Abstract
Soybean [Glycine max (L.) Merrill] is the most important leguminous crop in the world due to its high contents of high-quality protein and oil for human and animal consumption as well as for industrial uses. An accurate and saturated genetic linkage map of soybean is an essential tool for studies on modern soybean genomics. In order to update the linkage map of a F2 population derived from a cross between Misuzudaizu and Moshidou Gong 503 and to make it more informative and useful to the soybean genome research community, a total of 318 AFLP, 121 SSR, 108 RFLP, and 126 STS markers were newly developed and integrated into the framework of the previously described linkage map. The updated genetic map is composed of 509 RFLP, 318 SSR, 318 AFLP, 97 AFLP-derived STS, 29 BAC-end or EST-derived STS, 1 RAPD, and five morphological markers, covering a map distance of 3080 cM (Kosambi function) in 20 linkage groups (LGs). To our knowledge, this is presently the densest linkage map developed from a single F2 population in soybean. The average intermarker distance was reduced to 2.41 from 5.78 cM in the earlier version of the linkage map. Most SSR and RFLP markers were relatively evenly distributed among different LGs in contrast to the moderately clustered AFLP markers. The number of gaps of more than 25 cM was reduced to 6 from 19 in the earlier version of the linkage map. The coverage of the linkage map was extended since 17 markers were mapped beyond the distal ends of the previous linkage map. In particular, 17 markers were tagged in a 5.7 cM interval between CE47M5a and Satt100 on LG C2, where several important QTLs were clustered. This newly updated soybean linkage map will enable to streamline positional cloning of agronomically important trait locus genes, and promote the development of physical maps, genome sequencing, and other genomic research activities.
Key words: soybean, linkage map, SSR, RFLP, AFLP, STS
1. Introduction
Soybean, Glycine max (L.) Merr., supplies a large amount of high-quality protein and oil for food products and industrial materials. Recently, researchers have reported that various biochemical constituents of soybean seeds exert physiological functions beneficial to human health.1–3 The availability of numerous characteristics in soybean, such as symbiosis with root bacteroids, has set the stage for international efforts to explore soybean at the whole genome level.4,5 In modern genomics, the size of soybean genome (1.12–1.81 × 109 bp) has been considered to be moderate.6 Evolutionally, soybean is referred to as a recently diploidized tetraploid, and generally more than two copies are present for over 90% of the non-repetitive sequences in the soybean genome.7 In addition, 40–60% of the soybean sequences are repetitive.8,9 In the crop legumes, most crops belong to either the Hologalegina or the Phaseoloid lineage.10 Although two model legumes, Lotus and Medicago, belong to the Hologalegina lineage, it has been recently proposed that soybean genome could be used as a model for the Phaseoloid legumes due to the economic and biological importance of soybean, the moderate genome size, as well as the existing infrastructure for soybean research and commercial production.5,11
An accurate and saturated genetic linkage map of soybean is essential for studies on modern soybean genomics, i.e. identification of subtle or new trait loci including quantitative trait loci (QTLs), map-based cloning, and physical map construction or even whole-genome sequencing. The first soybean genetic map was constructed with 57 classical markers.12 Thereafter, molecular maps have been gradually integrated using restriction-fragment length polymorphism (RFLP) markers,13–16 random amplified polymorphic DNA (RAPD) markers,17 simple sequence repeat (SSR)18,19 and amplified-fragment length polymorphism (AFLP) markers.20,21 In recent years, integrated maps have been reported, each of which was merged from several maps derived from different mapping populations using JoinMap.22,23 More recently, an integrated map with sequence-based genic markers has also been constructed.24
Moshidou Gong503 (Glycine gracilis), which originated in Northeast China, is morphologically intermediate between the cultivated G. max and the wild form, G. soja.25 However, these three forms which are fully cross-compatible, effectively constitute a single species, G. max.25,26 Crosses between the cultivar (Misuzudaizu) and the intermediate form (Moshidou Gong503) would provide good genetic resources for linkage map construction and for the isolation of agronomically and biologically important genes. A framework of genetic linkage map had been previously constructed mainly with RFLP and SSR markers using a single F2 population of this combination.27–29 In addition, several agronomically and biologically important trait loci such as flowering time, growth habit, and seed quality were identified using this mapping population27,28 and its progenies (RILs).30,31 Further integration of this linkage map with a large number of SSR or RFLP markers and with other types of markers, i.e. AFLP or AFLP-derived sequence-tagged site (STS) markers, may enable to make this linkage map more informative and more useful for soybean genomics studies and particularly for the isolation of agronomically and biologically important QTL genes harbored by the parents, Misuzudaizu and Moshidou Gong503. Therefore, the objectives of the present study were threefold; (i) to develop AFLP and AFLP-derived STS markers; (ii) to develop a larger number of SSR and RFLP markers; and (iii) to integrate the newly developed markers into the framework of the previously described linkage map.28
2. Materials and methods
2.1. Plant materials and DNA extraction
A framework of the genetic linkage map had been previously constructed using an F2 population that was derived from a cross between the cultivar Misuzudaizu and a weedy form, Moshidou Gong 503, as ovule and pollen parents, respectively. This mapping population consisting of 190 F2 plants was used in the present study.27,28 However, the DNA was newly extracted for the present study from the leaves that had been preserved at −80°C, using the CTAB method32 with a slight modification.
2.2. AFLP marker development
The AFLP procedure was performed essentially as described by Vos et al.33 A total of 100–150 ng of genomic DNA was completely digested with EcoRI and MseI. Digested DNA was subjected to ligation with adapters that were compatible with the restriction sites (AFLP Core Reagent Kit, Life Technology, USA). After ligation, the reaction mixtures were diluted 10 times with TE. For the amplification of the restricted and ligated fragments, a two-step protocol was adopted. The first step included the selective pre-amplification of adapter-ligated DNA with primers with one additional selective nucleotide (+1/+1). In the second step, selective amplification of pre-amplified DNA was performed with adapter primers with two more additional selective nucleotides (+3/+3). All the amplification reactions were performed with TaKaRa EXTaq (TaKaRa, Japan). Electrophoresis was conducted by high-efficiency genome scanning (HEGS)34,35 with non-denaturing 11–13% polyacrylamide separating gels and 5% stacking gels. Gels were stained by Vistra Green (Amersham Pharmacia Biotech, UK) and were detected with FluorImager 585 (Amersham Pharmacia Biotech). Only clearly distinguishable polymorphic AFLP bands were scored for mapping in the present study.
Nomenclature for the AFLP markers includes the letter E for the EcoRI primer and the letter M for the MseI primer, each of which being followed by a number representing combinations of three selective nucleotides. The letter C was added as the prefix referring to the marker developed at Chiba University.
2.3. Development of STS markers from AFLP, BAC-end, or EST sequences
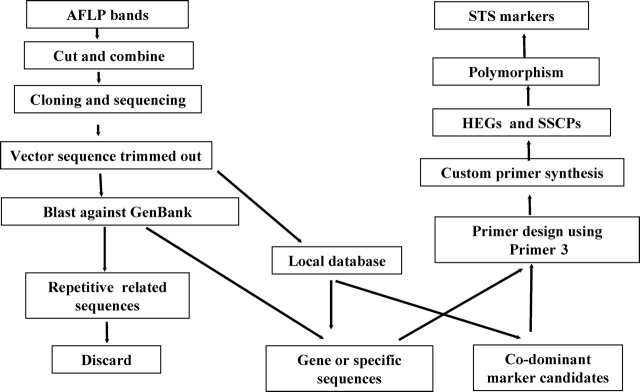
Compared with AFLP markers, STS markers are more valuable in marker-assisted selection (MAS) and more transferable between populations. Therefore, polymorphic AFLP fragments were converted into STS markers by cloning and sequencing. At first, polymorphic AFLP bands amplified from Misuzudaizu or Mashidou Gong 503 were excised from the polyacrylamide gel. DNA was extracted using a freeze-squeeze method (Xia et al., unpublished). These fragments were cloned using the pGEM®-T Easy Vector System (Promega, USA). Positive clones were confirmed by colony PCR.36 Plasmid DNA was isolated using the PI-200 Automatic DNA isolation system (Kurabo, Japan). Sequencing was performed using the ABI BigDye 3 system and analyzed using the ABI Prism3100 (Applied Biosystems, USA). Vector sequences were trimmed out using Chromas (version 2.23) (http://www.technelysium.com.au). After BLAST search against GenBank, all the retrotransposons or other repetitive sequences were discarded.37 A local sequence database was constructed by pooling the all sequences together using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Accordingly, all the sequences were searched over the local database to identify any orthologous sequences targeting for co-dominant marker development (Fig. 1). A total of 415 pairs of primers were designed to specific AFLP-derived sequences on line using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi).
Figure 1.
Strategy for developing AFLP-derived STS markers. See section 2 for details.
Furthermore, 150 primer pairs were designed to BAC-end sequences38,39 (http://www.soybeangenome.siu.edu). Among them, ∼75 primer pairs were kindly provided by D. A. Lightfoot, Southern Illinois University at Carbondale, Carbondale, IL 62901, USA. In addition, ∼50 and 60 primer pairs were designed to cDNAs from developing seeds and to expressed sequence tag (EST) homologs of flowering time-related genes,40 respectively.
For the mapping of new STS markers, all the primer pairs were initially tested for polymorphism between the two parents using HEGS34,35 and single-strand confirmation polymorphism (SSCP)41 techniques. The primer pairs showing a clear polymorphism between the two parents were mapped with HEGS, whereas the primer pairs with subtle polymorphisms were alternatively mapped with SSCP. The STS markers being developed at Chiba University were referred to as CSTS.
2.4. SSR marker development
In the early version of the linkage map, 96 SSR markers were mapped. Among them, 75 were developed at the USDA and DuPont Corporation and 21 SSR markers at Chiba University. In the present study, new SSR markers were mainly developed from genomic DNA or by surveying EST-SSR in the database. To isolate DNA fragments including SSRs with CA and CT repeats, a magnetic bead method was used for enrichment of the motif-containing sequences. The genomic DNA of Norin No.2 was digested with EcoRI and MseI. Digested DNA was ligated with adapters as described in AFLP marker development. After ligation, the fragments bearing CA and CT repeats were enriched with streptavidin-coated paramagnetic particles (Promega) probed with 3′-biotinylated (TG)8 and (AG)8 oligonucleotides, respectively. The enriched fraction was refined using SUPREC®-02 (TaKaRa), amplified by MseI and EcoRI primers and ligated to pGEM®-T Easy Vector System (Promega), and then transformed into Escherichia coli DH5α (Toyobo, Japan). The transformants were screened by blue–white selection. The positive clones were identified by colony hybridization using a DIG Luminescent Detection kit (Roche Diagnostics, USA) with DIG-labeled (TG)8 or (AG)8 probes. The PCR products of the positive clones were sequenced and the primers were designed using Primer 3 on line. In some cases, a dual-step method42,43 was used to isolate CA and CT-motif SSRs. The procedure was performed as previously described by Tamura et al.44 (AT)n(AC)n-motif SSRs were isolated using the streptavidin-coated magnetic beads described earlier, since this type of repeat is abundant in the soybean genome and AT repeats are difficult to screen directly due to the self-complementarity of the probe sequence. The SSR markers including AC, AG, AT, AAC, AAG, AAT, ACG, AGT, ATG, GGA, GGC, and GCT core-motifs were developed from motif-containing EST sequences. These sequences were identified by homology search of motif repeats against the EST data in DNA Data Bank of Japan (DDBJ) by FASTA. The minimum number of repeats for dinucleotide motif and trinucleotide motif SSRs was set to 10 and 7, respectively. The SSR markers developed at Chiba University were referred to as CSSRs in the present study.
2.5. RFLP analysis
On the basis of the earlier version of the linkage map,28 additional soybean cDNA clones derived from green leaves and clones of up-regulated genes in the nodules of Lotus japonicus45 were employed as probes to generate RFLP markers. The DNA was digested with eight restriction enzymes, ApaI, BamHI, BglII, DraI, EcoRI, EcoRV, HindIII, and KpnI. Electrophoresis, Southern blotting and hybridization procedures were performed as previously described.27
2.6. Linkage map construction
Most of the markers were mapped with F2 population consisting of 192 individuals. However, ∼200 markers, including newly developed RFLP and AFLP-derived STS markers, were mapped with 94 randomly selected F2 individuals. All the markers were checked against the expected 3:1 segregation by the χ2 test at a 5% significance level. The new marker data set was added to the original data set to produce the combined data set. Linkage analyses were performed using MAPMAKER (version 3) software.46 The commands ‘try’, ‘order’, and ‘build’ in MAPMAKER were used independently or in combination to insert new marker(s) into the framework of the previously described linkage map.28 Recombination frequencies were converted into map distance in centimorgans using the Kosambi mapping function.47 A LOD score of 3.0 and a maximum distance of 37.2 cM were used as linkage criteria for new marker insertion. The error detection function was set ‘on’ to detect any possible scoring errors. The linkage map was graphically visualized with MapChart.48
3. Results
3.1. AFLP marker development
Out of ∼800 primer pairs tested, 135 primer pairs that showed a clear polymorphism between the parents, Misuzudaizu and Moshidou Gong503, were selected for further analysis for the whole F2 population. Approximately 15–30 main bands were clearly amplified per primer combination. Each selected primer combination generated between 1 and 6 polymorphic bands (Fig. 2). The polymorphism rate of AFLPs was 4.8%, a value lower than the 11.3% value reported for barley49 and 14.8% for sorghum.50 The DNA quality, PCR, electrophoresis, and subsequent staining can all influence AFLP profiling. The HEGS system used in the present study generated clear and reproducible AFLP profiles within a range of 200–1200 bp, ensuring accurate genotype scoring (Fig. 2). The total number of bands generated and fragment intensity appeared to be negatively related to some extent. High GC content for both EcoRI + 3 and MseI + 3 selective nucleotides normally generated few but clear fragments, whereas a lower GC content led to a larger number of fragments with a lower quality. This phenomenon could be explained by the unusually high A + T nucleotide content in the soybean genome.51
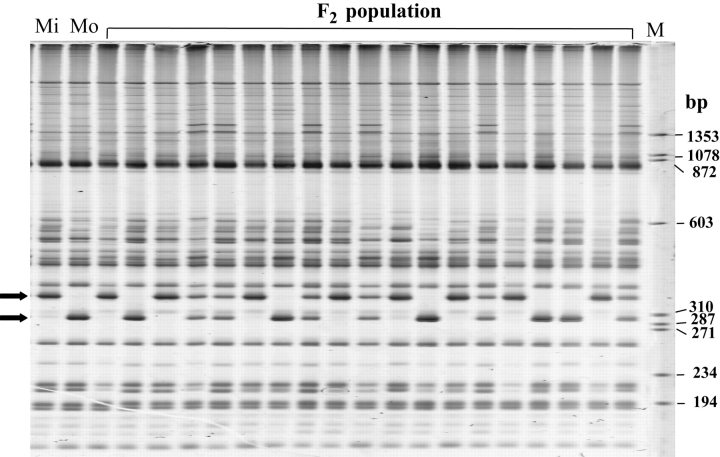
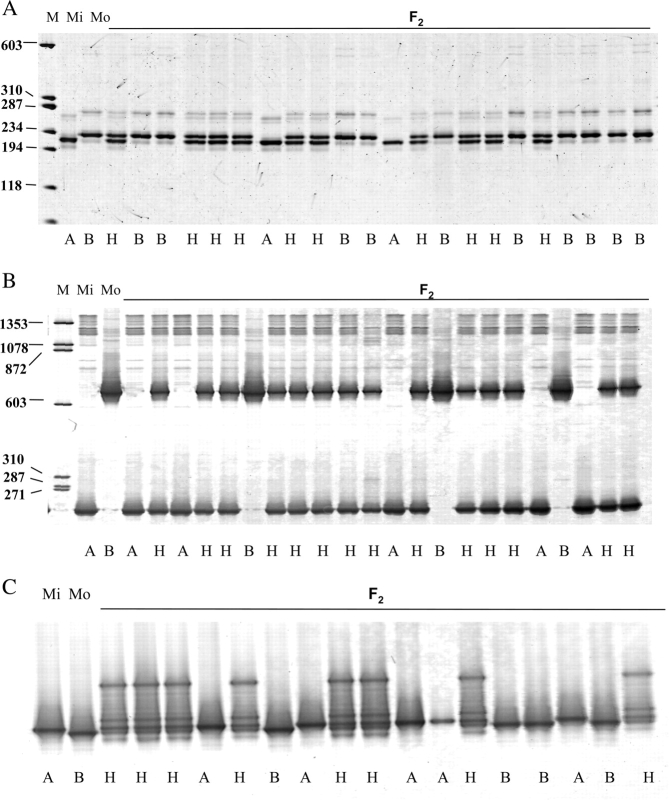
Figure 2.
AFLP marker analysis of the F2 mapping population. The left two lanes denoted by Mi and Mo were generated from the parents, Misuzudaizu and Moshidou Gong 503, respectively, with a combination of AFLP primers, E35(GAG) and M7(CTG). Lanes 3–21 were generated from the F2 population with the same primer combination. Arrows on the left side of the gel indicate mapped AFLP markers. Molecular weight marker ΦX174 HaeIII is shown in the lane denoted by M with the size (in bp) on the right side of the gel.
A total of 373 polymorphic bands were scored. However, 40 redundant markers, which were generated from the same combination and displayed the same genotype, were excluded. Apart from 10 unlinked markers and 5 unsuccessfully positioned markers, a total of 318 AFLP markers were successfully integrated into the framework of the previously described linkage map.28 Among the mapped markers, 164 markers showed a predominance for Misuzudaizu, whereas 154 markers for Moshidou Gong 503. Among the 164 markers with a predominance for Misuzudaizu, 149 (90.9%) markers segregated in a 3:1 ratio, whereas 9 and 6 markers segregated in 2:1 and 4:1 ratios, respectively. Among the 154 markers with a predominance for Moshidou Gong 503, 143 (92.9%) markers segregated in a 3:1, whereas 5 and 6 markers segregated in 2:1 and 4:1 ratios, respectively. The overall distortion rate of 8.2% was much lower than the 40% rate reported for two intraspecific crosses between two annual species of Medicago.52 Segregation distortion may be related to the differential parental genomes or to distorting factors such as sterility loci. Moreover, errors in genotyping scoring may also cause segregation distortion.23
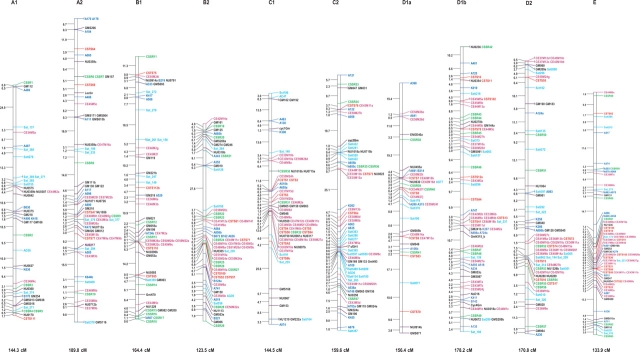
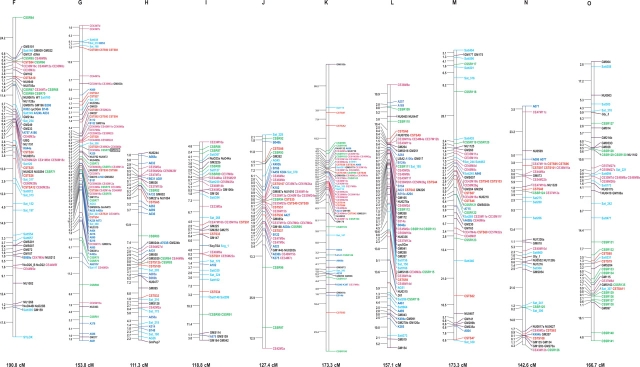
The 318 newly mapped markers were not uniformly distributed among the linkage groups (LGs) within a range of 3–31 per LG (Table 1). The number of new markers mapped to a given LG was not significantly correlated with the length of the LG (cM) [correlation coefficient, r = 0.1578 (P > 0.05)]. A certain degree of clustering of the AFLP markers was found in the putative centromeric or telomeric regions in LGs such as LGs B2, C1, D2, and E (Fig. 3). However, AFLP markers in the present study were not as strongly clustered as these reported by Qi et al.49 in barley and by Keim et al.21 in soybean. Although some researchers have reported a relatively uniform distribution of AFLP markers, it has been well documented in many crops, including soybean,21,49,50 that the strong clustering of AFLP markers is often associated with telomeric or centromeric regions. In the present study, the AFLP markers were generated using a restriction enzyme (EcoRI) that is insensitive to the methylation of CG dinucleotides. Thus, some particular regions, such as the heterochromatin regions around centromeres and telomeres, were accessible to EcoRI-based AFLP markers. Furthermore, in such regions, crossing-over during the meiosis was markedly reduced and the markers tended to cluster. In the present study, AFLP markers with a higher quality generated from a higher GC content in selective nucleotides may have to some extent reduced the level of clustering. The use of the enzymes PstI/MseI or TaqI/HindIII for AFLP marker generation might have further reduced the level of clustering of the AFLP markers, since either or both of the restriction enzymes are methylation sensitive.21,49,50 The AFLP markers presented here are accessible via the marker nomenclature (Supplementary Table S1).
Table 1.
Comparison of marker information in the newly constructed linkage map with that in the previous linkage map
| Previous linkage map(Yamanaka et al. 2001) | Newly constructed linkage map | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LG | Length (cM) | Marker nos. | SSR (CSSR1) | RFLP | Other types2 | Length (cM) | Marker nos. | AFLP | SSR | RFLP | STS | Other types2 | |||
| New CSSR | Public SSR3 | Total | New RFLP | Total | New CSTS | ||||||||||
| A1 | 132.9 | 18 | 3 | 15 | 0 | 144.3 | 42 | 3 | 5 | 10 | 15 | 8 | 23 | 1 | 0 |
| A2 | 202.2 | 27 | 3 (1) | 24 | 0 | 189.8 | 64 | 15 | 4 | 7 | 12 | 9 | 33 | 4 | 0 |
| B1 | 142.3 | 19 | 1 | 18 | 0 | 164.4 | 55 | 12 | 8 | 7 | 15 | 6 | 24 | 4 | 0 |
| B2 | 104.6 | 24 | 4 | 20 | 0 | 123.5 | 65 | 19 | 11 | 6 | 17 | 4 | 24 | 5 | 0 |
| C1 | 129.1 | 19 | 4 (1) | 15 | 0 | 144.5 | 64 | 21 | 3 | 6 | 9 | 10 | 25 | 9 | 0 |
| C2 | 158.2 | 32 | 6 | 25 | 1 | 159.6 | 71 | 14 | 5 | 17 | 22 | 6 | 31 | 3 | 1 |
| D1a | 166.8 | 16 | 5 | 11 | 0 | 156.4 | 38 | 10 | 4 | 5 | 9 | 3 | 14 | 5 | 0 |
| D1b | 164.4 | 23 | 3 | 20 | 0 | 178.2 | 75 | 23 | 8 | 9 | 17 | 7 | 27 | 8 | 0 |
| D2 | 159.3 | 22 | 6 | 16 | 0 | 170.8 | 72 | 22 | 8 | 16 | 24 | 4 | 20 | 6 | 0 |
| E | 118.0 | 27 | 3 | 24 | 0 | 133.9 | 73 | 23 | 6 | 7 | 13 | 2 | 26 | 11 | 0 |
| F | 195.4 | 41 | 9 (1) | 31 | 1 | 190.8 | 86 | 15 | 7 | 14 | 22 | 11 | 42 | 6 | 1 |
| G | 157.7 | 39 | 6 (3) | 32 | 1 | 153.8 | 107 | 31 | 7 | 9 | 19 | 8 | 40 | 16 | 1 |
| H | 107.1 | 26 | 1 | 25 | 0 | 111.3 | 50 | 8 | 4 | 6 | 10 | 3 | 28 | 4 | 0 |
| I | 113.5 | 24 | 8 (1) | 16 | 0 | 118.8 | 50 | 12 | 5 | 12 | 18 | 0 | 16 | 4 | 0 |
| J | 102.4 | 20 | 3 (2) | 17 | 0 | 127.4 | 54 | 18 | 4 | 3 | 9 | 2 | 19 | 8 | 0 |
| K | 181.6 | 26 | 6 (2) | 20 | 0 | 173.3 | 76 | 25 | 9 | 7 | 18 | 3 | 23 | 10 | 0 |
| L | 152.6 | 41 | 4 | 36 | 1 | 157.1 | 80 | 17 | 8 | 8 | 16 | 5 | 41 | 5 | 1 |
| M | 109.8 + 11.4 | 22 | 6 | 16 | 0 | 173.3 | 57 | 13 | 7 | 11 | 18 | 3 | 19 | 7 | 0 |
| N | 128.1 | 14 | 1 (1) | 12 | 1 | 142.6 | 48 | 10 | 2 | 7 | 10 | 8 | 20 | 7 | 1 |
| O | 171.3 | 23 | 14 (9) | 8 | 1 | 166.7 | 50 | 7 | 6 | 10 | 25 | 6 | 14 | 3 | 1 |
| Total | 2908.7 | 503 | 96 (21) | 401 | 6 | 3080.5 | 1277 | 318 | 121 | 177 | 318 | 108 | 509 | 126 | 6 |
1CSSR—SSR markers developed at Chiba University.
2Other types—including phenotypic markers and a RAPD marker.
3Public SSR—SSR markers developed at other institutes than Chiba University.
Figure 3.
Soybean genetic linkage map constructed with RFLP, SSR, STS, and AFLP markers. The linkage map was graphically visualized with MapChart. The name of each LG is indicated on the top of the bar. Distances between markers are indicated on the left side of each LG, as calculated by the Kosambi function. Total length of each LG is also indicated at the bottom of each group. Different colors represent the following markers: Violet red, AFLP; Red, CSTS; Green, CSSR; Light sky blue, SSR (Public); Dark blue, RFLP; Black, RFLP (Public); Italic Black, Phenotypic marker; Boxed Black, RAPD marker.
3.2. STS marker development
Over 500 AFLP polymorphic fragments, including ∼200 mapped AFLP markers were successfully sequenced. Approximately 15% of them were associated with repetitive sequences, such as Ty3/Gypsy and STR120.37 Interestingly, ∼10% were related to mitochondria or chloroplast gene-related sequences. Of 415 pairs of primers were designed to the non-repetitive sequences, a total of 97 AFLP-derived STS markers were successfully mapped and integrated into the framework of the previously described linkage map28 (Fig. 3). Among them, 64 markers with clear polymorphisms were mapped using HEGS (Fig. 4B), whereas 33 markers were mapped with SSCP (Fig. 4C). Furthermore, 58 markers were co-dominant with our mapping population.
Figure 4.
Analysis of segregation of CSSR and CSTS markers using HEGS (A, CSSR60; B, CSTS73) and SSCP techniques (C, CSTS48). Lanes denoted with Mi, Mo, and F2 were generated from the parents, Misuzudaizu and Moshidou Gong 503, and their F2 population, respectively. Molecular weight marker ΦX174 HaeIII is shown in the lane ‘M’ with the size (in bp) on the left side of the panels A and B. The genotypes for each lane were indicated at the bottom of each lane.
Initially, 30 AFLP-derived STS were converted from mapped AFLP markers, all of them being tagged to the same locus as the original AFLP markers being mapped. The other 67 markers were converted from randomly selected polymorphic AFLP bands. Among all the 97 AFLP-derived STS markers, 24 single, 7 double, 1 triple, 1 quadruple, and 1 quintuple markers were mapped to 34 loci, at which one or more AFLP markers had already resided. In addition, two double and two triple AFLP-derived STS markers were mapped to four loci at which no AFLP marker was tagged, suggesting that AFLP-derived STS markers also tended to be distributed in a clustering fashion as the AFLP markers do.
Additionally, 19 STS markers were developed from 150 primer pairs designed to BAC-end sequences at a polymorphism rate of 12.6%. Among the 110 PCR primer pairs designed to cDNA or flowering time gene homologs in soybean, only 10 markers were mapped at a polymorphism rate of only 9.1%. Taken together, a total of 126 CSTS markers were mapped within a range of 1 to 16 markers per LG (Table 1). The number of STS mapped to a given LG was not significantly correlated with the length of the LG (cM) [correlation coefficient, r = 0.0216 (P > 0.05)].
3.3. SSR marker development
Out of 702 new SSRs, 121 SSR markers were successfully mapped in the present study, including 41 markers from genomic DNAs and 80 from the EST database. Along with the 20 CSSR markers mapped in the earlier version of the linkage map, a total of 61 genomic DNA-derived SSR markers were classified with different motifs, i.e. 27 with CT, 3 with AC, 1 with GTG, and 30 with compound-motif repeats. An example of segregation of CSSR60 is shown in Fig. 4A. Polymorphism rates of genomic SSRs were 8, 18, and 53% for AC repeats, CT repeats, and (AT)n(AC)n motif, respectively.
Among the 80 EST-SSRs, 16 and 64 markers were developed from dinucleotide and trinucleotide motifs, respectively. Since the repeat numbers for the EST-SSRs are generally lower than those for genomic SSRs,22,39 we set the minimum repeat number for dinucleotides and trinucleotides to 10 and 7, respectively. The polymorphism was 25.15% (80/318) within a range of 15–50%, depending on the motifs, being slightly higher than the polymorphism rate of 18.0% reported by Song et al.22 Since we used HEGS and SSCP techniques for mapping, it was possible to detect subtle polymorphisms (Fig. 4B and C).
A total of 318 SSR markers were mapped in 20 different LGs, within a range of 9–25 markers per LG (Table 1). SSR distribution was significantly correlated with the length of the LG (r = 0.4449, P < 0.05). In contrast to AFLP markers, the SSR markers were relatively evenly distributed, although slight clustering was observed in some specific regions. This slight clustering phenomenon can be ascribed to the fact that SSR markers are significantly associated with the low-copy fractions of the plant genome.53
3.4. RFLP marker development
In addition to the 404 RFLP markers in the framework of the previously described linkage map,28 a total of 108 RFLP markers were newly generated with additional cDNA clones from green leaves and up-regulated cDNA clones in the nodules of L. japonicus as probes. These markers were successfully integrated into the existing linkage map framework. In total, 509 RFLP markers ware distributed among the LGs within a range of 13–44 markers per LG (Table 1). However, RFLP distribution was not significantly correlated with the length of the LG (r = 0.2905, P > 0.05).
3.5. The characteristics of the current linkage map
On the basis of the earlier version of the linkage map, a total of 318 AFLP, 121 SSR, 108 RFLP, and 126 STS markers were newly developed and integrated (Table 1, Fig. 3). The current genetic map is composed of 1277 loci at 2.41 cM intervals, covering a map distance of 3080 cM (Kosambi function) in 20 LGs. Most SSR and RFLP markers were relatively evenly distributed among the different LGs, although the AFLP markers were moderately clustered and several relatively large gaps still remained (Fig. 3). The coverage of the linkage map was extended since 17 markers were mapped beyond distal ends of the previous linkage map (Fig. 3). This is presently the densest linkage map developed from a single F2 population in soybean, although integrated maps, each of which was merged from several maps derived from different mapping populations, have been reported.23,24
3.6. Information about the developed markers
The information about the mapped markers regarding LG, map position, gene/accession numbers, and primer sequences and marker type is available in the online version of this article (Supplementary Table S1). In addition, primer information for about the STS and SSR markers, which were developed but not presented in Supplementary Table S1, is also accessible online-only (Supplementary Table S2).
4. Discussion
4.1. Marker order and position among different mapping populations
In our updated linkage map, 139 SSR markers were shared with the LGs described by Song et al.22 Most markers were in consensus order in both LGs, indicating a significant correlation (r = 0.6064, P < 0.01) between the length of the LGs in both maps.22 However, reversions occurred in some regions in LGs A1, D2, and G. In the LG G, the order of Sat_223 and Sat_260 was 0.2 cM apart in the present map, whereas 0.22 cM apart with a reversed order in the linkage map constructed by Song et al.22 Comparison of different linkage maps constructed from different populations with a different genetic background using different marker sets indicated that most markers showed the consensus order, although some intervals or regions always displayed some discrepancy in the marker order or positions. This phenomenon may be due to inversion, insertion, deletion, or transition of genomic regions as well as meiotic drive and gametic or zygotic selection.23 Also, possible errors in genotyping scoring may distort marker orders and segregation ratios.23 In soybean, some markers, especially RFLP markers, could be mapped on more than one LG. Because soybean is an allotetraploid, it has been shown that for over 90% of the non-repetitive sequences in the soybean genome, there were two closely related copies at different loci.7 As reported earlier, there was some inconsistency existed between physical map and genetic map regarding the marker order and positions.54–56 With the new progress made in genome sequencing and comparative mapping, it is likely that these discrepancies or inconsistencies will be reduced or eventually clarified.
4.2. AFLP-derived STS markers
Conversion of AFLP fragments into polymorphic STS markers would enable to achieve a high throughput scoring of genotypes in fine mapping and MAS in breeding.57 Development of AFLP-derived STS markers tend to be laborious and time-consuming due to the lower conversion efficiency. The lower polymorphism rate for STS or other markers22 may be due to the low sequence variation in soybean and its wild ancestor G. soja.24 Zhu et al.58 reported values of 0.5 and 4.7 SNPs/Kb in coding and non-coding perigenic DNA, respectively. As a result, the polymorphism rate was 10 times lower than that reported in maize.59,60 AFLP-derived STS markers developed in the present study displayed a high degree of transferability since most of them showed polymorphism in the RIL populations, Jack × Fukuyataka and Peking × Akita (Hwang et al., personal communication). Although 97 AFLP-derived STSs and 29 BAC and EST-derived STSs have been developed, the number is not necessarily large enough for a saturated map.
4.3. Comparison with the earlier version of the linkage map
As a total of 673 newly developed AFLP, SSR, RFLP, and STS markers in addition to 101 new public SSR markers were integrated, the average intermarker distance was reduced by more than twofold to 2.41 from 5.78 cM in the earlier version of the linkage map.28 In addition, the proportion of PCR-based markers was 34.8%, a much higher value than the 19.2% reported in the earlier version of the linkage map. A large gap of more than 37.5 cM in LG C1 was filled and two unlinked LGs for LG M were joined. The number of gaps of more than 25 cM was reduced to 6 from 19 in the earlier version of the linkage map.28 Similar large gaps were also present on the same or similar positions in a linkage map constructed from a RIL population derived from the current mapping population, using an other set of markers (Hayashi et al., unpublished result), indicating that some of these gaps may be partially associated with the nature of the genome structure of the parents. Some hot-spots of recombination may lead to enlarged gaps in the genetic linkage map, in spite of short physical distances. In addition, the degree of coverage of the newly constructed linkage map was improved, as 17 markers were mapped beyond the distal ends of the LGs in the previous linkage map.
4.4. Usefulness of the linkage map
Map-based cloning requires very fine resolution mapping in the target interval, since the highest marker density can shorten chromosome walking. MAS is most effective when the markers are tightly linked to the gene of interest since crossing-over between the gene and markers dramatically decreases. In general, accurate and consistent integrated genetic and physical maps55,56 of the soybean genome should enable to distinguish new or subtle QTL(s) from any of the more than a thousand identified QTLs, and thereafter to clone and functionally confirm cloned QTL genes. Several agronomically and biologically important trait loci such as flowering time, growth habit, and seed quality have been identified with this mapping population28 and its progeny.31 In particular, 17 markers were tagged in the 5.7 cM interval between CE47M5a and Satt100 on LG C2, where various important QTLs were clustered. The current soybean linkage map became more informative and useful for positional cloning of agronomically important genes for traits including QTLs that are harbored by the parents. On the basis of this linkage map, several residual heterozygous lines (RHLs) have been developed from the progeny of this mapping population for fine-mapping of several QTLs.61 More than 30 primer pairs targeting SSR motifs have been specifically developed from physical contigs of the flowering time QTLs (FT1, FT2, and FT3), 70% of which displaying polymorphism between the parents. These markers should enable to further narrow the QTL gene regions toward the cloning of candidate QTL(s) (Xia et al. and Watanabe et al., unpublished results).
Soybean originated in East Asia and the vast collection of wild species and landraces should provide useful genetic resources for studies on soybean genomics. Recent studies have revealed that a large number of wild species of soybean contain a wide range of secondary metabolite compounds, which have preliminarily been found to be beneficial to human health.1–3 Genetic differences in the secondary metabolite compounds between the cultivar Misuzudaizu and the intermediate weedy form Moshidou Gong 503 were also observed.
4.5. Future perspectives
Owing to the presence of relatively large gaps or marker-sparse regions, targeted marker development via BAC sequencing62 is a powerful tool. An accurate and consistent integrated genetic map is useful for physical map development and whole genome sequencing. Conversely, a large number of targeted SSR and STS markers can be generated from genome sequencing for saturation of the linkage map in soybean. Furthermore, due to the lower polymorphism rate in the soybean genome, new types of markers such as SNP-based markers need to be gradually incorporated due to their abundance in the soybean genome and technical applicability.24 Ideally, near or over 10 000 evenly distributed PCR-based markers could satisfy most applications including QTL gene isolation, evolution studies, and other field of genomics.
Acknowledgments
We wish to thank Professor J. Rees, University of Western Cape, South Africa, for the critical reading of the manuscript and Professor D. A. Lightfoot, Southern Illinois University, for providing the primer pairs.
Funding: Ministry of Agriculture, Forestry and Fisheries (Rice Genome Project DM-2109, Green Technology Project DM-1213); Japan International Research Center for Agricultural Sciences (Comprehensive Studies on Soybean Improvement, Production and Utilization in South America); Program for the Promotion of Basic Research Activities for Innovative Biosciences; Ministry of Education, Science, Sports, and Culture of Japan (935600).
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org
References
- 1.Liu K. Soy Isoflavones, In. In: Liu K., editor. Chemistry, Processing Effects, Health Benefits, and Functional Foods and Ingredients. Champaign, IL: AOCS Press; 2004. pp. 52–72. [Google Scholar]
- 2.Lin J., Wang C. Soybean Saponins, In. In: Liu K., editor. Chemistry, Processing Effects, Health Benefits, and Functional Foods and Ingredients. Champaign, IL: AOCS Press; 2004. pp. 73–100. [Google Scholar]
- 3.Messina M. J. Potential public health implications of the hypocholesterolemic effects of soy protein. Nutrition. 2005;19:280–281. doi: 10.1016/s0899-9007(02)00995-4. [DOI] [PubMed] [Google Scholar]
- 4.Stacey G., Vodkin L., Parrott W. A., Shoemaker R. C. National Science Foundation-sponsored workshop report. Draft plan for soybean genomics. Plant Physiol. 2004;135:59–70. doi: 10.1104/pp.103.037903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson S. A., Rokhsar D., Stacey G., Shoemaker R. C., Schmutz J., Grimwood J. Toward a reference sequence of the soybean genome: a multiagency effort. Crop. Sci. 2006;46:55–61. [Google Scholar]
- 6.Arumaganthan K., Earle E. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991;9:208–218. [Google Scholar]
- 7.Shoemaker R. C., Polzin K., Labate J., et al. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144:329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg R. B. DNA sequence organization in the soybean plant. Biochem. Genet. 1978;16:45–68. doi: 10.1007/BF00484384. [DOI] [PubMed] [Google Scholar]
- 9.Danesh D., Penuela S., Mudge J., et al. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor. Appl. Genet. 1998;96:196–202. [Google Scholar]
- 10.Lavin M., Herendeen P. S., Wojciechowski M. F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- 11.Gepts P., Beavis W. D., Brummer E. C., et al. Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol. 2005;137:1228–1235. doi: 10.1104/pp.105.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer R. G., Kilen T. C. Qualitative genetics and cytogenetics. Agronomy. 1987;16:135–209. [Google Scholar]
- 13.Keim P., Diers B. W., Olson T. C., Shoemaker R. C. RFLP mapping in soybean: association between marker loci and variation in quantitative traits. Genetics. 1990;126:735–742. doi: 10.1093/genetics/126.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lark K. G., Weisemann J. M., Mathews B. F., Palmer R., Chase K., Macalma T. A genetic map of soybean (Glycine max L.) using an intraspecific cross of two cultivars: ‘Minsoy’ and ‘Noir 1. Theor. Appl. Genet. 1993;86:901–906. doi: 10.1007/BF00211039. [DOI] [PubMed] [Google Scholar]
- 15.Shoemaker R. C., Specht J. E. Integration of the soybean molecular and classical genetic linkage groups. Crop. Sci. 1995;35:436–446. [Google Scholar]
- 16.Mansur L. M., Orf J. H., Chase K., Jarvik T., Cregan P. B., Lark K. G. Genetic mapping of agronomic traits using recombinant inbred lines of soybean. Crop. Sci. 1996;36:1327–1336. [Google Scholar]
- 17.Ferreira A. R., Foutz K. R., Keim P. Soybean genetic map of RAPD markers assigned to an existing scaffold RFLP map. J. Hered. 2000;91:392–396. doi: 10.1093/jhered/91.5.392. [DOI] [PubMed] [Google Scholar]
- 18.Akkaya M. S., Shoemaker R. C., Specht J. E., Bhagwat A. A., Cregan P. B. Integration of simple sequence repeat DNA markers into a soybean linkage map. Crop. Sci. 1995;35:1439–1445. [Google Scholar]
- 19.Cregan P. B., Jarvik T., Bush A. L., et al. An integrated genetic linkage map of the soybean. Crop. Sci. 1999;39:1464–1490. [Google Scholar]
- 20.Maughan P. J., Marouf M. A. S., Buss G. R., Huestis G. M. Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and near-isogenic line analysis. Theor. Appl. Genet. 1996;93:392–401. doi: 10.1007/BF00223181. [DOI] [PubMed] [Google Scholar]
- 21.Keim P., Schupp J. M., Travis S. E., et al. A high-density soybean genetic map based on AFLP markers. Crop. Sci. 1997;37:537–543. [Google Scholar]
- 22.Song Q. J., Marek L. F., Shoemaker R. C., et al. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- 23.Kassem M. A., Shultz J., Meksem K., et al. An updated ‘Essex’ by ‘Forrest’ linkage map and first composite interval map of QTL underlying six soybean traits. Theor. Appl. Genet. 2006;113:1015–1026. doi: 10.1007/s00122-006-0361-8. [DOI] [PubMed] [Google Scholar]
- 24.Choi I. Y., Hyten D. L., Matukumalli L. K., et al. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics. 2007;176:685–696. doi: 10.1534/genetics.107.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hymowitz T. Speciation and cytogenetics, In. In: Boerma H. R., Specht J. E., editors. Soybeans: Improvement, Production and Uses. 3rd Ed. Madison, WI: American Society of Agronomy-Crop Science Society of America-Soil Science Society of America; 2004. pp. 97–136. Agronomy Monograph No. 16. [Google Scholar]
- 26.Smartt J. Grain Legumes-Evolution and Genetic Resources. Cambridge: Cambridge University Press; 1990. pp. 246–257. [Google Scholar]
- 27.Yamanaka N., Nagamura Y., Tsubokura Y., et al. Quantitative trait locus analysis of flowering time in soybean using a RFLP linkage map. Breed. Sci. 2000;50:109–115. [Google Scholar]
- 28.Yamanaka N., Ninomiya S., Hoshi M., et al. An informative linkage map of soybean reveals QTLs for flowering time, leaflet morphology and regions of segregation distortion. DNA Res. 2001;8:61–72. doi: 10.1093/dnares/8.2.61. [DOI] [PubMed] [Google Scholar]
- 29.Hossain K., Kawai G., Hayashi H., Hoshi M., Yamanaka N., Harada K. Characterization and identification of (CT)n microsatellites in soybean using sheared genomic libraries. DNA Res. 2000;7:103–110. doi: 10.1093/dnares/7.2.103. [DOI] [PubMed] [Google Scholar]
- 30.Tajuddin T., Watanabe S., Yamanaka N., Harada K. Analysis of quantitative trait loci for protein and lipid contents in soybean seeds using recombinant inbred lines. Breed. Sci. 2003;53:133–140. [Google Scholar]
- 31.Watanabe S., Tadjuddin T., Yamanaka N., Hayashi M., Harada K. Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breed. Sci. 2004;54:399–407. [Google Scholar]
- 32.Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vos P., Hogers R., Bleeker M., et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawasaki S., Murakami Y. Genome analysis of Lotus japonicus. J. Plant Res. 2000;113:497–506. [Google Scholar]
- 35.Hayashi M., Miyahara A., Sato S., et al. Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res. 2001;8:301–310. doi: 10.1093/dnares/8.6.301. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J., Frietsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Laten H. M., Havecker E. R., Farmer L. M., Voytas D. F. SIRE1, an endogenous retrovirus family from Glycine max, is highly homogeneous and evolutionarily young. Mol. Biol. Evol. 2003;20:1222–1230. doi: 10.1093/molbev/msg142. [DOI] [PubMed] [Google Scholar]
- 38.Xia Z., Sato H., Watanabe S., Kawasaki S., Harada K. Construction and characterization of a BAC library of soybean. Euphytica. 2005;141:129–137. [Google Scholar]
- 39.Shultz J. L., Kazi S., Bashir R., Afzal J. A., Lightfoot D. A. The development of BAC-end sequence-based microsatellite markers and placement in the physical and genetic maps of soybean. Theor. Appl. Genet. 2007;114:1081–1090. doi: 10.1007/s00122-007-0501-9. [DOI] [PubMed] [Google Scholar]
- 40.Hecht V., Foucher F., Ferrandiz C., et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005;137:1420–1434. doi: 10.1104/pp.104.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirasawa K., Monna L., Kishitani S., Nishio T. Single nucleotide polymorphisms in randomly selected genes among japonica rice (Oryza sativa L.) varieties identified by PCR-RF-SSCP. DNA Res. 2004;11:275–283. doi: 10.1093/dnares/11.4.275. [DOI] [PubMed] [Google Scholar]
- 42.Lian C., Zhou Z., Hougetsu T. A simple method for developing microsatellite markers using amplified fragments of inter-simple sequence repeat (ISSR) J. Plant Res. 2001;114:381–385. [Google Scholar]
- 43.Lian C., Hougetsu T., Matsushit N. G., Alexis L., Suzuki K., Yamada A. Development of microsatellite markers from an ectomycorrhizal fungus, Tricholoma matsutake, by an ISSR-suppression-PCR method. Mycorrhiza. 2003;13:27–31. doi: 10.1007/s00572-002-0193-6. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K., Nishioka M., Hayashi M., et al. Development of microsatellite markers by ISSR-suppression-PCR method in Brassica rapa. Breed. Sci. 2005;55:247–252. [Google Scholar]
- 45.Kouchi H., Shimomura K., Hata S., et al. Large scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 2004;11:263–274. doi: 10.1093/dnares/11.4.263. [DOI] [PubMed] [Google Scholar]
- 46.Lander E. S., Green P., Abrahamson J., et al. MAPMAKER: an interactive computer package for constructing primary linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 47.Kosambi D. D. The estimation of map distances from recombination values. Ann. Eugen. 1944;12:172–175. [Google Scholar]
- 48.Voorrips R. E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 49.Qi X., Stam P., Lindhout P. Use of locus-specific AFLP markers to construct a high-density molecular map in barley. Theor. Appl. Genet. 1998;96:376–384. doi: 10.1007/s001220050752. [DOI] [PubMed] [Google Scholar]
- 50.Klein P. E., Klein R. R., Cartinhour S. W., et al. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genome Res. 2000;10:789–807. doi: 10.1101/gr.10.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu T., Oliphant A., Schupp J. M., Keim P. Hypomethylated DNA sequences: characterization of the duplicated soybean genome. Mol. Gen. Genet. 1994;244:638–645. doi: 10.1007/BF00282754. [DOI] [PubMed] [Google Scholar]
- 52.Jenczewski E., Ghererdi M., Bonnin I., Prosperi J. M., Olivieri I., Huguet T. Insight on segregation distortion in two intraspecific crosses between annual species of Medicago (Leguminosae) Theor. Appl. Genet. 1997;94:682–691. [Google Scholar]
- 53.Morgante M., Hanafey M., Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- 54.Shultz J. L, Meksem K., Lightfoot D. A. Evaluating physical maps by clone location comparison. Genome Lett. 2003;2:99–107. [Google Scholar]
- 55.Shultz J. L., Kurunam D. J., Shopinski K. L., et al. The soybean genome database (SoyGD): a browser for display of duplicated, polyploid, regions and sequence tagged sites on the integrated physical and genetic maps of Glycine max. Nucleic Acids Res. 2006;34:D758–D765. doi: 10.1093/nar/gkj050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu C. S., Sun P., Nimmakayala P., et al. A BAC- and BIBAC-based physical map of the soybean genome. Genome Res. 2004;14:319–326. doi: 10.1101/gr.1405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meksem K., Ruben E., Hyten D., Triwitayakorn K., Lightfoot D. A. Conversion of AFLP bands into high-throughput DNA markers. Mol. Genet. Genomics. 2001;265:207–214. doi: 10.1007/s004380000418. [DOI] [PubMed] [Google Scholar]
- 58.Zhu Y. L., Song Q. J., Hyten D. L., et al. Single-nucleotide polymorphisms in soybean. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tenaillon M. I., Sawkins M. C., Long A. D., Gaut R. L., Doebley J. F., Gaut B. S. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) Proc. Natl. Acad. Sci. USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ching A., Caldwell K. S., Jung M., et al. SNP, frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genetics. 2002;3:19. doi: 10.1186/1471-2156-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamanaka N., Watanabe S., Toda K., et al. Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor. Appl. Genet. 2005;110:634–639. doi: 10.1007/s00122-004-1886-3. [DOI] [PubMed] [Google Scholar]
- 62.Cregan P. B., Mudge J., Fickus E. W., et al. Targeted isolation of simple sequence repeat markers through the use of bacterial artificial chromosomes. Theor. Appl. Genet. 1999;98:919–928. [Google Scholar]