Abstract
A cattle database of candidate genes and genetic markers for milk production and mastitis has been developed to provide an integrated research tool incorporating different types of information supporting a genomic approach to study lactation, udder development and health. The database contains 943 genes and genetic markers involved in mammary gland development and function, representing candidates for further functional studies. The candidate loci were drawn on a genetic map to reveal positional overlaps. For identification of candidate loci, data from seven different research approaches were exploited: (i) gene knockouts or transgenes in mice that result in specific phenotypes associated with mammary gland (143 loci); (ii) cattle QTL for milk production (344) and mastitis related traits (71); (iii) loci with sequence variations that show specific allele-phenotype interactions associated with milk production (24) or mastitis (10) in cattle; (iv) genes with expression profiles associated with milk production (207) or mastitis (107) in cattle or mouse; (v) cattle milk protein genes that exist in different genetic variants (9); (vi) miRNAs expressed in bovine mammary gland (32) and (vii) epigenetically regulated cattle genes associated with mammary gland function (1). Fourty-four genes found by multiple independent analyses were suggested as the most promising candidates and were further in silico analysed for expression levels in lactating mammary gland, genetic variability and top biological functions in functional networks. A miRNA target search for mammary gland expressed miRNAs identified 359 putative binding sites in 3′UTRs of candidate genes.
Keywords: association study, candidate genes, gene linkage, knockout models, mammary gland, mastitis, methylation, micro RNA, milk traits, quantitative trait loci
Introduction
Association and quantitative trait locus (QTL) studies in large farm animals are typically performed in outbred populations, making the identification of robust QTL and candidate genes difficult and less reliable due to the variation of genetic background and population-specific interactions between loci. This situation differs very much from the situation in model and laboratory animal species, where highly inbred lines and targeted gene knock-outs are available. Therefore, the only applicable approach for QTL identification and candidate gene detection in large farm animals is the combination of different pieces of evidence supporting the functionality of identified genomic regions in relation to multigenic traits (Mackay 2004). Guidelines and standards for reporting quantitative trait nucleotide discovery in livestock species which allow incorporation of QTL in breeding programmes have been reviewed by Ron and Weller 2007.
A fair amount of genetic research related to lactation and udder health has already been performed due to its economic importance for milk production and manufacturing. This has led to considerable improvement of milk yield (MY); however, the progress in technological properties of milk and udder health has been relatively slow. Shook (2006) reported that somatic cell score (SCS) associated loci have been proposed to improve resistance to mastitis in dairy cattle. In addition, the expression of micro RNAs (miRNAs) in the bovine mammary gland could also play an important role in regulatory pathways in mammary gland development, milk production and resistance or susceptibility to mastitis (Silveri et al. 2006).
The recent developments in molecular biology have opened the possibility of exploiting heterologous animal models for comparative studies (Shook 2006). Targeted gene disruption in mice (gene knock-out experiments; KOs) revealed several mammary gland related phenotypes. The release of cattle genome sequence has enabled discovery of new markers and creation of synteny maps including data from other species. For example, Ron et al. (2007) utilized murine gene expression data from multiple analyses combined with bovine QTL mapping data to identify candidate genes for QTL for milk production traits in dairy cattle.
Functional traits of the mammary gland have been studied using different approaches, including the QTL approach, association studies and the candidate gene approach. However, information extracted from these methodologically focused studies is fragmented and often controversial. Therefore, there is an urgent need to integrate information from different sources and to allow complementation of different pieces of evidence based on holistic, map driven approach. The possibility of searching the database using animal trait ontology terms to select targets based on the mapping information or to search for indicated sequence similarities in primary databases opens up the possibility of introducing complex decision-making strategies which integrate multiple pieces of evidence supporting the candidate status of the selected region.
The classical forward genetics approaches which are typically focused on a single gene effect have been successful in the identification of a limited number of causal genes. In dairy cattle, two genes, DGAT1 (Grisart et al. 2002) and ABCG2 (Cohen-Zinder et al. 2005), have been reported to affect MY and milk composition. Therefore, the identification of key drivers related to complex traits needs a more holistic approach, based on integration of gene-to-gene interactions with DNA variation data. This approach has recently been developed to elucidate the complexity of common human diseases by intersecting genotypic, molecular profiling and clinical data in segregating populations (Schadt 2006).
Our attempt was to create a database which would take advantage of a multidisciplinary approach linking different types of data and supporting the evidence for involvement of candidate loci into the mammary gland development, milk production traits and resistance or susceptibility to mastitis. The database aims to serve as a tool for systematic development of markers for potential use in marker-assisted selection (MAS), which could be used in cattle breeding programmes to address the most relevant physiological pathways in the mammary gland.
Materials and methods
The database contains candidate loci involved in mammary gland development, milk production and resistance or susceptibility to mastitis. Candidate loci were collected considering seven different research approaches: (i) gene knock-outs and transgenes in mice that result in specific phenotypes associated with mammary gland; (ii) cattle QTL for milk production and mastitis traits; (iii) loci with sequence variations that show specific allele-phenotype interactions associated with milk production or mastitis in cattle; (iv) genes with expression profiles associated with milk production or mastitis in cattle or mouse; (v) cattle milk protein genes that exist in different genetic variants; (vi) miRNAs expressed in bovine mammary gland; (vii) epigenetically regulated cattle genes associated with mammary gland function.
Data mining and description of the database
We reviewed the literature published up to December 2008 searching for the relevant publications through PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and Web of Science (http://isiknowledge.com) using key phrases: genetics, gene candidates, mammary gland, miRNA, mastitis, milk, epigenetics, methylation, QTL, SNP, association. The data from animal experiments were retrieved from the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org) using the phenotype ontology terms listed in Table S1, representing ontology terms revealed by the literature review.
Quantitaive trait loci were extracted from Cattle QTL Database Release 7 (1/2009): http://www.animalgenome.org using ontology terms associated with mastitis [SCS, somatic cell count (SCC), clinical mastitis (CM)] and milk traits [MY, milking speed (MSPD), dairy capacity composite index (DCCI), protein yield (PY), protein percentage (PP), protein content (PC), energy yield (EY), fat percentage (FP), fat yield (FY), fat content (FC)]. Candidate genes from expression experiments for QTL for milk production traits in cattle were retrieved from cgQTL database (http://cowry.agri.huji.ac.il/QTLMAP/qtlmap.htm).
Putative target sites for mammary gland expressed miRNAs in candidate genes were obtained using Sanger’s mirBase Targets – Version 5 (http://microrna.sanger.ac.uk/). Ensembl transcript identifiers for candidate genes were obtained from Ensembl database – Release 52 (http://www.ensembl.org/) and matched to the list of identifiers with putative miRNA target sites for miRNAs experimentally confirmed in the mammary gland. Polymorphisms in bovine miRNA target octamers of candidate genes were obtained from the Patrocles database (http://www.patrocles.org/).
Candidate genes identified in multiple studies (using the same or different approaches) were considered as the most promising candidates and were analysed for expression level in lactating mammary gland using GNF BioGPS (http://biogps.gnf.org), considering mouse expression data (data for Bos taurus are not available yet). Gene variation data of the most promising candidate genes in the promoter region (5 kb), 5′UTR, exon, intron (100 bp flanking sequence) and 3′UTR were obtained from Ensembl database (http://www.ensembl.org/). The ingenuity pathway analysis program (http://www.ingenuity.com) was used to cluster the most promising candidate genes in functional networks.
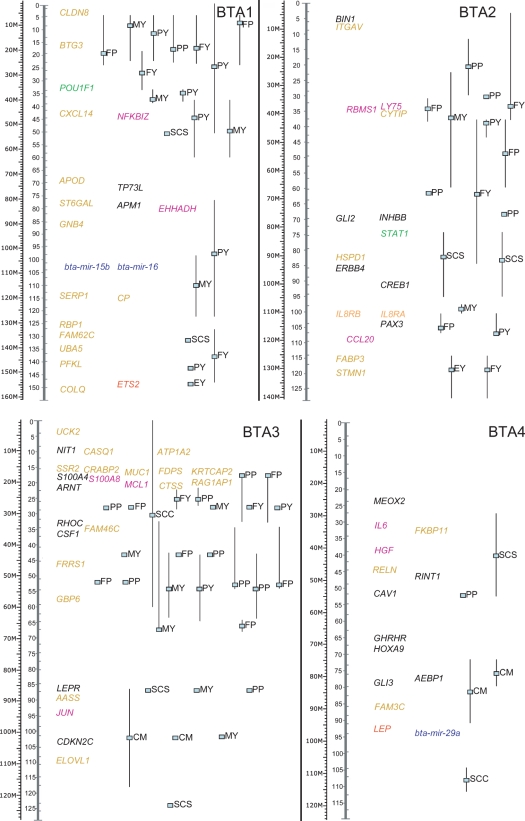
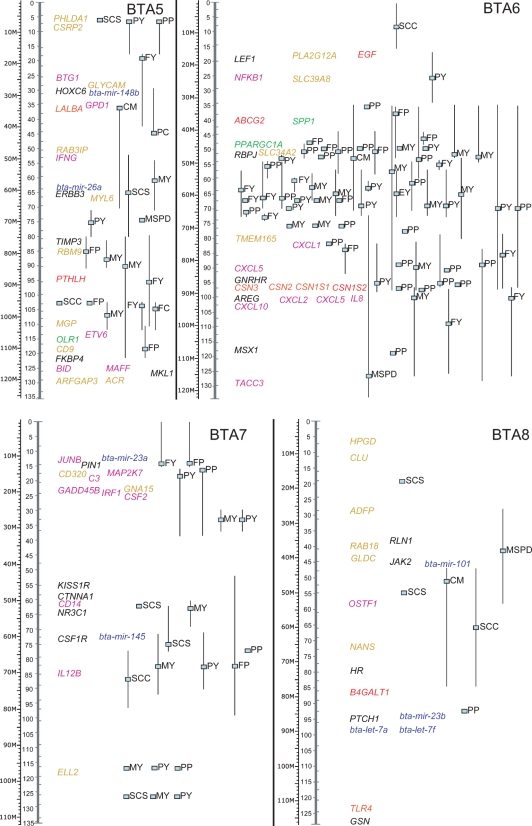
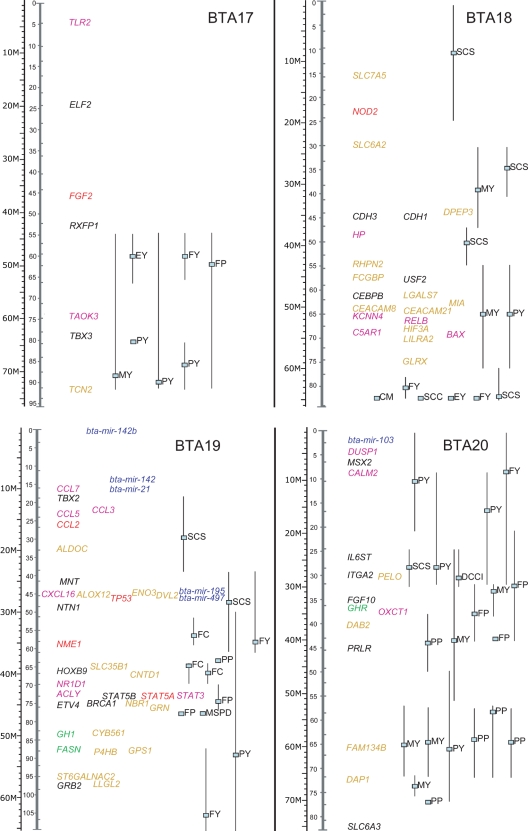
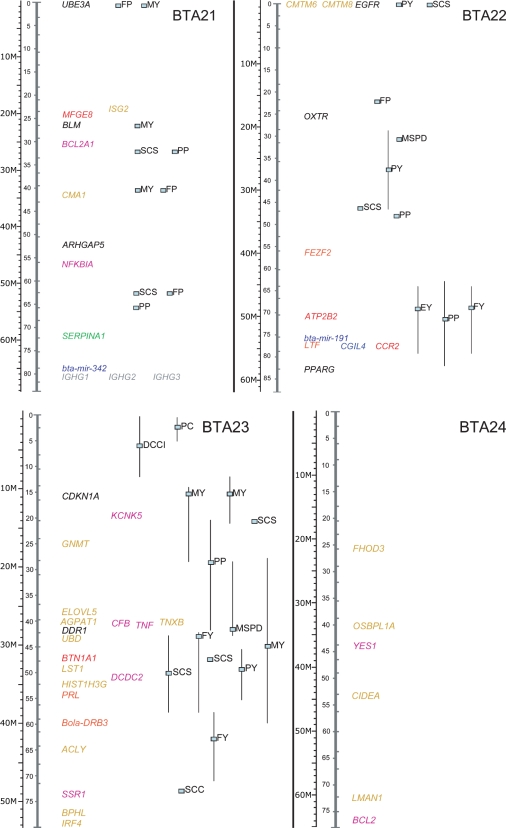
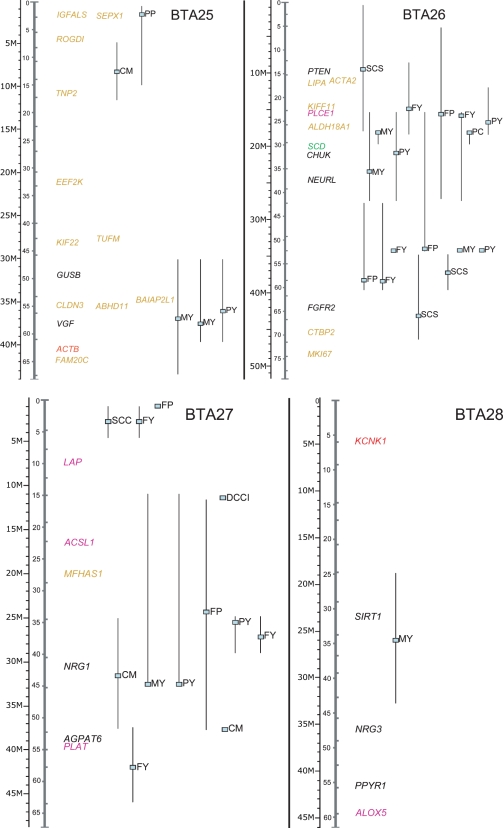
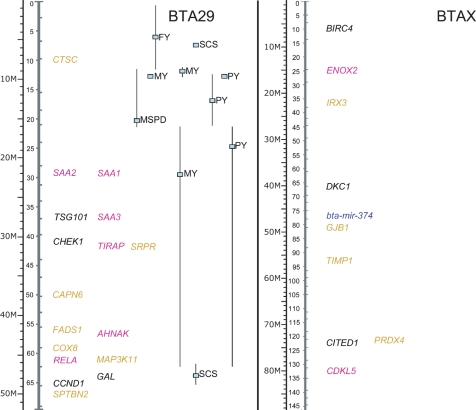
Our database was created in the Excel format and is available on-line: http://www.bfro.uni-lj.si/Kat_genet/genetika/mammary_gland.xls. Each gene from the mouse KO and gene transfer experiments is hyperlinked to phenotypic allele details in MGI database. Each QTL is hyperlinked to details in Cattle QTL database. The miRNAs are hyperlinked to details in the Sanger miRBase (http://microrna.sanger.ac.uk/) for miRNAs available in the database. Each gene from expression and association studies is hyperlinked to the Map Viewer –Bos taurus build (4.0) on NCBI (http://www.ncbi.nlm.nih.gov) or to MGI’s gene details, in cases when gene position for cattle was not available in the Map Viewer. Selected candidate genes and genomic loci were drawn on the genetic marker map.
Defining the map locations of the loci
The map location was retrieved from NCBI database Bos taurus build (4.0). If the map location was not available, we identified the location of the locus using the bovine–human synteny map. The bovine–human synteny map was constructed through BLASTing 8294 markers from MARC and RH maps (Everts-van der Wind et al. 2004; Itoh et al. 2005) with bovine contigs to obtain hits (defined as E < 10−19) with longer sequences. Hits were further BLASTed against the human genome; 6231 putative human bovine orthologs were found. Positions on the human physical map were obtained using Map Viewer on NCBI. The syntheny map was constructed using 6023 orthologs sorted in 213 blocks of synteny. Each synteny block with at least two markers (singletons were excluded) is described by its position on the physical human map and on the bovine cytogenetic map.
Results
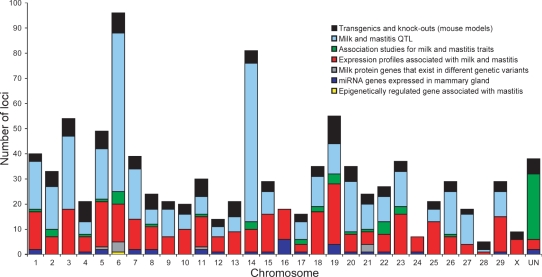
Genes, QTL, SNPs, AFLP markers and miRNAs representing 934 cattle loci involved in mammary gland development, milk production traits and resistance or susceptibility to mastitis were retrieved from different sources. The results are presented in the form of a genetic marker map (Fig. 1). The collected data include genetic as well as epigenetic background for mammary gland related traits. The database shows putative mammary gland related candidate loci on all chromosomes except on chromosome Y, with the highest number of candidate loci on chromosomes 6, 14 and 19 and the lowest on the chromosomes 28, 24, and X (Fig. 2). The Ingenuity Pathway Analysis identified that among the 44 candidate genes confirmed in multiple studies, 12 loci are involved in inflamatory response and antigen presentation and 10 loci are involved in development and function of connective tissue, muscle development and function as well as development and function of endocrine system. Eight loci are involved in cell mediated immune response and structure and development of lymphoid tissue and the other eight are involved in cellular development, movement and cancer. Three loci were associated with organ morphology, development of reproductive system and amino acid metabolism (Table 2). However, three genes could not be associated with physiological function using Ingenuity Pathway Analysis due to specificies cattle genome (LGB, BoLA-DRB3 and CSN1S2).
Figure 1.
Genetic map of cattle candidate genes and genetic markers for milk production and mastitis. The map includes mouse transgenic and knock-out experiments, QTL for milk and mastitis traits, genes and genetic markers tested for association with milk and mastitis traits, genes with expression patterns associated with milk and mastitis traits, milk protein genes that exist in different genetic variants, miRNAs expressed in mammary gland, and epigenetically regulated gene associated with mammary gland phenotype. The ruler to the extreme left of each figure represents mega-base pairs. The ruler next to the mega-base pairs scale represents distances in centimorgans. Loci are placed at approximate positions on both the sequence and the linkage map. Chromosomes are not drawn to scale. Legend: Transgenics and knock-outs (mouse models). 

Figure 2.
Number of candidate genes and genetic markers for mammary gland development, milk production traits and resistance or susceptibility to mastitis found with different approaches by chromosome.
Table 2.
The most promising candidate genes found by matching data sets from independent studies and their in silico analysis (level of expression in lactating mouse mammary gland, genetic variability in cattle and their functions).
| List of the most promising candidate genes and number of independent studies by approach |
In silico analysis |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Association studies |
Expression studies |
Number of SNPs |
|||||||||||||
| Gene | Gene name | Mouse KOs and transgenic experiments | Milk traits | Mastitis traits | Milk traits1 | Mastitis traits | Milk protein genetic variants | Epigenetic studies | Expression in lactating mammary gland2 (mouse) | Promoter (5 kb upstream) | 5′UTR | Exons3 | Introns (context 100 bp) | 3′UTR | Functions4 |
| Associated with milk production | |||||||||||||||
| ABCG2 | ATP-binding cassette, sub-family G (WHITE), member 2 | + + + | + | *** | 0 | 0 | 1 | 1 | 0 | E | |||||
| ATP2B2 | ATPase, Ca++ transporting, plasma membrane 2 | + | + | * | 1 | 0 | 1 | 3 | 0 | D | |||||
| B4GALT1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 | + | + | ** | 0 | 0 | 0 | 1 | 0 | A | |||||
| BTN1A1 | Butyrophilin, subfamily 1, member A1 | + | + | *** | 0 | 0 | 3 (2) | 0 | 0 | C | |||||
| CSN1S1 | Casein alpha s1 | + + + | + | + | + | *** | 0 | 0 | 1 (1) | 0 | 0 | C | |||
| CSN1S2 | Casein alpha s2 | + | + | NA | 0 | 0 | 0 | 0 | 0 | NA | |||||
| CSN2 | Casein beta | + | + | *** | 0 | 0 | 3 (3) | 0 | 0 | B | |||||
| CSN3 | Casein kappa | + | + + + | + | + | *** | 0 | 0 | 0 | 3 | 0 | B | |||
| DGAT1 | Diacylglycerol O-acyltransferase 1 | + | + + + + + + + | ** | NA | NA | NA | NA | NA | A | |||||
| EGF | Epidermal growth factor (beta urogastrone) | + | + | NA | 0 | 0 | 0 | 0 | 0 | B | |||||
| GHR | Growth hormone receptor | + + + + | ** | 0 | 0 | 0 | 0 | 0 | B | ||||||
| ID2 | Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | + | + | * | 0 | 0 | 0 | 1 | 0 | B | |||||
| LALBA | Lactalbumin, alpha | + | + | + | *** | 0 | 0 | 0 | 0 | 1 | A | ||||
| LEP | Leptin | + | + + + | + | ** | 0 | 0 | 8 (3) | 4 | 8 | B | ||||
| LGB | Lactoglobulin, beta | + + | + | NA | 0 | 1 | 1 | 0 | 0 | NA | |||||
| MFGE8 | Milk fat globule-EGF factor 8 protein | + | + | *** | 0 | 0 | 0 | 0 | 1 | A | |||||
| NME1 | Non-metastatic cells 1, protein (NM23A) expressed in | + | + | NA | 0 | 0 | 3 (2) | 0 | 1 | D | |||||
| PRL | Prolactin | + | + + | NA | 0 | 0 | 0 | 0 | 0 | B | |||||
| PTHLH | Parathyroid hormone-like peptide | + | + | ** | 0 | 0 | 0 | 0 | 0 | C | |||||
| STAT5A | Signal transducer and activator of transcription 5A | + | + + | ** | 0 | 0 | 0 | 0 | 0 | B | |||||
| XDH | Xanthine dehydrogenase | + | + | *** | 0 | 0 | 2 (1) | 1 | 0 | C | |||||
| Associated with mastitis resistance | |||||||||||||||
| ACTB | Actin, beta, cytoplasmic | + | + | * | 0 | 0 | 0 | 0 | 1 | D | |||||
| C5AR1 | Complement component 5a receptor 1 | + + | ** | 0 | 0 | 1 | 0 | 0 | B | ||||||
| CD14 | CD14 antigen | + + + + | *** | 0 | 1 | 1 | 0 | 0 | A | ||||||
| ETS2 | E26 avian leukaemia oncogene 2, 3′ domain | + | + | * | 0 | 0 | 0 | 1 | 1 | C | |||||
| FEZF2 | fez family zinc finger 2 | + | + | ** | 0 | 0 | 1 (1) | 1 | 0 | E | |||||
| IFNG | Interferon gamma | + + + | ** | 0 | 0 | 0 | 1 | 2 | D | ||||||
| IL1B | Interleukin 1 beta | + + + + | ** | 0 | 0 | 2 | 0 | 7 | D | ||||||
| IL6 | Interleukin 6 | + + | ** | 0 | 0 | 1 | 1 | 1 | B | ||||||
| IL8 | Interleukin 8 | + + + | NA | 0 | 0 | 0 | 5 | 7 | C | ||||||
| IL8RA | Interleukin 8 receptor, alpha | + + | ** | 0 | 0 | 0 | 0 | 0 | C | ||||||
| LBP | Lipopolysaccharide binding protein | + + | *** | 2 | 0 | 5 (2) | 0 | 0 | A | ||||||
| PTGS1 | Prostaglandin-endoperoxide synthase 1 | + | + | ** | 0 | 0 | 0 | 0 | 0 | A | |||||
| SAA3 | Serum amyloid A3 | + + | ** | 0 | 0 | 0 | 0 | 0 | A | ||||||
| TLR-2 | Toll-like receptor 2 | + + | ** | 0 | 0 | 3 (2) | 0 | 0 | A | ||||||
| TLR-4 | Toll-like receptor 4 | + + | + + | ** | 0 | 1 | 28 (8) | 4 | 2 | A | |||||
| TNF | Tumor necrosis factor | + + + + | ** | 0 | 0 | 0 | 0 | 0 | A | ||||||
| Associated with milk production and mastitis resistance | |||||||||||||||
| ACLY | ATP citrate lyase | + | + | ** | 0 | 0 | 2 | 3 | 0 | D | |||||
| BoLA-DRB3 | Major histocompatibility complex, class II, DRB3 | + + + | + + + | NA | 1 | 0 | 5 (5) | 0 | 0 | NA | |||||
| CCL2 | Chemokine (C-C motif) ligand 20 | + | + | * | 1 | 0 | 2 (2) | 1 | 0 | C | |||||
| KCNK1 | Potassium channel, subfamily K, member 1 | + | + | *** | 1 | 0 | 3 (1) | 1 | 0 | E | |||||
| LTF | Lactoferrin | + + | + | + | + | ** | 0 | 2 | 4 | 1 | 0 | A | |||
| RORA | RAR-related orphan receptor alpha | + | + | ** | 0 | 0 | 0 | 0 | 0 | D | |||||
| TP53 | Transformation related protein 53 | + | + | + | * | 0 | 0 | 1 | 1 | 0 | D | ||||
+, independent study.
Candidates suggested by Ron et al. (2007) on the basis of three independent expression experiments.
Mouse mammary gland expression score: *below median, **median to 10× median, ***above 10× median.
Number in brackets represents number of non-synonymous coding SNPs.
Top functions: A – inflammatory disease, inflammatory response, antigen presentation, B – connective tissue development and function, skeletal and muscular system development and function, endocrine system development and function, C – cancer, cellular movement and cellular development, D – cell-mediated immune response, lymphoid tissue structure and development, cancer, E – organ morphology, reproductive system development and function and amino acid metabolism.
Transgenics and knock-outs
Because of its numerous advantages (large amount of mutations, efficient techniques for targeted mutagenesis, precisely described phenotype changes), the mouse model has been used as a tool for identification of phenotype-genotype relationships. The availability of the complete mouse genome sequence allows comparisons with other species and identification of conserved regions (Guenet 2005). Currently, there are 143 genes that, when mutated or expressed as transgenes in mouse, result in phenotypes associated with mammary gland (Table S1).
Milk and mastitis QTL
There are 344 QTL associated with milk traits in cattle (MY, MSPD, DCCI, PY, PP, EY, FP) and 71 mastitis related traits (CM, SCS and SCC) available in AnimalQTL database. QTL are positioned on all chromosomes except on BTA16, BTA24 and BTAX. The reason why milk and mastitis QTL are spread over such a number of chromosomes might be in the numerous genetic and environmental factors that contribute to animal’s phenotype, including different traits and specific host-pathogen interactions. The highest density of QTL associated with milk traits was found on BTA6 and BTA14 and the highest density of mastitis related QTL on BTA3 and BTA14.
Association studies
SNPs associated with mastitis
Allele-phenotype association studies were performed for milk (MY, milk protein, PP, milk fat and FP) and mastitis (CM and SCS) traits. Association between DNA sequence variation and mammary gland phenotype has been demonstrated for twenty-four candidate genes (Sharif et al. 1999; Grisart et al. 2002; Blott et al. 2003;Kuss et al. 2003; Prinzenberg et al. 2003; Brym et al. 2004, 2005; Cohen-Zinder et al. 2005; Khatib et al. 2005; Kuss et al. 2005; Liefers et al. 2005;Leonard et al. 2005; Weikard et al. 2005; Zhou et al. 2005; Cobanoglu et al. 2006; do Nascimento et al. 2006; He et al. 2006; Kaminski et al. 2006; Khatib et al. 2006; Ron et al. 2006; Sanders et al. 2006; Kaupe et al. 2007; Leyva-Baca et al. 2007; Morris et al. 2007; Olsen et al. 2007; Pant et al. 2007; Robitaille et al. 2007;Rupp et al. 2007; Anton et al. 2008; Banos et al. 2008; Chebel et al. 2008; Hradecka et al. 2008; Ganai et al. 2009; Huang et al. 2008; Kaminski et al. 2008; Khatib et al. 2008; Macciotta et al. 2008; Wang et al. 2008). The association between DNA sequence variation and mastitis resistance or susceptibility has been found for ten candidate genes (Sharif et al. 1998; Youngerman et al. 2004;do Nascimento et al. 2006; Sharma et al. 2006b; Sugimoto et al. 2006; Wojdak-Maksymiec et al. 2006; Kaupe et al. 2007; Leyva-Baca et al. 2007; Pant et al. 2007; Rambeaud & Pighetti 2007; Rupp et al. 2007; Wang et al. 2007; Leyva-Baca et al. 2008) (Table S2). The evidence for the association of 11 genes (ABCG2, BoLA-DRB3, CSN1S1, CSN3, DGAT1, GHR, LGB, LEP, LTF, PRL and STAT5A) with mammary gland phenotype and three genes (IL8RA, TLR4 and BoLA-DRB3) with mastitis resistance or susceptibility has been reported more than once in different studies.
AFLP markers associated with mastitis
Genome screening for QTL is usually costly and highly laborious. Xiao et al. (2007) presented a simplified, inexpensive QTL mapping approach by integration of AFLP markers, DNA pooling and bioinformatics tools. Similarly, Sharma et al. (2006a) searched for genome-wide QTL-linked AFLP markers for mastitis resistance in Canadian Holsteins. Cows were screened by selective DNA pooling and AFLP technique. Twenty-seven AFLP markers associated with CM were found and the most promising marker named CGIL4 was then further characterized and mapped to BTA22 q24. However, due to their dominant character, the AFLPs are less informative than SNPs, which have become widely used with the progress of genome sequencing.
Expression profiles associated with milk production and mastitis
The high throughput technologies such as microarray analysis offer the possibility of studying changes in expression profiles of thousands of genes, in response to infection with a pathogen, simultaneously. Although microarray analysis has become an important tool in animal genomics, there is still the major problem that no clear consensus about the microarray data processing methods for detection of differentially expressed genes exists (Jaffrezic et al. 2007). Candidate genes with expression patterns associated with milk production in cattle were identified by Ron et al. (2007) by combining their mouse mammary gland gene expression experiments with two other expression experiments (Clarkson et al. 2004; Stein et al. 2004) using comparative mapping. The results are available as a web tool for candidate genes for QTL (cgQTL database). To date, twelve publications describing 107 genes with expression patterns associated with mastitis cases in cattle using microarrays (Pareek et al. 2005; Sugimoto et al. 2006; Zheng et al. 2006), real-time PCR (Long et al. 2001; Lee et al. 2003; Pfaffl et al. 2003; Schwerin et al. 2003; Goldammer et al. 2004; Swanson et al. 2004) and ELISA (Bannerman et al. 2004a,b; Lee et al. 2006) have been published (Table S3). The studies were performed in cattle and mouse using pathogens Streptococcus uberis, Streptococcus agalactiae, coliforms (i.e. Escherichia coli, Klebsiella pneumoniae), Staphylococcus spp. (i.e. aureus), Cornybacterium spp., and yeast. Differential expression of eleven genes (IL6, IL8, CD14, TLR4, IL1B, LBP, TLR2, C5AR1, TNF, IFNG and SAA3) during mastitis was confirmed in more than one (two to four) expression experiment, moreover, six genes (IL6, CD14, TLR4, IL1B, TLR2 and SAA3) were found to be differentially expressed in two species (cattle and mouse).
Milk protein genes
Farrell et al. (2004) reported 14 major proteins in bovine milk. Milk protein genes exist in different genetic variants that encode proteins that are slightly different chemically. Numerous investigatiors have focused on the association between certain genetic variants of milk proteins and yield traits, milk composition and technological properties of milk (Buchberger & Dovc 2000). However, the allele-specific effects are very much dependant on genetic background (breed) and experimental model (single locus vs. multi locus effects). Currently there are milk protein variants known for nine milk protein families in bovine milk (Table S4), but only a few of them affect milk traits significantly.
miRNA genes expressed in mammary gland
miRNAs are a new class of regulatory molecules and could also be involved in the regulation of gene expression in the mammary gland. To date, 32 miRNA genes have been reported to be expressed in the bovine mammary gland (Gu et al. 2007). Some of these miRNAs are located in overlapping regions with QTL for milk and/or mastitis traits (Table S5). Recently, several genes have been proven to be regulated via miRNAs, but so far none of them in mammary gland-related traits.
For miRNAs, experimentally proven to be expressed in mammary gland, we performed in silico searches for target sites and found 359 putative miRNA target sites in candidate genes. Using the Patrocles database, we found polymorphic miRNA target sites for bta-miR-199b, -miR-199a-5p, and -miR-361 in the IL1B gene and for –miR-126 in the CYP11B1 gene. Interestingly, the expression of -miR-199b, -miR-199a-5p and –miR-126 in the bovine mammary gland has already been experimentally confirmed.
Epigenetic factors
Epigenetic factors have also been demonstrated to be involved in CM (Vanselow et al. 2006). Principally, DNA-remethylation around the STAT5-binding enhancer in the CSN1S1 promoter was shown to be associated with shutdown of αS1-casein synthesis during acute mastitis. Interestingly, defensin genes BNBD5 and LAP are regulated in an opposite manner to the CSN1S1 promoter (Vanselow et al. 2006), which was also found to be associated with milk traits.
Discussion
The extensive literature and database search for mammary gland associated candidate genes and genome loci have been performed. We reviewed 934 loci involved in mammary gland development, milk production traits and resistance or susceptibility to mastitis in cattle (Table 1).
Table 1.
Summary of the data in the database of cattle candidate genes and genetic markers for mammary gland development, milk production traits and resistance or susceptibility to mastitis.
| Study approach | Number of loci |
|---|---|
| Knock-out and transgenic experiments | 143 |
| QTL | 415 |
| Association studies – milk traits | 24 |
| Association studies – mastitis | 10 |
| AFLP markers associated with mastitis | 27 |
| Expression studies – milk traits | 207 |
| Expression studies – mastitis | 107 |
| Milk protein genes that exist in different genetic variants | 9 |
| miRNAs expressed in mammary gland | 32 |
| Epigenetic factors | 1 |
| Total | 934* |
Unique loci (studies reporting individual gene more than once by different approaches were subtracted from sum).
Our criteria for inclusion of candidate regions into the database were association of the genetic marker with the animal trait, which mainly revealed functional candidates, and sequence similarity revealing structural candidates and map position (overlap with QTL of interest), which allowed identification of positional candidates.
As the data extracted from different sources are often fragmented and controversial, there is an urgent need to integrate information from different sources. Our database consists of cattle candidate loci for mammary gland development, milk production traits and resistance or susceptibility to mastitis comprising 934 loci. The database is available in Excel format and allows searching for loci by name, approach, reference and chromosomal location. The loci in the database are hyperlinked to the relevant public databases (NCBI, MGI, CattleQTLdb, and miRBase). Human and mouse homologs are available for all cattle genes, which represents an important advantage for a comparative approach. In cases when locations for bovine orthologs were unavailable on NCBI’s Map Viewer –Bos taurus build (4.0), we defined approximate locations of cattle orthologs by using a bovine-human synteny map. The cattle mammary gland database will serve as a source of candidates for functional studies and development of markers for the new generation of animal breeding tools.
Candidate loci were drawn to the genetic marker map (Fig. 1). The advantage of the map-based review is the identification of overlapping regions populated with candidate loci found by different approaches. A review of the genetic map approach has been previously published for obesity-related loci (Rankinen et al. 2006) and was heavily used as an important research tool in obesity studies.
We found 44 candidate genes identified in multiple independent studies using the same or different approaches, of which 22 were associated with milk production, 16 with mastitis and six with both (Table 2). Genes identified with multiple approaches or in multiple analyses using the same approach and/or in regions overlapping with QTL represent promising candidate genes for association with mammary gland development, lactation and resistance or susceptibility to mastitis.
The most promising candidates were further analysed in silico (Table 2). A search for expression in lactating mammary gland and polymorphisms (SNPs) in different regions (promoter, 5′UTR, exon, intron and 3′UTR) was performed. To date, there are 159 SNPs reported in 44 of the most promising candidate genes. We found 82 SNPs in exons, 34 in introns (100 bp flanking region), 32 in 3′UTRs, six in promoters and five in 5′UTRs of selected genes. The most polymorphic gene was TLR4, with 35 reported SNPs. Genes with a high number of reported polymorphisms were also LEP (20 SNPs), IL8 (12), IL1B (9) and LTF (7). Additionally, the pathway analysis was used to cluster the genes into five functional networks involved in a variety of biological functions.
Twenty-six genes (ABCG2, ACLY, ACTB, ATP2B2, B4GALT1, BoLA-DRB3, BTN1A1, CCL2, CSN1S2, CSN2, DGAT1, EGF, ETS2, FEZF2, ID2, KCNK1, MFGE8, NME1, LGB, PRL, PTGS1, PTHLH, RORA, STAT5A, TLR4 and XDH) were found to be associated with mammary gland phenotypes (milk and mastitis traits) using two different study approaches, LALBA, LEP, TP53 using three different approaches, and CSN3, CSN1S1 and LTF using four different approaches. Twenty-five genes were confirmed in multiple independent studies using the same approach; eleven of them in association studies for milk traits (ABCG2, BoLA-DRB3, CSN1S1, CSN3, DGAT1, GHR, LEP, LGB, LTF, PRL, and STAT5A), three in association studies for mastitis traits (BoLA-DRB3, IL8RA and TLR4) and 11 in mastitis expression experiments (C5AR1, CD14, IFNG, IL1B: IL6, IL8, LBP, SAA3, TLR2, TLR4 and TNF). Genes ABCG2, BoLA-DRB3, CSN1S1, CSN3, LEP, LTF and TLR4 were reported by at least two different approaches and confirmed in at least two independent studies for each approach. None of the miRNA genes overlapped with candidate gene locations, but 10 of them overlapped with QTL regions for different traits (Table S5). Moreover, we performed miRNA target searches in the collected candidate genes and found 359 putative target sites, of which two genes (IL1B and CYP11B) included polymorphic targets for miRNAs expressed in mammary gland. Those miRNA:mRNA pairs can now be experimentally tested for their possible involvement in the regulation of gene expression in the mammary gland.
The highest density of QTL was found on BTA6 and BTA14. As suggested by Khatkar et al. (2004), there are two distinct QTL regions on BTA6 at 49 ± 5.0 cM and 87 ± 7.9 cM. Genes PPARGC1 (Weikard et al. 2005), SPP1 (Leonard et al. 2005) and ABCG (Cohen-Zinder et al. 2005; Ron et al. 2006; Olsen et al. 2007), found in association studies for milk traits, are located in proximity of 49 ± 5.0 cM QTL region. In 87 ± 7.9 cM region, which overlaps with several PP QTL, casein genes (CSN1S1, CSN1S2, CSN2 and CSN3) are located. Khatkar et al. (2004) detected a genome-wide significant QTL for milk FP and yield close to the centromeric end of BTA14 where the DGAT1 gene is located. The DGAT1 gene that overlaps with several milk fat QTL was found in association studies for milk fat (Grisart et al. 2002; Kaminski et al. 2006; Kaupe et al. 2007, Anton et al. 2008, Banos et al. 2008; Hradecka et al. 2008; Kaminski et al. 2008) and in murine KO experiments, which resulted in the absence of milk production. As concluded by Grisart et al. (2002), the DGAT1 gene which is involved in triglyceride synthesis is the causative gene affecting milk fat on BTA14. Genes LEP (Liefers et al. 2005; Banos et al. 2008; Chebel et al. 2008), PRL (Brym et al. 2005; He et al. 2006), CSN3 (Kaminski et al. 2006, 2008; Robitaille et al. 2007), DGAT1 (Grisart et al. 2002; Kaminski et al. 2006; Kaupe et al. 2007; Anton et al. 2008; Banos et al. 2008; Hradecka et al. 2008; Kaminski et al. 2008) and STAT5A (Brym et al. 2004; Khatib et al. 2008) were reported in association studies for milk traits and in murine KO experiments.
The LTF gene on BTA22 was found in mastitis expression experiments (Pfaffl et al. 2003), association studies for milk phenotypes (Kaminski et al. 2006, 2008) and association studies for mastitis resistance or susceptibility (Wojdak-Maksymiec et al. 2006). Lactoferrin (LF), with its strong iron binding properties, is known to have several biological functions including host defence against microbial infection and anti-inflammatory activity. The multifunctional roles of LTF were reviewed by Ward et al. (2005). The finding that inflammation and involution of the mammary gland induces mammary expression of LF led to the suggestion that the LTF gene is a strong functional candidate for mastitis resistance or susceptibility (Kerr & Wellnitz 2003). As reported by Wojdak-Maksymiec et al. (2006), two alleles of LTF, A and B, were found in the studied population. The highest SCC was found in milk of the AB genotype, whereas the lowest one was found in cows of the AA genotype. The TLR4 gene on BTA8 was found in mastitis association and expression studies. Its differential expression was confirmed in two different experiments (Goldammer et al. 2004; Zheng et al. 2006) and association of its sequence polymorphisms with mastitis traits was found in two different studies (Sharma et al. 2006b; Wang et al. 2007). Therefore, TLR4 may be a strong candidate for functional studies to enhance mastitis resistance in cattle. The expression of the FEZF2 gene on BTA22 has been reported to be induced by mastitis and its sequence variation is associated with mastitis resistance or susceptibility (Sugimoto et al. 2006); cows susceptible to mastitis have a three-base insertion in a glycine-coding stretch of the gene. Sequence variation of the BTA23-located BoLA-DRB3 gene has also been reported to be associated with milk traits and mastitis resistance or susceptibility (Sharif et al. 1999; do Nascimento et al. 2006; Rupp et al. 2007). As suggested by do Nascimento et al. (2006), this might be due to a direct action of bovine major histocompatibility complex alleles on immune function, whereas effects on production traits might be only indirect and explained by better general health conditions of more productive animals. PTGS1 (Pfaffl et al. 2003), ACTB (Lee et al. 2006), TP53 (Schwerin et al. 2003) and ETS2 (Zheng et al. 2006) genes were found in mastitis expression studies and murine KO experiments that resulted in increased tumorigenesis of mammary gland and abnormal lactation. To address the developmental-specific expression profiles in the mammary gland, a new specialized microarray containing about 6000 highly enriched unique sequences from mouse mammary libraries (mammochip) has been developed and applied for expression profiling of the mouse mammary gland during development (Miyoshi et al. 2002). Comparison of gene expression in the wild type lactating and virgin mammary gland and in KO for the inhibitor of differentiation 2 (Id2) gene revealed four distinct groups of genes showing different expression profiles.
Major lactoprotein genes (CSN1S1, CSN1S2, CSN2, CSN3, LALBA and LGB) exist in different genetic variants that code for chemically different protein variants. The genetic variants of milk proteins have diverse effects on milk composition and cheese making ability. It is possible that effects on milk composition and cheese making ability are not the direct consequence of polymorphisms at lactoprotein gene loci but rather the consequence of QTL linked to the different genetic variants of these genes. Besides protein variant studies, milk protein genes have been also identified by other study approaches: (i) CSN1S1 in an epigenetic study (Vanselow et al. 2006), expression experiments for milk traits (Ron et al. 2007), and in association studies for milk traits (Prinzenberg et al. 2003; Kuss et al. 2005; Sanders et al. 2006), (ii) CSN2 in knock-out experiments in mice which resulted in abnormal lactation and abnormal milk composition, (iii) CSN3 gene in association studies for milk traits (Kaminski et al. 2006; Robitaille et al. 2007), expression experiments for milk traits (Ron et al. 2007) and in KO experiments in mice which resulted in abnormal lactation and abnormal milk composition, (iv) LALBA in KO experiments in mice which resulted in abnormal mammary gland morphology and in abnormal milk composition, expression experiments for milk traits (Ron et al. 2007) and (v) a LGB in association study for milk traits (Kuss et al. 2003).
Epigenetic modifications to the DNA sequence and associated chromatin are also known to regulate gene expression and contribute significantly to the phenotype. Variation in the epigenotype between genetically identical individuals can be associated with phenotypic differences. Moreover, the recent evidence suggests that the epigenome can be affected by environmental factors and that these changes can last a lifetime (Whitelaw & Whitelaw 2006). The CSN1S1 gene has been reported to be epigenetically regulated during mastitis (Vanselow et al. 2006).
Some of the genes which we identified on the cross-cut between different approaches or which were reported in multiple independent studies using the same approach were already identified and verified to affect QTL in cattle (i.e. ABCG2 and DGAT1), while others represent background for subsequent functional studies. Possible criterion to determine priority for further candidate gene analysis can be differential expression of the gene in the target organ, known physiological role to the trait (Ron et al. 2007) and positional overlapping with QTL of interest. The current database of cattle candidate genes and genetic markers for mammary gland development, milk production traits and resistance or susceptibility to mastitis consists of 934 unique loci. The project is ongoing and we plan to update the database periodically with further publications.
Acknowledgments
This work was supported by the Slovenian Research Agency (ARRS) (programme P4-0220, J.O. is financed through ARRS) and by the EU FP6 Network of Excellence EADGENE.
Supporting information
Additional supporting information may be found in the online version of this article.
Table S1 Transgenic and knockout murine models related to mammary gland phenotypes.
Table S2 Genes tested for association with milk and mastitis traits.
Table S3 Genes with expression patterns associated with mastitis.
Table S4 Milk protein genes and their variants (adapted from Farrell et al. 2004 and Buchberger & Dovc 2000).
Table S5 miRNAs expressed in mammary gland (adapted from Gu et al. 2007).
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Anton I, Kovacs K, Fesus L, Varhegyi J, Lehel L, Hajda Z, Polgar JP, Szabo F, Zsolnai A. Effect of DGAT1 and TG gene polymorphisms on intramuscular fat on milk production traits in different cattle breeds in Ilungary. Acta Veterinaria Hungarica. 2008;56:181–6. doi: 10.1556/AVet.56.2008.2.5. [DOI] [PubMed] [Google Scholar]
- Bannerman DD, Paape MJ, Hare WR, Hope JC. Characterization of the bovine innate immune response to intramammary infection with Klebsiella pneumoniae. Journal of Dairy Science. 2004a;87:2420–32. doi: 10.3168/jds.S0022-0302(04)73365-2. [DOI] [PubMed] [Google Scholar]
- Bannerman DD, Paape MJ, Lee JW, Zhao X, Hope JC, Rainard P. Escherichia coli and Staphylococcus aureus elicit differential innate immune responses following intramammary infection. Clinical and Diagnostic Laboratory Immunology. 2004b;11:463–72. doi: 10.1128/CDLI.11.3.463-472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banos G, Woolliams JA, Woodward BW, Forbes AB, Coffey MP. Impact of single nucleotide polymorphisms in leptin, leptin receptor, growth hormone receptor, and diacylglycerol acyltransferase (DGAT1) gene loci on milk production, feed, and body energy traits of UK dairy cows. Journal of Dairy Science. 2008;91:3190–200. doi: 10.3168/jds.2007-0930. [DOI] [PubMed] [Google Scholar]
- Blott S, Kim JJ, Moisio S, et al. Molecular dissection of a quantitative trait locus: A phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics. 2003;163:253–66. doi: 10.1093/genetics/163.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brym P, Kaminski S, Rusc A. New SSCP polymorphism within bovine STAT5A gene and its associations with milk performance traits in Black-and-White and Jersey cattle. Journal of Applied Genetics. 2004;45:445–52. [PubMed] [Google Scholar]
- Brym P, Kaminski S, Wojcik E. Nucleotide sequence polymorphism within exon 4 of the bovine prolactin gene and its associations with milk performance traits. Journal of Applied Genetics. 2005;46:179–85. [PubMed] [Google Scholar]
- Buchberger J, Dovc P. Lactoprotein genetic variants in cattle and cheese making ability. Food Technology and Biotechnology. 2000;38:91–8. [Google Scholar]
- Chebel RC, Susca F, Santos JEP. Leptin genotype is associated with lactation performance and health of Holstein cows. Journal of Dairy Science. 2008;91:2893–900. doi: 10.3168/jds.2007-0891. [DOI] [PubMed] [Google Scholar]
- Clarkson RWE, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Research. 2004;6:R92–109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobanoglu O, Zaitoun I, Chang YM, Shook GE, Khatib H. Effects of the signal transducer and activator of transcription 1 (STAT1) gene on milk production traits in Holstein dairy cattle. Journal of Dairy Science. 2006;89:4433–7. doi: 10.3168/jds.S0022-0302(06)72491-2. [DOI] [PubMed] [Google Scholar]
- Cohen-Zinder M, Seroussi E, Larkin DM, et al. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Research. 2005;15:936–44. doi: 10.1101/gr.3806705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts-van der Wind A, Kata SR, Band MR, et al. A 1463 gene cattle-human comparative map with anchor points defined by human genome sequence coordinates. Genome Research. 2004;14:1424–37. doi: 10.1101/gr.2554404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell HM, Jr, Jimenez-Flores R, Bleck GT, Brown EM, Butler JE, Creamer LK, Hicks CL, Hollar CM, Ng-Kwai-Hang KF, Swaisgood HE. Nomenclature of the proteins of cows’ milk--sixth revision. Journal of Dairy Science. 2004;87:1641–74. doi: 10.3168/jds.S0022-0302(04)73319-6. [DOI] [PubMed] [Google Scholar]
- Ganai NA, Bovenhuis H, van Arendonk JA, Visker MH. Novel polymorphisms in the bovine beta-lactoglobulin gene and their effects on beta-lactoglobulin protein concentration in milk. Animal Genetics. 2009;40:127–33. doi: 10.1111/j.1365-2052.2008.01806.x. [DOI] [PubMed] [Google Scholar]
- Goldammer T, Zerbe H, Molenaar A, Schuberth HJ, Brunner RM, Kata SR, Seyfert HM. Mastitis increases mammary mRNA abundance of beta-defensin 5, toll-like-receptor 2 (TLR2), and TLR4 but not TLR9 in cattle. Clinical and Diagnostic Laboratory Immunology. 2004;11:174–85. doi: 10.1128/CDLI.11.1.174-185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisart B, Coppieters W, Farnir F, et al. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Research. 2002;12:222–31. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eleswarapu S, Jiang H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Letters. 2007;581:981–8. doi: 10.1016/j.febslet.2007.01.081. [DOI] [PubMed] [Google Scholar]
- Guenet JL. The mouse genome. Genome Research. 2005;15:1729–40. doi: 10.1101/gr.3728305. [DOI] [PubMed] [Google Scholar]
- He F, Sun DX, Yu Y, Wang YC, Zhang Y. Association between SNPs within prolactin gene and milk performance traits in Holstein dairy cattle. Asian-Australasian Journal of Animal Sciences. 2006;19:1384–9. [Google Scholar]
- Hradecka E, Citek J, Panicke L, Rehout V, Hanusova L. The relation of GH1, GHR and DGAT1 polymorphisms with estimated breeding values for milk production traits of German Holstein sires. Czech Journal of Animal Science. 2008;53:238–45. [Google Scholar]
- Huang W, Maltecca C, Khatib H. A proline-to-histidine mutation in POU1F1 is associated with production traits in dairy cattle. Animal Genetics. 2008;39:554–7. doi: 10.1111/j.1365-2052.2008.01749.x. [DOI] [PubMed] [Google Scholar]
- Itoh T, Watanabe T, Ihara N, Mariani P, Beattie CW, Sugimoto Y, Takasuga A. A comprehensive radiation hybrid map of the bovine genome comprising 5593 loci. Genomics. 2005;85:413–24. doi: 10.1016/j.ygeno.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Jaffrezic F, De Koning DJ, Boettcher PJ, et al. Analysis of the real EADGENE data set: Comparison of methods and guidelines for data normalisation and selection of differentially expressed genes (Open Access publication) Genetics Selection Evolution. 2007;39:633–50. doi: 10.1186/1297-9686-39-6-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski S, Brym P, Rusc A, Wojcik E, Ahman A, Magi R. Associations between milk performance traits in Holstein cows and 16 candidate SNPs identified by arrayed primer extension (APEX) microarray. Animal Biotechnology. 2006;17:1–11. doi: 10.1080/08941920500460906. [DOI] [PubMed] [Google Scholar]
- Kaminski S, Malewski T, Ahman A, Wojcik E, Rusc A, Olenski K, Jakubczak A, Sazanov AA. Towards an integrated approach to study SNPs and expression of candidate genes associated with milk protein biosynthesis. Russian Journal of Genetics. 2008;44:459–65. [PubMed] [Google Scholar]
- Kaupe B, Brandt H, Prinzenberg EM, Erhardt G. Joint analysis of the influence of CYP11B1 and DGAT1 genetic variation on milk production, somatic cell score, conformation, reproduction, and productive lifespan in German Holstein cattle. Journal of Animal Science. 2007;85:11–21. doi: 10.2527/jas.2005-753. [DOI] [PubMed] [Google Scholar]
- Kerr DE, Wellnitz O. Mammary expression of new genes to combat mastitis. Journal of Animal Science. 2003;81(Suppl 3):38–47. doi: 10.2527/2003.81suppl_338x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib H, Heifetz E, Dekkers JC. Association of the protease inhibitor gene with production traits in Holstein dairy cattle. Journal of Dairy Science. 2005;88:1208–13. doi: 10.3168/jds.S0022-0302(05)72787-9. [DOI] [PubMed] [Google Scholar]
- Khatib H, Leonard SD, Schutzkus V, Luo W, Chang YM. Association of the OLR1 gene with milk composition in Holstein dairy cattle. Journal of Dairy Science. 2006;89:1753–60. doi: 10.3168/jds.S0022-0302(06)72243-3. [DOI] [PubMed] [Google Scholar]
- Khatib H, Monson RL, Schutzkus V, Kohl DM, Rosa GJM, Rutledge JJ. Mutations in the STAT5A gene are associated with embryonic survival and milk composition in cattle. Journal of Dairy Science. 2008;91:784–93. doi: 10.3168/jds.2007-0669. [DOI] [PubMed] [Google Scholar]
- Khatkar MS, Thomson PC, Tammen I, Raadsma HW. Quantitative trait loci mapping in dairy cattle: review and meta-analysis. Genetics Selection Evolution. 2004;36:163–90. doi: 10.1186/1297-9686-36-2-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss AW, Gogol J, Geidermann H. Associations of a polymorphic AP-2 binding site in the 5’-flanking region of the bovine beta-lactoglobulin gene with milk proteins. Journal of Dairy Science. 2003;86:2213–8. doi: 10.3168/jds.s0022-0302(03)73811-9. [DOI] [PubMed] [Google Scholar]
- Kuss AW, Gogol J, Bartenschlager H, Geldermann H. Polymorphic AP-1 binding site in bovine CSN1S1 shows quantitative differences in protein binding associated with milk protein expression. Journal of Dairy Science. 2005;88:2246–52. doi: 10.3168/jds.S0022-0302(05)72900-3. [DOI] [PubMed] [Google Scholar]
- Lee JW, Paape MJ, Elsasser TH, Zhao X. Elevated milk soluble CD14 in bovine mammary glands challenged with Escherichia coli lipopolysaccharide. Journal of Dairy Science. 2003;86:2382–9. doi: 10.3168/jds.S0022-0302(03)73832-6. [DOI] [PubMed] [Google Scholar]
- Lee JW, Bannerman DD, Paape MJ, Huang MK, Zhao X. Characterization of cytokine expression in milk somatic cells during intramammary infections with Escherichia coli or Staphylococcus aureus by real-time PCR. Veterinary Research. 2006;37:219–29. doi: 10.1051/vetres:2005051. [DOI] [PubMed] [Google Scholar]
- Leonard S, Khatib H, Schutzkus V, Chang YM, Maltecca C. Effects of the osteopontin gene variants on milk production traits in dairy cattle. Journal of Dairy Science. 2005;88:4083–6. doi: 10.3168/jds.S0022-0302(05)73092-7. [DOI] [PubMed] [Google Scholar]
- Leyva-Baca I, Schenkel F, Sharma BS, Jansen GB, Karrow NA. Identification of single nucleotide polymorphisms in the bovine CCL2, IL8, CCR2 and IL8RA genes and their association with health and production in Canadian Holsteins. Animal Genetics. 2007;38:198–202. doi: 10.1111/j.1365-2052.2007.01588.x. [DOI] [PubMed] [Google Scholar]
- Leyva-Baca I, Schenkel F, Martin J, Karrow NA. Polymorphisms in the 5’ upstream region of the CXCR1 chemokine receptor gene, and their association with somatic cell score in Holstein cattle in Canada. Journal of Dairy Science. 2008;91:407–17. doi: 10.3168/jds.2007-0142. [DOI] [PubMed] [Google Scholar]
- Liefers SC, Veerkamp RF, Te Pas MF, Chilliard Y, Van der Lende T. Genetics and physiology of leptin in periparturient dairy cows. Domestic Animal Endocrinology. 2005;29:227–38. doi: 10.1016/j.domaniend.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Long E, Capuco AV, Wood DL, Sonstegard T, Tomita G, Paape MJ, Zhao X. Escherichia coli induces apoptosis and proliferation of mammary cells. Cell Death and Differentiation. 2001;8:808–16. doi: 10.1038/sj.cdd.4400878. [DOI] [PubMed] [Google Scholar]
- Macciotta NPP, Mele M, Conte G, Serra A, Cassandro M, Dal Zotto R, Borlino AC, Pagnacco G, Secchiari P. Association between a polymorphism at the stearoyl CoA desaturase locus and milk production traits in Italian Holsteins. Journal of Dairy Science. 2008;91:3184–9. doi: 10.3168/jds.2007-0947. [DOI] [PubMed] [Google Scholar]
- Mackay TF. The genetic architecture of quantitative traits. Annual Review of Genetics. 2004;35:303–39. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Meyer B, Gruss P, et al. Mammary epithelial cells are not able to undergo pregnancy-dependent differentiation in the absence of the helix-loop-helix inhibitor Id2. Molecular Endocrinology. 2002;16:2892–901. doi: 10.1210/me.2002-0128. [DOI] [PubMed] [Google Scholar]
- Morris CA, Cullen NG, Glass BC, Hyndman DL, Manley TR, Hickey SM, McEwan JC, Pitchford WS, Bottema CD, Lee MA. Fatty acid synthase effects on bovine adipose fat and milk fat. Mammalian Genome. 2007;18:64–74. doi: 10.1007/s00335-006-0102-y. [DOI] [PubMed] [Google Scholar]
- do Nascimento CS, Machado MA, Martinez ML, et al. Association of the bovine major histocompatibility complex (BoLA) BoLA-DRB3 gene with fat and protein production and somatic cell score in Brazilian Gyr dairy cattle (Bos indicus) Genetics and Molecular Biology. 2006;29:641–7. [Google Scholar]
- Olsen HG, Nilsen H, Hayes B, Berg PR, Svendsen M, Lien S, Meuwissen T. Genetic support for a quantitative trait nucleotide in the ABCG2 gene affecting milk composition of dairy cattle. BMC Genetics. 2007;8:32. doi: 10.1186/1471-2156-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant SD, Schenkel FS, Leyva-Baca I, Sharma BS, Karrow NA. Identification of single nucleotide polymorphisms in bovine CARD15 and their associations with health and production traits in Canadian Holsteins. BMC Genomics. 2007;8:421. doi: 10.1186/1471-2164-8-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek R, Wellnitz O, Van Dorp R, Burton J, Kerr D. Immunorelevant gene expression in LPS-challenged bovine mammary epithelial cells. Journal of Applied Genetics. 2005;46:171–7. [PubMed] [Google Scholar]
- Pfaffl MW, Wittmann SL, Meyer HH, Bruckmaier RM. Gene expression of immunologically important factors in blood cells, milk cells, and mammary tissue of cows. Journal of Dairy Science. 2003;86:538–45. doi: 10.3168/jds.S0022-0302(03)73632-7. [DOI] [PubMed] [Google Scholar]
- Prinzenberg EM, Weimann C, Brandt H, Bennewitz J, Kalm E, Schwerin M, Erhardt G. Polymorphism of the bovine CSN1S1 promoter: Linkage mapping, intragenic haplotypes, and effects on milk production traits. Journal of Dairy Science. 2003;86:2696–705. doi: 10.3168/jds.S0022-0302(03)73865-X. [DOI] [PubMed] [Google Scholar]
- Rambeaud M, Pighetti GM. Differential calcium signaling in dairy cows with specific CXCR1 genotypes potentially related to interleukin-8 receptor functionality. Immunogenetics. 2007;59:53–8. doi: 10.1007/s00251-006-0170-x. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Robitaille G, Britten M, Methot S, Petitclerc D. Polymorphisms within the 5′-flanking region of bovine K-casein gene (CSN3) and milk production-related traits. Milchwissenschaft-Milk Science International. 2007;62:243–5. [Google Scholar]
- Ron M, Weller JI. From QTL to QTN identification in livestock - winning by points rather than knock-out: a review. Animal Genetics. 2007;38:429–39. doi: 10.1111/j.1365-2052.2007.01640.x. [DOI] [PubMed] [Google Scholar]
- Ron M, Cohen-Zinder M, Peter C, Weller JI, Erhardt G. Short communication: a polymorphism in ABCG2 in Bos indicus and Bos taurus cattle breeds. Journal of Dairy Science. 2006;89:4921–3. doi: 10.3168/jds.S0022-0302(06)72542-5. [DOI] [PubMed] [Google Scholar]
- Ron M, Israeli G, Seroussi E, Weller JI, Gregg JP, Shani M, Medrano JF. Combining mouse mammary gland gene expression and comparative mapping for the identification of candidate genes for QTL of milk production traits in cattle. BMC Genomics. 2007;8:183. doi: 10.1186/1471-2164-8-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp R, Hernandez A, Mallard BA. Association of bovine leukocyte antigen (BoLA) DRB3.2 with immune response, mastitis, and production and type traits in Canadian Holsteins. Journal of Dairy Science. 2007;90:1029–38. doi: 10.3168/jds.S0022-0302(07)71589-8. [DOI] [PubMed] [Google Scholar]
- Sanders K, Bennewitz J, Reinsch N, Thaller G, Prinzenberg EM, Kuhn C, Kalm E. Characterization of the DGAT1 mutations and the CSN1S1 promoter in the German Angeln dairy cattle population. Journal of Dairy Science. 2006;89:3164–74. doi: 10.3168/jds.S0022-0302(06)72590-5. [DOI] [PubMed] [Google Scholar]
- Schadt EE. Novel integrative genomics strategies to identify genes for complex traits. Animal Genetics. 2006;37:18–23. doi: 10.1111/j.1365-2052.2006.01473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerin M, Czernek-Schafer D, Goldammer T, Kata SR, Womack JE, Pareek R, Pareek C, Walawski K, Brunner RM. Application of disease-associated differentially expressed genes--mining for functional candidate genes for mastitis resistance in cattle. Genetics Selection Evolution. 2003;35(Suppl 1):S19–34. doi: 10.1186/1297-9686-35-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S, Mallard BA, Wilkie BN, Sargeant JM, Scott HM, Dekkers JC, Leslie KE. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with occurrence of disease and milk somatic cell score in Canadian dairy cattle. Animal Genetics. 1998;29:185–93. doi: 10.1046/j.1365-2052.1998.00318.x. [DOI] [PubMed] [Google Scholar]
- Sharif S, Mallard BA, Wilkie BN, Sargeant JM, Scott HM, Dekkers JC, Leslie KE. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) with production traits in Canadian dairy cattle. Animal Genetics. 1999;30:157–60. doi: 10.1046/j.1365-2052.1999.00459.x. [DOI] [PubMed] [Google Scholar]
- Sharma BS, Jansen GB, Karrow NA, Kelton D, Jiang Z. Detection and characterization of amplified fragment length polymorphism markers for clinical mastitis in Canadian Holsteins. Journal of Dairy Science. 2006a;89:3653–63. doi: 10.3168/jds.S0022-0302(06)72405-5. [DOI] [PubMed] [Google Scholar]
- Sharma BS, Leyva I, Schenkel F, Karrow NA. Association of toll-like receptor 4 polymorphisms with somatic cell score and lactation persistency in Holstein bulls. Journal of Dairy Science. 2006b;89:3626–35. doi: 10.3168/jds.S0022-0302(06)72402-X. [DOI] [PubMed] [Google Scholar]
- Shook GE. Major advances in determining appropriate selection goals. Journal of Dairy Science. 2006;89:1349–61. doi: 10.3168/jds.S0022-0302(06)72202-0. [DOI] [PubMed] [Google Scholar]
- Silveri L, Tilly G, Vilotte JL, Le Provost F. MicroRNA involvement in mammary gland development and breast cancer. Reproduction Nutrition Development. 2006;46:549–56. doi: 10.1051/rnd:2006026. [DOI] [PubMed] [Google Scholar]
- Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy MA, Heath VJ, Bell AK, Ferrier RK, Sandilands GP, Gusterson BA. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Research. 2004;6:R75–91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Fujikawa A, Womack JE, Sugimoto Y. Evidence that bovine forebrain embryonic zinc finger-like gene influences immune response associated with mastitis resistance. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6454–9. doi: 10.1073/pnas.0601015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K, Gorodetsky S, Good L, Davis S, Musgrave D, Stelwagen K, Farr V, Molenaar A. Expression of a beta-defensin mRNA, lingual antimicrobial peptide, in bovine mammary epithelial tissue is induced by mastitis. Infection and Immunity. 2004;72:7311–4. doi: 10.1128/IAI.72.12.7311-7314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanselow J, Yang W, Herrmann J, Zerbe H, Schuberth HJ, Petzl W, Tomek W, Seyfert HM. DNA-remethylation around a STAT5-binding enhancer in the alphaS1-casein promoter is associated with abrupt shutdown of alphaS1-casein synthesis during acute mastitis. Journal of Molecular Endocrinology. 2006;37:463–77. doi: 10.1677/jme.1.02131. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu S, Gao X, Ren H, Chen J. Genetic polymorphism of TLR4 gene and correlation with mastitis in cattle. Journal of Genetics and Genomics. 2007;34:406–12. doi: 10.1016/S1673-8527(07)60044-7. [DOI] [PubMed] [Google Scholar]
- Wang X, Maltecca C, Tal-Stein R, Lipkin E, Khatib H. Association of bovine fibroblast growth factor 2 (FGF2) gene with milk fat and productive life: an example of the ability of the candidate pathway strategy to identify quantitative trait genes. Journal of Dairy Science. 2008;91:2475–80. doi: 10.3168/jds.2007-0877. [DOI] [PubMed] [Google Scholar]
- Ward PP, Paz E, Conneely OM. Multifunctional roles of lactoferrin: a critical overview. Cellular and Molecular Life Sciences. 2005;62:2540–8. doi: 10.1007/s00018-005-5369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikard R, Kuhn C, Goldammer T, Freyer G, Schwerin M. The bovine PPARGC1A gene: molecular characterization and association of a SNP with variation of milk fat synthesis. Physiological Genomics. 2005;21:1–13. doi: 10.1152/physiolgenomics.00103.2004. [DOI] [PubMed] [Google Scholar]
- Whitelaw NC, Whitelaw E. How lifetimes shape epigenotype within and across generations. Human Molecular Genetics. 2006;15:R131–7. doi: 10.1093/hmg/ddl200. Spec No 2. [DOI] [PubMed] [Google Scholar]
- Wojdak-Maksymiec K, Kmiec M, Ziemak J. Associations between bovine lactoferrin gene polymorphism and somatic cell count in milk. Veterinarni Medicina. 2006;51:14–20. doi: 10.1111/j.1439-0442.2006.00899.x. [DOI] [PubMed] [Google Scholar]
- Xiao QJ, Wibowo TA, Wu XL, Michal JJ, Reeves JJ, Busboom JR, Thorgaard GH, Jiang ZH. A simplified QTL mapping approach for screening and mapping of novel AFLP markers associated with beef marbling. Journal of Biotechnology. 2007;127:177–87. doi: 10.1016/j.jbiotec.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Youngerman SM, Saxton AM, Oliver SP, Pighetti GM. Association of CXCR2 polymorphisms with subclinical and clinical mastitis in dairy cattle. Journal of Dairy Science. 2004;87:2442–8. doi: 10.3168/jds.S0022-0302(04)73367-6. [DOI] [PubMed] [Google Scholar]
- Zheng J, Watson AD, Kerr DE. Genome-wide expression analysis of lipopolysaccharide-induced mastitis in a mouse model. Infection and Immunity. 2006;74:1907–15. doi: 10.1128/IAI.74.3.1907-1915.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GL, Liu HG, Liu C, Guo SL, Zhu Q, Wu YH. Association of genetic polymorphism in GH gene with milk production traits in Beijing Holstein cows. Journal of Biosciences. 2005;30:595–8. doi: 10.1007/BF02703558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.