Abstract
Ecotropic viral integration site 1 (EVI1) plays important roles in leukaemia and development, and its expression is temporally and spatially highly restricted during the latter process. Nevertheless, the only physiological agent that to date has been shown to regulate transcription of this gene in mammalian cells is all-trans retinoic acid. Here we describe the identification of a retinoic acid response element that was located in the most distal of several alternative first exons of the human EVI1 gene and was constitutively bound by canonical retinoid receptors in NTERA-2 teratocarcinoma cells. Furthermore, it was the target of negative feedback by EVI1 on the induction of its own promoter by retinoic acid. This process required a previously described transcription repression domain of EVI1. Extending its role as a modulator of the retinoic acid response, EVI1 had the opposite effect on the RARβ retinoic acid response element, whose induction by all-trans retinoic acid it enhanced through a mechanism that involved almost all of its known functional domains. Augmentation of the retinoic acid response by EVI1 was also observed for the endogenous RARβ gene. Thus, we have established EVI1 as a novel type of modulator of the retinoic acid response, which can both enhance and repress induction by this agent in a promoter-specific manner.
Keywords: all-trans retinoic acid, EVI1, feedback inhibition, RARE, transcription regulation
Introduction
Ecotropic viral integration site 1 (EVI1) is an evolutionarily conserved gene with important roles both in malignant diseases and in normal development [1]. Its homozygous disruption in mice led to multiple malformations, including small or missing limb buds, defects in the development of the central and peripheral nervous systems and an immature heart. Evi1−/− mice died before 11.5 days postcoitum due to heart failure and massive haemorrhaging [2]. EVI1 is thought to exert its biological effects mainly by acting as a regulator of gene transcription [1]. It codes for a 1051 amino acid nuclear protein with two sets of zinc finger domains that contain seven and three zinc finger motifs, respectively, are able to bind to DNA independently of each other, and are separated from each other by an intervening region (IR) and a transcription repression domain (RD). At the C-terminus of EVI1, an acidic region (AR) involved in transcription activation is present [1]. EVI1 interacts with transcriptional coactivators and corepressors, and is able to both activate and repress gene transcription [3–7]. However, so far only very few direct EVI1 target genes have been described [7,8] and EVI1 may exert its effects in part by modifying the activity of other sequence-specific transcription factors [9–12].
The human EVI1 gene has several different first exons, whose alternative use gives rise to the transcript variants EVI1_1a, EVI1_1b, EVI1_1c, EVI1_1d, and EVI1_3L (Fig. 1A) [1,13–15]. The transcriptional start sites and the putative regulatory regions of these mRNA variants are located in close vicinity to each other, and they are all predicted to be translated into the same 1051 amino acid protein [14]. In contrast, the mRNA variant MDS1/EVI1 gives rise to an EVI1 protein with an extension of 188 amino acids at its N-terminus [16,17], which, at least in some experimental systems, acted differently from, and even in an opposite manner to, the 1051 amino acid EVI1 protein [3,18–20]. The MDS1/EVI1 transcript consists of parts of the mRNA from the MDS1 gene, joined to the second exon of the EVI1 mRNA [16,17]. The first exon of MDS1, and therefore also its presumptive regulatory regions, are located more than 500 kb upstream of EVI1 exon 1a. Despite this, the expression patterns of MDS1/EVI1 resembled those of the other EVI1 mRNA variants in almost all investigated human and murine tissues [14,17].
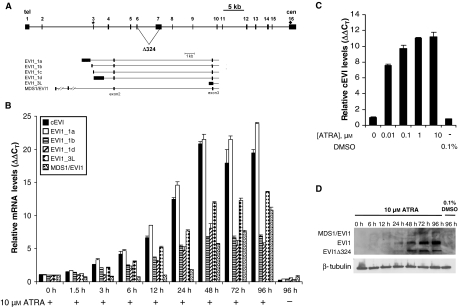
Fig. 1.
Induction of different EVI1 mRNA and protein variants in response to ATRA in NTERA-2 cells. (A) Schematic of the human EVI1 gene and its mRNA splice and 5′-end variants. Boxes, exons; lines, introns. The upper panel shows the entire EVI1 gene, with alternative splicing producing the Δ324 mRNA variant indicated by triangular lines. Asterisk and diamond, positions of the start codon in exon 3 and of the stop codon in exon 16, respectively. In the lower panel, only the 5′-end of the gene with several alternative first exons is shown. The alternative first exon contained within EVI1_1a is exon 1a, etc. Reproduced with permission from [1]. (B) NTERA-2 cells were treated with 10 μm ATRA or an equivalent amount of dimethylsulfoxide for the indicated periods of time. RNA was extracted and reverse transcribed, and the mRNA levels of cEVI1 (i.e. the sum of all EVI1 transcripts, amplified using forward and reverse primers located in exons 7 and 8/9, respectively; black columns), EVI1_1a (white columns), EVI1_1b (horizontally striped columns), EVI1_1d (diagonally striped columns), EVI1_3L (checked columns) and MDS1/EVI1 (stippled columns) were assayed by RTQ-RT-PCR. The expression of each EVI1 mRNA variant relative to the housekeeping gene cyclophilinD and the 0 h time point, as well as standard deviations between replicate measurements, were calculated using the ΔΔCT method [45]. Please note that because the ΔΔCT method relates expression values to the 0 h time point for each amplicon, only induction factors, but not expression levels, can be compared between different amplicons. (C) NTERA-2 cells were treated with the indicated amounts of ATRA or dimethylsulfoxide for 15 h. RTQ-RT-PCR for cEVI1 was performed as described above. (D) NTERA-2 cells were treated with 10 μm ATRA or an equivalent amount of dimethylsulfoxide for the indicated periods of time. Proteins were extracted and subjected to immunoblot analysis using an antibody against EVI1, or a β-tubulin antibody as a loading control. The EVI1 antibody detected three bands whose molecular masses corresponded to those of MDS1/EVI1 (∼ 170 kDa), EVI1 (∼ 140 kDa) and of the protein product of an internal splice variant of EVI1, Δ324 (∼ 100 kDa).
Although EVI1 was expressed in a temporally and spatially highly restricted manner during mammalian development [2,21], the only physiological agent that to date has been identified as potentially contributing to this regulation is all-trans retinoic acid (ATRA) [14,22,23]. Interestingly, mice with a homozygous disruption of the retinaldehyde dehydrogenase-2 gene, which is responsible for embryonic retinoic acid (RA) synthesis, die on the same day of embryonic development as Evi1−/− mice, with a partially overlapping phenotype [2,24]. Like EVI1 [2,22], ATRA also regulates many aspects of neuronal differentiation [25]. Together, these observations suggest that EVI1 may be involved in mediating some of the biological effects of ATRA.
RA regulates gene expression by binding to nuclear receptors that directly act as transcription factors at the promoters of target genes. Two classes of retinoid receptors have been described: retinoic acid receptors (RARs) and retinoid X receptors (RXRs). Each receptor class has three paralogous members in human cells, designated α, β and γ [26,27]. Gene regulation by RA is usually mediated by heterodimers between one member of each of the RAR and the RXR families. These bind to retinoic acid response elements (RAREs) that consist of two direct repeats (DR) of the consensus sequence PuG(G/T)TCA, separated by one, two or, most commonly, five spacer nucleotides, and accordingly termed DR1, DR2 or DR5 elements. RAR and RXR bind to their cognate response elements in a constitutive manner. In the absence of RA, they repress target gene transcription, whereas ligand binding activates target promoters by inducing conformational changes in the receptors that lead to replacement of corepressors by coactivators [26,27]. Numerous genes have been reported to be regulated by retinoid receptors in a direct manner [28]. In addition, RARs and RXRs have featured both synergistic and antagonistic interactions with other sequence-specific transcription factors [26].
EVI1 was induced by ATRA in several human and murine cell lines, and this induction was at least in part due to an increased rate of transcription [14,22,23]. Nevertheless, so far no RARE has been found in the regulatory regions of the EVI1 gene [22]. Here we describe the identification of a functional RARE that is located in exon 1a of the human EVI1 gene, that binds to retinoid receptors in a constitutive manner, and whose induction by ATRA is inhibited by EVI1. Furthermore, we show that EVI1 augments the induction of the RARβ gene by ATRA.
Results
Induction of different EVI1 mRNA and protein variants by ATRA in NTERA-2 cells
To characterize the regulation of the EVI1 gene by ATRA, NTERA-2 cells were incubated with 10 μm of this agent for various periods of time, and the expression of EVI1 and its mRNA 5′-end variants (except for EVI1_1c, whose levels are too low for reliable detection [13,14]) was measured by real-time quantitative RT-PCR (RTQ-RT-PCR). EVI1_1a, EVI1_1b, EVI1_1d and EVI1_3L (Fig. 1A), as well as the cEVI1 amplicon (which extends from the 3′-end of exon 7 to the 5′-end of exon 9, and thus represents all EVI1 transcript variants, Fig. 1A) were induced by ATRA to varying extents, but with similar kinetics: their levels were elevated as early as 3 h after the addition of ATRA, continued to rise until after 48 h, and then remained constant until at least 96 h after ATRA addition (Fig. 1B). The MDS1/EVI1 mRNA, in contrast, began to slightly increase in abundance 24 h after the addition of ATRA, was distinctly induced at 48 h and further accumulated up to the 96 h time point (Fig. 1B).
In a dose–response experiment, cEVI1 was strongly induced by as little as 10 nm ATRA and reached maximal levels at 1 μm ATRA (Fig. 1C).
Immunoblot analyses of extracts prepared from NTERA-2 cells treated with 10 μm ATRA for various periods of time revealed three bands specifically recognized by an EVI1 antibody (Fig. 1D). The molecular masses of these bands were consistent with those reported for MDS1/EVI1 (∼ 170 kDa), EVI1 (∼ 140 kDa) and the protein product of an alternatively spliced EVI1 mRNA, Δ324 (∼ 100 kDa) [16,29]. The 140 kDa band, which gave the strongest signal, began to appear 12–24 h after the addition of ATRA and increased in intensity up to the 96 h time point. The 100 kDa band was considerably fainter, but exhibited similar induction kinetics. The 170 kDa band was the faintest, and was detectable only at the 72 and 96 h time points (Fig. 1D). The immunoblot and RTQ-RT-PCR analyses consistently showed that EVI1 was induced strongly and rapidly upon incubation of NTERA-2 cells with ATRA, whereas MDS1/EVI1 was upregulated with delayed kinetics.
An inverted DR5 RARE in EVI1 exon 1a confers ATRA responsiveness to the EVI1 promoter and binds canonical retinoid receptors in a constitutive manner
To search for a RARE in the putative regulatory regions of the human EVI1 gene, three fragments that together covered most of the genomic region from positions −2.6 to +3.9 kb relative to the 5′-end of EVI1 exon 1a were cloned into the luciferase reporter vectors pGL3basic and pGL3promoter (Fig. 2A). Only EVI1(+15/+1106)/pGL3, whose insert largely corresponds to EVI1 exon 1a, but not EVI1(−2629/−72)/pGL3 or EVI1(+1258/+3903)/pGL3, responded to ATRA in reporter gene assays (Fig. 2A, and data not shown). Because pGL3basic- and pGL3promoter-based constructs yielded comparable results, subsequent experiments were performed only with pGL3basic. Deletion of the first 240 nucleotides of EVI1(+15/+1106)/pGL3 completely eliminated the ATRA response (Fig. 2B), suggesting that it was mediated by the inverted DR5 RARE consensus sequence CGACCTTTTTGTGACCT present between nucleotide positions +182 and +198 relative to the beginning of exon 1a. Indeed, EVI1(+86/+274)/pGL3 was still responsive to ATRA. The importance of the RARE consensus sequence in conferring ATRA responsiveness to the EVI1 promoter was corroborated through mutation of the six residues comprising its second half-site, which yielded EVI1(+86/+1106)mut/pGL3. This construct did not exhibit any ATRA response (Fig. 2B). However, its basal activity was elevated as compared with that of the corresponding wild-type construct (Fig. 2B), probably due to the loss of repression by unliganded RARs and RXRs [27].
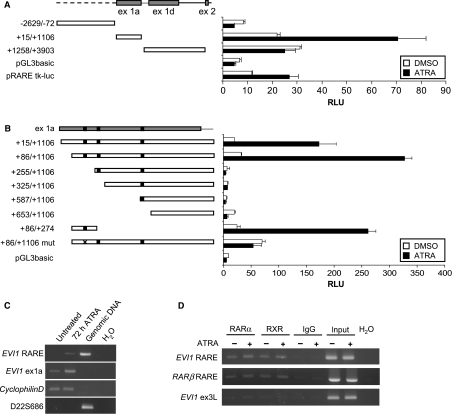
Fig. 2.
A functional RARE is located in exon 1a of the human EVI1 gene. (A) Schematic of the genomic region at the 5′-end of the human EVI1 gene and of the fragments cloned into pGL3basic, and luciferase reporter assays. Stippled line, EVI1 upstream region; grey boxes, exons (exons 1b and 1c are too small to be depicted in this schematic); solid lines, introns. The positions of cloned fragments are indicated relative to the transcriptional start site of EVI1_1a (GenBank accession no. BX640908). pRARE-tk-luc contains two copies of the human RARβ RARE and the tk minimal promoter in pGL2, and was used as a positive control for the ATRA response. For luciferase assays, the indicated reporter plasmids were transfected into NTERA-2 cells, together with the renilla luciferase plasmid pRL; 10 μm ATRA or an equivalent amount of dimethylsulfoxide was added 1 day after transfection. Another ∼ 24 h later, cells were lysed and luciferase activities were measured. Relative luciferase units (RLU) were derived by normalizing firefly to renilla luciferase activities. Error bars represent the standard deviations between duplicate measurements. White bars, dimethylsulfoxide; black bars, ATRA. (B) Schematic of EVI1(+15/+1106)/pGL3 and its derivative constructs, and luciferase reporter assays. Grey box, exon 1a; black boxes, predicted RAREs. In EVI1(+86/+1106)mut/pGL3, the six nucelotides comprising the second half-site of the EVI1 RARE were mutated (indicated by a cross). Luciferase reporter gene assays were performed as in (A). (C) RT-PCR confirming that the RARE of the EVI1 gene is located within its transcribed region. NTERA-2 cells were incubated with ATRA for 0 or 72 h. RNA was extracted, treated with DNaseI, reverse transcribed and amplified using primers flanking the EVI1 RARE (EVI1_RARE-F, EVI1_RARE-R; Table S1B), an intron-spanning primer pair located in a more proximal region of EVI1_1a (EVI1_1a-fwd, EVI1_1a-rev; Table S1A) and a primer pair for the housekeeping gene cyclophilinD (cycD-F, cycD-R; Table S1A). The effectiveness of the DNaseI treatment was verified with primers for the microsatellite marker D22S686 (D22S686-F, D22S686-R; Table S1B). (D) ChIP was performed on NTERA-2 cells treated with ATRA or dimethylsulfoxide for 24 h using an antibody specific to RARα, a pan-RXR antibody, or unspecific rabbit IgG as a negative control. Immunoprecipitated chromatin and input DNA as a positive control were amplified with primers flanking the EVI1 RARE (EVI1_RARE-F, EVI1_RARE-R; Table S1B), primers for the RARβ RARE (RARβ_RARE-F, RARβ_RARE-R; Table S1B) or with negative control primers located in EVI1 exon 3L (EVI1_ex3L-F; EVI1_ex3L-R; Table S1B).
To confirm that the EVI1 RARE was located within the transcribed region of the EVI1 gene, RT-PCR with primers surrounding the RARE (Table S1B) was performed on DNAse-treated RNA from NTERA-2 cells that had been incubated with ATRA or vehicle for 72 h. The RARE amplicon was elevated in response to ATRA (Fig. 2C), demonstrating that ATRA-induced transcription of the EVI1 gene indeed initiates in part or entirely upstream of its RARE.
Chromatin immunoprecipitation (ChIP) experiments revealed that RARα and RXR were associated with the EVI1 RARE both in the absence and presence of ATRA (Fig. 2D). Confirming the specificity of these interactions, only faint bands were observed when negative control immunoprecipitates or primers were used (Fig. 2D). Similar results were obtained for the RARE of the RARβ gene promoter, which was employed as a positive control (Fig. 2D). In summary, the first exon of the EVI1 gene contains a consensus RARE motif that is bound by canonical RARs and RXRs, which repress it in the absence and activate it in the presence of ligand.
ATRA regulates the MDS1/EVI1 promoter neither directly, nor through EVI1
Because ATRA induced the MDS1/EVI1 mRNA only slowly (Fig. 1B), its action on the MDS1/EVI1 promoter seemed unlikely to be direct. Indeed, none of four overlapping fragments, which together covered the genomic region from position −3.0 to +2.5 kb relative to the transcriptional start site of MDS1/EVI1, conferred responsiveness to treatment with ATRA for 24 or 48 h to a luciferase reporter (Fig. 3A, and data not shown). We therefore asked whether ATRA might regulate the slowly responding MDS1/EVI1 promoter in an indirect manner through the rapidly induced EVI1 protein. However, none of the MDS1/EVI1 promoter fragments was induced by exogenously expressed EVI1, either in the absence or in the presence of ATRA (Fig. 3A). Therefore, even though ATRA clearly induces MDS1/EVI1, the mechanism through which it does so remains unclear at present.
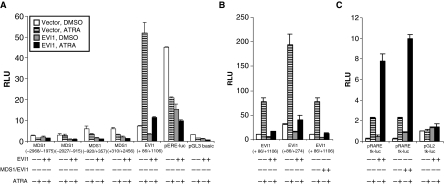
Fig. 3.
EVI1 does not mediate ATRA regulation of the MDS1/EVI1 promoter, but decreases the ATRA response of its own RARE and enhances that of the RARβ RARE. (A) Four genomic fragments, whose positions are indicated relative to the transcriptional start site of the MDS1/EVI1 gene (assumed to be identical to that of the MDS1 gene; GenBank accession no. NM_004991), were cloned into the luciferase reporter vector pGL3basic. NTERA-2 cells were cotransfected with these reporter constructs, HA-EVI1/pEFzeo or empty vector, and the renilla luciferase vector pRL-SV40. EVI1(+86/+1106)/pGL3 served as a positive control for the ATRA response; pEREluc, which contains two copies of an EVI1 DNA binding site, as a positive control for the effects of EVI1; and empty pGL3basic as a negative control. ATRA or dimethylsulfoxide was added 1 day after transfection, and cells were lysed and luciferase activities determined another ∼ 24 h later. Relative luciferase units (RLU) were derived by normalizing firefly to renilla luciferase activities. Error bars represent the standard deviations between duplicate measurements. (B, C) Luciferase assays after transfection of NTERA-2 cells with the indicated reporter plasmids and the expression vectors HA-EVI1/pEFzeo or HA-MDS1/EVI1/pEFzeo were performed as described in (A). pRARE-tk-luc contains two copies of the RARE of the human RARβ gene promoter and the tk minimal promoter in pGL2; pGL2-tk-luc contains only the tk minimal promoter in pGL2.
pERE/luc served as a positive control for the effects of EVI1 in the reporter assays, and showed the expected decrease in luciferase activity upon ectopic expression of EVI1 (Fig. 3A). Notably, ATRA also reduced the activity of pERE/luc (Fig. 3A), probably through its ability to upregulate the expression of endogenous EVI1. This supports the assumption that EVI1 protein induced by ATRA in NTERA-2 cells is functional as a transcriptional regulator. Interestingly, ATRA induction of EVI1(+86/+1106)/pGL3, which was included in the experiment as a positive control for the ATRA response, was strongly reduced in the presence of exogenously expressed EVI1 (Fig. 3A), suggesting negative feedback by EVI1 on its own promoter.
Both EVI1 and MDS1/EVI1 counteract the ATRA response of the EVI1 promoter, but enhance that of the RARβ RARE
As with EVI1(+86/+1106)/pGL3, the ATRA response of EVI1(+86/+274)/pGL3 was strongly reduced by exogenously expressed EVI1 (Fig. 3B). This indicated that EVI1 may interfere directly with the action of RAR, RXR and/or closely associated proteins at its own promoter. We next asked whether the repressive effects of EVI1 would extend to other ATRA responsive genes. Surprisingly, ATRA induction of pRARE-tk-luc, which contains two copies of the RARE that is present in the human RARβ gene promoter [30], was not diminished, but rather enhanced, by EVI1 (Fig. 3C). Like EVI1, MDS1/EVI1 also counteracted ATRA induction of the EVI1 promoter and enhanced that of the RARβ RARE (Fig. 3B,C).
Protein domains involved in modulation of the ATRA response by EVI1
In order to identify regions in the EVI1 protein that contribute to positive and negative modulation of the ATRA response, a series of EVI1 deletion constructs was prepared (Fig. 4A). All constructs had an N-terminal hemagglutinin (HA) epitope tag, and either retained a predicted nuclear localization sequence (NLS) in the IR, or were engineered to contain an NLS. The expression and nuclear location of the truncated proteins was confirmed by immunoblot and indirect immunofluorescence analyses (Fig. S1).
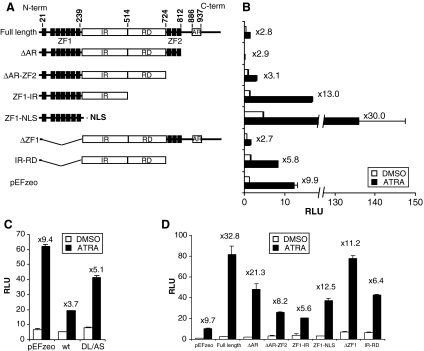
Fig. 4.
Protein domains involved in negative and positive modulation of the ATRA response by EVI1. (A) Schematic of the EVI1 deletion constructs. Black boxes, zinc finger motifs; ZF1, ZF2, zinc finger domains 1 and 2, respectively; IR, intervening region; RD, repression domain; AR, acidic region. Amino acid positions delimiting these domains are indicated. All constructs are based on the pEFzeo vector backbone, and contain an N-terminal HA epitope tag. The SV40 large T antigen NLS was engineered onto the ZF1 construct; all other constructs include a predicted NLS contained in the IR. (B–D) Luciferase assays using the indicated EVI1 deletion constructs along with the reporter vectors EVI1(+86/+1106)/pGL3 (B, C) or pRARE-tk-luc (D) were performed as described in Fig. 3. wt, wild-type EVI1; DL/AS, CtBP binding site mutant. White bars, dimethylsulfoxide; black bars, ATRA. Fold induction by ATRA in the presence of each construct is indicated. The effects of all mutations were highly reproducible; only deletion of the AR diminished the activity of EVI1 on pRARE-tk-luc more modestly in an independent experiment.
Deletion of the first zinc finger domain (ZF1), of the C-terminal AR, or of the AR and the second zinc finger domain (ZF2) (Fig. 4A, B: ΔZF1, ΔAR and ΔAR-ZF2, respectively) did not affect the repressive effects of EVI1 on the ATRA response of EVI1(+86/+1106)/pGL3 (Fig. 4B). However, a mutant with a C-terminal deletion that removed the RD in addition to the AR and the ZF2 (Fig. 4A, B: ZF1-IR) was inactive. Further deletion of the IR to yield a construct consisting only of the ZF1 (Fig. 4A, B: ZF1-NLS) even enhanced the ATRA response of the EVI1 promoter (Fig. 4B). A truncated EVI1 protein comprising only the IR and the RD (Fig. 4A, B: IR-RD) still had repressive activity, albeit at a reduced level (Fig. 4B). The RD was thus clearly important for the negative feedback by EVI1 on its own promoter. It contains two closely spaced binding sites for the corepressor protein C-terminal binding protein (CtBP), the more C-terminal one of which was reported to be functionally more important [4,5,31]. Mutation of this site on the background of the full-length EVI1 protein only moderately decreased the ability of EVI1 to counteract the ATRA response, suggesting that CtBP was not primarily responsible for transcriptional repression by EVI1 in this context (Fig. 4C).
To test which domains of EVI1 were involved in enhancing the ATRA response of the RARβ RARE, similar experiments were performed using the pRARE-tk-luc reporter plasmid. Deletion of the AR (Fig. 4A, D: ΔAR) had only a modest effect on the enhancement of the ATRA response by EVI1, but deletion of both the AR and the ZF2 (Fig. 4A, D: ΔAR-ZF2) eliminated it (Fig. 4D). Likewise, an EVI1 protein devoid of the ZF1 (Fig. 4A, D: ΔZF1), or constructs encoding only the ZF1 (Fig. 4A, D: ZF1-NLS), the ZF1 and IR (Fig. 4A, D: ZF1-IR), or the IR and RD (Fig. 4A, D: IR-RD), were incompetent to increase the ATRA response of pRARE-tk-luc (Fig. 4D). Taken together, several or all EVI1 protein domains contributed to the enhancement of the ATRA response by this transcription factor.
EVI1 increases the ATRA response of the endogenous RARβ gene in U937T cells
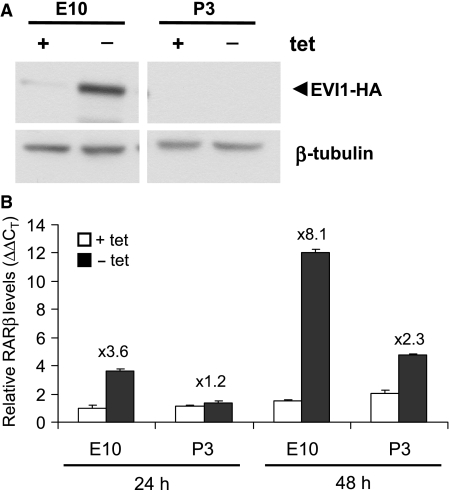
Finally, we asked whether EVI1 would be able to modulate induction of an endogenous gene by ATRA. In NTERA-2 cells, endogenous EVI1 was likely to obscure the effects of experimentally expressed EVI1. To circumvent this problem, the cell line U937T_EVI1-HA E10, a U937 derivative that does not express its own EVI1 gene, but has been engineered to express exogenous EVI1 in a tetracycline repressible manner (Fig. 5A) [32], was used. U937T_EP P3 cells, which contain the corresponding empty plasmid instead of the EVI1 expression plasmid, served as a negative control. In U937T_EVI1-HA E10 cells, the RARβ mRNA was undetectable by RTQ-RT-PCR in the absence of ATRA, but was strongly upregulated by this agent. Induction of EVI1 by removal of tetracycline led to an enhancement of this response (Fig. 5B), demonstrating that EVI1 was indeed able to modulate the ATRA response of an endogenous gene.
Fig. 5.
EVI1 enhances the ATRA response of the endogenous RARβ gene in U937T_EVI1-HA cells. (A) Immunoblot analysis demonstrating induction of EVI1-HA in U937T_EVI1-HA E10 cells (E10) 48 h after removal of tetracycline (tet) from the culture media. U937T cells transfected with empty plasmid (U937T_EP P3 cells; P3) were used as a negative control. EVI1-HA was detected with an HA antibody; hybridization with an antibody against β-tubulin was used as a loading control. (B) U937T_EVI1-HA E10 cells (E10) and U937T_EP P3 cells (P3) were maintained in the presence or absence of tetracycline and of ATRA for 24 and 48 h. RARβ mRNA levels were determined by RTQ-RT-PCR and related to those of the housekeeping gene cyclophilinD using the ΔΔCT method [45]. Because RARβ expression was undetectable in the absence of ATRA, the corresponding data are not shown. White and black columns, cells cultured in the presence or absence of tetracycline, respectively.
Discussion
Expression of the EVI1 gene is highly regulated during normal development [2,21,33] and is deregulated in certain types of cancer [13,15,34–36]. Nevertheless, little is known about the control of its transcription. In the present report, we have confirmed induction of the human EVI1 gene by ATRA, and demonstrated that a classical inverted DR5 RARE is located within its transcribed region, near the beginning of exon 1a. This element was necessary and sufficient to confer ATRA responsiveness to a luciferase reporter gene. Like other RAREs [26,27], it was bound by RAR and RXR in a constitutive manner, and repressed in the absence and activated in the presence of ligand. In silico analysis also predicted a DR2 RARE in exon 1 of the MDS1/EVI1 gene. However, neither this element nor any other sequences in a 5.5 kb region surrounding the transcriptional start site of MDS1/EVI1 were able to confer ATRA responsiveness to a luciferase reporter gene. Also, MDS1/EVI1 was not regulated by ATRA indirectly through the rapidly induced EVI1 protein. It remains possible, however, that regulatory elements outside the region investigated in this study do indeed confer direct responsiveness to ATRA and/or EVI1 to the MDS1/EVI1 promoter.
Interestingly, we found that the EVI1 RARE not only mediated transcriptional induction in response to ATRA, but in addition conferred negative feedback by EVI1 on its own promoter. On the other hand, EVI1 enhanced the ATRA response of a reporter gene driven by the RARβ RARE, as well as that of the endogenous RARβ gene. This not only argues against a nonspecific squelching effect as an explanation for the negative feedback of EVI1 on its own promoter, but also establishes EVI1 as a dual regulator of the ATRA response, which is able to repress or enhance gene induction by this agent in a promoter-specific manner. Notably, in silico analysis using tfsearch (http://www.cbrc.jp/research/db/TFSEARCH.html) did not reveal any EVI1 consensus DNA binding sites overlapping with, or located in the vicinity of, the EVI1 or the RARβ RAREs. Accordingly, ChIP experiments using either an EVI1 or an HA antibody failed to reveal an interaction between the EVI1 protein and the EVI1 or RARβ RAREs in native or transfected NTERA-2 cells, or in U937T_EVI1-HA cells (data not shown). This may be due to technical problems related to, for example, antibody quality, epitope accessibility or the positioning of side chains reactive with cross-linking reagents. Alternatively, modulation of the ATRA response by EVI1 may not involve interactions between EVI1 and DNA. Instead, the induction of EVI1 and RARβ by ATRA may require different cofactors, and EVI1 may differentially interact with, and affect the activity of, these cofactors in the nucleoplasm.
Not surprisingly, the positive and negative effects of EVI1 on the ATRA response were mediated through different protein domains. For the inhibitory effects of EVI1 on the induction of its own promoter by ATRA, the region between the two zinc finger domains was both necessary and sufficient. This region consists of a previously established RD [4,5,37] and an IR, which may also be able to mediate transcriptional repression [38]. Even though expression of the isolated IR+RD considerably diminished the ATRA response of the EVI1 promoter, full repression required the additional presence of either N-terminally or C-terminally adjacent domains. These may contribute in a partially redundant manner to pertinent interactions between EVI1 and other proteins, and/or be required to maintain the IR and RD in the conformation they adopt in the context of the full-length protein. A point mutation (DL/AS) disrupting the functionally more important of two consensus binding sites for the corepressor CtBP in the EVI1 RD [4,5] reduced the negative modulation of the ATRA response by EVI1. However, EVI1 DL/AS also exhibited a diminished ability to enhance the ATRA response of the RARβ RARE (data not shown), an activity unlikely to be mediated by a corepressor. This suggested that the DL/AS mutation may not only affect CtBP binding, but may also alter the structure of EVI1 and/or its interactions with other proteins. Other corepressors shown to functionally interact with EVI1, such as histone deacetylase 1 [6] or the histone methyl transferase SUV39H1 [39,40], may therefore contribute to, or mediate, repression of the ATRA response by EVI1. As for the ability of EVI1 to enhance ATRA induction of the RARβ gene, several of its protein domains appeared to take part in it. Deletion of the AR, which had previously been implicated in transcriptional activation by EVI1 [41,42], only moderately reduced the activating effect of EVI1 on the RARβ RARE. However, further deletion of the ZF2, or deletion of only the ZF1, abolished this activity, which therefore appears to require multiple and complex macromolecular interactions.
It remains to be determined whether EVI1 modulates more of the several hundred reported ATRA responsive genes [28] in a similar manner as the EVI1 or RARβ genes. EVI1 probably indirectly affects the expression of at least some of them through the increased levels of RARβ it contributes to. Because ATRA-induced transcription of the EVI1 gene itself would probably also be augmented through this mechanism, a potentially detrimental self-amplifying feed-forward loop could ensue. The negative feedback of EVI1 on the ATRA induction of its own promoter plausibly serves to prevent such a self-enhancing regulatory loop.
Our results also shed new light on previous observations that ectopic expression of EVI1, possibly in concert with low amounts of ATRA naturally present in sera used for cell culture, mimicked neuronal differentiation of P19 cells in response to exogenously added ATRA [22], and that targeted disruption of the retinaldehyde dehydrogenase-2 and Evi1 genes in mice caused partially overlapping phenotypes [2,24]: EVI1 appears to act both as a downstream target of the ATRA response and as an upstream regulator of it. It may therefore contribute to the biological effects of ATRA in a dual manner: first, by activating or repressing its own, direct target genes; and second, by modulating the expression of retinoid receptor-regulated genes. Another exciting possibility raised by our results is that EVI1 overexpressing leukaemias may exhibit increased sensitivity to ATRA-induced differentiation. If this presently speculative assumption can be substantiated by further experiments, new treatment possibilities may emerge for myeloid leukaemia with EVI1 overexpression, a disease entity that is notoriously resistant to currently used therapeutic regimens [15,36,43].
Materials and methods
Cell culture
The human teratocarcinoma cell line NTERA-2 (ACC 527) was obtained from the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany. It was cultivated in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Invitrogen) and 5% horse serum (Invitrogen) in a humidified 37 °C incubator at 5% CO2. U937T cells, kindly provided by G. Grosveld, St Jude Hospital, Memphis, TN, USA, had been derived from U937 human histiocytic lymphoma cells by stable transfection with a construct driving tetracycline regulable expression of the tetVP16 fusion protein [44]. Transfection with an expression plasmid coding for a tetracycline regulable, HA epitope tagged EVI1 protein or with empty plasmid as a control yielded U937T_EVI1-HA E10 and U937T_EP P3 cells, respectively [32]. These were maintained at 37 °C and 5% CO2 in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum, 500 μg·mL−1 hygromycin (PAA, Pasching, Austria), 0.5 μg·mL−1 puromycin (Sigma, St Louis, MO, USA) and 1 μg·mL−1 tetracycline (Serva, Heidelberg, Germany). To induce the expression of EVI1-HA, cells were washed three times with phosphate-buffered saline and resuspended in growth media without tetracycline.
ATRA stock solutions contained 10 mm ATRA in dimethylsulfoxide. Unless indicated otherwise, ATRA was used at final concentrations of 10 and 1 μm for NTERA-2 cells and U937T derivative cell lines, respectively.
RNA isolation, cDNA synthesis and RTQ-RT-PCR
Total RNA was extracted using Trizol (Invitrogen), treated with RNase-free DNaseI (Invitrogen), and reverse transcribed using random hexamer primers (Invitrogen) and M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. RTQ-RT-PCR was carried out in an ABI Prism 7700 Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA) using the Power SYBR® GREEN PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. Primers are shown in Table S1A. All primers exhibited optimal amplification efficiencies in serial dilution experiments. Assays for the housekeeping gene cyclophilinD were carried out in duplicate; all other assays were carried out in triplicate. Expression values for the genes of interest relative to cyclophilinD and to a reference value were determined using the ΔΔCT method [45].
Plasmids
EVI1 promoter fragments −2629/−72, +15/+1106 and +1258/+3903 (relative to the transcription start site of EVI1_1a, GenBank accession no. BX640908) were amplified from BAC clone RP11-33A1 using Dynazyme (Finnzyme, Espoo, Finland; a DNA polymerase with proofreading activity) and primers containing engineered restriction sites. Similarly, MDS1/EVI1 promoter fragments −2968/−1975, −2027/−915, −920/+357 and +310/+2456 (relative to the transcription start site of MDS1, GenBank accession no. NM_004991) were amplified from BAC clone RP11-816J6. PCR products were cloned into pCR® 2.1-TOPO® (Invitrogen) and then transferred into the luciferase reporter plasmid pGL3basic (Promega, Madison, WI, USA). Initial experiments with the EVI1 promoter fragments were also carried out with analogous constructs in pGL3promoter, but as both types of plasmid yielded similar results, subsequent analyses were restricted to pGL3basic. Deletion subclones of EVI1(+15/+1106)/pGL3 were generated through restriction digestion and religation or through PCR. A mutation that changed the sequence of the second half-site of the EVI1 RARE from TGACCT to CTTTAG [46] was introduced into EVI1(+86/+1106)/pGL3 using overlap extension PCR. The resulting PCR product was cloned into pCR® 2.1-TOPO® (Invitrogen) and then transferred into pGL3basic. Primers and restriction sites used to generate promoter constructs are available upon request. pRARE-tk-luc contains two copies of the RARE of the human RARβ gene promoter [30] and a tk minimal promoter in pGL2 (Promega); the corresponding empty control vector pGL2-tk-luc is identical except that it lacks the RAREs. Both plasmids were kindly provided by H. Harant, Ingenetix, Vienna, Austria. pERE/luc consists of two copies of the recognition sequence for the N-terminal zinc finger domain of EVI1, the tk minimal promoter and the pGL3basic vector backbone [6].
HA-EVI1/pEFzeo and HA-MDS1/EVI1/pEFzeo contain the human EVI1 and MDS1/EVI1 cDNAs, respectively, both with an engineered Kozak consensus for translation initiation and an N-terminal HA epitope tag, under the control of the eIF1α promoter [6]. To obtain EVI1 deletion constructs, internal restriction fragments of HA-EVI1/pEFzeo were replaced by suitable PCR products in a manner that left the Kozak sequence and the HA tag intact. To construct EVI1 ZF1-NLS, a double-stranded oligonucleotide containing a tandem repeat of the SV40 large T antigen NLS [47], followed by a stop codon, and a PCR product covering the EVI1 ZF1 region and part of the NLS were used as templates in a fusion PCR. The resulting ZF1-NLS PCR product was cloned into HA-EVI1/pEFzeo as described above. All other deletion constructs retained a predicted NLS in the IR of EVI1. Primers and restriction sites used to generate EVI1 deletion constructs are available upon request. The human EVI1 cDNA with the CtBP binding site mutation, DL/AS, and the corresponding wild-type cDNA had been cloned into pME18S, and were kindly provided by M. Kurokawa, Tokyo University, Japan [5].
Transient transfections and reporter gene assays
One day before transfection, NTERA-2 cells were seeded into 24-well plates at a density of 8 × 104 cells per well. Transfections were carried out using DAC-30 (Eurogentec, Seraing, Belgium; production discontinued) or FuGene6 (Roche, Basel, Switzerland) according to the manufacturers’ instructions, employing 0.6 μg DNA and 2 μL DAC-30, or 0.6 μg DNA and 3 μL FuGene6, per well. Transfection efficiencies were > 25%.
To assess the ATRA responsiveness of the EVI1 promoter fragments, 540 ng of reporter plasmid were cotransfected with 60 ng of the renilla luciferase plasmid pRL-TK (Promega). To determine the responsiveness of reporter plasmids to EVI1, its deletion derivatives or MDS1/EVI1, 300 ng of reporter plasmid was cotransfected with 300 ng of the respective expression vector and 6 ng of the renilla luciferase plasmid pRL-SV40 (Promega). 10 μm ATRA, or an equivalent amount of dimethylsulfoxide, was added to each well 1 day after transfection. On day two after transfection, the cells were lysed, and firefly and renilla luciferase activities were assayed in an Aureon PhL luminometer (Aureon Biosystems, Vienna, Austria) using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activities were normalized to renilla luciferase activities to adjust for variability in transfection efficiencies. Duplicate samples were assayed for each data point.
ChIP
NTERA-2 cells were cultivated in 15 cm dishes to ∼ 80% confluence and treated with 10 μm ATRA, or an equivalent amount of dimethylsulfoxide, for 24 h. Proteins were cross-linked to DNA by incubating the cells with 1% formaldehyde (Sigma) for 10 min at 37 °C. After quenching with 125 mm glycine (Sigma) for 5 min at room temperature, the cells were washed with ice-cold phosphate-buffered saline containing a complete protease inhibitor cocktail (Roche), scraped off the dishes and lysed in a buffer containing 50 mm Tris pH 8.1, 10 mm EDTA, 1% SDS and a complete protease inhibitor cocktail (Roche). Chromatin was sheared with eight cycles of 15 s each at 90% duty cycle on a Branson sonifier 450 (Branson, Danbury, CT, USA). Sample concentrations were normalized based on their A280. Chromatin was diluted 1 : 10 into ChIP buffer (OneDay ChIP kit, Diagenode, Liege, Belgium) and agitated overnight at 4 °C with 8 μg of RARα antibody (sc-551, Santa Cruz, CA, USA), pan-RXR antibody (sc-774, Santa Cruz) or rabbit IgG. Further processing was performed using the OneDay ChIP kit according to the manufacturer’s instructions.
In an aliquot of the sheared chromatin, the cross-link was reversed by overnight incubation at 65 °C with 2% SDS, 100 mm NaHCO3, 10 mm dithiothreitol and 200 mm NaCl. After proteinase K digestion, DNA was recovered by phenol/chloroform extraction and ethanol precipitation. Agarose gel electrophoresis showed a DNA smear ranging in size from 100 bp to 2 kb, with maximal intensity at 500 bp. Immunoprecipitated DNA and de-crosslinked (‘input’) DNA as a positive control were subjected to PCR with primers flanking the EVI1 RARE (EVI1_RARE-F, EVI1_RARE-R; Table S1B), primers flanking the RARβ RARE (RARβ_RARE-F, RARβ_RARE-R; Table S1B; [48]), and with negative control primers located in EVI1 exon 3L (EVI1_ex3L-F, EVI1_ex3L-R; Table S1B).
Immunoblot analysis
For protein extraction, cells were subjected to repeated freeze–thawing in a buffer containing 20 mm Tris pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% NP-40, 0.5 mm phenylmethanesulfonyl fluoride and a complete protease inhibitor cocktail (Roche). Denaturing PAGE, tank blotting onto Bio Trace NT nitrocellulose membranes (Pall Corporation, Port Washington, NY, USA) and antibody hybridizations were performed using standard procedures. Primary antibodies directed against the HA-tag (mouse anti-HA.11 clone 16B12; Covance, Berkeley, CA, USA), EVI1 (rabbit anti-EVI1 C50E12; Cell Signaling Technology, Danvers, MA, USA) or β-tubulin (mouse anti-β-tubulin clone TUB 2.1; Sigma) were used at dilutions of 1 : 10000, 1 : 1000 and 1 : 2500, respectively. Horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit secondary IgGs were used at dilutions of 1 : 40000–1 : 100000, and detected using the Super Signal West Femto kit (Pierce, Rockford, IL, USA).
Acknowledgments
This work was supported by the Austrian Science Foundation, grants P17896-B14 and P19795-B12 to RW. The authors gratefully acknowledge Dr Hanna Harant, Ingenetix, Vienna, Austria, Dr Mineo Kurokawa, University of Tokyo, Japan and Dr Gerard Grosveld, St Jude Children’s Research Hospital, Memphis, TN, USA, for providing pRARE-tk-luc, the EVI1 CtBP mutant and corresponding wild-type plasmids, and U937T cells, respectively. Lilli Simböck from the laboratory of Dr Christian Seiser, Medical University of Vienna, Austria, was of invaluable help in establishing the ChIP assay. Dr Herbert Stangl and Dr Witta Monika Strobl of the Department of Medical Chemistry of the Medical University of Vienna generously allowed access to their luminometer.
Glossary
Abbreviations
- AR
acidic region
- ATRA
all-trans retinoic acid
- ChIP
chromatin immunoprecipitation
- CtBP
C-terminal binding protein
- DR
direct repeat
- EVI1
ecotropic viral integration site 1
- HA
hemagglutinin
- IR
intervening region
- NLS
nuclear localization sequence
- RA
retinoic acid
- RAR
retinoic acid receptor
- RARE
retinoic acid response element
- RD
repression domain
- RTQ-RT-PCR
real-time quantitative RT-PCR
- RXR
retinoid X receptor
- ZF1
zinc finger domain 1
- ZF2
zinc finger domain 2
Supporting information
The following supplementary material is available:
Fig. S1. Expression and nuclear location of protein products derived from EVI1 deletion constructs.
Doc. S1. Immunofluorescence analysis.
Table S1. Primers used for RTQ-RT-PCR, RT-PCR, genomic PCR and ChIP PCR.
This supplementary material can be found in the online version of this article.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- 1.Wieser R. The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions. Gene. 2007;396:346–357. doi: 10.1016/j.gene.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Hoyt P, Bartholomew C, Davis A, Yutzey K, Gamer L, Potter S, Ihle J, Mucenski M. The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech Dev. 1997;65:55–70. doi: 10.1016/s0925-4773(97)00057-9. [DOI] [PubMed] [Google Scholar]
- 3.Soderholm J, Kobayashi H, Mathieu C, Rowley J, Nucifora G. The leukemia-associated gene MDS1/EVI1 is a new type of GATA-binding transactivator. Leukemia. 1997;11:352–358. doi: 10.1038/sj.leu.2400584. [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty S, Senyuk V, Sitailo S, Chi Y, Nucifora G. Interaction of EVI1 with CBP and P/CAF results in reversible acetylation of EVI1 and in colocalization in nuclear speckles. J Biol Chem. 2001;276:44936–44943. doi: 10.1074/jbc.M106733200. [DOI] [PubMed] [Google Scholar]
- 5.Izutsu K, Kurokawa M, Imai Y, Maki K, Mitani K, Hirai H. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood. 2001;97:2815–2822. doi: 10.1182/blood.v97.9.2815. [DOI] [PubMed] [Google Scholar]
- 6.Vinatzer U, Taplick J, Seiser C, Fonatsch C, Wieser R. The leukaemia-associated transcription factors EVI-1 and MDS1/EVI1 repress transcription and interact with histone deacetylase. Br J Haematol. 2001;114:566–573. doi: 10.1046/j.1365-2141.2001.02987.x. [DOI] [PubMed] [Google Scholar]
- 7.Yuasa H, Oike Y, Iwama A, Nishikata I, Sugiyama D, Perkins A, Mucenski M, Suda T, Morishita K. Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J. 2005;24:1976–1987. doi: 10.1038/sj.emboj.7600679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yatsula B, Lin S, Read A, Poholek A, Yates K, Yue D, Hui P, Perkins A. Identification of binding sites of EVI1 in mammalian cells. J Biol Chem. 2005;280:30712–30722. doi: 10.1074/jbc.M504293200. [DOI] [PubMed] [Google Scholar]
- 9.Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, Yazaki Y, Matsumoto K, Hirai H. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature. 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- 10.Buonamici S, Li D, Mikhail F, Sassano A, Platanias L, Colamonici O, Anastasi J, Nucifora G. EVI1 abrogates interferon-alpha response by selectively blocking PML induction. J Biol Chem. 2005;280:428–436. doi: 10.1074/jbc.M410836200. [DOI] [PubMed] [Google Scholar]
- 11.Laricchia-Robbio L, Fazzina R, Li D, Rinaldi C, Sinha K, Chakraborty S, Nucifora G. Point mutations in two EVI1 Zn fingers abolish EVI1-GATA1 interaction and allow erythroid differentiation of murine bone marrow cells. Mol Cell Biol. 2006;26:7658–7666. doi: 10.1128/MCB.00363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senyuk V, Sinha K, Li D, Rinaldi C, Yanamandra S, Nucifora G. Repression of RUNX1 activity by EVI1: a new role of EVI1 in leukemogenesis. Cancer Res. 2007;67:5658–5666. doi: 10.1158/0008-5472.CAN-06-3962. [DOI] [PubMed] [Google Scholar]
- 13.Vinatzer U, Mannhalter C, Mitterbauer M, Gruener H, Greinix H, Schmidt H, Fonatsch C, Wieser R. Quantitative comparison of the expression of EVI1 and its presumptive antagonist, MDS1/EVI1, in patients with myeloid leukemia. Genes Chromosomes Cancer. 2003;36:80–89. doi: 10.1002/gcc.10144. [DOI] [PubMed] [Google Scholar]
- 14.Aytekin M, Vinatzer U, Musteanu M, Raynaud S, Wieser R. Regulation of the expression of the oncogene EVI1 through the use of alternative mRNA 5’-ends. Gene. 2005;356:160–168. doi: 10.1016/j.gene.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 15.Haas K, Kundi M, Sperr W, Esterbauer H, Ludwig W, Ratei R, Koller E, Gruener H, Sauerland C, Fonatsch C, et al. Expression and prognostic significance of different mRNA 5′-end variants of the oncogene EVI1 in 266 patients with de novo AML: EVI1 and MDS1/EVI1 overexpression both predict short remission duration. Genes Chromosomes Cancer. 2008;47:288–298. doi: 10.1002/gcc.20532. [DOI] [PubMed] [Google Scholar]
- 16.Fears S, Mathieu C, Zeleznik Le N, Huang S, Rowley J, Nucifora G. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc Natl Acad Sci USA. 1996;93:1642–1647. doi: 10.1073/pnas.93.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wimmer K, Vinatzer U, Zwirn P, Fonatsch C, Wieser R. Comparative expression analysis of the antagonistic transcription factors EVI1 and MDS1-EVI1 in murine tissues and during in vitro hematopoietic differentiation. Biochem Biophys Res Commun. 1998;252:691–696. doi: 10.1006/bbrc.1998.9588. [DOI] [PubMed] [Google Scholar]
- 18.Sitailo S, Sood R, Barton K, Nucifora C. Forced expression of the leukemia-associated gene EVI1 in ES cells: a model for myeloid leukemia with 3q26 rearrangements. Leukemia. 1999;13:1639–1645. doi: 10.1038/sj.leu.2401585. [DOI] [PubMed] [Google Scholar]
- 19.Sood R, Talwar TrikhaA, Chakrabarti S, Nucifora G. MDS1/EVI1 enhances TGF-beta 1 signaling and strengthens its growth-inhibitory effect, but the leukemia-associated fusion protein AML1/MDS1/EVI1, product of the t(3;21), abrogates growth-inhibition in response to TGF-beta 1. Leukemia. 1999;13:348–357. doi: 10.1038/sj.leu.2401360. [DOI] [PubMed] [Google Scholar]
- 20.Cuenco G, Ren R. Both AML1 and EVI1 oncogenic components are required for the cooperation of AML1/MDS1/EVI1 with BCR/ABL in the induction of acute myelogenous leukemia in mice. Oncogene. 2004;23:569–579. doi: 10.1038/sj.onc.1207143. [DOI] [PubMed] [Google Scholar]
- 21.Perkins A, Mercer J, Jenkins N, Copeland N. Patterns of Evi-1 expression in embryonic and adult tissues suggest that Evi-1 plays an important regulatory role in mouse development. Development. 1991;111:479–487. doi: 10.1242/dev.111.2.479. [DOI] [PubMed] [Google Scholar]
- 22.Kazama H, Kodera T, Shimizu S, Mizoguchi H, Morishita K. Ecotropic viral integration site-1 is activated during, and is sufficient for, neuroectodermal P19 cell differentiation. Cell Growth Differ. 1999;10:565–573. [PubMed] [Google Scholar]
- 23.Xi Z, Russell M, Woodward S, Thompson F, Wagner L, Taetle R. Expression of the Zn finger gene, EVI-1, in acute promyelocytic leukemia. Leukemia. 1997;11:212–220. doi: 10.1038/sj.leu.2400547. [DOI] [PubMed] [Google Scholar]
- 24.Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 25.McCaffery P, Adams J, Maden M, Rosa-Molinar E. Too much of a good thing: retinoic acid as an endogenous regulator of neural differentiation and exogenous teratogen. Eur J Neurosci. 2003;18:457–472. doi: 10.1046/j.1460-9568.2003.02765.x. [DOI] [PubMed] [Google Scholar]
- 26.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 27.McGrane M. Vitamin A regulation of gene expression: molecular mechanism of a prototype gene. J Nutr Biochem. 2007;18:497–508. doi: 10.1016/j.jnutbio.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Balmer J, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 29.Bordereaux D, Fichelson S, Tambourin P, Gisselbrecht S. Alternative splicing of the Evi-1 zinc finger gene generates mRNAs which differ by the number of zinc finger motifs. Oncogene. 1990;5:925–927. [PubMed] [Google Scholar]
- 30.de The H, Vivanco-Ruiz M, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- 31.Palmer S, Brouillet J, Kilbey A, Fulton R, Walker M, Crossley M, Bartholomew C. Evi-1 transforming and repressor activities are mediated by CtBP co-repressor proteins. J Biol Chem. 2001;276:25834–25840. doi: 10.1074/jbc.M102343200. [DOI] [PubMed] [Google Scholar]
- 32.Konrad T, Karger A, Hackl H, Schwarzinger I, Herbacek I, Wieser R. Inducible expression of EVI1 in human myeloid cells causes phenotypes consistent with its role in myelodysplastic syndromes. J Leukoc Biol. 2009;86:813–822. doi: 10.1189/jlb.0109042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Campenhout C, Nichane M, Antoniou A, Pendeville H, Bronchain O, Marine J, Mazabraud A, Voz M, Bellefroid E. Evi1 is specifically expressed in the distal tubule and duct of the Xenopus pronephros and plays a role in its formation. Dev Biol. 2006;294:203–219. doi: 10.1016/j.ydbio.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 34.Brooks D, Woodward S, Thompson F, Dos Santos B, Russell M, Yang J, Gua X, Trent J, Alberts D, Taetle R. Expression of the zinc finger gene EVI-1 in ovarian and other cancers. Br J Cancer. 1996;74:1518–1525. doi: 10.1038/bjc.1996.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poppe B, Dastugue N, Vandesompele J, Cauwelier B, De Smet B, Yigit N, De Paepe A, Cervera J, Recher C, De Mas V, et al. EVI1 is consistently expressed as principal transcript in common and rare recurrent 3q26 rearrangements. Genes Chromosomes Cancer. 2006;45:349–356. doi: 10.1002/gcc.20295. [DOI] [PubMed] [Google Scholar]
- 36.Lugthart S, van Drunen E, van Norden Y, van Hoven A, Erpelinck C, Valk P, Beverloo H, Lowenberg B, Delwel R. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111:4329–4337. doi: 10.1182/blood-2007-10-119230. [DOI] [PubMed] [Google Scholar]
- 37.Bartholomew C, Kilbey A, Clark A, Walker M. The Evi-1 proto-oncogene encodes a transcriptional repressor activity associated with transformation. Oncogene. 1997;14:569–577. doi: 10.1038/sj.onc.1200864. [DOI] [PubMed] [Google Scholar]
- 38.Kilbey A, Bartholomew C. Evi-1 ZF1 DNA binding activity and a second distinct transcriptional repressor region are both required for optimal transformation of Rat1 fibroblasts. Oncogene. 1998;16:2287–2291. doi: 10.1038/sj.onc.1201732. [DOI] [PubMed] [Google Scholar]
- 39.Cattaneo F, Nucifora G. EVI1 recruits the histone methyltransferase SUV39H1 for transcription repression. J Cell Biochem. 2008;105:344–352. doi: 10.1002/jcb.21869. [DOI] [PubMed] [Google Scholar]
- 40.Spensberger D, Delwel R. A novel interaction between the proto-oncogene Evi1 and histone methyltransferases, SUV39H1 and G9a. FEBS Lett. 2008;582:2761–2767. doi: 10.1016/j.febslet.2008.06.056. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka T, Nishida J, Mitani K, Ogawa S, Yazaki Y, Hirai H. Evi-1 raises AP-1 activity and stimulates c-fos promoter transactivation with dependence on the second zinc finger domain. J Biol Chem. 1994;269:24020–24026. [PubMed] [Google Scholar]
- 42.Sato T, Goyama S, Nitta E, Takeshita M, Yoshimi M, Nakagawa M, Kawazu M, Ichikawa M, Kurokawa M. Evi-1 promotes para-aortic splanchnopleural hematopoiesis through up-regulation of GATA-2 and repression of TGF-b signaling. Cancer Sci. 2008;99:1407–1413. doi: 10.1111/j.1349-7006.2008.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 44.Boer J, Bonten-Surtel J, Grosveld G. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol Cell Biol. 1998;18:1236–1247. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Jiang S, Wu M, Chen L, Hung M, Lin H, Chang G, Chang T. Identification and characterization of the retinoic acid response elements in the human RIG1 gene promoter. Biochem Biophys Res Commun. 2005;331:630–639. doi: 10.1016/j.bbrc.2005.03.214. [DOI] [PubMed] [Google Scholar]
- 47.Fischer-Fantuzzi L, Vesco C. Cell-dependent efficiency of reiterated nuclear signals in a mutant simian virus 40 oncoprotein targeted to the nucleus. Mol Cell Biol. 1988;8:5495–5503. doi: 10.1128/mcb.8.12.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H, Park U, Kim E, Um S. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 2007;26:3545–3557. doi: 10.1038/sj.emboj.7601797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.