Abstract
The midbrain-hindbrain boundary (MHB) is a highly conserved fold in the vertebrate embryonic brain. We have termed the deepest point of this fold the MHB constriction (MHBC) and have begun to define the mechanisms by which it develops. In the zebrafish, the MHBC is formed soon after neural tube closure, concomitant with inflation of the brain ventricles. The MHBC is unusual, as it forms by bending the basal side of the neuroepithelium. At single cell resolution, we show that zebrafish MHBC formation involves two steps. The first is a shortening of MHB cells to approximately 75% of the length of surrounding cells. The second is basal constriction, and apical expansion, of a small group of cells that contribute to the MHBC. In the absence of inflated brain ventricles, basal constriction still occurs, indicating that the MHBC is not formed as a passive consequence of ventricle inflation. In laminin mutants, basal constriction does not occur, indicating an active role for the basement membrane in this process. Apical expansion also fails to occur in laminin mutants, suggesting that apical expansion may be dependent on basal constriction. This study demonstrates laminin-dependent basal constriction as a previously undescribed molecular mechanism for brain morphogenesis.
Keywords: basal constriction, apical expansion, zebrafish morphogenesis, laminin, brain ventricle, midbrain-hindbrain boundary, midbrain-hindbrain boundary constriction (MHBC), cell shape
1. Introduction
During development of the vertebrate brain, the neural tube assumes a complex structure that includes formation of the brain ventricles and the appearance of conserved folds and bends. These folds and bends delineate functional units of the brain and are likely to shape the brain such that it can pack into the skull. The midbrain-hindbrain boundary (MHB) is the site of one of the earliest bends in the developing brain. In the embryo, the MHB functions as an embryonic organizing center (Brand et al., 1996; Joyner, 1996; Puelles and Martinez-de-la-Torre, 1987; Sato et al., 2004) and later becomes the cerebellum and part of the tectum (Louvi et al., 2003).
We have called the deepest point in the MHB the “midbrain-hindbrain boundary constriction” (MHBC). In the present study, we ask what processes are necessary for MHBC morphogenesis, using the zebrafish as a model. In the zebrafish, the MHBC forms between 17 and 24 hours post fertilization (hpf), concomitant with formation of the brain ventricles. At this stage of development, the neuroepithelium is a pseudostratified-columnar epithelium where apical cell surfaces face the brain ventricle lumen, and basal cell surfaces, on the outside of the tube, abut the basement membrane.
Thus, interestingly, the MHBC forms by bending the basal side of the neuroepithelium. This is unusual, since essentially all epithelial bends have been described at the apical surface, via apical constriction. A single report mentions that Drosophila salivary gland morphogenesis may involve bending of the basal side of the epithelium (Fristrom, 1988). The organization of the neuroepithelium, and correlation with brain ventricle inflation led us to consider three factors that may drive MHBC morphogenesis: (1) fluid pressure on the inside of the neural tube as the brain ventricles inflate (Lowery and Sive, 2005), (2) changes in cell shape during bending, and (3) interactions of the basal side of the neuroepithelium with the basement membrane.
We show here that MHBC morphogenesis involves two processes, cell shortening at the MHB and basal constriction of the neuroepithelial cells at the MHBC. Basal constriction is dependent upon laminin function, but not upon inflation of the brain ventricles. These data indicate that the MHBC forms through changes in cell shape, dependent on the extracellular matrix, which have not previously been described during brain morphogenesis.
2. Results and Discussion
2.1 Zebrafish MHBC morphogenesis occurs soon after neural tube closure
In the zebrafish, brain morphogenesis begins after neural tube closure at 17 hpf (Kimmel et al., 1995; Lowery and Sive, 2005). At this stage, a slight indentation, visible on the outside of the tube at the MHB anlage (Fig. 1A), corresponds to the basal side of the neuroepithelium. Beginning at 18 hpf, the opposing apical sides of the neuroepithelium separate along the midline and inflate to form the fore-, mid-, and hindbrain ventricles (Lowery and Sive, 2005). However, cells at the MHB remain closely apposed at the midline. At 21 hpf, after the midbrain and hindbrain ventricles have opened further, the indentation at the MHB outside the tube is more prominent, but still shallow (Fig. 1B). By 24 hpf, the MHB is bent acutely at the basal surface creating a sharp point on the outside of the tube (Fig. 1C). This was clearly visible in all wild type embryos by staining the outside of the neural tube with a laminin antibody (Fig. 1D). We have called this sharp point, at the deepest point of this fold, the midbrain-hindbrain boundary constriction (MHBC). This constriction is highly conserved amongst the vertebrates (Rhinn and Brand, 2001).
Fig. 1. Zebrafish MHB morphogenesis occurs between 17 and 24 hpf, and requires laminin but not ventricle inflation.
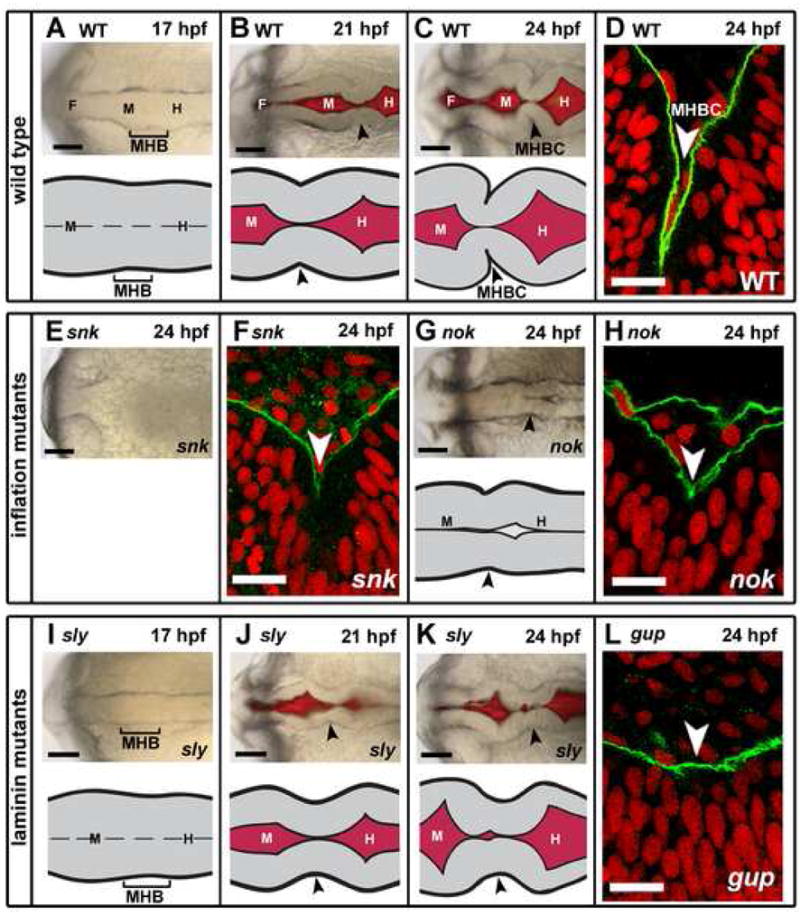
(A–C) Brightfield and fluorescent images and schematics of wild type (WT) MHBC formation. (D) WT embryo at 24 hpf was stained with anti-laminin 1 antibody (green); nuclei were stained with propidium idodide (red). Laminin lines the basal surface of the neuroepithelium. (E) Brightfield image of snk, a ventricle inflation mutant, at 24 hpf. (F) snk embryo at 24 hpf stained as in D. (G) Brightfield image and schematic of nok, a ventricle inflation mutant, at 24 hpf. (H) nok embryo at 24 hpf stained as in as D. (I–K) Brightfield and fluorescent images and schematics of MHBC formation in the laminin mutant, sly. (L) gup embryo at 24 hpf stained as in D. Arrowheads indicate MHB at 21 hpf and MHBC at 24 hpf. F, forebrain; M, midbrain; H, hindbrain. Scale bars: A–C, E, G, I–K = 100 μM, D, F, H, L = 6 μM.
2.2 A sharp MHBC forms in ventricle inflation mutants
In order to determine the mechanisms regulating MHBC morphogenesis, we asked whether brain ventricle inflation plays a role in this process. We hypothesized that pressure from the embryonic cerebrospinal fluid (eCSF) within the brain ventricles is required to form the MHBC, through a passive “pushing” mechanism (Lowery and Sive, 2005). Supporting this hypothesis, blood flow modifies heart chamber morphology and stimulates valve morphogenesis (Berdougo et al., 2003; Hove et al., 2003; Seidman and Seidman, 2001).
We analyzed MHBC morphogenesis in two zebrafish mutants lacking inflated brain ventricles, snakehead (snk), with a mutation in atp1a1 encoding a Na+K+ ATPase (Lowery and Sive, 2005) and nagie oko (nok), a mutant allele of the MAGUK scaffolding protein, Mpp5 (Wei and Malicki, 2002). snk and nok embryos were imaged at 24 hpf to examine the overall outline of the neural tube and shape of the MHBC. The abnormal refractility in snk embryos prevented visualization of the MHBC by brightfield microscopy (Fig. 1E). However, laminin staining of all snk embryos analyzed revealed that the MHBC does define a sharp point, although the angle at the MHB is less acute than that of wild type embryos (Fig. 1F). In nok mutants laminin staining indicated that the MHBC also defined a sharp point in all embryos observed (Fig. 1G,H).
The acuteness of the MHBC in the tissue in both snk and nok is clearly reduced compared to wild type embryos. Thus, ventricle inflation may be required to push together the neuroepithelium to form an extremely acute angle, but it is not required to form a sharp point, at the MHBC.
2.3 Laminin is required for MHBC formation
We also hypothesized that the basement membrane, which lies adjacent to the MHBC on the outside of the brain primordium, may play a role in its formation. Laminin is a major component of the basement membrane, that interacts with integrins, to mediate adhesion to the cytoskeleton of overlying cells (Miner and Yurchenco, 2004). A role for laminin has been demonstrated during mouse salivary gland branching, axon pathfinding in multiple organisms, and zebrafish notochord development (Bernfield et al., 1984; Garcia-Alonso et al., 1996; Karlstrom et al., 1996; Parsons et al., 2002; Paulus and Halloran, 2006). Laminin has not previously been implicated in brain morphogenesis in any system, although it has been shown to play a role in development of the eye, which is derived from neuroepithelium (Svoboda and O’Shea, 1987).
We tested the requirement for laminin by examining the MHBC in the sleepy mutant (slym86) that has a mutation in the gamma1 laminin gene (lamc1) (Parsons et al., 2002) and in the grumpy mutant (guphi1113b), which has a viral insertion in the first intron of the laminin beta1 gene (lamb1), (Amsterdam et al., 2004 and A. Amsterdam, personal communication). By brightfield imaging, sly mutants showed an initially normally shaped neural tube (Fig. 1I,J), but by 24 hpf, the MHBC remained a shallow indentation (Fig. 1K). Similar results were observed with gup mutants (data not shown). Consistent with brightfield imaging, at 24 hpf, a shallow MHBC was observed in gup mutant embryos stained with the laminin 1 antibody (Fig. 1L). This angle was consistently shallow in all embryos, observed either by brightfield imaging or by laminin 1 staining. We used the guphi1113b viral insertion mutants for laminin 1 antibody staining, because the laminin 1 antibody is not immunoreactive in the allele of sly used in this study (slym86), nor in the other gup allele previously described (gupm186) (Parsons et al., 2002). Although the mechanism by which this antibody recognizes laminin 1 in guphi1113 is not known, the viral insertion may result in a recognizable, but non-functional protein, whereas point mutation alleles of slym86 and gupm186 result in the introduction of a premature stop codon and likely severely truncated proteins (Parsons et al., 2002). These data showed that laminin function is essential for the sharp point normally seen at the MHBC and define a new role for laminin in brain morphogenesis.
2.4 Cells shorten and basally constrict at the MHBC
Bends or folds in epithelial sheets are driven by changes in cell length and formation of wedge-shaped cells, such as the cell shortening and apical constriction during neurulation in Xenopus, optic vesicle formation in mice, and ventral furrow invagination in Drosophila (Lee et al., 2007; Smith et al., 1994; Svoboda and O’Shea, 1987; Sweeton et al., 1991). We therefore hypothesized that wedge-shaped cells would be required to form the MHBC. However, based on the orientation of the MHBC, we hypothesized that such wedge-shaped cells would be basally, rather than apically, constricted.
In order to test this hypothesis, we analyzed cell shape at the MHBC in wild type embryos by expressing membrane-localized green fluorescent protein (memGFP) and imaging live embryos by laser-scanning confocal microscopy. At 17 hpf, cells in the midbrain, hindbrain, and MHB are uniform in length and are both spindle and columnar-shaped, with some rounded dividing cells visible (Fig. 2A,A′). In contrast, by 21 hpf, MHB cells are shorter in length (0.76 the apical-basal length) than those in either the midbrain or hindbrain (Fig. 2B,B′,J). Do these MHB cells shorten relative to surrounding cells, or do they fail to lengthen in concert with the rest of the neuroepithelium? By imaging wild type embryos, using spinning-disk confocal microscopy, to generate a live time-lapse data series, between 17 and 21 hpf, we showed that single cells at the MHB shorten relative to surrounding cells (Fig. 2D–I). While the shortening event appears to be graded along the MHB, the uneven nature of the pseudostratified neuroepithelium makes quantification of subtle changes in cell length in regions flanking the future MHBC, difficult to measure. In conclusion, a first step in MHBC formation is the shortening of cells at the MHB.
Fig. 2. MHBC formation requires cell shortening and basal constriction.
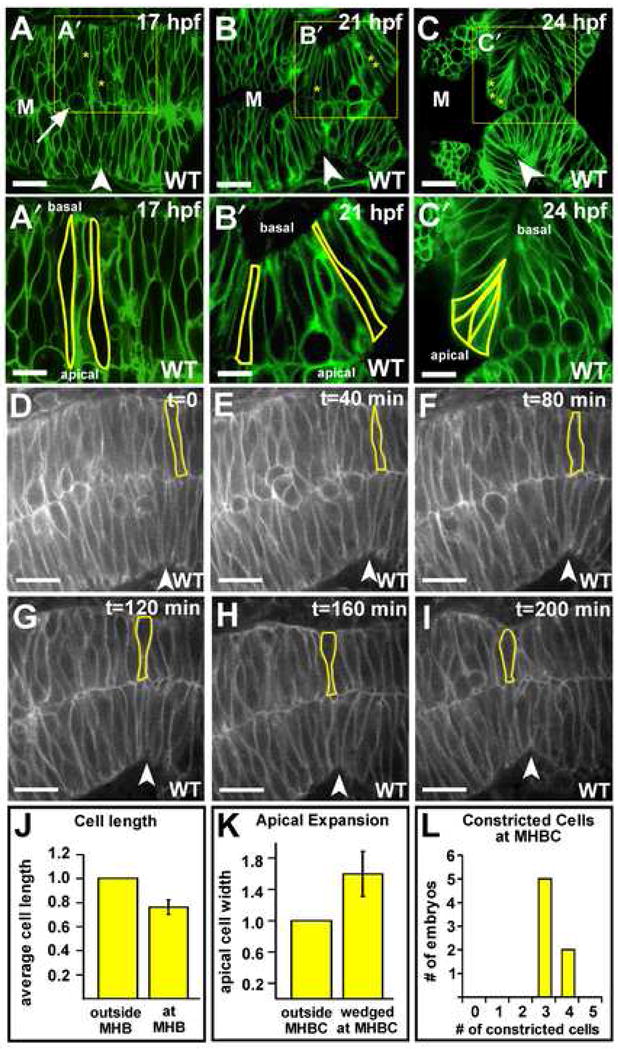
(A–C′) Live laser-scanning confocal imaging of wild type embryos injected with memGFP mRNA at the one cell stage and imaged at 17, 21, and 24 hpf. Boxed regions from A–C are enlarged for panels A′–C′. Individual cells in the MHB are outlined, and a dividing cell is indicated by an arrow in B. Asterisks in A–C mark cells outlined in A′–C′. Cells with two asterisks are outside the MHB. (A′) At 17 hpf, cells at the MHB are similar in length to the cells in the surrounding tissue. (B′) At 21 hpf, cells at the MHB are shorter than the cells in the surrounding tissue. (C′) At 24 hpf, cells at the MHBC are constricted basally and expanded apically. In A– C′, some green fluorescence is visible within outlined cells since the plane of section contains the surface of the cell membrane. (D–I) Time-course of MHB morphogenesis beginning at 17 hpf. A single cell is outlined and followed through the time course. Cells at the MHB shorten relative to those surrounding (n = 6 embryos). (J) Relative cell lengths at and outside the MHB in 21 hpf wild type embryos. Cells at the MHB were 0.76 times the length of those outside the MHB (+/− 0.06 s.d.) (n = 8 embryos, 3 cells at the MHB and 4 cells outside the MHB were measured per embryo). (K) Relative apical width of unwedged cells (those outside the MHBC) and basally constricted cells (at the MHBC) in wild type embryos at 24 hpf. Cells at the MHBC had 1.6 times the apical width of those outside the MHBC (+/− 0.29 s.d.) (n = 6 embryos, 2 cells at the MHBC and 3 cells outside the MHBC were measured per embryo). (L) Numbers of basally constricted cells at the MHBC in wild type embryos at 24 hpf (n = 9 embryos). Arrowheads indicate the MHBC. M, midbrain. Scale bars: A–C = 20 μM, A′–C′ = 9 μM, D–I = 30 μM.
Subsequent to cell shortening, we found that, by 24 hpf, a group of cells at the MHBC had become wedge-shaped, with constriction at their basal surface (Fig. 2C,C′). Within a single plane (Z-section) three to four wedge-shaped cells meet at a sharp point to form the MHBC in wild type (Fig. 2C′,L). Basally constricted cells were defined as those with a clear wedge-shaped morphology such that their basal surface was constricted to a point. We found that the apical width of the wedge-shaped cells at the MHBC had expanded to 1.6 times that of cells outside the MHBC (outlined cells in Fig. 2C′, Fig. 2K). Interestingly, although the midline in the MHB does not separate, we found that the basally constricted MHBC cells were not apposed at the midline, but instead were oriented with their apical surfaces exposed to the midbrain ventricle lumen (Fig. 2C,C′). These data demonstrate that cells at the MHBC undergo basal constriction and apical expansion.
2.5 Basal constriction at the MHBC occurs without ventricle inflation, but requires laminin
Since the MHBC forms a sharp point in the ventricle inflation mutants snk and nok, we asked whether basally constricted cells formed in these mutants. Confocal imaging indicated that cells at the MHBC in both mutants demonstrated basal constriction (Fig. 3A–C′). However, unlike wild type, the basally constricted cells in these mutants apparently did not show apical expansion, relative to adjacent cells in the same embryo (Fig. 3H). This may be because apical expansion requires that cells have an unconstrained apical surface, which occurs when wild type MHBC cells abut the midbrain lumen. Where the ventricles do not inflate and the midline of the brain primordium does not separate, the mutant cells may be constrained in their ability to expand apically. Therefore, the reduced bend angle formed at the MHBC in nok and snk may be due to failure of the cells at the MHBC to expand apically, in response to ventricle inflation. These data show that the basal constriction in the MHBC can occur independent of brain ventricle inflation, and moreover, that basal constriction is independent of apical expansion.
Fig. 3. Basal constriction at the MHBC is laminin-dependent and not dependent on ventricle inflation.
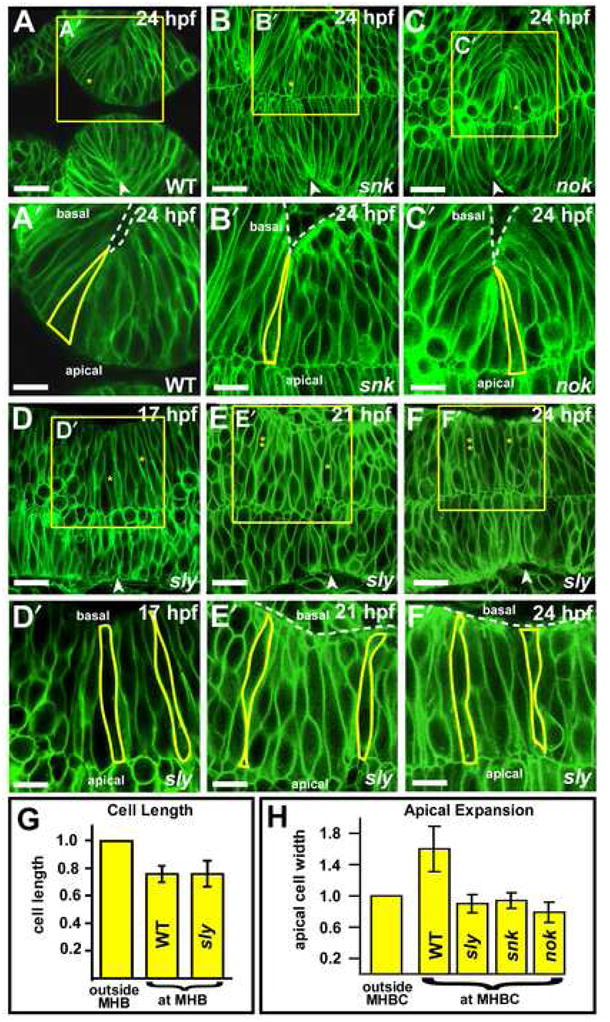
(A–C′) Live laser-confocal imaging of wild type, snk and nok embryos at 24 hpf after injection with memGFP. Boxed regions in A–C are enlarged in panels A′–C′. Cells at the MHBC in (A–A′) wild type, (B–B′) snk and (C–C′) nok undergo basal constriction (see cell outlines). (D–F′) Imaging of sly mutants injected with memGFP mRNA at the one cell stage and imaged at 17, 21, and 24 hpf. Boxed regions in D–F are enlarged for panels D′–F′. (D′) At 17 hpf, MHB and surrounding cells are similar in length (see outlined cells). (E′) At 21 hpf, cells at the MHB are shorter than those surrounding. One cell at and one cell outside the MHB are outlined in yellow. Some cells are visible outside the neural tube. (F′) At 24 hpf, cells at the MHBC fail to basally constrict. For panels A–C and D–F asterisks indicate the cell that is outlined in the image below. Cells with two asterisks are outside MHB. Arrowheads indicate the MHBC. Dotted lines delineate the outside of the neural tube. Some green fluorescence is visible within outlined cells since the plane of section contains the surface of the cell membrane. Anterior is to the left in all images. Scale bars: A–C = 22 μM, A′–C′ = 12 μM, D–F = 18 μM, D′–F′ = 9 μM. (G) Length of cells at the MHB relative to those outside the MHB in WT and sly mutants. At 21 hpf, cells at the MHB (at MHB) in sly mutants are 0.76 (+/− 0.094 s.d.) the length of those outside the MHB (outside MHB), as in WT embryos (WT data is the same as from Figure 2) (n = 6 embryos, 2 cells at MHB, 3 cells outside MHB were measured per embryo). (H) Graph compares the apical width of cells outside MHBC to cells at MHBC in WT, sly, snk, and nok embryos at 24 hpf. Basally constricted cells at the MHBC do not apparently show corresponding apical expansion snk and nok (n = 3 embryos each mutant, 2 cells at MHBC, 3 cells outside MHBC were measured per embryo). Cells at the MHBC in sly mutants that have shortened, but are not constricted basally, are also not expanded apically (n = 6 embryos, 3 cells at MHBC, 4 cells outside MHBC were measured per embryo).
In order to determine what aspect of MHBC formation is disrupted in laminin mutants, we analyzed sly embryos for changes in cell length and shape (Fig. 3D–F′). At 17 hpf, the cells at the MHB of sly mutants appeared similar to wild type (compare Fig. 2A′ with Fig. 3D′). By 21 hpf, cells at the MHBC in sly mutants were 0.76 the length of cells on either side, similar to wild type (Fig. 3E,E′,G). However, by 24 hpf, cells at the MHBC in sly mutants had not basally constricted (Fig. 3F′). Neither was apical expansion observed in the MHBC of these mutants (Fig. 3H). Based on somite number, and marker gene expression (Supplemental Fig 1), development of laminin mutant embryos was not retarded relative to wild type. These data show that laminin is necessary for basal constriction, defining a new role for this protein in morphogenesis. The data also show that basal constriction is required for formation of the sharp point at the MHBC and that cell shortening is insufficient for this process. Further, the absence of apical expansion in the laminin mutants indicates that this process may be dependent on basal constriction.
Together with results from the ventricle inflation mutants, the data show that apical expansion occurs only in the presence of, but is not required for, basal constriction, and suggest that basal constriction, and not apical expansion, is the active process driving MHBC morphogenesis.
2.6 Other process involved in MHBC morphogenesis
What other processes might be involved in MHBC formation? One possibility is that differential cell proliferation or apoptosis contributes to the MHBC. We have investigated cell death and proliferation in the brain during MHBC development, and find no significant differences between wild type and sly or gup embryos in the MHB region (Supplemental Fig. 2).
The role of the actin cytoskeleton during apical constriction is well established (Haigo et al., 2003; Pilot and Lecuit, 2005). Therefore, we hypothesized that actin may also localize basally in cells that are undergoing basal constriction at the MHBC. We addressed this by analyzing fixed wild type and sly embryos, at 24hpf, for actin localization using phalloidin staining. In wild type embryos, actin localized basally and apparently, at highest intensity in the basally constricted cells at the MHBC (Fig. 4A,C). In sly embryos, where cells are not constricted basally, actin lined the basal side of the neuroepithelium, but did not concentrate in any cells in this region (Fig. 4B,D). Although these data suggest differences in actin localization in the MHBC region in wild type versus sly embryos, further experiments, in live embryos, are required to more accurately investigate actin dynamics and localization at the MHBC.
Fig. 4. Actin is localized basally at the MHBC in wild type embryos.
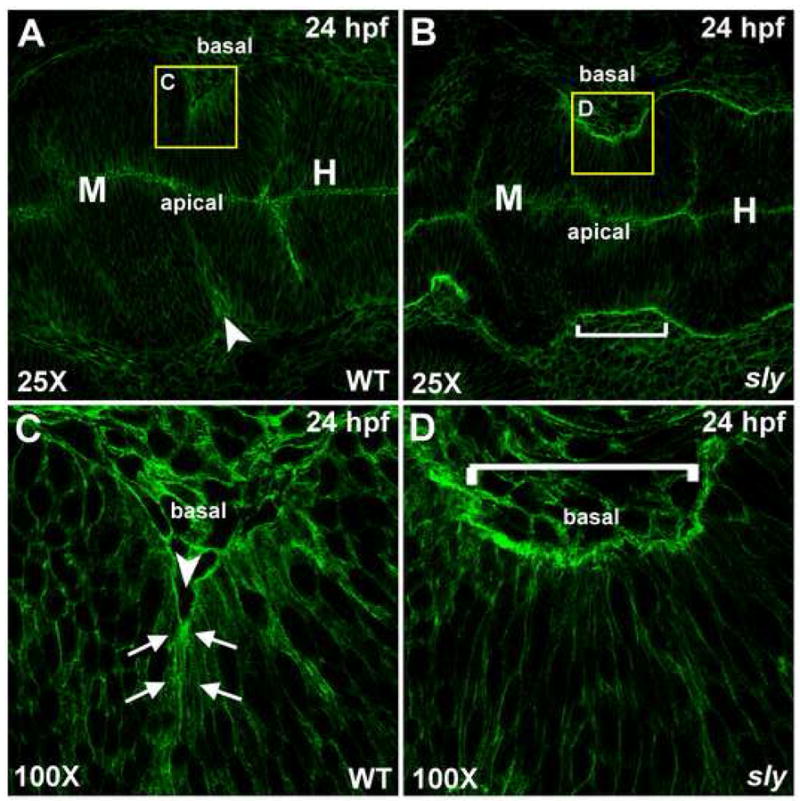
(A–D) Phalloidin stained wild type (A,C) and sly (B,D) embryos that were fixed at 24 hpf. Stained embryos were flat mounted in glycerol and imaged by confocal microscopy at 25X (A,B) and 100X (C,D). Arrows indicate points of basal actin accumulation in wild type embryos. Anterior is to the left in all images. Arrowhead indicates the MHBC in wild type and a bracket indicates the MHBC region in sly mutants. M, midbrain; H, hindbrain.
2.7 Model for MHBC morphogenesis
The model presented in Fig. 5 summarizes our data indicating that two steps are involved in formation of the MHBC in the embryonic brain. In the first step, cells of the MHB shorten relative to the surrounding cells. This is followed by laminin-dependent basal constriction and coordinate apical expansion of a small group of cells that contribute to the sharp bend of the MHBC. Basal constriction is an active process, and not a passive consequence of brain ventricle inflation or other aspects of brain morphogenesis, as it occurs in mutants lacking ventricle inflation. An active role for the basal side of the neuroepithelium is supported by failure of basal constriction in laminin mutants. Additionally, since apical expansion does not occur when the basement membrane is disrupted, information from the apical side of the neuroepithelium does not drive basal constriction, but rather basal constriction and apical expansion may occur coordinately.
Fig. 5. Schematic of the two steps required in MHBC formation.
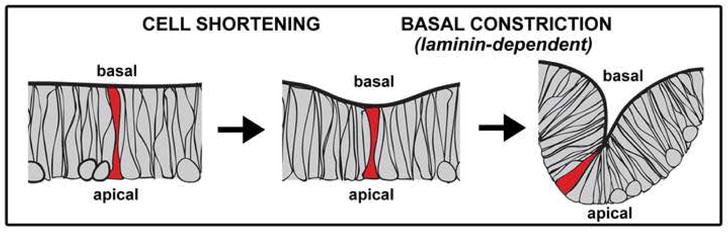
In step one, cells shorten at the MHB. In step two, 3 to 4 cells (in a plane of section) basally constrict, and expand apically, to form a sharp point at the MHBC. This second step is laminin dependent.
The mechanisms underlying basal constriction are not known; however, our data show that laminin provides a crucial function. Laminin is a secreted protein and can act on both the cells from which it is secreted and those surrounding (Parsons et al., 2002). Since laminin lines the entire neural tube, it is unlikely to play an instructive role in MHBC formation. Rather, laminin is likely to modulate basal constriction through its interaction with integrins in the basal cell membrane and subsequent regulation of cytoskeletal function. Although a difference in actin localization at the MHBC region may be present between wild type and sly mutants, we do not know whether actin is directly involved in driving basal constriction in these cells.
One clear requirement for MHBC formation is its precise positioning in the brain. While Fgf and Wnt signaling pathways are necessary to form the MHBC (Brand et al., 1996; Sato et al., 2004), it is not clear whether these genes are solely responsible for specifying MHB fate, or whether they also play a more direct role in MHBC morphogenesis. Laminin acts downstream of MHB specification, since genes indicative of specified MHB, including engrailed 3 and pax2a are normally expressed in laminin mutants (Supplemental Fig 1). Separating the effects of region-specific signaling pathways in positioning the MHB from their possible role in later MHBC morphogenesis, as well as determining the cell biology underlying MHBC morphogenesis will be the focus of future analyses.
3. Experimental Procedures
3.1 Fish lines and Maintenance
Zebrafish lines were maintained and stages determined as previously described (Kimmel et al., 1995; Westerfield, 1995). Strains used include wild-type AB, slym86 (Schier et al., 1996), guphi1113b (Amsterdam et al., 2004), nokm227 (Malicki et al., 1996), and snkto273a (Jiang et al., 1996).
3.2 Live imaging
Brain ventricle imaging was carried out as previously described (Lowery and Sive, 2005). For confocal imaging, single cell embryos were micro-injected with CAAX-eGFP mRNA (memGFP) (kindly provided by J. B. Green, Dana-Farber Cancer Institute Boston, MA) transcribed with the mMessage mMachine kit (Ambion). The embryos were mounted inverted in 0.7% agarose (Sigma) and imaged by fluorescent, laser-scanning confocal microscopy (Zeiss LSM510) or with spinning disk confocal microscopy (Perkin Elmer Ultraview RS). Time-lapse data was analyzed using Imaris (Bitplane).
3.3 Quantitation of cell length and apical cell width
Slices for measurement were chosen based on the ability to outline the entire extent of a cell from the apical to basal surface of the neuroepithelium and by following the cell through a full Z-series. The length of three cells at the MHBC and four cells outside the MHBC were measured using Imaris (Bitplane) software, and the ratio between cell lengths at and outside the MHBC were calculated for each embryo and averaged. The width of two wedge shaped cells at the MHBC and three unconstricted cells outside the MHBC at 24 hpf in each embryo were measured using Imaris (Bitplane). The error bars in Fig. 2 indicate the standard deviation between the ratios found for each embryo.
3.4 Immunohistochemistry
Embryos were fixed in 4% paraformaldehyde and dehydrated in methanol. After rehydrating in PBT, embryos were permeablized with 2.5 μg/ml proteinase K for 1 minute, and blocked in PBT, 0.1% Triton X, 1% BSA, and 1% NGS. Embryos were incubated overnight at room temperature in laminin antibody (laminin rabbit anti-mouse, Sigma L-9393, 1:150), washed, and incubated in secondary antibody, (goat anti-rabbit IgG Alexa Flour 488, Invitrogen, 1:500) in combination with propidium iodide (PI) (Invitrogen, 1:1000). Embryos were flat mounted in glycerol, imaged using a Zeiss LSM510 laser-scanning confocal microscope, and images analyzed with LSM software (Zeiss) and Photoshop.
3.5 Actin staining
Embryos were fixed in 4% paraformaldehyde for 2 hrs at room temperature and washed in PBT. Embryos were incubated at 4 degrees C overnight in Alexa Fluor 488 phalloidin (Invitrogen A12379) 1:40 in PBT, washed overnight at 4 degrees C in PBT, mounted in glycerol, imaged using a Zeiss LSM510 laser-scanning confocal microscope and images analyzed with LSM software (Zeiss) and Photoshop.
Supplementary Material
Fig. S1. Patterning gene expression is normal in sly mutants during MHBC formation. (A–H) Wild type and sly mutants were analyzed by in situ hybridization for pax2a and krox20 at 17, 20, and 24 hpf. Gene expression at 17 hpf was normal in sly compared to wild type shown in panels A and B. At 20 hpf gene expression remained normal, shown dorsally C, D and laterally E, F. At 24 hpf pax2a and krox20 are still expressed normally in sly mutants compared to wild type panels G, H. (I–J) Wild type and sly mutants were analyzed for expression of engrailed 3 (eng3) and krox20 at 24 hpf. In sly mutants, the eng3 expression domain appeared to be extended along the neural tube around the MHB relative to wild type. This is because the MHB tissue in the mutant is also elongated due to the lack of a sharp MHBC and MHB fold, indicating that gene expression in the MHB at the time of MHBC formation is the same in wild type embryos and in sly mutants. The bracket marks the MHB region in each embryo. Anterior is to the left in all images.
Fig. S2. Levels of proliferation and cell death in laminin mutants are normal. (A–C) Wild type and laminin mutant embryos were fixed and stained with PH3 antibody as a marker for cell proliferation at 22 hpf and the cells in the MHB were counted and compared between wild type and laminin mutants. Representative images of PH3 stained wild type and sly embryos are seen in panels A and B respectively. Quantitation of PH3 positive cells indicated that there was no difference in the number of positive cells in the MHB of laminin mutants compared to wild type controls, panel C. (n = 3 control, n = 5 laminin mutants; 3 sly, 2 gup) (D–E) Wild type and laminin mutants were assayed by TUNEL to determine amount of apoptosis at 22 hpf. Cells in the MHB were counted and compared between wild type and laminin mutants. Representative images of TUNEL stained wild type and sly mutants are seen in panels D and E respectively. Differences in refractility between D and E are due to microscope settings. Quantitation of TUNEL positive cells in the MHB of laminin mutants compared to wild type controls indicated that there was no difference in the number of positive cells, panel F. (n = 7 control, n = 5 laminin mutants, 3 sly, 2 gup). Anterior is to the left in all images. M, midbrain; H, hindbrain.
Acknowledgments
We thank the members of the Sive lab for helpful comments and Olivier Paugois for fish husbandry. Thanks to Nancy Hopkins and Adam Amsterdam for the guphi1113b mutant and to James Evans and Paul Matsudaira for advice and expertise in live imaging. This work was conducted using the Whitehead Institute-MIT Bioimaging Center at MIT and the W. M. Keck Foundation Biological Imaging Facility at the Whitehead Institute. HLS is supported by NIH MH70926 and MH077253. JHG is supported by a MIT CSBi/Merck postdoctoral fellowship. EGG is supported by an NSF pre-doctoral fellowship. LAL is supported by a NIH NRSA pre-doctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–9. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Bernfield M, Banerjee SD, Koda JE, Rapraeger AC. Remodelling of the basement membrane: morphogenesis and maturation. Ciba Found Symp. 1984;108:179–96. doi: 10.1002/9780470720899.ch12. [DOI] [PubMed] [Google Scholar]
- Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Lun K, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, van Eeden FJ, Nusslein-Volhard C. Mutations in zebrafish genes affecting the formation of the boundary between midbrain and hindbrain. Development. 1996;123:179–90. doi: 10.1242/dev.123.1.179. [DOI] [PubMed] [Google Scholar]
- Fristrom D. The cellular basis of epithelial morphogenesis. A review. Tissue Cell. 1988;20:645–90. doi: 10.1016/0040-8166(88)90015-8. [DOI] [PubMed] [Google Scholar]
- Garcia-Alonso L, Fetter RD, Goodman CS. Genetic analysis of Laminin A in Drosophila: extracellular matrix containing laminin A is required for ocellar axon pathfinding. Development. 1996;122:2611–21. doi: 10.1242/dev.122.9.2611. [DOI] [PubMed] [Google Scholar]
- Haigo SL, Hildebrand JD, Harland RM, Wallingford JB. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr Biol. 2003;13:2125–37. doi: 10.1016/j.cub.2003.11.054. [DOI] [PubMed] [Google Scholar]
- Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421:172–7. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Brand M, Heisenberg CP, Beuchle D, Furutani-Seiki M, Kelsh RN, Warga RM, Granato M, Haffter P, Hammerschmidt M, Kane DA, Mullins MC, Odenthal J, van Eeden FJ, Nusslein-Volhard C. Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development. 1996;123:205–16. doi: 10.1242/dev.123.1.205. [DOI] [PubMed] [Google Scholar]
- Joyner AL. Engrailed, Wnt and Pax genes regulate midbrain--hindbrain development. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- Karlstrom RO, Trowe T, Klostermann S, Baier H, Brand M, Crawford AD, Grunewald B, Haffter P, Hoffmann H, Meyer SU, Muller BK, Richter S, van Eeden FJ, Nusslein-Volhard C, Bonhoeffer F. Zebrafish mutations affecting retinotectal axon pathfinding. Development. 1996;123:427–38. doi: 10.1242/dev.123.1.427. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lee C, Scherr HM, Wallingford JB. Shroom family proteins regulate gamma-tubulin distribution and microtubule architecture during epithelial cell shape change. Development. 2007;134:1431–41. doi: 10.1242/dev.02828. [DOI] [PubMed] [Google Scholar]
- Louvi A, Alexandre P, Metin C, Wurst W, Wassef M. The isthmic neuroepithelium is essential for cerebellar midline fusion. Development. 2003;130:5319–30. doi: 10.1242/dev.00736. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development. 2005;132:2057–67. doi: 10.1242/dev.01791. [DOI] [PubMed] [Google Scholar]
- Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–73. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- Miner JH, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004;20:255–84. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Pollard SM, Saude L, Feldman B, Coutinho P, Hirst EM, Stemple DL. Zebrafish mutants identify an essential role for laminins in notochord formation. Development. 2002;129:3137–46. doi: 10.1242/dev.129.13.3137. [DOI] [PubMed] [Google Scholar]
- Paulus JD, Halloran MC. Zebrafish bashful/laminin-alpha 1 mutants exhibit multiple axon guidance defects. Dev Dyn. 2006;235:213–24. doi: 10.1002/dvdy.20604. [DOI] [PubMed] [Google Scholar]
- Pilot F, Lecuit T. Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev Dyn. 2005;232:685–94. doi: 10.1002/dvdy.20334. [DOI] [PubMed] [Google Scholar]
- Puelles L, Martinez-de-la-Torre M. Autoradiographic and Golgi study on the early development of n. isthmi principalis and adjacent grisea in the chick embryo: a tridimensional viewpoint. Anat Embryol (Berl) 1987;176:19–34. doi: 10.1007/BF00309748. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Brand M. The midbrain--hindbrain boundary organizer. Curr Opin Neurobiol. 2001;11:34–42. doi: 10.1016/s0959-4388(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Sato T, Joyner AL, Nakamura H. How does Fgf signaling from the isthmic organizer induce midbrain and cerebellum development? Dev Growth Differ. 2004;46:487–94. doi: 10.1111/j.1440-169x.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Harvey M, Malicki J, Solnica-Krezel L, Stainier DY, Zwartkruis F, Abdelilah S, Stemple DL, Rangini Z, Yang H, Driever W. Mutations affecting the development of the embryonic zebrafish brain. Development. 1996;123:165–78. doi: 10.1242/dev.123.1.165. [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–67. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- Smith JL, Schoenwolf GC, Quan J. Quantitative analyses of neuroepithelial cell shapes during bending of the mouse neural plate. J Comp Neurol. 1994;342:144–51. doi: 10.1002/cne.903420113. [DOI] [PubMed] [Google Scholar]
- Svoboda KK, O’Shea KS. An analysis of cell shape and the neuroepithelial basal lamina during optic vesicle formation in the mouse embryo. Development. 1987;100:185–200. doi: 10.1242/dev.100.2.185. [DOI] [PubMed] [Google Scholar]
- Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112:775–89. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31:150–7. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish. University of Oregon Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Patterning gene expression is normal in sly mutants during MHBC formation. (A–H) Wild type and sly mutants were analyzed by in situ hybridization for pax2a and krox20 at 17, 20, and 24 hpf. Gene expression at 17 hpf was normal in sly compared to wild type shown in panels A and B. At 20 hpf gene expression remained normal, shown dorsally C, D and laterally E, F. At 24 hpf pax2a and krox20 are still expressed normally in sly mutants compared to wild type panels G, H. (I–J) Wild type and sly mutants were analyzed for expression of engrailed 3 (eng3) and krox20 at 24 hpf. In sly mutants, the eng3 expression domain appeared to be extended along the neural tube around the MHB relative to wild type. This is because the MHB tissue in the mutant is also elongated due to the lack of a sharp MHBC and MHB fold, indicating that gene expression in the MHB at the time of MHBC formation is the same in wild type embryos and in sly mutants. The bracket marks the MHB region in each embryo. Anterior is to the left in all images.
Fig. S2. Levels of proliferation and cell death in laminin mutants are normal. (A–C) Wild type and laminin mutant embryos were fixed and stained with PH3 antibody as a marker for cell proliferation at 22 hpf and the cells in the MHB were counted and compared between wild type and laminin mutants. Representative images of PH3 stained wild type and sly embryos are seen in panels A and B respectively. Quantitation of PH3 positive cells indicated that there was no difference in the number of positive cells in the MHB of laminin mutants compared to wild type controls, panel C. (n = 3 control, n = 5 laminin mutants; 3 sly, 2 gup) (D–E) Wild type and laminin mutants were assayed by TUNEL to determine amount of apoptosis at 22 hpf. Cells in the MHB were counted and compared between wild type and laminin mutants. Representative images of TUNEL stained wild type and sly mutants are seen in panels D and E respectively. Differences in refractility between D and E are due to microscope settings. Quantitation of TUNEL positive cells in the MHB of laminin mutants compared to wild type controls indicated that there was no difference in the number of positive cells, panel F. (n = 7 control, n = 5 laminin mutants, 3 sly, 2 gup). Anterior is to the left in all images. M, midbrain; H, hindbrain.


