Abstract
The innate immune system is a two-edged sword; it is absolutely required for host defense against infection but, uncontrolled, can trigger a plethora of inflammatory diseases. Here we used systems biology approaches to predict and validate a gene regulatory network involving a dynamic interplay between the transcription factors NF-κB, C/EBPδ, and ATF3 that controls inflammatory responses. We mathematically modeled transcriptional regulation of Il6 and Cebpd genes and experimentally validated the prediction that the combination of an initiator (NF-κB), an amplifier (C/EBPδ) and an attenuator (ATF3) forms a regulatory circuit that discriminates between transient and persistent Toll-like receptor 4-induced signals. Our results suggest a mechanism that enables the innate immune system to detect the duration of infection and to respond appropriately.
Introduction
The innate immune system must provide stable, specific, and protective responses in a diverse pathogenic environment, while at the same time attenuating the collateral damage inflicted by the inflammation associated with such responses1-8. Much has been learned about the recognition mechanisms that facilitate the specificity of innate immune responses. In general, pattern recognition receptors such as the Toll-like receptors (TLRs) recognize microbial components9-11 and activate intracellular signaling pathways leading to the transcriptional induction of genes that are critical for protective inflammatory responses12,13.
Bacterial lipopolysaccharide (LPS) is a major surface component of Gram negative bacteria and is detected by TLR4 (http://www.signaling-gateway.org/molecule/query?afcsid=A002296)14. LPS-stimulation leads to macrophage activation as characterized by changes in extracellular and intracellular microbial killing systems, production and secretion of pro-inflammatory cytokines and chemokines, enhanced expression of co-stimulatory receptors that are essential for efficient T-cell activation and enhanced production of arachidonic acid metabolites15,16. These and other inflammatory responses in macrophages are largely driven at the level of transcription17,18. However, the gene regulatory program of TLR-induced macrophage activation is not well understood. It is known that macrophages express more than 500 transcription factors19, of which approximately 100 are induced by LPS; this suggests a high degree of complexity in the regulation of TLR4-induced responses.
In this report we use the tools of systems biology20-24 to unravel a transcriptional circuit leading to the TLR4-activated state in macrophages. Briefly, temporal activation of macrophages by LPS was analyzed using microarrays and these data were then clustered to reveal regulated ‘waves’ of transcription. It is well established that genes which are co-regulated often share cis-regulatory elements, and that transcriptional programs are propagated by sequential cascades of transcription factors25,26. We therefore identified transcription factors in the first cluster of expressed genes (cluster 1) and used computational motif scanning to predict which genes in cluster 2 contained promoter binding sites for cluster 1 transcription factors. These predictions were then validated using chromatin immunoprecipitation (ChIP), a method which also permitted us to establish the kinetics of promoter occupancy. These kinetic data allowed mathematical modeling of the transcriptional circuitry, which, in turn, enabled the prediction of novel functions that are not easily identified using conventional approaches. The functional predictions were then tested in cell culture systems and in mice.
We used this strategy to identify a new regulatory circuit involving the transcription factors NF-κB (http://www.signaling-gateway.org/molecule/query?afcsid=A002052), ATF3 (http://www.signaling-gateway.org/molecule/query?afcsid=A003217) and C/EBPδ. We predicted and validated that C/EBPδ acts as an amplifier of NF-κB responses, and that it discriminates between transient and persistent TLR4 signals. Using ChIP-on-chip analysis we identified 63 LPS-induced C/EBPδ-targets of which a large of fraction was implicated in host defense to bacterial infection. Integration of the kinetic and functional data strongly suggests a mechanism by which C/EBPδ participates in the control of persistent bacterial infections.
Results
NF-κB and ATF3 control C/EBPδ expression
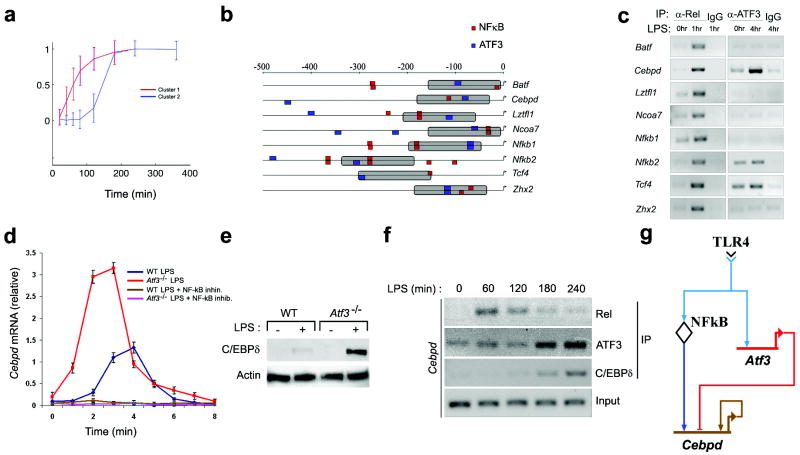
A schematic of the experimental design used here is depicted in Supplementary Fig.1 online. Transcriptome analysis demonstrated that LPS induced the expression of two temporal clusters of transcription factors within 3 hrs: the early cluster (cluster 1) was comprised of 23 transcription factors and the intermediate cluster (cluster 2) contained 55 transcription factors (Fig. 1a, and Supplementary Table 1). We decided to focus on the transcriptional circuitry involving two cluster 1 transcription factors, Rel (NF-κB) and ATF3, as we previously demonstrated that Rel activates and ATF3 attenuates a subset of LPS-induced genes27. We therefore scanned the promoters of the 55 cluster 2 transcription factors for ATF3 and NF-κB binding sites, and identified 8 genes whose promoters contained candidate binding sites for both ATF3 and NF-κB within 1500 bp of the transcriptional start site and within 150 bp of each other (Supplementary Table 2). This subset included Batf, Cebpd, Lztfl1, Ncoa7, Nfkb1, Nfkb2, Tcf4, and Zhx2 (Fig. 1b). ChIP analysis demonstrated that LPS induced the binding of NF-κB (at 1hr) and ATF3 (at 4hr) to the promoters of Cebpd, Nfkb2 and Tcf4 (Fig. 1c). We focused our subsequent experiments on Cebpd as LPS stimulated NF-κB and ATF3 binding to the Cebpd promoter (Fig. 1c). Transcription of Cebpd was induced by LPS (Fig. 1d). Pharmacological inhibition of NF-κB blocked LPS-induced Cebpd transcription (Fig. 1d), and LPS-induced Cebpd mRNA (Fig. 1d) and protein (Fig. 1e) quantities were substantially increased in Atf3-null macrophages. Taken together, these data demonstrate that LPS-induced transcription of Cebpd is absolutely dependent on NFκB, and that NFκB-dependent Cebpd mRNA production is attenuated by ATF3. ChIP analysis demonstrated rapid and transient recruitment of Rel to the promoter of the Cebpd gene; maximal binding was seen 1 hr after stimulation with LPS (Fig. 1f). By contrast, LPS-stimulated ATF3 binding to the promoter of the Cebpd gene (over basal amounts) occurred after 3 h and was sustained (Fig. 1f). Interestingly, Motif scanning of the 5’ cis-regulatory region of the Cebpd gene predicted that this transcription factor can bind to its own promoter (Supplementary Table 3); this prediction was confirmed by ChIP (Fig. 1f). The kinetics of C/EBPδ binding to its own promoter paralleled that of ATF3.
Figure 1. Prediction and validation of LPS-induced transcription factor network involving NF-κB, C/EBPδ and ATF3.
a, Macrophages from wild-type mice were stimulated with 10 ng/ml LPS for the indicated times. mRNA was isolated and subjected to microarray analysis. A total of 78 TFs were detected in cluster 1 (red line) and cluster 2 (blue line) kinetic clusters. Shown are normalized log fold-change gene expression data over time. Each profile shows the cluster-average expression for a single cluster over 6 h after LPS-stimulation. Data represent the average of three independent experiments.
b, ATF3 and NF-κB binding sites were identified in the cis-regulatory regions of transcription factor genes (cluster 2). Predicted targets were filtered using the additional constraint of a 150bp proximity limit (gray bars) between putative ATF3 and NF-κB binding sites.
c, Macrophages from wild-type mice were stimulated with 10ng/ml LPS, for the indicated time periods. Nuclear Rel and ATF3 were immunoprecipitated and the indicated genes were amplified by via PCR from transcription factor-bound DNA. IgG immunoprecipitates, negative control. Data are representative of two independent experiments.
d, WT and Atf3-/- macrophages were stimulated with 10 ng/ml LPS in the presence or absence of the NF-κB inhibitor sc-514 (25μM) for the indicated times. Data represent the average of three independent experimental values ± standard error.
e, Macrophages from wild-type and Atf3-/- mice were stimulated with 10ng/ml LPS. Cells were harvested 4 h after LPS stimulation and the lysates were subjected to immunoblotting with the indicated antibodies. Actin, loading control. Data are representative of three independent experiments.
f, Macrophages from wild-type mice were stimulated with 10 ng/ml LPS for the indicated times. Kinetics of Rel, C/EBPδ, and ATF3 recruitment to the Cebpd promoter were assayed by ChIP followed by PCR amplification, as in c. Data are representative of three independent experiments.
g, Transcriptional factor transcriptional network model depicted as a BioTapestry diagram36.
These observations suggested a model wherein TLR4 activates NF-κB, which then binds to the promoter of Cebpd and activates it (Fig. 1g). TLR4 also activates the transcription of Atf327 (data not shown), and stimulates its later recruitment to the Cebpd promoter. The binding of ATF3 to the promoter of Cebpd inhibits its NF-κB-dependent transcription. C/EBPδ is also recruited to its own promoter in a TLR4-dependent manner, suggesting auto-regulation.
Regulatory circuit involving NF-κB, C/EBPδ and ATF3
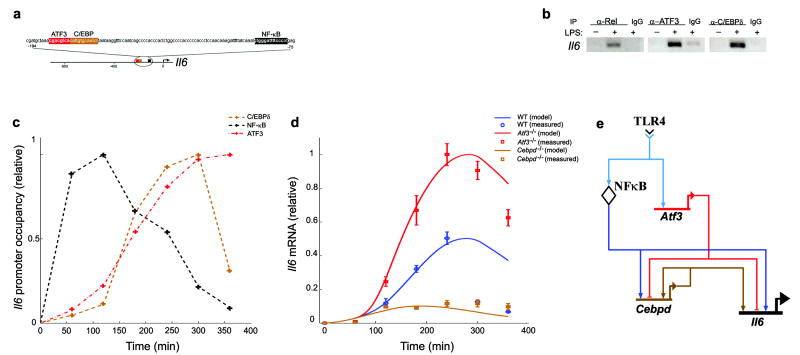
Motif scanning analysis predicted the presence of binding sites for NF-κB, C/EBPδ and ATF3 in the cis-regulatory regions of 146 LPS-induced genes (Supplementary Table 4). Many of these genes play well-established roles in regulating the immune response; Il6 was selected for further study because we had previously explored its regulation by NF-κB and ATF327. Motif scanning predicted the existence of NF-κB, ATF3 and C/EBPδ binding sites in the cis-regulatory region of Il6 (Fig. 2a) and this prediction was confirmed using ChIP (Fig. 2b). Notably, the binding of NF-κB, ATF3 and C/EBPδ to the Il6 promoter occurred in an LPS-dependent manner (Fig. 2b). The Rel subunit of NF-κB was transiently recruited to the Il6 promoter with maximal binding at 2 h after LPS stimulation (Fig. 2c), as was RelA (data not shown). ATF3 and C/EBPδ demonstrated slower kinetics of recruitment with maximal binding at 4-5 h after LPS stimulation (Fig. 2c).
Figure 2. Mathematical model characterizing Il6 transcriptional regulation in TLR4-stimulated macrophages.
a, Shown are NF-κB, ATF3 and C/EBPδ binding sites locations in the Il6 gene promoter relative to transcription start site, as predicted by motif scanning.
b, Macrophages from wild-type mice were stimulated with 10 ng/ml LPS for 6 h and processed for ChIP assays as in Fig. 1c. The binding of immunoprecipitated NF-κB (Rel), ATF3 and C/EBPδ to the Il6 promoter was measured by PCR. InG, negative control immunoprecipitation. Data are representative of three independent experiments.
c, Macrophages from wild-type mice were stimulated with 10 ng/ml LPS for the indicated times and processed for ChIP assays as in Fig. 1c. The binding of immunoprecipitated NFκB (Rel), C/EBPδ and ATF3 to the Il6 promoter was measured by quantitative real-time RT-PCR. Transcription factor binding was normalized to the amount of PCR product loaded. Data represent the average of three independent experiments.
d, Predicted Il6 expression amounts in wild-type, Cebpd-/- and Atf3-/- cases of the kinetic model are shown as lines and measured Il6 mRNA quantities in wild-type, Cebpd-/- and Atf3-/- macrophages are shown as points. Data represent the average of three independent experimental values ± standard error.
e, Extended transcriptional network model depicted as a BioTapestry diagram36: Il6 gene expression is controlled by superposition of three network motifs: first, positive auto-regulation which is mediated by C/EBPδ binding its own promoter; second, feed-forward transcriptional activation of Il6, mediated by NF-κB and C/EBPδ; third, feed-forward transcriptional inhibition mediated by ATF3 binding to Cebpd and Il6 promoters.
To explore relative roles of each transcription factor in LPS-induced IL-6 production, we examined Cebpd-/- and Atf3-/- macrophages (Fig. 2d), as well as macrophages treated with NF-κB inhibitors (Supplementary Fig. 4). LPS-induced Il6 mRNA production was significantly increased in Atf3-/- macrophages and substantially decreased in Cebpd-/- cells relative to the wild-type macrophages (Fig. 2d). Notably, LPS-induced NF-κB activation and ATF3 expression were unaltered in Cebpd-/- macrophages (data not shown). Taken together, these data suggested a model in which Il6 production is initiated by NF-κB, amplified by CEBPδ, and attenuated by ATF3 (Fig. 2e).
We developed a mathematical model of this regulatory network. We assumed that the rate of Il6 transcriptional initiation depends on the fractional promoter occupancy by the transcription factors NF-κB, ATF3, and CEBPδ, that NF-κB acts as an activator of Cebpd (Fig. 1d) and Il6 transcription (Supplementary Fig. 4), that ATF3 attenuates transcription of Cebpd (Fig. 1d) and Il6 (Fig. 2d), and that C/EBPδ acts only in cooperation with NF-κB (Supplementary Fig. 5). A detailed description of the kinetic modeling is provided in Supplementary data. This model was fit to the LPS-induced Il6 expression measurements in wild-type, Atf3-/-, and Cebpd-/- macrophages described above (Fig. 2d). Seven parameters of the kinetic model were determined by minimizing the prediction error for recapitulating time-course gene and protein expression data in LPS-stimulated macrophages in three genotypes (wild-type, Atf3-/-, and Cebpd-/-). The model parametric complexity, with a ratio of 1.17 fit parameters per dynamic variable and a ratio of approximately six measured data points per fit parameter, is comparable to previously published kinetic models28,29. Consistent with a model that is not over fitted, the predicted Il6 transcriptional response was found to be robust with respect to simultaneous variation of the seven parameters (Supplementary Fig. 3).
The salient properties of the model are depicted in Fig. 2e. TLR4 stimulates the translocation of NF-κB to the nucleus where it activates a low degree transcription of Il6. Concomitantly NF-κB induces expression of C/EBPδ, which then binds to the Il6 promoter and cooperates with NF-κB to stimulate maximal transcription of the cytokine gene; this is known as coherent feed-forward type I regulation30. Additional features of the model include autoregulation of Cebpd (positive feedback) and ATF3-mediated attenuation of Cebpd and Il6 transcription.
C/EBPδ discriminates transient from persistent TLR4 signals
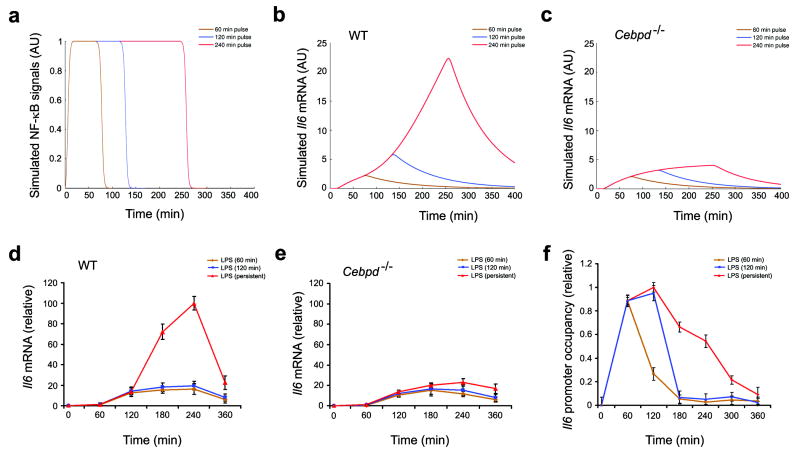
Coherent feed-forward type I regulation protects biological systems from unwanted responses to fluctuating inputs30. Given the double-edged nature of inflammation it is critical that the macrophage be capable of discerning a persistent from a transient insult; this would enable the cell to discriminate between real and spurious threats. For this reason, we hypothesized that the architecture of the NF-κB, ATF3 and C/EBPδ regulatory circuit may have evolved to serve the function of discriminating between transient and persistent innate immune stimuli. To test this hypothesis we simulated Il6 transcriptional activation under LPS pulsing by computationally varying the NF-κB activation signal as described in Supplementary data (Fig. 3a). Short-duration pulses were predicted by the model to induce only weak Il6 mRNA production while persistent stimulation was predicted to super-induce Il6 transcription (Fig. 3b). Furthermore, the super-induction of Il6 induced by persistent stimulation was predicted to be absent in Cebpd-/- macrophages (Fig. 3c). Measurements of Il6 expression in wild-type and Cebpd-/- macrophages were in qualitative agreement with these model predictions (Fig. 3d,e). Notably, the measured Rel binding dynamics to the Il6 promoter (Fig. 3f) were in qualitative agreement with computationally simulated NF-κB inputs (Fig. 3a). We re-confirmed our model predictions using measured Rel binding (Fig. 3f) as an input for computational simulation of Il6 mRNA production in wild-type and Cebpd-/-macrophages (Supplementary Fig. 2). Taken together, these results suggest that the overall function of the feed-forward motifs involving NF-κB, C/EBPδ and ATF3 is to detect and to respond to persistent signals while filtering out brief inputs. It is also formally possible that C/EBPδ mediates a mechanism to sense the dose rather than the duration of the response. This possibility is less likely, as the LPS concentration required for half-maximal induction of Il6 transcription (with continuous stimulation) was similar in both WT and Cebpd-/- macrophages (half-maximal concentration ≈ 0.4 ng/ml).
Figure 3. Computational simulations of the transcriptional response of Il6 to TLR4 signals of varying duration reveal a threshold effect.
a, Computational simulation of transient and persistent NF-κB signals. To simulate LPS pulsing, the NFκB signal was computationally varied over time. NF-κB signals of the same amplitude but different duration are shown.
b,c Outputs of computationally simulated Il6 transcriptional response to transient LPS signals in WT (b) and Cebpd-/- (c) macrophages.
d,e Macrophages from wild-type (d) and Cebpd-/- (e) mice were stimulated for 1 or 2 h or persistently with 10ng/ml LPS. Cells were harvested at the indicated time points and Il6 mRNA was measured by quantitative real-time RT-PCR. Data points represent the average of three independent experimental values and error bars indicate ± standard error.
f, Macrophages from wild-type mice were stimulated for 1 h or 2 h or persistently with 10 ng/ml LPS. Cells were harvested at the indicated times and processed for ChIP assays as in Fig. 1c. Presence of immunoprecipitated Rel, on the Il6 promoter was measured by quantitative real-time RT-PCR. Transcription factor binding was normalized to the amount of PCR product loaded. Data points represent the average of three independent experimental values and error bars indicate ± standard error.
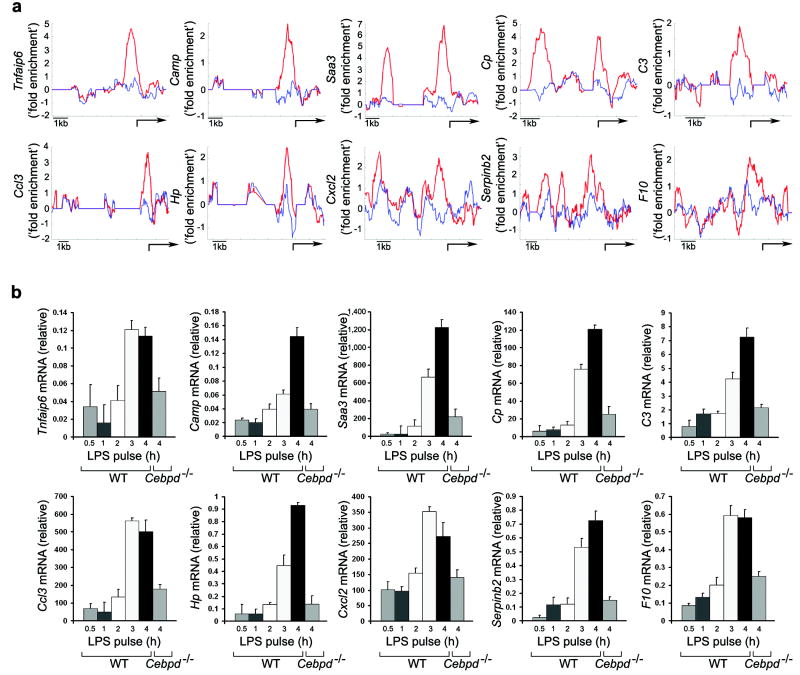
Given the important functional role played by the NF-κB, C/EBPδ and ATF3 circuit, it is likely that other innate immune genes in addition to Il6 are regulated in this manner. To begin to define this set of genes we performed whole genome location analysis and observed that TLR4 activation stimulated the recruitment of C/EBPδ to the promoters of 63 LPS-induced genes at 6 h after LPS stimulation, including Serpinb2, Cp, Saa3, Hp, Camp, C3, Tnfaip6, Ccl3, Cxcl2, and F10 (Fig. 4a, Supplementary Table 5). Transcription genes in response to persistent LPS stimulation was significantly blunted in Cebpd-/- macrophages, confirming the notion that C/EBPδ regulates these genes (Fig. 4b).
Figure 4. Identification of C/EBPδ-direct targets.
a, Macrophages from wild-type mice were stimulated for 6 h with 10 ng/ml LPS. Cells were harvested and processed for immunoprecipitation with polyclonal antibodies against C/EBPδ. The binding of immunoprecipitated C/EBPδ to the promoters of target genes was detected using the Affymetrix GeneChip Mouse Promoter 1.0R Array. Averaged and normalized C/EBPδ binding across indicated promoters are shown for LPS-stimulated (red line) and unstimulated (blue line) cells. Arrow, transcription start site. Data represent the average of two independent experimental values.
b, Macrophages from wild-type and Cebpd-/- mice were stimulated with 1ng/ml LPS for the indicated time periods. Four hours after LPS stimulation cells were harvested and indicated mRNA transcripts were measured by quantitative real-time RT-PCR. Data points represent the average of three independent experimental values and error bars indicate ± the standard error.
However, transcription induced in response to short-duration TLR4 stimulation induced was similar in wild-type and Cebpd-/- macrophages (data not shown). Overall these results suggest that, as in the case of Il6, C/EBPδ discriminated between transient and persistent signals leading to the activation of these genes. A large number of the C/EBPδ-regulated genes are known to be associated with host defense to infection (Supplementary Table 5).
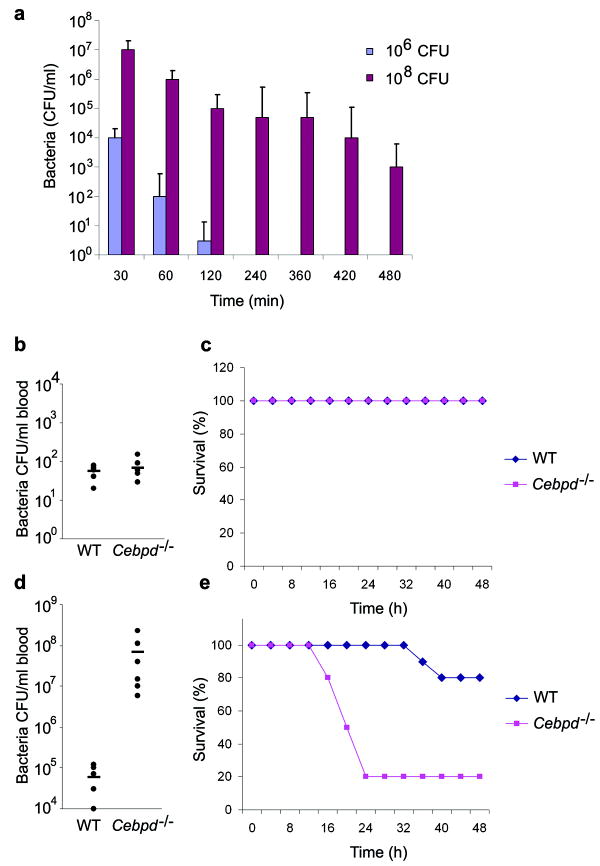
We therefore tested whether C/EBPδ could discriminate between transient and persistent infection in vivo. We established a Gram negative bacterial peritoneal infection model in mice in which a low dose of Escherichia coli, H904931 (1×106 colony-forming units (cfu)) was cleared rapidly, whereas a high dose of bacteria (1×108 cfu) resulted in a persistent infection (Fig. 5a). Next we compared the capacity of wild-type and Cebpd-/- mice to clear low and high doses of E. coli; the bacterial burden in the blood was examined 18 h post-infection. Wild-type and Cebpd-/- mice cleared the low dose bacterial infection with similar efficiency (Fig. 5b, c). However, high dose infection of Cebpd-/- mice resulted in severe bacteremia; Cebpd-/- mice had a 1,000-fold higher bacterial load in the blood than wild-type mice (Fig. 5d). In addition, whereas either wild-type nor Cebpd-/- mice succumbed following low dose infection (Fig. 5c), 80% of Cebpd-/- mice, but no wild-type mice, succumbed within 24 h with a high dose of bacteria (5×108 cfu.) (Fig. 5e).
Figure 5. The role of C/EBPδ in the restriction of transient and persistent bacterial infections.
a, Wild-type mice were challenged intraperitoneally with low (1×106 colony-forming units (cfu)) or high (1×108 cfu) doses of Escherichia coli, H9049. Shown are averaged peritoneal bacterial counts (± standard error) at the indicated time points after infection (n=6 mice for each data point).
b, Bacterial burden in the blood was measured 18 h after intraperitoneal (i.p.) infection of wild-type and Cebpd-/- mice with 1×106 c.f.u. E. coli H9049. Individual c.f.u. values and the average (horizontal line) from one representative experiment out of three are shown (n=6 mice for each group)
c, Survival curve comparing wild type and Cebpd-/- mice infected i.p. with 1×106 c.f.u. E. coli H9049 (n =10 for each group). Data are representative of three independent experiments.
d, Bacterial burden in the blood was measured 18 h after i.p. infection of wild-type and Cebpd-/- mice with 1×108 c.f.u. E. coli H9049. Individual c.f.u. values and the average (horizontal line) from one representative experiment out of three are shown (n=6 mice for each group)
e, Survival curve comparing wild type and Cebpd-/- after i.p. challenge with 5×108 c.f.u. E. coli H9049 per mouse (n =10 for each group). Data are representative of three independent experiments.
Discussion
It is becoming increasingly apparent that the tools of systems biology are invaluable in deciphering the complexity of the immune system and in predicting novel drug targets20-24. TLR4 stimulation results in the induction of a complex gene regulatory network that programs macrophage activation resulting in an effective host response to pathogens12,13,15,16. We have shown that the TLR4 agonist, LPS, regulates the transcription of approximately 2000 genes within 24 hrs in macrophages32. It is well established that transcriptional programs are propagated by sequential cascades of transcription factors25,26. We showed here that LPS-stimulation of macrophages induces the transcription of two clusters of transcription factors within 3 hrs; the first cluster contains 23 TFs and the second cluster contains 55 TFs. Next we used a combination of mathematical and biological experiments to predict and validate a transcriptional network involved in TLR4 activation. The power of the approach lies in its ability to rapidly uncover complex interactions between transcription factors, and to define the functional emergent properties of the system which, in turn, suggest the molecular underpinnings of the biological response. An analysis of the transcription factors in clusters 1 and 2 predicted a number of networks involved in the TLR4 response.
We focused on an NF-κB(Rel)/ATF3/C/EBPδ sub-network; each of these transcription factors had previously been shown to participate in host defense27,33,34, but their interaction, and the consequences of this interaction in the innate immune response, was not previously described. High density temporal measurements of LPS-induced binding of these transcription factors to the Il6 promoter, combined with gene deletion studies, enabled us to construct a model of a regulatory circuit that participates in the transcription of this cytokine gene. In this model TLR4 stimulates the translocation of NF-κB to the nucleus where it activates weak transcription of Il6. Concomitantly NF-κB induces C/EBPδ, which then binds to the Il6 promoter and cooperates with NF-κB to stimulate maximal transcription of the cytokine gene. At a later time point ATF3 attenuates Cebpd and Il6 transcription. We previously demonstrated that ATF3 recruits histone deacetylase 1 (HDAC1) to the Il6 promoter in an LPS-dependent manner. The ATF3-associated HDAC1 then deacetylates histones, resulting in the closure of chromatin and the inhibition of Il6 transcription27. It is known that C/EBPδ binds to and recruits the histone acetylase CBP to its target promoters, thus leading to the increased histone acetylation and to the opening of chromatin35. It is therefore possible that the NF-κB (Rel)-ATF3-C/EBPδ regulatory network is regulated by epigenetic chromatin remodeling. The relationship between NFkB and C/EBPδ suggests coherent feed-forward type I regulation30. This type of regulation has been suggested to protect biological systems from unwanted responses to fluctuating inputs30. The inflammatory response is a two-edged sword and it is therefore critical that inflammatory cells be able to discriminate between real and perceived threats. The coherent feed-forward type I regulatory circuit described above could, in principle, enable immune cells to filter transient insults from more dangerous persistent attacks. Exploration of this concept necessitates computational simulation of the system; therefore we used time-delay differential equations to simulate pulses of NF-κB activation and to examine transcriptional responses in silico. These simulations demonstrated a threshold effect in the transcriptional regulation of Il6 and a critical role for C/EBPδ in a regulatory circuit that discriminates transient and persistent TLR4-stimulation. The predictions were validated in LPS-stimulated macrophages and in an in vivo model of bacterial infection.
We used a combination of motif scanning, microarray and ChIP-to-chip analysis and identified a large number of LPS-induced C/EBPδ targets. These genes demonstrated differential transcriptional responsiveness to persistent and transient LPS-dependent stimulation of macrophages in vitro, and many have ascribed roles in host defense to bacterial infection. Consistent with the in vitro studies, Cebpd-null mice were able to resist low dose, transient, infection with E. coli H9049, but were highly susceptible to higher dose, persistent infection. In summary, we have used the tools of systems biology to demonstrate that TLR4-induced inflammatory responses are regulated by the integration of transcriptional “on” and “off” switches with “amplifiers” and “attenuators”. In addition, we have demonstrated a mechanism by which the macrophages are able to discriminate between real and perceived threats. Collectively these regulatory elements may facilitate the maintenance of effective host defense and the prevention of inflammatory disease.
Methods
Mouse BM-derived macrophages
BM derived macrophages (BMDM) were isolated from C57BL/6, Atf3-/- and Cebpd-/- mice essentially as described27. Briefly, BM cells collected from femurs were plated on non-tissue culture-treated plastic in complete RPMI containing 10% FBS (Hyclone Laboratories), 2mM L-glutamine, 100 IU/mL penicillin and 100ug/mL streptomycin,(all from Cellgro, Mediatech), and supplemented with recombinant human M-CSF (rhM-CSF) at 50 ng/mL (Chiron). BMDM were stimulated with high purity 10 ng/mL LPS (S. Minnesota, List Biologicals) for the indicated times. LPS-induced NF-κB activation was inhibited with 25μM sc-514 (Calbiochem).
Microarray analysis
Total RNA was isolated using the TRIzol Reagent (Invitrogen) and overall quality was analyzed using an Agilent 2100 Bioanalyzer. Sample mRNA was amplified and labeled using the Affymetrix One-Cycle Eukaryotic Target Labeling Assay protocol and reagents. Biotinylated cRNA was hybridized to an Affymetrix GeneChip® Mouse Genome 430 2.0 array using standard protocols and reagents from Affymetrix. Probe intensities were measured using the Affymetrix GeneChip Scanner 3000 and processed into CEL files using Affymetrix GeneChip Operating Software. Probe intensities were background-adjusted, normalized, and probeset-summarized using the Robust Multi-chip Average (RMA) method using the software Bioconductor, then exported to Matlab® (MathWorks) for further analysis.
Raw data can be downloaded from the Gene Expression Omnibus (GEO) accession number GSE14769.
Quantitative real-time PCR
To measure mRNA transcript expression in macrophages total RNA was isolated using Trizol (Invitrogen), reverse-transcribed and subjected to real-time PCR, using TaqMan® Gene Expression Assays (Applied Biosystems). Data acquisition was performed on an 7900HT fast real-time PCR system (Applied Biosystems). Data were normalized to the expression of Eif1a mRNA transcripts in individual samples. A comprehensive listing of Taqman Gene Expression Assays used here is provided in Supplementary Methods.
Chromatin immunoprecipitation (ChIP) assay and immunoblotting
For ChIP binding analysis formalin-fixed cells were sonicated and processed for immunoprecipitation, using anti-Rel (C), anti-ATF3 (C-19) and anti-C/EBPδ (M-17) antibodies (Santa Cruz), essentially as described previously27. Immunoprecipitated DNA samples were amplified using target promoter-specific primers. A list of promoter-specific primers is provided in Supplementary Methods.
For immunoblotting, macrophages were lysed and processed for immunoblots, as described previously 27.
ChIP-on-chip analysis
For ChIP-on-chip binding analysis formalin-fixed cells were sonicated and processed for immunoprecipitation with polyclonal antibodies specific for C/EBPδ, essentially as described previously 27. Immunoprecipitated DNA samples were amplified and labeled using the Affymetrix Chromatin Immunoprecipitation Assay protocol and hybridized to GeneChip Mouse Promoter 1.0R Array. Analysis of the ChIP-on-chip data was performed using Model-based Analysis of Tiling-arrays37. Raw data can be downloaded from the Gene Expression Omnibus (GEO) accession number GSE14812.
Supplementary Material
Acknowledgments
We acknowledge M. Gilchrist, E. Gold and C. Rosenberger for discussions and critical reading of the manuscript. We thank A. Nachman, I. Podolsky, C. Lorang and T. Stolyar for technical assistance. This work was supported by Irvington Institute Fellowship Program of the Cancer Research Institute (to V.L.) and the NIH (to A.A.)
Footnotes
Competing interests’ statement: The authors declare that they have no competing financial interests.
Motif scanning, gene expression profiling, mice, analysis of robustness of model preduction and kinetic modeling are described in Supplementary Methods.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev of Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 4.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi KS, Flavell RA. Shielding the double-edged sword: negative regulation of the innate immune system. J Leukoc Biol. 2004;75:428–433. doi: 10.1189/jlb.0703321. [DOI] [PubMed] [Google Scholar]
- 6.Barnes PJ, Karin M. Nuclear factor-κB - A pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336(15):1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 7.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 8.Liew FY, Xu D, Brint EK, O’Neill LAJ. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 9.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Royet J, Reichhart JM, Hoffmann JA. Sensing and signaling during infection in Drosophila. Curr Opin Immunol. 2005;17:11–17. doi: 10.1016/j.coi.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 14.Beutler B. Tlr4: Central component of the sole mammalian LPS sensor. Curr Opin Immunol. 2000;12:20–26. doi: 10.1016/s0952-7915(99)00046-1. [DOI] [PubMed] [Google Scholar]
- 15.Taylor PR, et al. Macrophage receptors and immune recognition. Annu Rev of Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 16.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 17.Boldrick JC, et al. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci U S A. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nau GJ, et al. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci U S A. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roach JC, et al. Transcription factor expression in lipopolysaccharide-activated peripheral-blood-derived mononuclear cells. Proc Natl Acad Sci USA. 2007;104:16245–16250. doi: 10.1073/pnas.0707757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aderem A. Systems biology: its practice and challenges. Cell. 2005;121:511–513. doi: 10.1016/j.cell.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 22.Suthram S, Sittler T, Ideker T. The plasmodium protein network diverges from those of other eukaryotes. Nature. 2005;438:108–112. doi: 10.1038/nature04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ideker T, Galitski T, Hood L. A new approach to decoding life: Systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 24.Aderem A, Smith KD. A systems approach to dissecting immunity and inflammation. Semin Immunol. 2004;16:55–67. doi: 10.1016/j.smim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci U S A. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- 27.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 29.Ramsey SA, et al. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat Genet. 2006;38:1082–1087. doi: 10.1038/ng1869. [DOI] [PubMed] [Google Scholar]
- 30.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 31.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 32.Ramsey SA, et al. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput Biol. 2008;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 34.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 35.Kovács KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J Biol Chem. 2003;278:36959–36965. doi: 10.1074/jbc.M303147200. [DOI] [PubMed] [Google Scholar]
- 36.Longabaugh WJR, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 37.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.