Abstract
Motivational states are important determinants of behavior. In fruit flies appetitive memory expression is constrained by satiety and promoted by hunger. Here we identify a neural mechanism that integrates the motivational state of hunger and memory. We show that stimulation of neurons that express Neuropeptide F (dNPF), an ortholog of mammalian NPY, mimicks food-deprivation and promotes memory performance in satiated flies. Robust appetitive memory performance requires the dNPF receptor in six dopaminergic neurons that innervate a distinct region of the mushroom bodies. Blocking these dopaminergic neurons releases memory performance in satiated flies whereas stimulation suppresses memory performance in hungry flies. Therefore dNPF and dopamine provide a motivational switch in the mushroom body that controls the output of appetitive memory.
Introduction
Motivation provides behavior with purpose and intensity and ensures that particular motor actions are expressed at the appropriate time. Although the concept of motivation has interested psychologists and ethologists for decades (Hull, 1951; Tolman, 1932; Thorpe, 1956; Bindra, 1959; Hinde, 1966; Lorenz, 1950; Dethier, 1976; Toates, 1986; Kennedy, 1987), a detailed neurobiological perspective of the mechanisms underlying state-dependent changes in behavior is lacking. Understanding how motivational systems are organized in the brain and how they impact neural circuits that direct behavior is a major question in neurobiology and addresses the functional connection between body and mind.
Hunger is perhaps the most heavily studied of the regulatory, or homeostatic, motivational drive states because food availability is easily manipulated in the laboratory. Hunger results from internally generated metabolic deficit signals and these signals in turn, increase the likelihood that the animal initiates food-seeking behavior (Dethier, 1976; Saper et al., 2002; Abizaid and Horvath, 2008). Models of motivation include learned representations of cues associated with food, such as smell and taste, that provide additional incentive and direction to locate a particular food source (Hull, 1951; Toates, 1986). When the food is located and consumed, the homeostatic process comes full-circle and the motivational drive to feed is neutralized. However, it is unclear how neural systems representing hunger and satiety are integrated with those of memory.
The idea that motivation could be approached experimentally in insects followed seminal studies of food-seeking behavior in the blowfly Phormia regina (Dethier, 1976). It was noted that although exposing gustatory receptor neurons on the proboscis to sugar always generated an electrophysiological response, the blowfly did not consistently respond by extending the proboscis. However, a food-deprived blowfly was more likely to respond with proboscis extension. A sophisticated genetic tool-kit for manipulating neural circuits (Keene and Waddell, 2007) coupled with robust behaviors makes the fruit fly Drosophila melanogaster ideal to understand the physiological mechanism that underlies such state-dependent behavior.
Drosophila can be efficiently trained to associate odorants with sucrose reward (Tempel et al., 1983; Krashes and Waddell, 2008). Importantly, fruit flies have to be hungry to effectively express appetitive memory performance (Krashes and Waddell, 2008). Therefore motivated decision-making and appetitive memory performance emerges in Drosophila when the incentive of the conditioned odor, the learned representation of that odor, and the internal motivational drive state of hunger are positively integrated. This apparent state-dependence implies that signals for hunger and satiety may interact with memory circuitry to regulate the behavioral expression of learned food-seeking behavior. The mushroom body (MB) in the fly brain is a critical site for appetitive memory (Schwaerzel et al., 2003; Keene et al., 2006; Krashes and Waddell, 2008). Synaptic output from the MB α´ β´ neurons is required to consolidate appetitive memory whereas output from the αβ subset is specifically required for memory retrieval (Krashes et al., 2007; Krashes and Waddell, 2008). This anatomy provides a foundation for understanding neural circuit integration between systems representing a motivational state and those for memory.
Neuropeptide Y (NPY) is a highly conserved 36 amino acid neuromodulator that stimulates food-seeking behavior in mammals (Tatemoto et al., 1982; Clark et al., 1984; Kalra, 1997). NPY mRNA levels are elevated in neurons in the arcuate nucleus of the hypothalamus of food-deprived mice (Sahu et al., 1988; Sanacora et al., 1990) and injection of NPY into the paraventricular nucleus increases feeding (Stanley and Leibowitz, 1985). Most impressively, ablating NPY expressing neurons from adult mice leads to starvation (Bewick et al., 2005; Gropp et al., 2005; Luquet et al., 2005). NPY exerts its effects through a family of NPY receptors and appears to have inhibitory function (Colmers et al., 1988; Colmers et al., 1991; Klapstein and Colmers, 1993; Qian et al., 1997; Rhim et al., 1997; Sun et al., 2003; Browning and Travagli, 2003; Lin et al., 2004). NPY therefore must repress the action of inhibitory pathways in order to promote feeding behavior. Drosophila Neuropeptide F is an ortholog of NPY, which has a C-terminal amidated phenylalanine instead of the amidated tyrosine in vertebrates (Brown et al., 1999). Evidence suggests that dNPF plays a similar role in appetitive behavior in flies. dNPF overexpression prolongs feeding in larvae and delays the developmental transition from foraging to pupariation (Wu et al., 2003). Furthermore, overexpressing a dNPF receptor gene, npfr1 (Garczynski et al., 2002), causes well-fed larvae to eat bitter-tasting food that wild-type larvae will only consume if they are hungry (Wu et al., 2005b).
In this study we exploited dNPF to identify a neural circuit that participates in motivational control of appetitive memory behavior in adult fruit flies. We show that stimulating dNPF neurons promotes appetitive memory performance in fed flies, mimicking the hungry state. npfr1 is required in dopaminergic (DA) neurons that innervate the MB for satiety to suppress appetitive memory performance. Directly blocking the DA neurons during memory testing reveals performance in fed flies whereas stimulating them suppresses performance in hungry flies. These data suggest that six DA neurons are a key module of dNPF-regulated circuitry, through which the internal motivational states of hunger and satiety are represented in the MB.
Results
Stimulating dNPF neurons promotes memory retrieval in fed flies
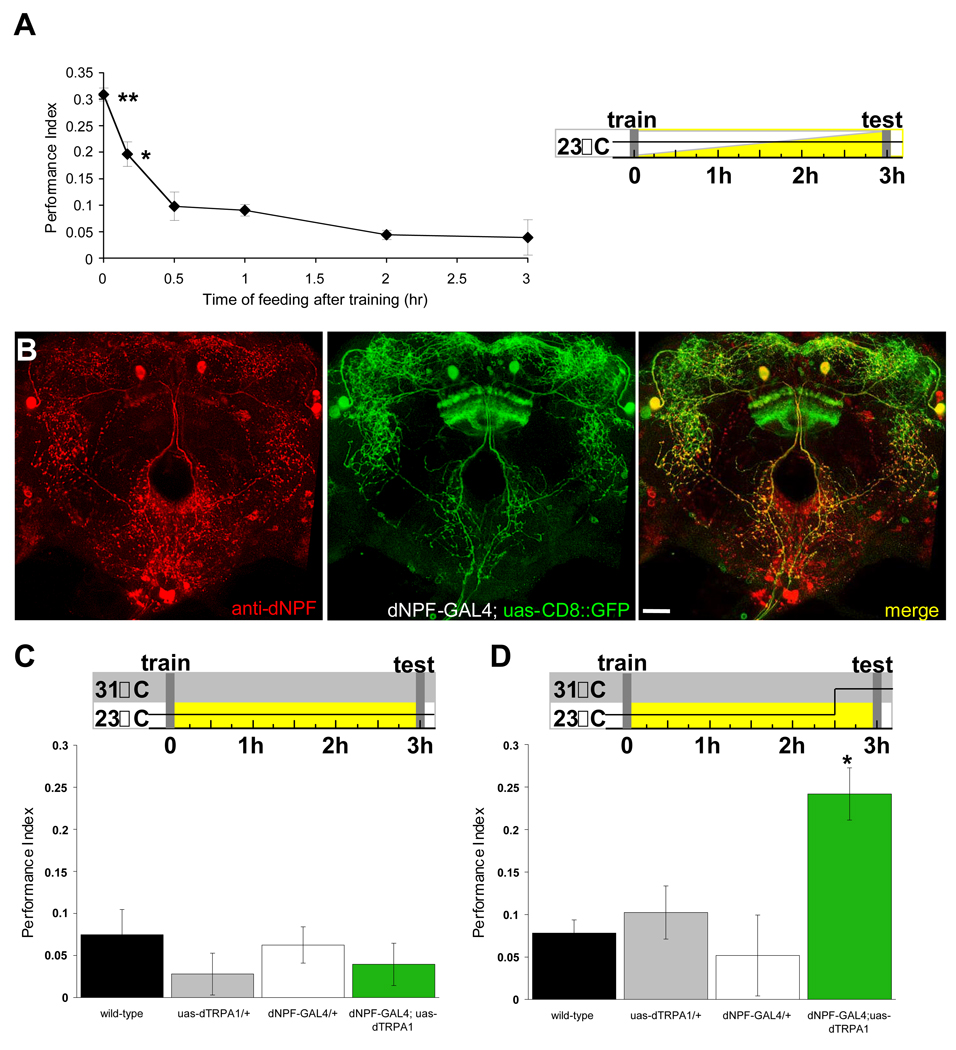
Feeding flies after appetitive conditioning suppresses memory performance (Figure 1A) and the suppression is reversed by re-starving flies (Krashes and Waddell, 2008). Food-deprivation is also required for efficient appetitive learning but a learning defect could simply result from satiated flies failing to ingest the reinforcing sucrose. In this study we specifically manipulated memory retrieval and in all experiments we ensured that flies were efficiently trained, by food-depriving them for 18hr before training. Immediately after training we transferred flies to vials with, or without, food for 3hr before testing appetitive memory. Flies starved before and after training display robust appetitive memory but memory performance steadily declines following 10–30min of feeding (Figure 1A) indicating a continuum of performance relative to the satiety state of the flies.
Figure 1. Stimulating dNPF neurons promotes appetitive memory expression in satiated flies.
A. Feeding after training suppresses appetitive memory performance. Double asterisk, significant difference (P<0.007); single asterisk (P<0.03) from all other groups. Temperature shift protocols are shown. White bar represents fly storage in empty vials, while yellow bar indicates flies stored with food -figure format used throughout this study. B. dNPF is expressed in neurons that innervate the dorsal and lateral protocerebrum, the SOG and the CC. Immunostaining with anti-dNPF antibody (red), partially overlaps (yellow, merge) with dNPF-GAL4 driven CD8::GFP (green). dNPF stained cells in the SOG are not labeled by dNPF-GAL4. The dNPF antibody only labels the upper layer of the fan-shaped body, consistent with processes in the ellipsoid body and lower fan-shaped body being post-synaptic regions. Scale bar represents 20µm. C. Feeding flies after training suppresses memory performance. All flies were food-deprived, trained, fed and tested at 23°C. D. Stimulating dNPF neurons before testing produces memory performance in fed flies. All flies were food-deprived, trained, and fed for 150min at 23°C. All flies were then transferred to 31°C for 30min and tested for appetitive memory. Asterisk denotes significant difference (P<0.05, ANOVA) from unmarked groups. Data are mean ± standard error of the mean (SEM).
Immunostaining for dNPF in adult fly brains reveals neurons in the subesophageal ganglion (SOG), the dorsal and lateral protocerebrum and the central complex (CC) (Wen et al., 2005; and Figure 1B). One can control some of these neurons using a dNPF promoter-driven GAL4 to express GAL4-uas promoter driven transgenes (Wen et al., 2005). dNPF-GAL4 driven uas-CD8::GFP labels most of the dNPF-immunoreactive neurons whose cell bodies reside in the dorsal protocerebrum but not those whose somata are clustered in the SOG (Figure 1B and S1).
We reasoned that dNPF release might represent the food-deprived state in the brain and so tested whether stimulating dNPF-expressing neurons could over-ride the suppression of memory performance by feeding. We expressed the heat-sensitive uas-dTrpA1 transgene (Hamada et al., 2008) with dNPF-GAL4. dTrpA1 encodes a Transient Receptor Potential (TRP) channel that is required in a small number of neurons in the brain for temperature preference in Drosophila (Hamada et al., 2008). Ectopically expressed dTRPA1 conducts Ca2+ and depolarizes neurons when flies are exposed to >25°C allowing one to stimulate specific neurons. We first food-deprived and trained wild-type, dNPF-GAL4, uas-dTrpA1 and dNPF-GAL4;uas-dTrpA1 flies, fed them ad libitum for 3hr and tested appetitive memory at the permissive 23°C. No group showed robust appetitive memory under these conditions (Figure 1C) and no statistical differences were apparent between groups (P>0.57). However, stimulating dNPF neurons for 30min before and during testing by shifting the flies to 31°C revealed memory performance in dNPF-GAL4;uas-dTrpA1 flies that was statistically different from all other groups (P<0.006)(Figure 1D). Therefore stimulating dNPF neurons mimics food-deprivation consistent with dNPF being a key factor in the internal state of hunger in the brain.
Localizing the relevant dNPF modulated circuit
We used a uas-RNA interference (RNAi) transgene against the dNPF receptor, uas-npfr1RNAi (Wu et al., 2003; Wu et al., 2005b) to localize the relevant dNPF-modulated neurons, reasoning that npfr1 disruption would impair appetitive memory in hungry flies. We verified the efficacy of the uas-npfr1RNAi transgene for our purpose by expressing it in all neurons using n-synaptobrevin-GAL4 and testing appetitive memory performance. As expected the memory performance of uas-npfr1RNAi;n-syb-GAL4 flies was impaired and was statistically different from all other control groups (P<0.04). However, uas-npfr1RNAi;n-syb-GAL4 flies were normal for aversive olfactory conditioning (Tully and Quinn, 1985)(Figure S2).
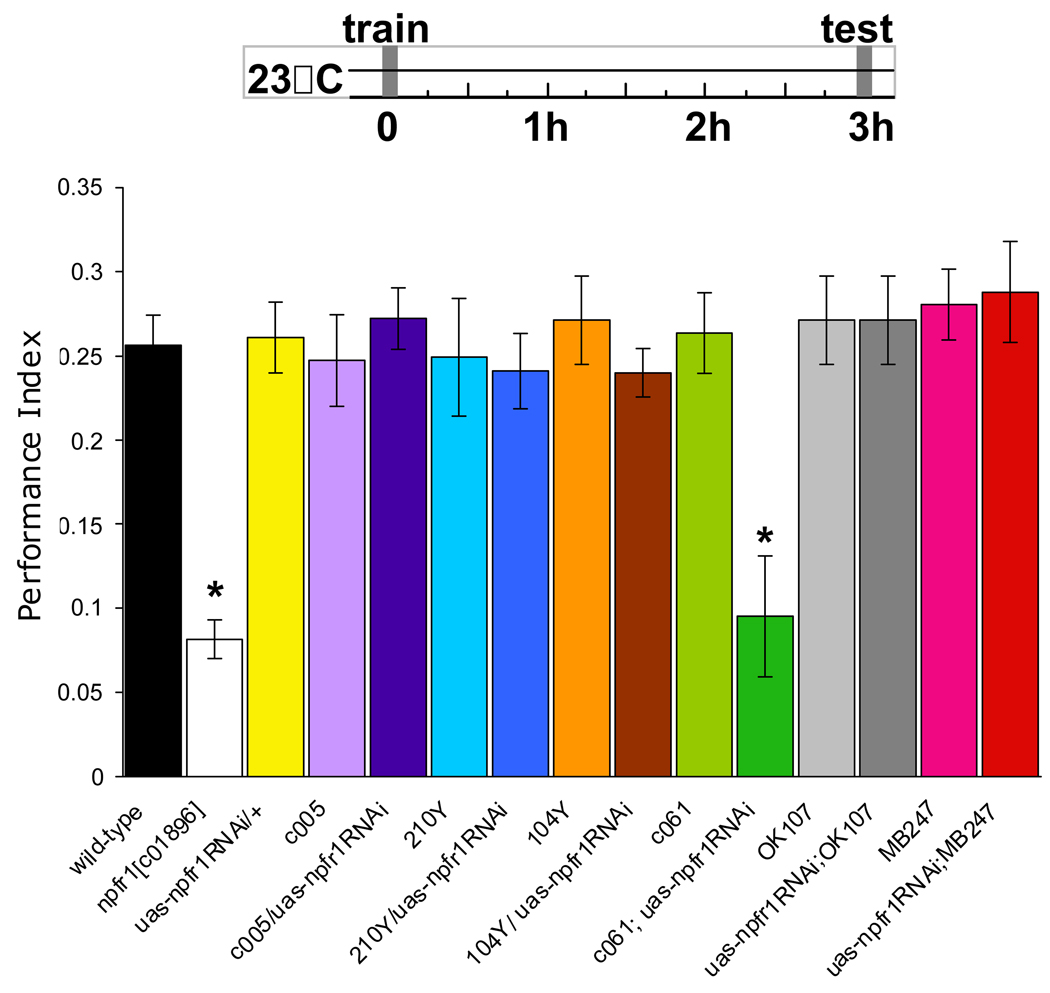
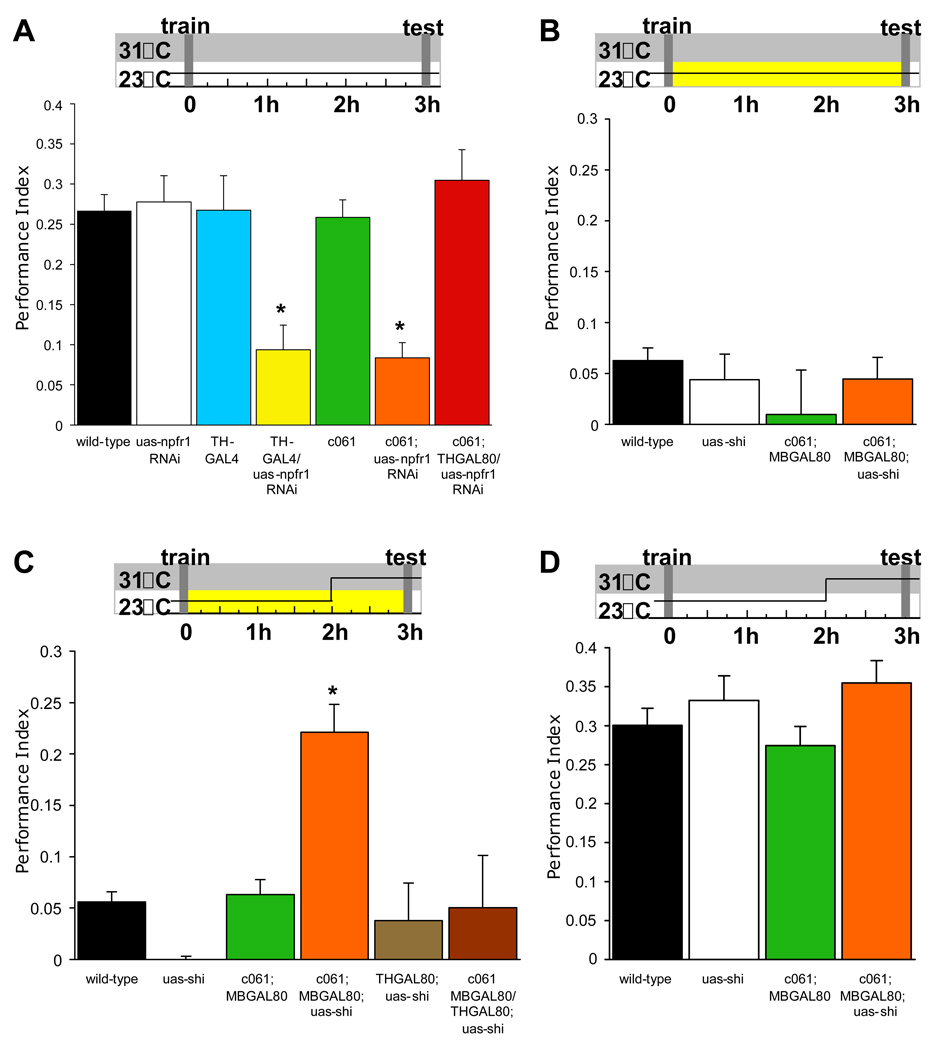
We next drove uas-npfr1RNAi with GAL4 drivers that express in the dorsal protocerebrum and CC - c005, 210Y, 104Y and c061 and in all MB neurons or the MB αβ and γ neurons - OK107 and MB247. We food-deprived wild-type flies, flies with a piggyBac element in the npfr1 locus (Bellen et al., 2004), flies expressing uas-npfr1RNAi in specific neurons, and flies harboring GAL4 or uas-npfr1 RNAi alone, and tested appetitive memory 3hr after training. The performance of npfr1[c01896] and c061;uas-npfr1RNAi flies was statistically different (both P<0.01) from all other flies (Figure 2). These data suggest c061 neurons mediate the effects of dNPF on appetitive memory expression.
Figure 2. Disruption of npfr1 expression impairs appetitive memory in food-deprived flies.
A. Food-deprived npfr1[c01896] flies and those expressing uas-npfr1RNAi with c061 have impaired 3hr appetitive memory whereas those expressing uas-npfr1RNAi with c005, 210Y, 104Y, OK107 or MB247 are normal. Asterisk denotes significant difference (P<0.01, ANOVA) from other unmarked groups. Data are mean ± SEM.
Some c061 neurons innervate the MB
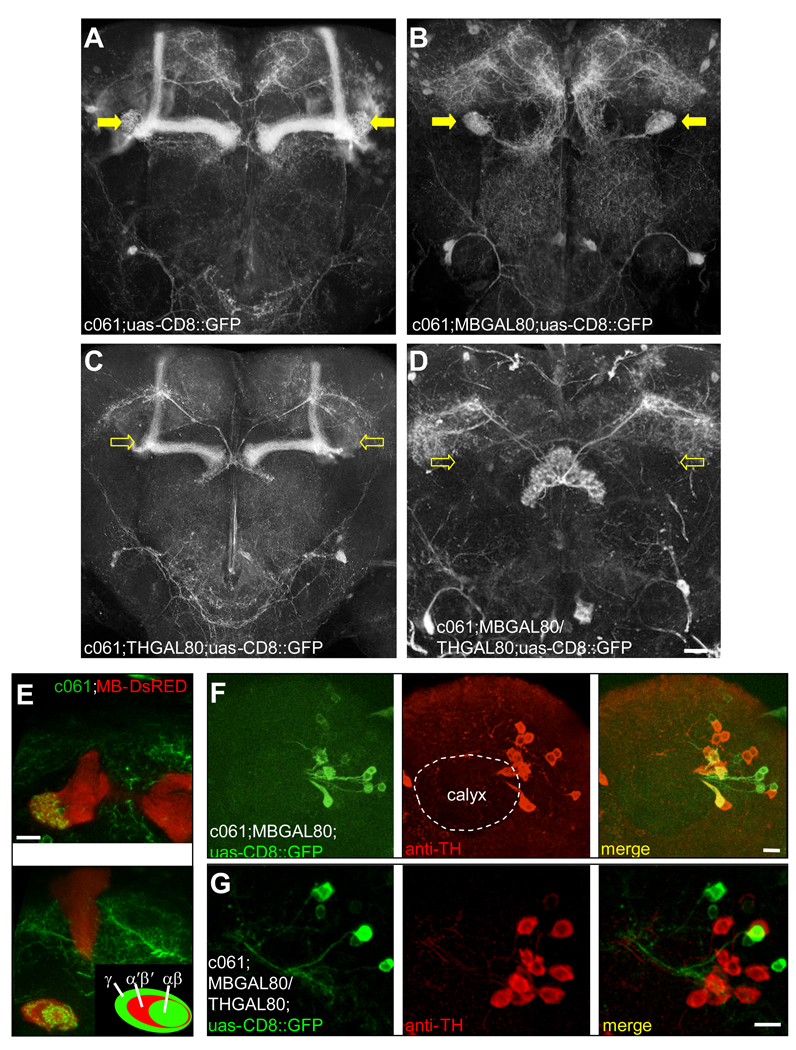
We visualized c061 neurons with uas-CD8::GFP. Confocal analysis revealed expression including intrinsic neurons of the MBs (Figure 3A). Since MB expression of uas-npfr1RNAi did not disrupt memory (Figure 2), we crossed in a GAL80 transgene that blocks GAL4 activity in all MB neurons (MBGAL80; Krashes et al., 2007). MBGAL80 abolished MB expression but prominent expression remained in three neurons per hemisphere whose projections densely innervate the MB heel and peduncle. Another cluster of five neurons per hemisphere innervate a specific layer in the fan-shaped body of the CC (Figure 3B, 5A and Movie S1 and Movie S2). Higher resolution imaging revealed innervation of the MB peduncle occupied by αβ but not α´β´ neurons (Figure 3E). Output from α´β´ neurons is required to consolidate appetitive memory whereas output from αβ neurons is required for appetitive memory retrieval (Krashes et al., 2007; Krashes and Waddell, 2008). Finding neurons that innervate the MB heel and αβ neurons is consistent with a model where satiety affects memory retrieval by modulating MB αβ and γ neurons.
Figure 3. c061 labels six DA neurons that innervate the MB.
A. Projection view of a c061;uas-CD8::GFP brain. Filled yellow arrows mark the heel region of the MB resembling that in TH-GAL4 labeled brains (Figure S4A). B. Combining MBGAL80 with c061;uas-CD8::GFP eliminates MB expression and reveals neurons that innervate the heel of the MB (filled yellow arrows). Also see Movie S1 and Movie S2. C. Combining THGAL80 with c061;uas-CD8::GFP eliminates expression in the neurons innervating the region between the MB lobes and MB heel (hollow arrows) but leaves expression elsewhere intact. D. Projection view of a c061;MBGAL80/THGAL80;uas-CD8::GFP brain. THGAL80 removes expression from the neurons innervating the dorsal protocerebrum and MB heel (hollow arrows) and leaves expression in the fan-shaped body and elsewhere intact. Scale bar represents 20 µm. E. Higher magnification single confocal section views of the MB heel and peduncle region in a c061; uas-CD8::GFP brain. Top panel shows innervation of the MB heel. Bottom panel, innervation in the base of the peduncle. Inset, section through the peduncle showing zones occupied by the αβ,α´β´ and γ MB neurons. The MB is co-labeled with MB-DsRED. F. Confocal section through a c061;MBGAL80;uas-CD8::GFP brain at the level of the MB calyx (outlined). GFP (green) labels 3 cell bodies at the side of the calyx and 5 more lateral cell bodies. Counter staining with anti-TH antibody (red) labels 12 cell bodies in the PPL1 cluster and 3 of them overlap (merge, yellow) with c061;MBGAL80 driven GFP. Scale bar represents 10 µm. G. Confocal section through a PPL1 cluster in a c061;MBGAL80/THGAL80;uas-CD8::GFP brain. GFP (green) labels 5 cell bodies and none overlap with anti-TH staining. Scale bar represents 10 µm.
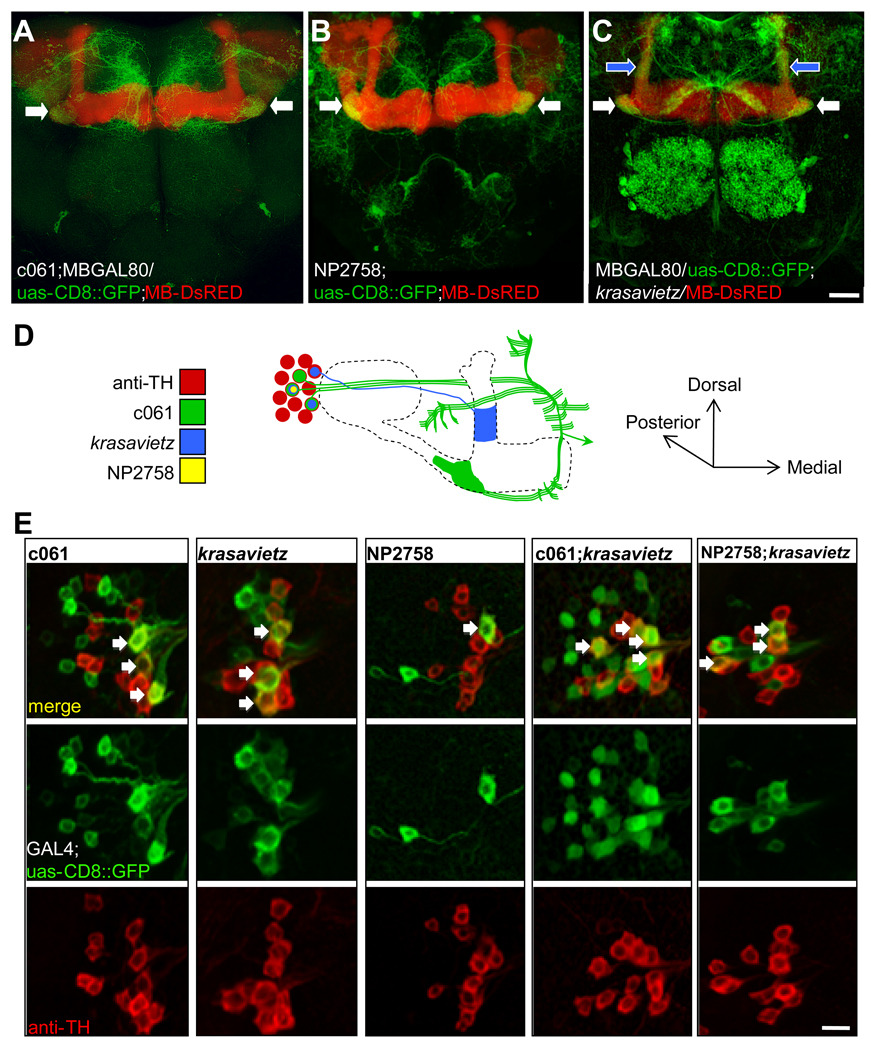
Figure 5. The c061 DA neurons are MB-MP neurons.
A. Projection view of a c061;MBGAL80;uas-CD8::GFP brain showing MB-MP neurons (white arrows) and neurons in the subesophageal ganglion (also see Movie S1 and Movie S2). B. Projection view of a NP2758;uas-CD8::GFP brain showing MB-MP neurons (white arrows) and neurons in the subesophageal ganglion (also see Movie S3 and Movie S4). C. Projection view of a MBGAL80; krasavietz/uas-CD8::GFP brain showing MB-MP neurons (white arrows). DA neurons innervating the MB α-stalk (blue arrows, also see Figure S4C), and neurons in the fan-shaped body and local neurons in the antennal lobe are also labeled (also see Movie S5 and Movie S6). The MB is labeled with MB-DsRED. D. Cartoon illustrating the gross structure of MB-MP neurons and the expression pattern of each GAL4. The MB is shown as an outline. DA neuron cell bodies (red, anti-TH) of a single PPL1 cluster are shown with the labeling of each GAL4 overlayed. The cell body organization is not stereotyped and it is difficult to distinguish the projections of each MB-MP neuron. No order or detail is inferred here. At least one MB-MP neuron sends a contralateral projection to the other MB (green arrow head). E. krasavietz and NP2758 label a subset of c061 MB-MP neurons. Each column shows the separate and merged channels from confocal images of a PPL1 cluster in brains co-labeled with GAL4 driven GFP (green) and anti-TH (red). Double-labeled neurons are marked with an arrow in the merged images. The quantification of neurons is shown in Figure S4B. Scale bar represents 20 µm (A, B, C) or 10 µm (E).
The MB-innervating neurons are dopaminergic
Some DA neurons innervate the MB heel and base of the peduncle (Friggi-Grelin et al., 2003; Riemensperger et al., 2005; Tanaka et al., 2008; Figure S4). We therefore immunostained c061;MBGAL80;uas-CD8::GFP brains with anti-tyrosine hydroxylase (TH) antibody. TH specifically labels DA neurons in flies because they do not produce epinephrine or norepinephrine. This analysis revealed that the three c061 MB-innervating neurons double label with GFP and anti-TH (Figure 3F) consistent with them releasing dopamine. Their position by the MB calyx defines them as belonging to the protocerebral posterior lateral 1 (PPL1) DA neuron cluster (Friggi-Grelin et al., 2003; Riemensperger et al., 2005).
Finding the MB-innervating neurons label for TH allowed us to use a TH-promoter driven GAL80 (THGAL80) to remove DA neuron expression (Sitaraman et al., 2008). We combined c061 and c061;MBGAL80 with THGAL80 and uas-CD8::GFP and visualized brains co-labeled with anti-TH. THGAL80 suppressed expression in DA neuron somata (Figure 3C, D and G) and eliminated expression in processes innervating the heel and peduncle region of the MB (Figure 3C and 3D). Expression remained in c061;THGAL80 brains in MB, fan-shaped body and SOG (Figure 3C). In c061;MBGAL80/THGAL80 brains expression remained in the fan-shaped body and SOG (Figure 3D). Therefore c061 DA neurons innervate the dorsal protocerebrum and MB heel and peduncle. Transgenic markers of neural polarity suggest DA processes in the dorsal protocerebrum are postsynaptic while those in the MB heel and peduncle are presynaptic (Zhang et al., 2007; and data not shown).
npfr1 expression in DA neurons is required for appetitive memory
We tested the importance of npfr1 in DA neurons by expressing uas-npfr1RNAi with TH-GAL4. We food-deprived flies before and after training and tested 3hr appetitive memory. Performance of TH-GAL4;uas-npfr1RNAi flies was statistically different from that of wild-type, TH-GAL4 and uas-npfr1RNAi control flies (P<0.01; Figure 4A). We also used THGAL80 to test whether DA neuron expression was required for the appetitive memory defect of c061;uas-npfr1RNAi flies. Memory of c061;THGAL80;uas-npfr1RNAi flies was statistically indistinguishable from controls (P>0.9) and was statistically different from that of c061;uas-npfr1RNAi and THGAL4; uas-npfr1RNAi flies (Figure 4A). Therefore npfr1 expression is required in DA neurons that innervate the MB for appetitive memory performance in hungry flies.
Figure 4. c061 DA neurons regulate appetitive memory performance.
Temperature shift protocols are shown above each graph. A. Expressing uas-npfr1RNAi in all DA neurons with TH-GAL4 or in a subset of DA neurons with c061 impairs 3hr appetitive memory in hungry flies. Expressing uas-npfr1RNAi in c061 neurons except the DA neurons (c061;THGAL80;uas-npfr1RNAi) does not affect appetitive memory. B. Feeding flies after training suppresses 3hr memory. All genotypes were food-deprived, trained, fed and tested at the permissive temperature of 23°C. C. Blocking synaptic output from c061;MBGAL80 neurons before testing reveals memory performance in fed flies. Removing uas-shits1 expression from the DA neurons reverses the memory promoting effect (c061;MBGAL80/THGAL80;uas-shits1 flies). All genotypes were food-deprived, trained and stored in food vials for 120min at 23°C. Flies were then shifted to 31°C for 60min and tested for memory. D. Blocking output from c061;MBGAL80 neurons does not enhance performance in food-deprived flies. All genotypes were food-deprived, trained and stored in empty vials for 120min at 23°C. Flies were then shifted to 31°C for 60min before flies were tested for appetitive memory at 31°C. Asterisks denote significant difference (P<0.05, ANOVA) from other unmarked groups. Data are mean ± SEM.
Blocking DA neurons promotes memory retrieval in fed flies
We used c061;MBGAL80 and THGAL80 to test whether DA neurons were responsible for inhibiting memory performance in fed flies. We directly blocked their output during memory testing with the dominant temperature-sensitive uas-shibire ts1 (shits1) transgene (Kitamoto, 2001). shits1 blocks membrane recycling and thus synaptic vesicle release at the restrictive temperature of 31°C and this blockade is reversible by returning flies to <25°C.
Flies were food deprived, trained and immediately transferred to vials containing food before testing 3hr memory. We performed this experiment at 23°C throughout (Figure 4B), or we blocked the neurons prior to, and during memory retrieval by shifting flies to 31°C for 1hr before testing (Figure 4C). We tested wild-type and single transgene GAL4 and uas-shits1 flies in parallel. At 23°C performance was suppressed by feeding and there were no significant differences between groups (P>0.77; Figure 4B). However, when c061;MBGAL80;uas-shits1 neurons were blocked prior to and during retrieval appetitive memory performance was statistically different from all other groups (all P<0.04) (Figure 4C). Expressing uas-shits1 in c061;MBGAL80 neurons except the DA neurons did not enhance performance (Figure 4C). Memory of c061;MBGAL80/THGAL80;uas-shits1 flies was statistically indistinguishable from the control groups (P>0.99). Importantly, blocking DA neurons did not further enhance hungry fly performance (all P>0.17; Figure 4D). Therefore these data are consistent with the DA neurons limiting memory performance in fed flies. It is likely that dopamine provides the inhibition because the DA neurons do not label for the inhibitory transmitter gamma-aminobutyric acid, GABA (Figure S3).
The DA neurons are MB-MP neurons
Similar neurons that innervate the MB have been described (Tanaka et al., 2008). NP2758 labels a single pair of MB-MP neurons, named according to the regions of the MB that they innervate: medial lobe and pedunculus (MP) (Figure 5B and Movie S3 and Movie S4). From here we refer to MB-innervating DA neurons as MB-MP neurons. We also found that krasavietz-GAL4 (Dubnau et al., 2003; Shang et al., 2007) combined with MBGAL80 (Krashes et al., 2007) expresses in MB-MP neurons (Figure 5C and Movie S5 and Movie S6).
We counted the TH positive neurons in the PPL1 cluster in each GAL4 (Figure 5E and S4B). Three TH positive cells are labeled by GFP in each PPL1 cluster in c061;MBGAL80;uas-CD8::GFP flies. MBGAL80;krasavietz/uas-CD8::GFP also labels three TH neurons but two are MB-MP neurons and the other innervates the vertical MB α lobe (Figure 5C and S4C). Lastly, we confirmed that NP2758;uas-CD8::GFP labels one MB-MP neuron per hemisphere. We combined the lines in pairs and counted cells co-labeled with GFP and anti-TH to determine whether c061, krasavietz and NP2758 label overlapping MB-MP neurons. Four cell bodies are labeled in PPL1 in c061;MBGAL80;krasavietz flies. One of these is the α lobe projecting krasavietz neuron (Figure 5C, 5D and S4), so MBGAL80;krasavietz labels two of the three c061 MB-MP neurons. Three cell bodies are labeled in PPL1 in NP2758;MBGAL80;krasavietz flies showing that NP2758 labels one of the two MBGAL80;krasavietz MB-MP neurons. Therefore c061;MBGAL80 labels three MB-MP neurons, MBGAL80;krasavietz labels two of these and NP2758 labels one of the MB-MP neurons that is common to c061;MBGAL80 and MBGAL80;krasavietz (Figure 5D). We did not observe more than three MB-MP neurons on each side of the brain.
Blocking NP2758 or krasavietz;MBGAL80 neurons prior to, and during memory retrieval did not reveal performance in fed flies (Figure S5). Therefore it is either necessary to block all six MB-MP neurons to release appetitive memory in fed flies or the two MB-MP neurons uniquely labeled by c061 could be responsible.
MB-MP stimulation inhibits appetitive memory expression in hungry flies
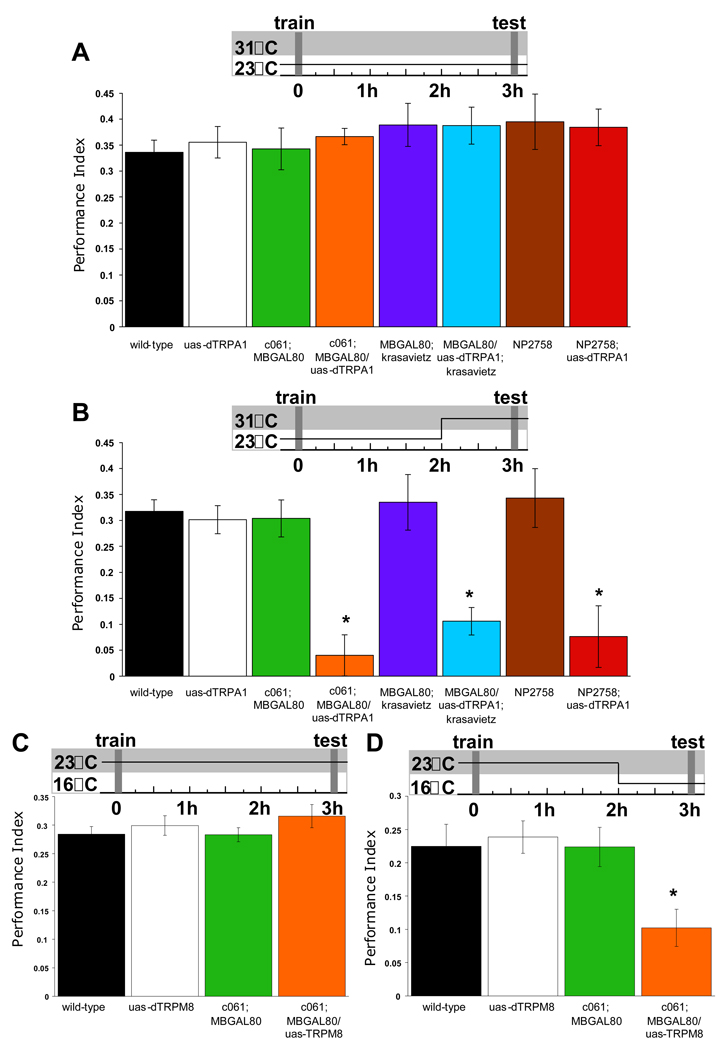
To further assess whether MB-MP neurons limit appetitive memory expression, we tested whether MB-MP neuron stimulation suppressed memory in hungry flies. We tested wild-type flies, flies expressing uas-dTrpA1 in MB-MP neurons and GAL4 and uas-dTrpA1 flies in parallel using two different temperature regimens; permissive 23°C throughout (Figure 6A), or we stimulated neurons prior to, and during memory retrieval by shifting flies to 31°C (Figure 6B). We starved flies, trained them and transferred them to empty vials before testing 3hr memory. All groups displayed robust appetitive memory at 23°C and there was no statistical difference between groups (P>0.96) (Figure 6A). However, acute MB-MP neuron stimulation prior to, and during memory retrieval severely impaired memory (Figure 6B). The performance of c061; MBGAL80/uas-dTrpA1 flies, MBGAL80/uas-dTrpA1;krasavietz flies and NP2758;uas-dTrpA1 flies was statistically different from all other groups (P<0.04). These data suggest that stimulating two MB-MP neurons is sufficient to block appetitive memory performance.
Figure 6. MB-MP stimulation suppresses appetitive memory expression in hungry flies.
Temperature shift protocols are shown above each graph. A. The permissive 23°C does not disrupt 3hr appetitive memory. All genotypes were starved, trained, stored for 3hr in empty vials and tested for memory at 23°C. B. Stimulating 6, 4 or 2 MB-MP neurons with uas-dTrpA1 before and during testing attenuates memory performance in starved flies. All genotypes were food-deprived, trained and stored in empty vials for 120min at 23°C and were then shifted to 31°C for 60min before and during testing. C. The permissive temperature of 23°C does not disrupt 3hr appetitive memory. All genotypes were starved, trained, stored in empty vials for 3 hr and tested for appetitive memory at 23°C. D. Stimulating 6 MB-MP neurons with uas-TRPM8 before and during testing attenuates memory performance in starved flies. All genotypes were food-deprived, trained and stored in empty food vials for 120min at 23°C and were then shifted to 16°C for 60min before and during testing. Asterisk denotes significant difference (P<0.05, ANOVA) from other unmarked groups. Data are mean ± SEM.
The suppression of performance with MB-MP activation is not due to irreversible MB damage. Food-deprived c061;MBGAL80/uas-dTrpA1 flies stimulated during acquisition (Figure S6A) or for 1hr after training (Figure S6B) showed normal 3hr memory (P>0.58 and P>0.70 respectively). Furthermore, brief stimulation during testing is sufficient to suppress performance (P<0.005; Figure S6C).
We also stimulated MB-MP neurons with the cold-sensitive uas-TRPM8 transgene (Peabody et al., 2009). The mammalian TRPM8 channel is activated below 18°C (McKemy et al., 2002; Peier et al., 2002). We starved and trained flies and put them in empty food vials for 3hr before testing appetitive memory. No statistical difference was apparent between the performance of flies at the permissive 23°C (P>0.50) (Figure 6C). However, stimulating c061-MB-MP neurons by shifting flies to 16°C for 1hr before testing impaired memory (Figure 6D). Performance of c061;MBGAL80;uas-TRPM8 flies was statistically different from all other groups (P<0.03). Therefore stimulating MB-MP neurons with dTRPA1 or TRPM8 suppresses performance in hungry flies (Figure 6B and D) and mimics feeding (Figure 1A).
To exclude the possibility that manipulations with uas-shits1 and uas-dTrpA1 interfere with olfaction or gustation, we tested the acuity of all flies used in this study. No significant differences were found between the relevant groups for either odor or sucrose acuity (Table S1). Therefore blocking output from MB-MP neurons reveals appetitive memory performance in satiated flies whereas stimulating them suppresses appetitive memory expression in hungry flies. These data are consistent with MB-MP neurons being a neural mechanism through which satiety suppresses appetitive memory performance.
Discussion
Drosophila as a model for motivational systems
It is critical to an animal’s survival that behaviors are expressed at the appropriate time. Motivational systems provide some of this behavioral control. Apart from the observation that motivational states are often regulated by hormones or neuromodulatory factors (Toates, 1986; Watts, 2003), we know little about how motivational states modulate specific neural circuitry. Hungry fruit flies form appetitive long-term memory, following a 2min pairing of odorant and sucrose and memory performance is only robust if the flies remain hungry (Krashes and Waddell, 2008). Therefore this paradigm includes key features of models for motivational systems (Toates, 1986): the conditioned odor provides the incentive cue predictive of food, there is a learned representation of the goal object (odorant/sucrose), and the expression of learned behavior depends on the internal physiological state (hunger and not satiety). In this study we identified a neural circuit mechanism that integrates hunger/satiety and appetitive memory.
What normally regulates dNPF-expressing neurons?
We do not know the signals that ordinarily control dNPF-releasing neurons. In mammals NPY-expressing neurons are a critical part of a complex hypothalamic network that regulates food-intake and metabolism (Saper et al., 2002). In times of adequate nutrition, NPY-expressing neurons are inhibited by high levels of leptin and insulin that are transported into the brain following release from adipose tissue and the pancreas (Figlewicz and Benoit, 2009). In hungry mice, leptin and insulin levels fall leading to loss of inhibition of NPY neurons. Flies do not have leptin but they have several insulin-like peptides (Arquier et al., 2008), that may regulate dNPF neurons. Some NPY expressing neurons are directly inhibited by glucose (Levin et al., 2006). Fly neurons could sense glucose with the Bride of Sevenless receptor (Kohyama-Koganeya et al., 2008). In blowflies satiety involves mechanical tension of the gut and abdomen (Gelperin, 1967; Gelperin, 1971). Lastly, it will be interesting to test the role of other extracellular signals implicated in fruit fly feeding behavior including the hugin (Melcher and Pankratz, 2005) and take-out neuropeptides (Sarov-Blat et al., 2000; Meunier et al., 2007).
A model for the role of MB-MP neurons
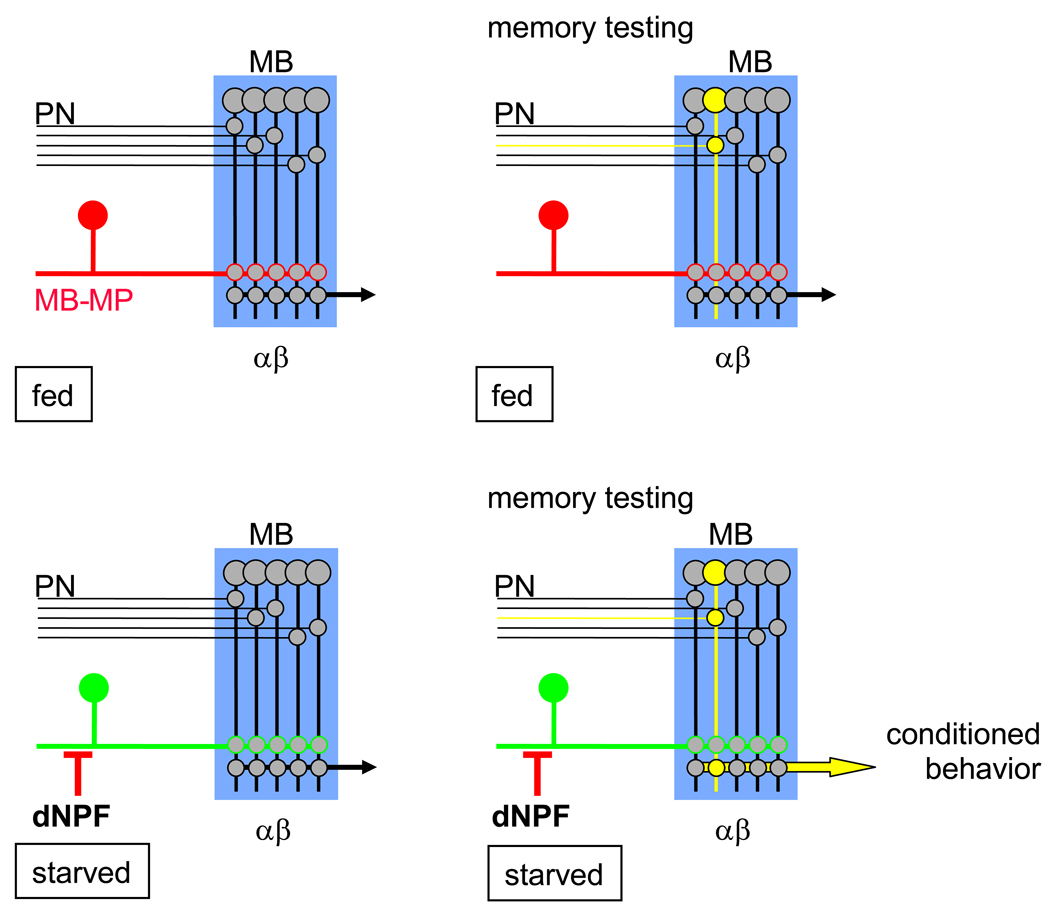
NPY inhibits synaptic function in mammals (Colmers et al., 1988; Colmers et al., 1991; Klapstein and Colmers, 1993; Qian et al., 1997; Rhim et al., 1997; Sun et al., 2003; Browning and Travagli, 2003; Lin et al., 2004) and our data suggest that dNPF promotes appetitive memory performance by suppressing inhibitory MB-MP neurons. We propose a model where MB-MP neurons gate MB output (Figure 7). Appetitive memory performance is low in fed flies because the MB αβ and γ neurons are inhibited by tonic dopamine release from MB-MP neurons. Hence, when the fly encounters the conditioned odorant during memory testing, the MB neurons encoding that olfactory memory respond, but the signal is not propagated beyond the MB due to the inhibitory influence of MB-MP neurons. However, when the flies are food-deprived dNPF levels rise and dNPF disinhibits MB-MP neurons, and other circuits, through the action of NPFR1. dNPF disinhibition of the MB-MP neurons opens the gate on the MB. Therefore, when hungry flies encounter the conditioned odorant during memory testing, the relevant MB neurons are activated and the signal propagates to downstream neurons, leading to expression of the conditioned behavior.
Figure 7. Model for the role of MB-MP neurons.
Left panels illustrate the state of the inhibitory control exerted upon the MB in the fed state (top) and starved state (bottom). When fed flies are exposed to the conditioned odor during memory testing (right panels) the appropriate projection neurons and MB neurons are activated (yellow). However, the signal only propagates beyond the MB neurons in hungry flies when the MB-MP neuron ‘gate’ is open. Red lines denote inhibition and green lines relief from inhibition. See discussion for more detail.
Satiety and hunger are not absolute states. We sometimes observe above chance performance scores in fed flies and shorter periods of feeding after training suggest that inhibition of performance is graded. This could be accounted for by a competitive push-pull inhibitory mechanism between dNPF and MB-MP neurons.
By gating the MB through the MB-MP neurons, hunger and satiety are likely affecting the relative salience of learned odor cues in the fly brain. However, MB-MP neurons are unlikely to change the sensory representation of odor in the MB because flies trained with stimulated MB-MP neurons perform normally when tested for memory without stimulation (Figure S6A). Therefore odors are likely perceived the same irrespective of MB-MP neuron activity. Furthermore, the MB-MP neurons did not affect naïve responses to the specific odorants used. It will be interesting to test whether MB-MP neurons change responses to other odorants and/or modulate arousal (Andretic et al., 2005; Kume et al., 2005; Seugnet et al., 2008), visual stimulus salience (Zhang et al., 2007) and attention-like phenomena (van Swinderen, 2007).
Structural and functional subdivision of DA neurons
There are eight different morphological classes of DA neurons that innervate the MB (Mao and Davis, 2009) and our data imply functional subdivision. Previous studies concluded that DA neurons convey aversive reinforcement (Schwaerzel et al., 2003; Schroll et al., 2006; Riemensperger et al., 2005 and see Figure S7).
We specifically manipulated the MB-MP DA neurons. MB-MP neurons are not required for acquisition of aversive olfactory memory (P>0.94)(Figure S7) consistent with a distinct function in controlling the expression of appetitive memory. Since several studies have implicated the MB α lobe in memory (Pascual and Preat, 2001; Yu et al., 2005; Yu et al., 2006), other DA neurons in PPL1 that innervate the α lobe (like those labeled in MBGAL80;krasavietz, Figure 5C and S4) may provide reinforcement. The MB-MP neurons may also be functionally divisible and independently regulated to gate MB function. The idea that a specific DA circuit restricts stimulus-evoked behavior is reminiscent of literature tying dopamine to impulse control in mammals (Weintraub, 2008; Blum et al., 1996). Previous studies of DA neurons in Drosophila (Schwaerzel et al., 2003; Schroll et al., 2006; Seugnet et al., 2008; Andretic et al., 2005; Kume et al., 2005; Seugnet et al., 2008; Zhang et al., 2007) have simultaneously manipulated all, or large numbers of DA neurons. Our data suggest that the DA neurons should be considered as individuals, or small groups.
Motivation and learning in flies
Flies have to be hungry to efficiently acquire appetitive memory but whether this reflects a state-dependent neural mechanism or results from the failure to ingest enough sugar is unclear. Stimulating MB-MP neurons in hungry flies did not impair appetitive memory formation (Figure S6A) and therefore MB-MP neurons are unlikely to constrain learning in fed flies. Other dNPF-regulated neurons may provide this control since NPY has been implicated in learning (Redrobe et al., 2004).
Hunger simultaneously regulates discrete neural circuit modules
The dNPF-expressing neurons innervate broad regions of the brain and may simultaneously modulate distinct neural circuits to promote food-seeking. MB-MP neurons represent a circuit through which the salience of learned food-relevant odorant cues is regulated by relative nutritional state. Given the apparent role of the MB as a locomotor regulator (Huber, 1967; Martin et al., 1998; Pitman et al., 2006; Joiner et al., 2006), MB-MP neurons may also generally promote exploratory behavior. There are likely to be independent circuits for other elements of food-seeking behavior including those that potentiate gustatory pathway sensitivity and promote ingestion.
NPY stimulates feeding but inhibits sexual behavior in rats (Clark et al., 1985). Modulators exerting differential effects could provide a neural mechanism to establish a hierarchy of motivated states and coordinate behavioral control. dNPF may potentiate activity in food-seeking related circuits while suppressing circuits required for other potentially competing behaviors, eg. sexual pursuit.
Regulating behavior with inhibitory control
In this study we provide the first multi-level neural circuit perspective for a learned motivated behavior in fruit flies. Our work demonstrates a clear state-dependence for the expression of appetitive memory. Odorants that evoke conditioned appetitive behavior in hungry flies are ineffective at evoking appetitive behavior in satiated flies. Therefore the fly brain is not simply a collection of input-output reflex units and includes neural circuits through which the internal physiological state of the animal establishes the appropriate context for behavioral expression.
Dethier (1976) proposed that ‘a satiated fly receives maximum inhibitory feedback so that sensory input is behaviorally ineffective. As deprivation increases inhibition wanes and sensory input becomes increasingly effective in initiating feeding’. Our data provide experimental evidence that this prediction is also likely to be accurate for expression of appetitive memory in the fruit fly where the mechanism involves neuromodulation in the central brain. The DA MB-MP neurons inhibit the expression of appetitive memory performance in satiated flies whereas dNPF disinhibits the MB-MP neurons in food-deprived flies. The likelihood that appetitive behavior is triggered by the conditioned odorant is therefore determined by the competition between inhibitory systems in the brain. The concept that continuously active inhibitory forces in the insect brain control behavioral expression was also proposed many years ago (Roeder, 1955). Here we provide evidence that these neurons exist and that their hierarchical arrangement is a key determinant of behavioral control.
Experimental Procedures
Fly strains
See supplemental information for fly stock source. To express dTRPA1 in dNPF neurons we crossed uas-dTrpA1 females to dNPF-GAL4 male flies. To screen for neurons that required npfr1 we crossed female uas-npfr1RNAi flies to c061, c061; THGAL80, 210Y, c005, 104Y, OK107, MB247, TH-GAL4, n-syb or n-syb; uas-dcr2 males. c061 is located on the X-chromosome so female c061;MBGAL80 flies were crossed to uas-shits1 males. Similarly, we crossed c061;MBGAL80 females with THGAL80; uas-shits1 males. To express uas-shits1 in MB-MP neurons female uas-shits1 flies were crossed to NP2758 or MBGAL80; krasavietz males. Since NP2758 is on the X-chromosome, only female flies were assayed from the NP2758 cross. We expressed dTRPA1 in the MB-MP neurons by crossing female uas-dTrpA1 flies to NP2758 or MBGAL80;krasavietz males or c061;MBGAL80 females with uas-dTrpA1 males. All GAL4 and uas-transgene flies were crossed with wild-type females to create heterozygous controls. We visualized GAL4 expression by crossing to uas-mCD8::GFP or uas-mCD8::GFP; MB-DsRED flies (Lee and Luo, 1999; Riemensperger et al., 2005).
Behavioral analysis
All flies were food deprived for 16–20 hr before training in milk bottles containing a 10×6cm filter paper soaked with water. The olfactory appetitive paradigm was performed as described (Krashes and Waddell, 2008). Following training, flies were stored for 3hr in vials with food or containing only a water-damp filter paper. All experiments performed after feeding included a control group of food-deprived flies. The performance index (PI) was calculated as the number of flies running toward the conditioned odor minus the number of flies running toward the unconditioned odor divided by the total number of flies in the experiment. A single PI value is the average score from flies of the identical genotype tested with each odor (3-Octanol or 4-Methylcyclohexanol). Olfactory and gustatory acuity was performed according to Keene et al. (2006).
Statistical analyses were performed using KaleidaGraph (Synergy Software). Overall analyses of variance (ANOVA) were followed by planned pairwise comparisons between the relevant groups with a Tukey HSD post-hoc test. Unless stated otherwise, all experiments are n≥8.
Immunohistochemistry
Adult female flies were collected 3–5 days after eclosion, brains or entire central nervous systems were dissected in ice-cold PBS [1.86mM NaH2PO4, 8.41mM Na2HPO4, 175mM NaCl] and fixed in 2% paraformaldehyde solution in PBS for 10min at room temperature (RT). Samples were then washed 5A for 15min with PBS containing 0.25% Triton-X100 (PBT), blocked for 1hr with PBT containing 5% NGS (all at RT) and incubated with primary antibody in blocking solution for 2 days at 4°C. Samples were washed 5× for 15min in PBT, incubated with secondary antibody in PBT for 12hr at 4°C and washed 10× for 15min with PBT, 2× in PBS for 15min and mounted in Vectashield (Vector Labs) for confocal microscopy. Imaging was performed on a Zeiss LSM 5 Pascal confocal microscope and images were processed in ImageJ and Adobe Photoshop. In some cases, debris on the brain surface was manually deleted from the relevant confocal sections to permit construction of a clear projection view of the z-stack. Antibodies were diluted: mouse IgG2a anti-GFP (Invitrogen 1:200); rabbit anti-Tyrosine hydroxylase (Chemicon 1:100); rabbit anti-dNPF (gift from P. Shen 1:2000); rabbit anti-GABA (Sigma 1:100); mouse monoclonal 4B1 anti-Drosophila ChAT (Hybridoma Bank, University of Iowa 1:100), FITC conjugated anti-Mouse IgG2a(Jackson Laboratory 1:200); Cy3 conjugated anti-Rabbit (Jackson Laboratory 1:200); Cy5 conjugated anti-Mouse IgG1γ (Jackson Laboratory 1:200).
Supplementary Material
Acknowledgements
We thank Andre Fiala, Paul Garrity, Julie Simpson, Ping Shen, Tim Tully and Troy Zars for flies. We also thank Geraldine Wright, Hans Hoffman, Tzumin Lee and Stanley Heinze for their input. This work was supported by NIH MH09883 and MH081982 to S.W., and NRSA DA024499 to M. J. K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abizaid A, Horvath TL. Brain circuits regulating energy homeostasis. Regul Pept. 2008;149:3–10. doi: 10.1016/j.regpep.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Arquier N, Geminard C, Bourouis M, Jarretou G, Honegger B, Paix A, Leopold P. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 2008;7:333–338. doi: 10.1016/j.cmet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick GA, Gardiner JV, Dhillo WS, Kent AS, White NE, Webster Z, Ghatei MA, Bloom SR. Post-embryonic ablation of AgRP neurons in mice leads to a lean, hypophagic phenotype. FASEB J. 2005;19:1680–1682. doi: 10.1096/fj.04-3434fje. [DOI] [PubMed] [Google Scholar]
- Bindra D. Motivation: A Systematic Reinterpretation. New York: Ronald Press Company; 1959. [Google Scholar]
- Blum K, Sheridan PJ, Wood RC, Braverman ER, Chen TJ, Cull JG, Comings DE. The D2 dopamine receptor gene as a determinant of reward deficiency syndrome. J R Soc Med. 1996;89:396–400. doi: 10.1177/014107689608900711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Crim JW, Arata RC, Cai HN, Chun C, Shen P. Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides. 1999;20:1035–1042. doi: 10.1016/s0196-9781(99)00097-2. [DOI] [PubMed] [Google Scholar]
- Browning KN, Travagli RA. Neuropeptide Y and peptide YY inhibit excitatory synaptic transmission in the rat dorsal motor nucleus of the vagus. J Physiol. 2003;549:775–785. doi: 10.1113/jphysiol.2003.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JT, Kalra PS, Crowley WR, Kalra SP. Neuropeptide Y and human pancreatic polypeptide stimulate feeding behavior in rats. Endocrinology. 1984;115:427–429. doi: 10.1210/endo-115-1-427. [DOI] [PubMed] [Google Scholar]
- Clark JT, Kalra PS, Kalra SP. Neuropeptide Y stimulates feeding but inhibits sexual behavior in rats. Endocrinology. 1985;117:2435–2442. doi: 10.1210/endo-117-6-2435. [DOI] [PubMed] [Google Scholar]
- Colmers WF, Klapstein GJ, Fournier A, St-Pierre S, Treherne KA. Presynaptic inhibition by neuropeptide Y in rat hippocampal slice in vitro is mediated by a Y2 receptor. Br J Pharmacol. 1991;102:41–44. doi: 10.1111/j.1476-5381.1991.tb12129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmers WF, Lukowiak K, Pittman QJ. Neuropeptide Y action in the rat hippocampal slice: site and mechanism of presynaptic inhibition. J Neurosci. 1988;8:3827–3837. doi: 10.1523/JNEUROSCI.08-10-03827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Dethier VG. The Hungry Fly - a Physiological Study of the Behaviour. Cambridge: Harvard University Press; 1976. [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Figlewicz DP, Benoit SC. Insulin, leptin, and food reward: update 2008. Am J Physiol Regul Integr Comp Physiol. 2009;296:R9–R19. doi: 10.1152/ajpregu.90725.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides. 2002;23:773–780. doi: 10.1016/s0196-9781(01)00647-7. [DOI] [PubMed] [Google Scholar]
- Gelperin A. Stretch receptors in the foregut of the blowfly. Science. 1967;157:208. doi: 10.1126/science.157.3785.208. [DOI] [PubMed] [Google Scholar]
- Gelperin A. Abdominal sensory neurons providing negative feedback to the feeding behavior of the blowfly. Journal of Comparative Physiology A. 1971;72:17–31. [Google Scholar]
- Gropp E, Shanabrough M, Borok E, Xu AW, Janoschek R, Buch T, Plum L, Balthasar N, Hampel B, Waisman A, Barsh GS, Horvath TL, Bruning JC. Agouti-related peptide-expressing neurons are mandatory for feeding. Nat Neurosci. 2005;8:1289–1291. doi: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinde RA. Animal behaviour: a synthesis of ethology and comparative psychology. New York: McGraw-Hill Book Company; 1966. [Google Scholar]
- Huber F. Central control of movements and behavior of invertebrates. In: Wiersma CAG, editor. Invertebrate Nervous Systems. University of Chicago Press; 1967. pp. 333–351. [Google Scholar]
- Hull CL. Essentials of Behavior. New Haven: Yale University Press; 1951. [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kalra SP. Appetite and body weight regulation: is it all in the brain? Neuron. 1997;19:227–230. doi: 10.1016/s0896-6273(00)80934-4. [DOI] [PubMed] [Google Scholar]
- Keene AC, Krashes MJ, Leung B, Bernard JA, Waddell S. Drosophila dorsal paired medial neurons provide a general mechanism for memory consolidation. Curr Biol. 2006;16:1524–1530. doi: 10.1016/j.cub.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- Kennedy JS. Animal Motivation: The Beginning of the End? In: Jermy T, Chapman RF, Bernays EA, Stoffolano JG, editors. Perspectives in Chemoreception and Behavior. New York: Springer; 1987. [Google Scholar]
- Kim YC, Lee HG, Han KA. D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci. 2007;27:7640–7647. doi: 10.1523/JNEUROSCI.1167-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Lee HG, Seong CS, Han KA. Expression of a D1 dopamine receptor dDA1/DmDOP1 in the central nervous system of Drosophila melanogaster. Gene Expr Patterns. 2003;3:237–245. doi: 10.1016/s1567-133x(02)00098-4. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Klapstein GJ, Colmers WF. On the sites of presynaptic inhibition by neuropeptide Y in rat hippocampus in vitro. Hippocampus. 1993;3:103–111. doi: 10.1002/hipo.450030111. [DOI] [PubMed] [Google Scholar]
- Kohyama-Koganeya A, Kim YJ, Miura M, Hirabayashi Y. A Drosophila orphan G protein-coupled receptor BOSS functions as a glucose-responding receptor: loss of boss causes abnormal energy metabolism. Proc Natl Acad Sci U S A. 2008;105:15328–15333. doi: 10.1073/pnas.0807833105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Waddell S. Rapid consolidation to a radish and protein synthesis-dependent long-term memory after single-session appetitive olfactory conditioning in Drosophila. J Neurosci. 2008;28:3103–3113. doi: 10.1523/JNEUROSCI.5333-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Levin BE, Kang L, Sanders NM, Dunn-Meynell AA. Role of neuronal glucosensing in the regulation of energy homeostasis. Diabetes. 2006;55:122–130. [Google Scholar]
- Lin S, Boey D, Herzog H. NPY and Y receptors: lessons from transgenic and knockout models. Neuropeptides. 2004;38:189–200. doi: 10.1016/j.npep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Liu G, Seiler H, Wen A, Zars T, Ito K, Wolf R, Heisenberg M, Liu L. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- Lorenz KZ. Physiological Mechanisms in Animal Behaviour. Company of Biologists: Cambridge University Press; 1950. The comparative method in studying innate behaviour patterns. [Google Scholar]
- Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JR, Ernst R, Heisenberg M. Mushroom bodies suppress locomotor activity in Drosophila melanogaster. Learn Mem. 1998;5:179–191. [PMC free article] [PubMed] [Google Scholar]
- McKemy DD, Neuhausser WM, Julius D. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 2002;416:52–58. doi: 10.1038/nature719. [DOI] [PubMed] [Google Scholar]
- Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 2005;3:e305. doi: 10.1371/journal.pbio.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier N, Belgacem YH, Martin JR. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol. 2007;210:1424–1434. doi: 10.1242/jeb.02755. [DOI] [PubMed] [Google Scholar]
- Pascual A, Preat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- Peabody NC, Pohl JB, Diao F, Vreede AP, Sandstrom DJ, Wang H, Zelensky PK, White BH. Characterization of the decision network for wing expansion in Drosophila using targeted expression of the TRPM8 channel. J Neurosci. 2009;29:3343–3353. doi: 10.1523/JNEUROSCI.4241-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peier AM, Moqrich A, Hergarden AC, Reeve AJ, Andersson DA, Story GM, Earley TJ, Dragoni I, McIntyre P, Bevan S, Patapoutian A. A TRP channel that senses cold stimuli and menthol. Cell. 2002;108:705–715. doi: 10.1016/s0092-8674(02)00652-9. [DOI] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Qian J, Colmers WF, Saggau P. Inhibition of synaptic transmission by neuropeptide Y in rat hippocampal area CA1: modulation of presynaptic Ca2+ entry. J Neurosci. 1997;17:8169–8177. doi: 10.1523/JNEUROSCI.17-21-08169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrobe JP, Dumont Y, Herzog H, Quirion R. Characterization of neuropeptide Y, Y(2) receptor knockout mice in two animal models of learning and memory processing. J Mol Neurosci. 2004;22:159–166. doi: 10.1385/JMN:22:3:159. [DOI] [PubMed] [Google Scholar]
- Rhim H, Kinney GA, Emmerson PJ, Miller RJ. Regulation of neurotransmission in the arcuate nucleus of the rat by different neuropeptide Y receptors. J Neurosci. 1997;17:2980–2989. doi: 10.1523/JNEUROSCI.17-09-02980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T, Voller T, Stock P, Buchner E, Fiala A. Punishment prediction by dopaminergic neurons in Drosophila. Curr Biol. 2005;15:1953–1960. doi: 10.1016/j.cub.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Roeder KD. Spontaneous activity and behavior. Sci. Monthly. 1955;80:362–370. [Google Scholar]
- Sahu A, Kalra PS, Kalra SP. Food deprivation and ingestion induce reciprocal changes in neuropeptide Y concentrations in the paraventricular nucleus. Peptides. 1988;9:83–86. doi: 10.1016/0196-9781(88)90013-7. [DOI] [PubMed] [Google Scholar]
- Sanacora G, Kershaw M, Finkelstein JA, White JD. Increased hypothalamic content of preproneuropeptide Y messenger ribonucleic acid in genetically obese Zucker rats and its regulation by food deprivation. Endocrinology. 1990;127:730–737. doi: 10.1210/endo-127-2-730. [DOI] [PubMed] [Google Scholar]
- Saper CB, Chou TC, Elmquist JK. The need to feed: homeostatic and hedonic control of eating. Neuron. 2002;36:199–211. doi: 10.1016/s0896-6273(02)00969-8. [DOI] [PubMed] [Google Scholar]
- Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101:647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, Fiala A. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol. 2008;18:1110–1117. doi: 10.1016/j.cub.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Claridge-Chang A, Sjulson L, Pypaert M, Miesenbock G. Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell. 2007;128:601–612. doi: 10.1016/j.cell.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D, Zars M, Laferriere H, Chen YC, Sable-Smith A, Kitamoto T, Rottinghaus GE, Zars T. Serotonin is necessary for place memory in Drosophila. Proc Natl Acad Sci U S A. 2008;105:5579–5584. doi: 10.1073/pnas.0710168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley BG, Leibowitz SF. Neuropeptide Y injected in the paraventricular hypothalamus: a powerful stimulant of feeding behavior. Proc Natl Acad Sci U S A. 1985;82:3940–3943. doi: 10.1073/pnas.82.11.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QQ, Baraban SC, Prince DA, Huguenard JR. Target-specific neuropeptide Y-ergic synaptic inhibition and its network consequences within the mammalian thalamus. J Neurosci. 2003;23:9639–9649. doi: 10.1523/JNEUROSCI.23-29-09639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- Tatemoto K, Carlquist M, Mutt V. Neuropeptide Y--a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature. 1982;296:659–660. doi: 10.1038/296659a0. [DOI] [PubMed] [Google Scholar]
- Tempel BL, Bonini N, Dawson DR, Quinn WG. Reward learning in normal and mutant Drosophila. Proc Natl Acad Sci U S A. 1983;80:1482–1486. doi: 10.1073/pnas.80.5.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe WH. Learning and Instinct in Animals. Cambridge: Harvard University Press; 1956. [Google Scholar]
- Toates FM. Motivational Systems. Cambridge: Cambridge University Press; 1986. [Google Scholar]
- Tolman EC. Purposive behavior in animals and men. New York: The Century Co.; 1932. [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol [A] 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- van Swinderen B. Attention-like processes in Drosophila require short-term memory genes. Science. 2007;315:1590–1593. doi: 10.1126/science.1137931. [DOI] [PubMed] [Google Scholar]
- Watts AG. Motivation. In: Arbib MA, editor. The Handbook of Brain Theory and Neural Networks. Cambridge: Bradford Books, MIT press; 2003. [Google Scholar]
- Weintraub D. Dopamine and impulse control disorders in Parkinson's disease. Ann Neurol. 2008;64 Suppl 2:S93–S100. doi: 10.1002/ana.21454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39:147–161. doi: 10.1016/s0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci U S A. 2005a;102:13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. 2005b;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- Yu D, Akalal DB, Davis RL. Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Keene AC, Srivatsan A, Waddell S, Davis RL. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123:945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Yuan N, Lee D. Suppression of excitatory cholinergic synaptic transmission by Drosophila dopamine D1-like receptors. Eur J Neurosci. 2007;26:2417–2427. doi: 10.1111/j.1460-9568.2007.05870.x. [DOI] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- Zhang K, Guo JZ, Peng Y, Xi W, Guo A. Dopamine-mushroom body circuit regulates saliency-based decision-making in Drosophila. Science. 2007;316:1901–1904. doi: 10.1126/science.1137357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.