Abstract
Many bacterial proteins, including most secretory proteins, are translocated across the plasma membrane by the interplay of the cytoplasmic SecA ATPase and a protein-conducting channel formed by the SecY complex. SecA catalyzes the sequential movement of polypeptide segments through the SecY channel. How SecA interacts with a broad range of polypeptide segments is unclear, but structural data raise the possibility that translocation substrates bind into a “clamp” of SecA. Here, we have used disulfide bridge cross-linking to test this hypothesis. To analyze polypeptide interactions of SecA during translocation, two cysteines were introduced into a translocation intermediate: one that cross-links to the SecY channel and the other one for cross-linking to a cysteine placed at various positions in SecA. Our results show that a translocating polypeptide is indeed captured inside SecA's clamp and moves in an extended conformation through the clamp into the SecY channel. These results define the polypeptide path during SecA-mediated protein translocation and suggest a mechanism by which ATP hydrolysis by SecA is used to move a polypeptide chain through the SecY channel.
Keywords: disulfide bridge cross-linking, protein translocation, SecY, secretion, SecA clamp
Many bacterial polypeptides, including most secretory proteins, are transported across the plasma membrane by a process that is similar to protein translocation across the endoplasmic reticulum membrane in eukaryotes (for review, see ref. 1). The polypeptide substrates contain hydrophobic signal sequences that are usually located at the N terminus and cleaved off after membrane transfer. Translocation occurs through a hydrophilic channel formed by a conserved heterotrimeric membrane protein complex, called the SecY complex in bacteria and archaea and the Sec61 complex in eukaryotes. The complex consists of a large α-subunit (SecY or Sec61p) that spans the membrane ten times, and two smaller β- and γ-subunits (called SecG and SecE in bacteria). In bacteria, the SecY channel can either associate with the ribosome to translocate proteins during their synthesis (cotranslational translocation), or it can cooperate with the cytosolic ATPase SecA to transport polypeptides after completion of their synthesis (posttranslational translocation).
SecY forms an hourglass shaped translocation pathway with water-filled funnels toward both sides of the membrane (2–4). The constriction of the pore is located approximately halfway across the membrane and consists of a pore ring of amino acids whose hydrophobic, bulky side chains project radially toward the interior of the channel. The cytoplasmic funnel is empty whereas the external funnel is plugged by a short helix. The channel is opened by displacement of the plug helix, which is triggered by the binding of the signal sequence of a substrate (5, 6).
The SecA ATPase uses the energy of ATP hydrolysis to push polypeptides through the SecY channel (7). SecA is a multidomain protein (8). It contains two RecA-like nucleotide-binding domains (NBD1 and NBD2), which bind the nucleotide at their interface and move relative to one another during the ATP hydrolysis cycle. SecA also contains a polypeptide-cross-linking domain (PPXD), a helical wing domain (HWD), and a helical scaffold domain (HSD). The latter consists of a long helix and two shorter ones that form a two-helix finger (3).
A recent crystal structure of the SecA – SecYEG complex revealed that SecA uses its PPXD and the long helix of the HSD to interact with the SecY channel and that SecA's two- helix finger inserts deeply into the cytoplasmic funnel of SecY (3). Disulfide cross-linking experiments indicate that the fingertip contacts a translocating polypeptide chain right above the entrance into the translocation pore, and mutagenesis experiments show that a tyrosine (or another bulky, hydrophobic residue) at the fingertip is essential for protein translocation (9). These data suggest that motions of the two-helix finger move a polypeptide chain into the SecY channel with the tyrosine providing the major contact site. However, interactions of the fingertip with a polypeptide chain cannot explain how a translocation substrate is recognized and bound by SecA.
The mechanism by which SecA interacts with its translocation substrates is indeed an interesting problem. The initial binding likely involves the signal sequence, but ultimately, SecA must interact with a broad range of polypeptide segments to move them sequentially into the SecY channel (10). One possible polypeptide-binding site is the “clamp” of SecA (3), which is formed by the PPXD, the NBD2, and parts of the HSD (Fig. S1). Structures of SecA in isolation show the clamp in various open conformations (8, 11–13), which would allow a polypeptide chain to enter it. Structures of SecA bound to the SecY channel show the clamp in a closed conformation, in which the PPXD has rotated all of the way toward the NBD2 (3) (Fig. S1). This motion might capture a polypeptide chain and position it above the SecY pore. Although this model is attractive, other polypeptide binding sites in SecA have been proposed (8, 12, 14, 15). In addition, the SecA-SecYEG structure shows that the closed clamp leaves only little space for a translocating polypeptide chain (3), which emphasizes the need for experimental testing of the proposed model. Recent spin-label perturbation experiments provide evidence that a polypeptide chain can indeed bind to the clamp of nontranslocating SecA in solution, but several positions outside the clamp also showed interactions (16). In fact, our disulfide bridge cross-linking experiments with single cysteines in SecA and in the substrate indicate that SecA interacts rather promiscuously with polypeptides when not engaged in translocation (see Results). It is therefore important to define substrate interactions by analyzing translocating SecA molecules.
Here, we have tested the postulated role of SecA's clamp using disulfide bridge cross-linking to analyze interactions of SecA that is engaged in translocation. We show that a polypeptide chain is captured inside the clamp and moves through it into the SecY pore. Our data delineate the entire path of a translocating polypeptide chain during SecA-mediated protein translocation and shed light on the mechanism by which polypeptides are translocated by SecA.
Results
Experimental Strategy.
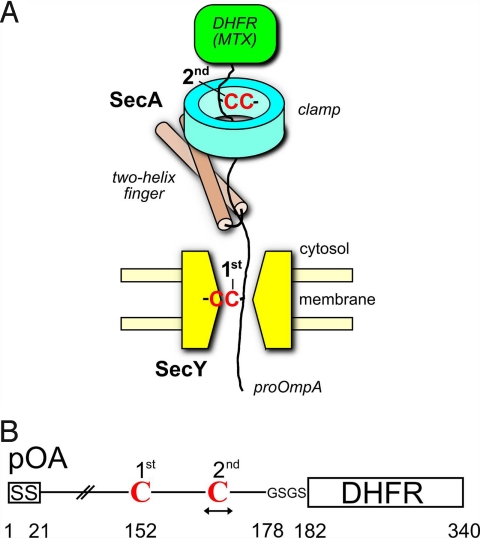
We used the following disulfide bridge cross-linking strategy to test interactions of translocating SecA with a polypeptide chain. Two cysteines were introduced into a translocation intermediate of the substrate proOmpA (pOA), one cysteine that would cross-link to the pore ring of the SecY channel and another one further toward the C terminus that would cross-link to a cysteine in SecA (Fig. 1 A and B). Double cross-links of pOA to both SecY and SecA would indicate that the polypeptide chain has engaged both translocation components. By changing the position of the second cysteine in pOA and placing the corresponding partner cysteine at various positions in SecA, we should be able to identify SecA residues that are in the path of a translocating polypeptide chain. Specifically, this strategy should allow us to test whether a translocating polypeptide chain moves through SecA's clamp.
Fig. 1.
Cross-linking strategy employing a translocation intermediate. (A) Scheme of the experimental strategy. Translocation of the C terminus of proOmpA was blocked by a fused dihydrofolate reductase (DHFR) domain that is folded in the presence of methotrexate (MTX). The fusion protein contains two cysteines (C), one for cross-linking to the SecY pore and a second at various positions for cross-linking to SecA mutants containing single cysteines (shown is a SecA with a cysteine in the clamp). (B) Scheme of the proOmpA (pOA)-DHFR construct. The position of the first cysteine (position 152) is kept constant whereas the position of the second is varied. GSGS is a linker sequence. SS, signal sequence.
To generate a translocation intermediate, the polypeptide chain must be trapped in the channel by preventing its complete translocation with a bulky group at the C terminus. In previous experiments, we used a tRNA at the C terminus of the translocation substrate pOA (pOA-tRNA) (9). The substrate was generated by in vitro translation of a truncated pOA mRNA, followed by release of the peptidyl-tRNA from the ribosome by urea treatment. Cross-linking experiments showed that the C-terminal residues immediately preceding the tRNA moiety were close to the tip of SecA's two-helix finger, indicating that most of the polypeptide chain had moved into the channel. Thus, the tRNA moiety had passed through SecA, perhaps because it was denatured by the urea treatment, and blocked further translocation only at the entrance into the SecY pore. Obviously, this translocation intermediate is inappropriate to identify interactions of SecA's clamp with a translocating polypeptide.
To generate a translocation intermediate in which the C terminus cannot pass SecA and move all of the way into the SecY pore, we fused the first 177 residues of pOA and four linker residues to dihydrofolate reductase (DHFR) (pOA-DHFR) (Fig. 1B) (17). DHFR folds well in the presence of its substrate analog methotrexate and provides an efficient block for the movement of the C terminus of the substrate through the membrane.
pOA-DHFR Is Translocated into the SecY Channel.
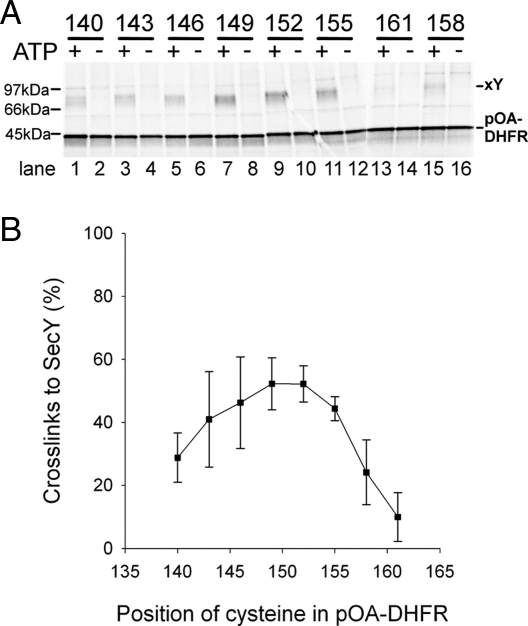
We first determined whether the pOA-DHFR translocation intermediate is captured inside the SecY channel. Single cysteines were introduced into pOA-DHFR and tested for cross-linking to a cysteine in the pore ring of SecY (position 282). These experiments were performed with E. coli SecA lacking endogenous cysteines. Disulfide bridge formation was induced by the addition of the oxidant copper phenanthroline, and the products were analyzed by nonreducing SDS/PAGE followed by autoradiography. As expected, disulfide bridge formation between pOA-DHFR and SecY was only observed in the presence of ATP, when SecA is functional (Fig. 2A, odd numbered lanes). In addition, the cross-links were dependent on the addition of the oxidant. Essentially, all of the tested positions in pOA-DHFR (positions 140 to 161) gave cross-links to SecY, although the efficiency was highest with positions 149–155 (Fig. 2 A and B). These results show that the polypeptide chain is indeed located inside the SecY channel. The broad range of cross-linking positions also suggests that the polypeptide chain can slide back and forth inside the pore. For further experiments we used pOA-DHFR substrates with a cysteine at position 152, which gave prominent cross-links to SecY (Fig. 2A, lane 9).
Fig. 2.
Interaction of a translocation intermediate with the SecY pore. (A) pOA-DHFR containing a single cysteine at the indicated positions was synthesized in vitro in the presence of [35S]methionine. These substrates were incubated in the presence or absence of ATP with SecA lacking cysteines and proteoliposomes containing SecY with a single cysteine at position 282 in the pore ring. Disulfide bridge formation was induced with an oxidant. The samples were separated by SDS/PAGE and analyzed by autoradiography. Note that the samples 158 and 161 were loaded in the wrong order. The positions of free and SecY-cross-linked substrate (pOA-DHFR and xY) are indicated. (B) Quantification of three experiments performed as in A (mean and standard deviation). Shown is the percentage of substrate cross-linked to SecY [xY/(xY + pOA-DHFR)].
Probing Interactions of pOA-DHFR by Double Cross-Linking to SecA and SecY.
To determine whether a translocating polypeptide moves through SecA's clamp, we used pOA-DHFR constructs that contained a cysteine at position 152 for cross-linking to the pore residue 282 in SecY and a second cysteine placed at more C-terminal positions (positions 157–184) to probe for interaction with SecA's clamp. The pOA-DHFR substrates were incubated in the presence or absence of ATP with proteoliposomes containing purified SecY (I282C) complex and SecA mutants that contained a single cysteine at different positions. Disulfide bridge formation was induced by addition of an oxidant.
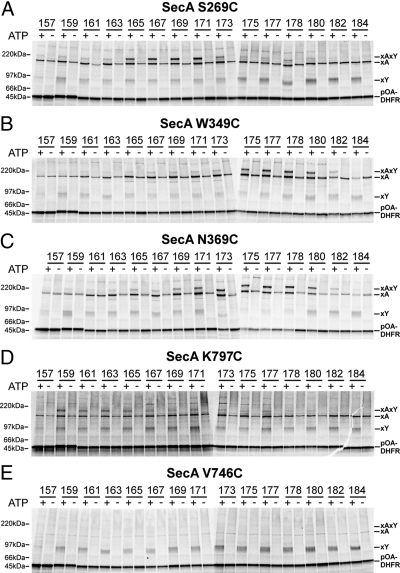
We first tested a SecA mutant that carried a cysteine at position 269 (SecA-S269C), a position in the PPXD located inside the clamp (Fig. 3A). Several positions indeed gave prominent double cross-links of pOA-DHFR to both SecY and SecA (xAxY). The double cross-links had the expected size and could be immunoprecipitated by SecY or SecA antibodies (Fig. S2). In contrast, the SecY single-cross-links were recognized only by SecY antibodies and the SecA-single cross-links by SecA antibodies, although a minor proportion of these cross-links was also precipitated nonspecifically by SecY antibodies (Fig. S2). The double cross-links and the single cross-links to SecY (xY) were only seen in the presence of ATP (Fig. 3A), whereas the cross-links to SecA (xA) were also seen in the absence of ATP, reflecting the promiscuous interaction of SecA with nontranslocating substrate.
Fig. 3.
Probing interactions of a translocation intermediate with SecA. (A) Interaction of a substrate with the SecA clamp. pOA-DHFR containing a cysteine at positions 152 and a second cysteine at the indicated positions was synthesized in vitro in the presence of [35S]methionine. The substrate was incubated in the presence or absence of ATP with SecA containing a single cysteine in the clamp at position 269 and with proteoliposomes containing SecY with a cysteine at position 282. The samples were treated with an oxidant and analyzed by nonreducing SDS/PAGE and autoradiography. The positions of free, SecA-, SecY-, and double cross-linked substrate (pOA-DHFR, xA, xY, and xAxY) are indicated. (B) As in A but with a cysteine in the SecA clamp at position 349. (C) As in A but with a cysteine in the SecA clamp at position 369. (D) Interaction of a substrate with the two-helix finger of SecA. The experiments were performed as in A but with a cysteine at the SecA fingertip (position 797). (E) A cysteine randomly placed into SecA does not interact with the translocation intermediate. The experiments were performed as in A but with a cysteine at position 746 of SecA.
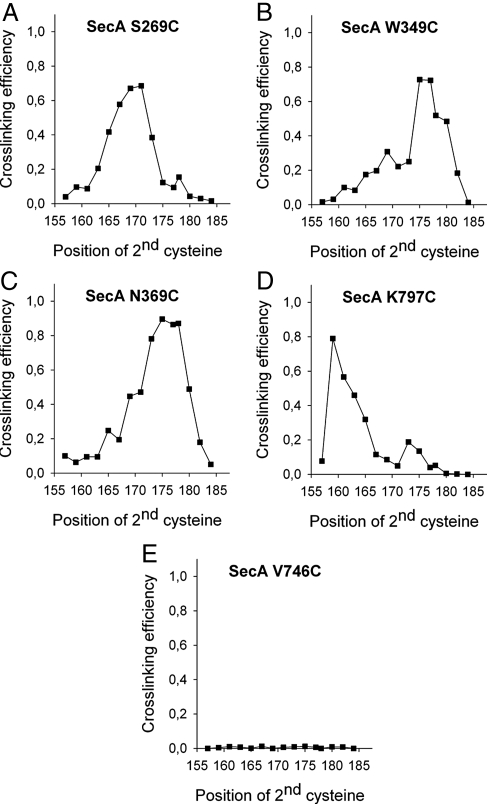
The analysis showed that positions 165 to 171 of the substrate gave the most prominent double cross-links to SecA-S269C (Fig. 3A); >70% of the pOA-DHFR molecules that cross-linked to SecY also cross-linked to SecA (Fig. 4A). At other positions of the substrate, the double cross-links were weaker, and instead, the SecY single cross-links were more intense (Figs. 3A and 4A). These results indicate that position 269 in the clamp of SecA is in the path of a translocating polypeptide, contacting a region in the substrate that is centered around residue 170.
Fig. 4.
Quantification of the interactions of a translocation intermediate with SecA. (A–E) Quantified experiments from Fig. 3 A–E. The cross-linking efficiency is expressed as the ratio of double cross-links over the total SecY cross-links [xAxY/(xAxY + xY)].
We next tested other positions in the clamp. Position 349 in the PPXD and position 369, which is located near the β-strands that connect the NBD1 with the PPXD, also gave efficient double cross-links (Fig. 3 B and C); quantification indicated cross-linking yields of up to 90% at the optimum cysteine positions in the substrate (Fig. 4 B and C). Other positions chosen inside the clamp also gave cross-linking yields exceeding 70% (Figs. S3 A–D and S5 A–D), with the exception of position 339 (≈55% yield) (Figs. S3E and S5E). In each case, the intensities of the double cross-links were dependent on the position of the second cysteine in the pOA-DHFR construct and displayed pronounced maxima. These results indicate that each position in the clamp contacts a certain region of the substrate. Because a substrate region, rather than a single residue, contacts a given SecA position, these data also indicate that the polypeptide chain is caught at different stages during its movement through the clamp.
In agreement with previous results (9), prominent double cross-links were also seen with a cysteine placed at the tip of the two-helix finger (position 797) (Figs. 3D and 4D). However, in contrast to the previous experiments in which the substrate carried a tRNA at the C terminus, the cross-links to the fingertip disappeared when the second cysteine was placed further toward the C terminus of pOA (Figs. 3D and 4D). These data are consistent with the assumption that the DHFR domain prevents the C terminus of the substrate from entering SecA's clamp whereas the tRNA only blocks translocation at a later point (at the entrance into the SecY pore).
Ten positions chosen outside the predicted path of a translocating polypeptide chain either did not give double cross-links at all or cross-linked only weakly (Figs. 3E and 4E; the other positions are shown in Figs. S4 A–I and S5 F–N).
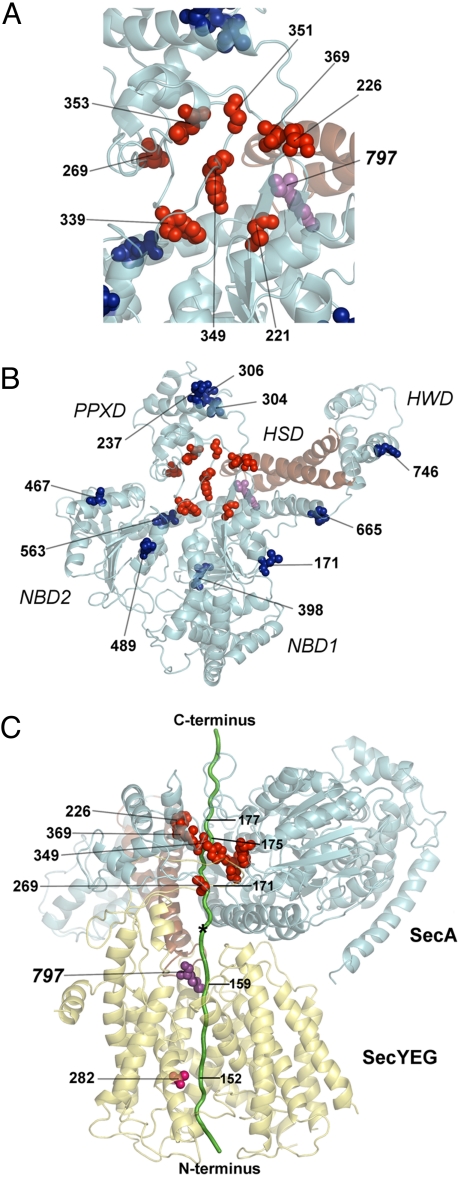
We mapped the tested positions onto SecA in the recently determined SecA-SecYEG structure (3). Viewed from the cytoplasm, the substrate-interacting positions are all localized inside the central cavity of the clamp (shown in red in Figs. 5 A and B), including positions in the PPXD and the two β-strands that connect the NBD1 with the PPXD. In a side view, these positions and the cross-linking position 797 of the two-helix finger delineate the path of the polypeptide chain from the fused DHFR domain all of the way into the SecY channel (Fig. 5C). The most C-terminal residues of pOA (approximately position 177), which immediately precede the DHFR block (beginning at position 182), contact the clamp on its membrane-distal side (position 226) (Fig. 5C). The preceding pOA residues around residue 173–175 contact positions 349 and 369 located approximately half-way inside the clamp, and the region around residue 171 is close to position 269 at the membrane-proximal side of SecA (Fig. 5C). Finally, pOA positions around residue 159 contact the two-helix finger (position 797) that is inserted into the SecY channel (Fig. 5C). In each case, multiple positions of the polypeptide chain contact a given SecA position, indicating that the substrate has some flexibility and is caught in different conformations. By measuring the distance between the pore ring of SecY and the tested positions in SecA, it appears that the translocating polypeptide chains can adopt rather extended conformations. For example, at least seven residues of pOA (residues 152 to 159) are required to span the distance of 25Å between SecY's pore ring and the tip of the two-helix finger (3.6Å per residue), which is close to a fully extended conformation (18). Similarly, most of the pOA positions that contact residue 226 in the clamp (≈66Å away from the pore ring) are 20–25 residues away from the position that cross-links to the pore ring (residues 171–177 versus 152; 2.6–3.3Å per residue), and a minor population of translocating chains appears to be entirely stretched out.
Fig. 5.
Interaction sites with a translocating polypeptide mapped onto the T. maritima SecA structure (PDB accession 3DIN). (A) View from the cytosol of SecA positions that were tested for cross-linking to a translocating polypeptide chain. Cross-linking positions in the clamp and two-helix finger are shown as red and magenta balls, respectively. Positions that gave weak or no cross-links are shown in blue. The two-helix finger is shown in brown. (B) As in A but with a zoomed-out view. (C) Side view of the SecA-SecY structure with a modeled translocating pOA-DHFR substrate (pOA is shown in green; the DHFR domain was omitted for clarity). Positions in SecA's clamp and the two-helix finger, which could be cross-linked to the substrate, are shown as red and magenta balls, respectively. The cross-linking SecY pore residue is shown as pink balls. The star indicates an opening toward the cytosol.
Discussion
Our results demonstrate that a translocating polypeptide chain is captured inside SecA's clamp and moves through it into the SecY channel. These results define the polypeptide path during SecA-mediated translocation and have important implications for the mechanism of translocation.
The identification of the clamp as the polypeptide-binding site is strongly supported by the fact that many other SecA positions showed only weak or no interactions with a translocating chain. This includes positions on the external surface of the clamp (positions 237, 304, and 306), which are located in a groove that NMR experiments implicated in signal sequence interaction (15). This suggests that SecA has distinct binding sites for signal sequences and polypeptide segments that follow them. How a signal sequence would be transferred from the outside of the clamp into the SecY channel is unclear, particularly because the SecA-SecYEG structure shows that a direct path is blocked by the interaction of the PPXD with loops of SecY (3). Perhaps, the signal sequence is released from its binding site before SecA interacts with SecY, or the hydrophobic interior of the clamp provides an additional or alternative binding site for the signal sequence.
At the beginning of translocation, SecA's clamp must be open to allow the entry of a substrate. This requires that the PPXD rotates away from the NBD2, a conformation represented by x-ray structures of SecA in isolation (8, 12) (see Fig. S1). We propose that the open clamp interacts through the two β-strands that link the NBD1 and PPXD with the backbone of a polypeptide chain. The interacting polypeptide segment would be induced to form a short β-strand that extends this β-sheet, a mechanism termed “β-strand augmentation” (19). This sequence-independent mode of interaction is supported by a recent structure of a SecA-peptide complex (20), as well as by two other SecA structures in which a polypeptide segment formed an additional short β-strand next to the β-sheet connecting NBD1 and PPXD (8, 21).
Once the polypeptide chain is bound, the PPXD would rotate toward the NBD2 and close the clamp, a conformation that is stabilized by an interaction of the PPXD with SecY (3). The capture of the polypeptide chain inside the clamp would contribute to the sequence-independent interaction of SecA with a substrate. This mechanism of polypeptide binding is in fact reminiscent of how many chaperones, such as Hsp70s, interact with a broad range of peptide substrates (22); in these cases, extended polypeptide chains are embraced by the walls of a deep groove, making side chain interactions less important. In the recently determined SecA-SecY structure, the interior of the clamp is partially occluded by a loop at the membrane-distal side (3). Our data now suggest that the loop is flexible and likely moves out of the way when a translocation substrate is present. In fact, positions 349 and 351 in this loop give efficient cross-links. Accommodation of a substrate in the clamp is facilitated by the fact that the polypeptide chain is in a rather extended conformation. Our results indicate that a translocating polypeptide chain is in an unfolded conformation all of the way from its entry into the SecA clamp to the pore ring of the SecY channel. Thus, any folding can only occur when the polypeptide chain emerges on the extracellular side of the SecY channel.
The closed clamp might simply position the polypeptide chain above the channel, allowing a translocating polypeptide chain to slide back and forth within its central cavity. However, we consider it more likely that the clamp tightens and widens during the ATP hydrolysis cycle through movements of the PPXD relative to the NBD2. The β-strand augmentation mechanism may still be important because it might prevent complete dissociation of the polypeptide. In this context, it should be noted that cysteines placed close to the two β-strands connecting the NBD1 and PPXD gave the highest cross-linking yields (positions 221, 226, 369).
The translocating polypeptide chain is fully surrounded by clamp residues over a distance of ≈26Å but has to cross a significant gap of ≈12Å from the clamp into the SecY pore. This opening toward the cytosol is located underneath the mouth of the clamp, right above the lateral gate of the SecY channel (indicated by a star in Fig. 5C) and would be of sufficient size to accommodate two polypeptide strands in an extended conformation. A polypeptide chain could therefore loop out sideways into the cytosol, rather than move into the SecY channel. However, this would be energetically unfavorable and would not normally occur during the translocation of a secretory protein. Also, because SecA is required for the biosynthesis of some membrane proteins (23–25), the cytosolic exit pathway may allow segments following transmembrane domains to emerge into the cytosol, similar to what has been proposed for membrane protein integration during cotranslational translocation (26).
Our results show that a polypeptide chain emerging from the clamp contacts the two-helix finger of SecA before it enters the SecY pore. The determined polypeptide path is consistent with a model in which, upon ATP binding by SecA, the finger would move toward the SecY pore and drag the polypeptide chain with it. At the same time, the clamp would loosen its grip on the polypeptide chain. Upon ATP hydrolysis, the clamp would tighten, the finger would disengage from the polypeptide and reset to “grab” the next polypeptide segment. This process would be repeated until the polypeptide chain is all of the way through the SecY channel.
Materials and Methods
Cloning, Mutagenesis, and Protein Purification.
Full-length E. coli proOmpA with a C-terminal hexa-histidine tag was cloned into pMAl p4E (NEB) using NdeI and HindIII restriction sites. A SalI site was introduced after position 175 and a NcoI site in front of the His-tag by site directed mutagenesis (Stratagene). These sites were used to insert E.coli DHFR with N- and C-terminal GSGS-linkers. The two endogenous cysteines of DHFR were removed, and single cysteines were introduced into the new construct at positions 152 and 157–184 by site-directed mutagenesis. All constructs were confirmed by sequencing.
For translocation and cross-linking assays, a linear fragment of pOA-DHFR containing a 5′ SP6 promoter and a 3′ stop-codon was generated by PCR. After in vitro transcription (Promega), the mRNA was translated in rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine. The crude lysate was then used for the experiments.
Point mutations in E. coli SecA and SecY were introduced by site directed mutagenesis. Both SecA and SecY complex were overexpressed in BL21 cells and purified using Ni- affinity chromatography (12). Purified SecY complex was reconstituted into liposomes made of E.coli polar lipids (27).
Cross-Linking.
Cross-linking was performed in 20 μl reactions containing 50 mM Hepes/KOH pH7.5, 50 mM KCl, 5 mM MgCl2, 2.5 mM ATP, 0.5 mg/ml BSA, 40 μg/ml SecA, 20 μg/ml SecY complex in proteoliposomes, and 1 μl lysate containing in vitro synthesized pOA-DHFR (9). For cross-linking in the absence of ATP, 0.5 units of hexokinase and 10 mM glucose were added to the reaction. After 15 min at 37 °C, cross-links were formed by the addition of 50 μM copper phenanthroline for 10min at 37 °C. Samples were treated with 10 mM N-ethylmaleimide (NEM) for 5 min on ice and then subjected to nonreducing SDS/PAGE on 4–20% Tris·HCl gels (Bio-Rad). The bands were visualized by phosphorimaging. QuantityOne (Bio-Rad) was used for local background subtraction and quantification. Cross-linking efficiencies were expressed as the ratio of counts in double cross-linked bands (xAxY) to the sum of counts in double cross-linked and single SecY cross-linked bands [xAxY/(xAxY + xY)].
Supplementary Material
Acknowledgments.
We thank K. Erlandson for providing protocols, mutants, and helpful hints; J. Zimmer for help with protein purification and modeling; E. Park for advice on cloning and mutagenesis; and J. Zimmer, A. Salic, Y. Chen, and A. Tripathi for critical reading of the manuscript. We also thank P. Pohl for generously providing the opportunity for B.W.B.'s stay in Boston. This work was supported by National Institutes of Health Grant GM052586. B.W.B. was supported by a fellowship from the Austrian Marshall Plan Foundation. T.A.R. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910550106/DCSupplemental.
References
- 1.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 2.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 3.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukazaki T, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris CR, Silhavy TJ. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J Bacteriol. 1999;181:3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 8.Hunt JF, et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 9.Erlandson KJ, et al. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiebel E, Driessen AJM, Hartl F-U, Wickner W. DμH+ and ATP Function at Different Steps of the Catalytic Cycle of Preprotein Translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 11.Sharma V, et al. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA. 2003;100:2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papanikolau Y, et al. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol. 2007;366:1545–1557. doi: 10.1016/j.jmb.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 14.Papanikou E, et al. Identification of the preprotein binding domain of SecA. J Biol Chem. 2005;280:43209–43217. doi: 10.1074/jbc.M509990200. [DOI] [PubMed] [Google Scholar]
- 15.Gelis I, et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper DB, et al. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arkowitz RA, Joly JC, Wickner W. Translocation Can Drive the Unfolding of a Preprotein Domain. EMBO J. 1993;12:243–253. doi: 10.1002/j.1460-2075.1993.tb05650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ainavarapu SR, et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remaut H, Waksman G. Protein–protein interaction through beta-strand addition. Trends Biochem Sci. 2006;31:436–444. doi: 10.1016/j.tibs.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Zimmer J, Rapoport TA. Conformational flexibility and peptide interaction of the translocation ATPase SecA. J Mol Biol. 2009 Oct 20; doi: 10.1016/j.jmb.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmer J, Li W, Rapoport TA. A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J Mol Biol. 2006;364:259–265. doi: 10.1016/j.jmb.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi HY, Bernstein HD. SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J Biol Chem. 1999;274:8993–8997. doi: 10.1074/jbc.274.13.8993. [DOI] [PubMed] [Google Scholar]
- 24.Neumann-Haefelin C, Schafer U, Muller M, Koch HG. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 2000;19:6419–6426. doi: 10.1093/emboj/19.23.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duong F, Wickner W. Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. EMBO J. 1998;17:696–705. doi: 10.1093/emboj/17.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapoport TA, Goder V, Heinrich SU, Matlack KE. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.