Abstract
The evolutionarily conserved Argonaute/PIWI (AGO/PIWI; a.k.a. PAZ-PIWI Domain, or PPD) family of proteins is crucial for the biogenesis and function of small non-coding RNAs (ncRNAs). This family can be divided into AGO and PIWI subfamilies. The AGO proteins are ubiquitously present in diverse tissues. They bind to small interfering RNAs (siRNAs) and microRNAs (miRNAs). In contrast, the PIWI proteins are predominantly present in the germline, and associate with a novel class of small RNAs known as PIWI-interacting RNAs (piRNAs). Tens of thousands of piRNA species, typically 24-32 nucleotide long, have been found in mammals, zebrafish, and Drosophila. Most piRNAs appear to be generated from a small number of long single-stranded RNA precursors that are often encoded by repetitive intergenic sequences in the genome. PIWI proteins play crucial roles during germline development and gametogenesis of many metazoan species, from germline determination and germline stem cell maintenance to meiosis, spermiogenesis, and transposon silencing. These diverse functions may involve piRNAs, and may be achieved via novel mechanisms of epigenetic and post-transcriptional regulation.
Keywords: epigenetic regulation, germ cell, stem cell, transposon silencing, translational regulation
I. Introduction
The past decade has witnessed rapid advancement in understanding how small non-coding RNAs (ncRNAs) regulate diverse developmental processes. Central to this regulation is the evolutionarily conserved piwi/argonaute (a.k.a. PAZ-PIWI Domain, or PPD) protein family, which was first discovered for their function for stem cell self-renewal and germline development (Cox et al. 1998). This protein family can be divided into AGO and PWI subfamilies, herein referred to as AGO and PIWI proteins, respectively. AGO proteins bind to small interfering RNAs (siRNAs) and micro RNAs (miRNAs), and have been shown to play crucial roles in the siRNA and miRNA pathways in many tissues; whereas PIWI proteins bind to novel class of ncRNAs called PIWI-interacting RNAS in 2006 (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006a; Lau et al. 2006; Watanabe et al. 2006) and play diverse function in germline development and gametogenesis.
In this review, we will summarize recent progress in understanding the biological function of PIWI proteins, the biogenesis and function of piRNAs, and the molecular mechanisms mediated by PIWI proteins and their piRNA partners.
II. Developmental Function of PIWI Proteins
The developmental functions of PIWI proteins have so far mostly inferred from analyzing the phenotype of the loss-of-function mutants of piwi genes. These analyses, mostly conducted in Drosophila, mice, C. elegans, zebrafish, and planarian, have offered definitive information about the function in PIWI proteins in both germline and somatic tissues. Table 1 summarizes frequently encountered PIWI proteins in diverse organisms.
A. Germline Function
PIWI proteins are mostly restricted to the germline. Different PIWI proteins exist and function in different stages of the germline cycle. Among different organisms, the crucial role of PIWI proteins can be found from the earliest stage of germline development (germline fate specification) to late stages of gametogenesis such spermiogenesis, egg activation, and fertilization. Current studies in different organisms point to the possibly conserved requirement of PIWI proteins throughout the germline cycle in diverse organisms.
a. Germ cell formation
The role of PIWI proteins in germ cell specification during early embryogenesis has been demonstrated in Drosophila. Germ cell formation in Drosophila requires a specialized form of cytoplasm known as the germ plasm. The germ plasm is found at the posterior pole of the embryo and is both necessary and sufficient for germ cell formation (Illmensee and Mahowald 1974). The germ plasm is morphologically distinct from the rest of the cytoplasm in the embryo in that it contains electron dense structures known as polar granules, which are often found in association with mitochondria (Mahowald 2001). Similar electron-dense structures are seen at different points in Drosophila germline development, most notably during oogenesis as the nuage, a perinuclear germline structure. Analogous germline structures are also seen in many metazoan species, including p granules in C. elegans, the mitochondrial cloud in Xenopus, and the chromatoid body in mammalian testes (Saffman and Lasko 1999). Although the role of polar granules and analogous structures are unknown, many proteins and RNAs known to participate in germ cell formation localize to these structures. In Drosophila, there is a strong correlation between polar granule formation and germ cell formation.
PIWI is a polar granule component and controls germ cell formation (Megosh et al. 2006). In embryos lacking maternal PIWI, there is a severe decrease in germ cell formation (Megosh et al. 2006). Although ectopic PIWI does not induce ectopic germ cell formation, increases in maternal piwi dosage lead to a proportional increase in primordial germ cell formation. It was not determined if all of these germ cells were functional, although they possessed physical characteristics of germ cells. Immuno-labeling of PIWI analyzed by confocal microscopy showed that PIWI co-localizes with polar granule markers such as VASA, a highly conserved DEAD box RNA helicase needed for germ cell formation (Hay et al. 1988; Lasko and Ashburner 1988; Hay et al. 1990; Lasko and Ashburner 1990), at the posterior pole. In addition, inducing ectopic localization of a polar granules component, OSKAR, at the anterior pole leads to PIWI localization to the OSKAR-containing granules at the anterior pole. Furthermore, PIWI interacts with VASA in a complex that can be isolated by reciprocal co-immunoprecipitation. These results strongly suggest that PIWI is a polar granule component.
PIWI was also found to associate with DICER1 and dFMRP (Megosh et al. 2006). DICER1 is integral to miRNA biogenesis and function, and dFMRP has direct roles in these processes as well (Bernstein et al. 2001; Caudy et al. 2002). The depletion of either dFMRP or DICER1 also leads to a decrease in germ cell formation, consistent with function of maternal PIWI and the knowledge of germ cell-specific miRNAs (Megosh et al. 2006). Moreover, PIWI protein associates with a small number of miRNAs in addition to a large number of piRNAs (Yin and Lin 2007). All these observations suggest that the function of PIWI in controlling the formation of primordial germ cells is via the miRNA mechanism.
The PIWI homolog AUBERGINE (AUB) is also a polar granule component, as shown by confocal immunofluorescence and immuno-electron microscopy, and is required for gem cell formation in Drosophila (Harris and Macdonald 2001; Thomson et al. 2008). The few fertilized embryos laid by aub-/- females contain no germ cell and display abdominal defects—typical phenotype of the grandchildless group of genes in Drosophila (Harris and Macdonald 2001; Mahowald 2001). Further localization experiments suggest that the role of AUB in OSK translation is independent of any role AUB plays in germ cell formation, as OSK localizes to the posterior before AUB (Harris and Macdonald 2001). Results from ectopic OSK experiments are consistent with this hypothesis; OSK can direct the formation of germ cells at ectopic sites (Ephrussi et al. 1991), and germ cells do not form in aub mutant embryos with ectopic OSK (Harris and Macdonald 2001). Taken together this shows AUB acts downstream of OSK in germ cell formation, while being upstream of OSK specifically osk mRNA translation during oogenesis.
b. Germline stem cell maintenance
The germline stem cell function of PIWI proteins was also first discovered for the Drosophila PIWI. Drosophila piwi gene was initially discovered in a mutagenesis screen for genes that control asymmetric stem cell division in the Drosophila germline (Lin and Spradling 1997). PIWI become localized to the nucleus after primordial germ cell formation, and is expressed in both germline and somatic cells in both male and female Drosophila (Cox et al. 1998). Piwi mutant flies fail to maintain germline stem cells in both males and females (Cox et al. 1998). Genetic mosaic and niche-cell specific expression analyses indicate that the expression of PIWI in the somatic niche cells is required for germline stem cell maintenance (Szakmary et al. 2005); whereas the expression of PIWI in germline stem cells promotes their division (Cox et al. 1998). Furthermore, overexpressing PIWI in the germarium somatic cells results in an increase in the number of GSC and rate of division. These experiments clearly established the function of PIWI in germline stem cell renewal.
There are three murine PIWI proteins, MIWI, MILI, and MIWI-2, all of which are expressed during spermatogenesis (Figure 1). Among them, Miwi2 expression is detected from day 15.5 dpc to 3 dpp in mitotically arrested prenatal germline stem cells (Aravin et al. 2008). In addition, Miwi2 is expressed in Sertoli cells, which are somatic supporting cells within seminiferous tubules. However the somatic expression of Miwi2 is not necessary for the germline function (Carmell et al., 2007). Mili is expressed from 3 dpp mitotically arrested prenatal germline stem cells to round spermatids (Kuramochi-Miyagawa et al 2004; Unhavaithaya et al, 2008; Wang et al, submitted). Miwi is expressed from meiotic spermatocytes to elongating spermatids (Deng and Lin 2001). In agreement with their expression pattern, both MILI and MIWI-2 show spermatogenic stem cell arrest as their first defect (Carmell et al. 2007; Unhavaithaya et al. 2008). The stem cell arrest phenotype is especially predominant in mili-deficient mice. Only occasionally the stem cells will escape the arrest and produce a few spermatocytes, which were then arrested at the prophase I, especially pachytene stage (Kuramochi-Miyagawa et al. 2004; Unhavaithaya et al. 2008). Somewhat different from Mili mutants, spermatogenesis in Miwi2-null mice was blocked predominantly at the leptotene stage of meiosis I (Carmell et al. 2007). There is massive degeneration of the spermatogonia population in these mice, implicating a function of MIWI-2 in germline stem cell maintenance.
Figure 1.
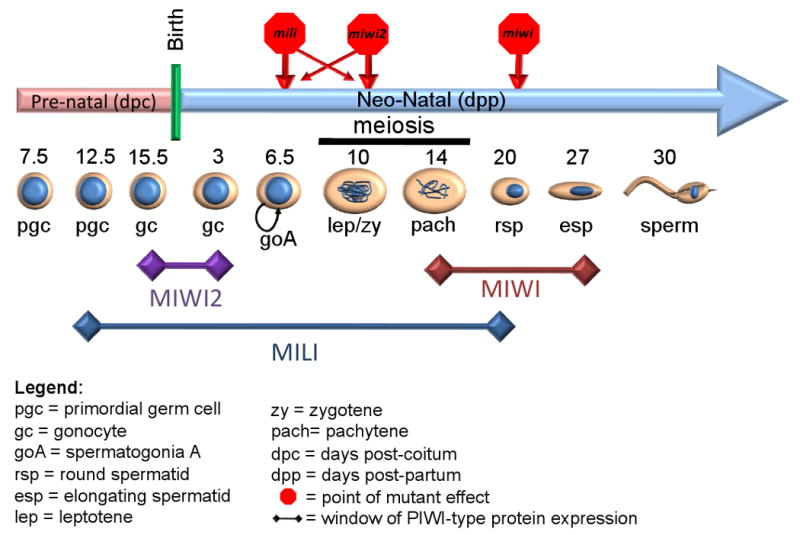
The role PIWI-like proteins during mammalian spermatogenesis. A schematic drawing of mouse spermatogenesis on a time coordinate, with MILI, MIWI, and MIWI2 expression periods indicated.
Expectedly, PIWI proteins have also been identified in other vertebrates such as zebrafish. Zebrafish have two PIWI proteins, Ziwi (zebrafish PIWI) and Zili (zebrafish PIWI like) (Houwing et al. 2007; Houwing et al. 2008). Ziwi is a cytoplasmic protein that is present in both the male and female gonads (Houwing et al. 2007). Ziwi is also expressed in very early stage embryos; this expression reflects the fact that Ziwi is maternally deposited. During subsequent embryogenesis, Ziwi become restricted to the germline, where it is expressed at highest levels during the mitotic and early meiotic stages of germ cell differentiation. Ziwi is found to co-localize with zebrafish VASA in granule-like structures in 2-4 cell stage, these structures are described as zebrafish nuage. The zebrafish nuage is analogous to the nuage in Drosophila or chromatoid body in mice, as described above. Gametogenesis occurs in ziwi mutant fish; there is no clear block of germ cell formation. However, germ cells undergo increased apoptosis, resulting in loss of all germ cells by the time fish reach adulthood (Houwing et al. 2007). Whether or not this is a direct role in GSC maintenance or a secondary effect is unknown.
Different from ZIWI, Zili expression starts 3 days post fertilization, long after zygotic transcription begins (Kane and Kimmel 1993). Zili is found in a granular pattern by 3 weeks post fertilization. In adults, Zili is expressed specifically in testis and ovaries (Houwing et al. 2008). During early spermatogenesis, Zili is found in cytoplasmic granules around spermatogonia and spermatocyte nuclei, but is not detectable in later stages of spermatogenesis. In the ovary, Zili is present at all stages of oogenesis in peri-nuclear granules, and is found in the oocyte nucleus during later stages of oogenesis. Like PIWI, Zili is re-localized to the nucleus during germline development, and also like PIWI, is not present at the sites of the most intense DAPI staining. zili null animals are agametic (Houwing et al. 2008). Strikingly like piwi mutant flies, there is a severe reduction in the number of germ cells in fish lacking Zili, accompanied by a lack of germ cell differentiation to mature oocytes or sperm. Hence, Zili might be required for germline stem cell maintenance.
c. Meiosis
A recurring theme of the role of Piwi proteins is during meiosis. In mice, miwi2 mutant show predominant arrest at the leptotene stage of meiosis (Carmell et al. 2007). The mutant spermatogenic cells show defects in double stranded DNA break repair, consistent with a role in proper meiotic recombination (Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007). Similarly, Mili-null mice are terminally blocked at the zygotene or early pachytene stages of meiotic prophase, (Kuramochi-Miyagawa et al. 2004). This phenotype is very similar to that displayed by mouse vasa homologue (mvh)-null mice (Tanaka et al. 2000). MVH interacts with Mili and Miwi directly in 293T cells, the expression of MVH in testis is not dependent on either Mili or Miwi. However, miwi deficiency does alter subcellular localization of MVH. However, due to the interaction between MVH and Miwi this suggests that Miwi may have a direct role in MVH localization and potential functions of MVH.
The role of PIWI-type proteins in meiosis appears to be conserved during evolution. In Zebrafish, in addition to a null allele, there is a hypomorphic allele of zili, ziliL590P. Males containing this allele are fertile and normal. Interestingly, females are sterile, showing an oogenic block in Meiosis I. Additionally, flies mutant for PIWI proteins and proteins involved in repeat associated siRNA (see later sections for their relationship to PIWI-proteins), also have defects in meiosis (Chen et al. 2007b; Klattenhoff et al. 2007; Lim and Kai 2007; Pane et al. 2007). Although a blockage of meiosis from flies to mammals is intriguing it should be noted that there are meiotic checkpoints operating in metazoan germlines. Therefore it is not clear at this time if PIWI-type proteins have a role in meiosis or whether PIWI-deficient germlines are simply too damaged due to other defects in gametogenesis.
d. Spermiogenesis
The first functional analysis of a mammalian PIWI protein was MIWI (Mouse PIWI). Miwi expression is largely restricted to the testis (Deng and Lin 2002), where it is a cytoplasmic protein expressed in the germline from meiosis I mid-pachytene stage of Meiosis I to the elongating spermatid stage. Consistent with its expression pattern, miwi knockout mice exhibit male sterility, characterized by a block at the early spermatid stage. This phenotype is similar to defects observed in CREM (cAMP-responsive element modulator) deficient mice. CREM is a key transcription activator of genes necessary for spermiogenesis (the morphogenetic process that transforms a round spermatid into a mature sperm). Genes that are normally expressed by post-meiotic cells are not expressed or severely down-regulated in CREM mutant mice. MIWI and CREM do not regulate each other's expression. However, a miwi complex with mRNAs of the Activator of CREM (ACT) and CREM target genes and is required for the stability of these mRNAs. Thus the Miwi and CREM pathway act together to initiate spermiogenesis.
e. Oogenesis
The oogeneic function of PIWI proteins are evident in Drosophila and zebrafish. In Drosophila, the complete female sterility of piwi mutants is not only due to germline stem cell depletion, but also defects in subsequent stages of oogenesis, such as aberrant number of nurse cells and abnormal egg chamber polarity (Lin and Spradling 1997). Mutations of the aubergine locus were discovered in mutagenic screens for defects in oogenesis (Schupbach and Wieschaus 1991). Most embryos display abnormalities in dorsal/ventral patterning, which is a hallmark. Epidermal growth factor receptor (Egfr) pathway defects in Drosophila. Consistent with this, it was found that levels of GURKEN (GRK) protein, the transforming growth factor α (TGFα) homolog, is reduced in aub mutant egg chambers (Wilson et al. 1996). Much less is known about the third PIWI-type protein in the Drosophila genome, AGO3. While no mutations of ago3 have been reported, AGO3 itself does associate with piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). Antibody staining generated against AGO3 show that it is a cytoplasmic protein found in ovaries and testis of mice (Brennecke et al. 2007; Gunawardane et al. 2007). There is evidence of a significant portion of AGO3 being in the soma of females flies, although its subcellular localization in somatic cells is unknown (Brennecke et al. 2007). Owing to its germline enrichment it will be quite probable that ago3 mutants will show defects in oogenesis, but there may be defects in the soma as well.
In zebrafish, both zili and ziwi mutant fish have defects in oogenesis, as mutations of both lead to female sterility, as previously discussed. In C. elegans, The PRG-1 protein is expressed throughout development and localizes to nuage-like structures called P granules. prg-1 mutant sperm exhibit extensive defects in egg activation and fertilization (Batista et al. 2008; Wang and Reinke 2008). There are no known defects for piwi mutants in mammalian oogenesis. This may reflect the difference in oogenesis between mammalian and non-mammalian systems.
B. Somatic Functions
Despite the common believe that PIWI proteins have only germline-restricted functions, the role of these proteins in somatic tissues have been increasingly explored, especially in the genetically amendable model systems.
a. Drosophila
The somatic function of PIWI proteins is evident during early embryogenesis. Embryos laid by piwi mutant mothers, are somatic lethal. (Cox et al. 1998; Cox et al. 2000; Megosh et al. 2006). Like PIWI, AUB has early embryonic functions. The few fertilized embryos laid by aub-/- females have abdominal deletions and no germ cells, as reviewed above (Mahowald 2001). In larval and adult flies, PIWI binds to chromosomes in somatic tissues such as salivary glands and eyes, and exhibit epigenetic effect at the binding sites (Pal-Bhadra et al. 2002; Pal-Bhadra et al. 2004; Brower-Toland et al. 2007; Yin and Lin 2007). The molecular mechanism and biological effects on such epigenetic regulation are under active exploration.
b. Planaria
The regeneration of planarians depends on adult stem cells known as neoblasts. Three-PIWI-type proteins are known to be expressed in neoblasts of the planaria Schmidtea mediterranea (Reddien et al. 2005; Palakodeti et al. 2008). Two of these PIWI proteins, SMEDWI-2 (S. mediterranea PIWI-2) and SMEDWI-3 are needed for neoblast maintenance, but not formation (Reddien et al. 2005; Palakodeti et al. 2008). This is consistent with a role in stem cell maintenance of PIWI proteins in Drosophila, zebrafish and mice.
c. Tetrahymena thermophila
The development of ciliates such as Tetrahymena thermophila depends on a somatic macronucleus and a germline micronucleus. During sexual reproduction known as conjunction specific DNA sequences are eliminated in the newly formed macronucleus. The reason for these specific regions to be deleted is not clear however the function of TWI1P a PIWI-type protein is needed for DNA elimination. There is no viable progeny produced by parental strains absent for TWI1P (Mochizuki et al. 2002).
d. Cancer
In humans, the overexpression of piwi genes is highly correlated with cancers in humans. The over- and ectopic expression of HIWI (Human Piwi) is associated with several cancer types (Qiao et al. 2002; Lee et al. 2006; Liu et al. 2006; Chen et al. 2007a). Striking evidence for a role of HIWI in cancer is the observation that repression of hiwi expression stops cancer growth in cell culture (Liu et al. 2006). As previously discussed PIWI-type proteins have a role in the maintenance of GCS or GSC formation. Keeping in mind the suspected relationship between stem cells and precancerous cells, it is therefore not surprising a family of proteins with roles in stem cell formation would also be involved in cancer development (Chen et al. 2007a; Leedham and Wright 2008).
III. Biochemical Characterization of PIWI proteins
Before we present a discussion on the function of PIWI proteins and piRNAs, an overview and definition of the PIWI proteins will be outlined here. The AGO and PIWI proteins determined by the similarity of a given candidate to either the AGO proteins of Arabidopsis or to Drosophila PIWI. AGO proteins been have found in nearly all eukaryotes, while PIWI proteins are restricted to animals and ciliates, organisms which undergo sexual reproduction (Beyret and Lin 2008). All AGO/PIWI are identified by the presence of two protein domains – the central PAZ (standing for Piwi/Argonaute/Zwille) domain and the C terminal 350-residue PIWI domain. Although the molecular function of each is only partially understood, the PAZ domain contains an OB (oligonucleotide/oligosaccharide binding) fold (Lingel et al. 2003; Yan et al. 2003). OB folds are a typical single stranded nucleic acid binding motif. The structure of the PIWI domain is similar to RNase H enzymes (Song et al. 2004). Three dimensional analyses of multiple AGO proteins is consistent with a model where the PAZ domain forms a pocket for target RNA while PIWI domain sits off to the side cutting bound RNA (Figure 2; Song et al. 2003; Yan et al. 2003; Song et al. 2004; Wang et al. 2008). Indeed, in vitro evidence shows that Drosophila PIWI proteins AGO3, PIWI and AUB can slice RNA targets (Gunawardane et al. 2007). Likewise, RIWI (Rat PIWI) containing complexes also show slicing activity (Lau et al. 2006). It is not clear if the PIWI-type proteins can cleave RNA targets in vivo, in fact other endonucleases have a function in piRNA cleavage (Pane et al. 2007). Although detailed structural analyses suggest potential functions of PIWI proteins, a detailed mutagenic analysis of conserved amino acids in the domains of PIWI proteins, and the impact of these mutations on ncRNA biology, remains to be performed to verify these proposed functions.
Figure 2.
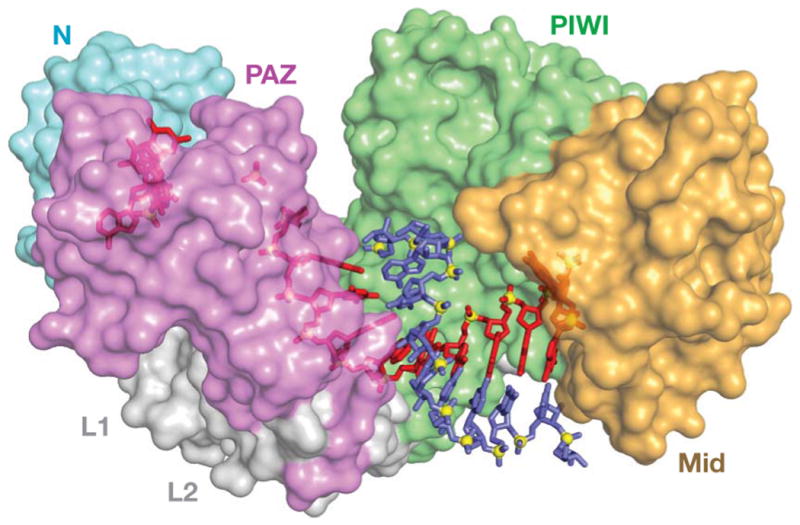
The 3-D structure of a ternary complex of the wild-type Thermus thermophilus AGO in complex with aDNA-piRNA heteroduplex. Red: DNA; blue: RNA. Adopted from Wang et al. 2008.
IV. Identification and Basic Features of piRNAs
Cloning of small RNAs associated with PIWI proteins lead to the discovery of piRNAs in mammalian systems in 2006. Subsequently, piRNAs were also discovered in other model systems by similar efforts.
A. piRNAs in mammals
Four independent efforts in searching for PIWI-interacting ncRNAs led to the identification of piRNAs in 2006 (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006a; Watanabe et al. 2006). Extracts of total RNAs from mouse testes contained an abundant species of approximately 30nt that are associated with MIWI, a mouse PIWI protein (Grivna et al. 2006a). Initial 40 piRNAs were initially cloned. The presence of these piRNAs was almost completely depleted in miwi mutant mice (Grivna et al. 2006a). From the 40 piRNAs initially cloned it was determined that small RNAs from at least one other isolation contained at least in part piRNAs (Grivna et al. 2006a). Using this data set it was observed that piRNAs were not evenly distributed across the mouse genome, but rather cluster at defined loci throughout the genome. This larger species of small known as piRNAs were only present in testicular RNA extracts from 20 day post partum (dpp) mice, and not present in 14 dpp mouse testes. Upon sucrose gradient fractionation, it was observed that piRNAs were found in mRNPs, monosomes, and polysomes in equal abundance. The presence of piRNAs in the polysomal fraction is indicative of a potential role of piRNAs in translational control, to be discussed further in later sections.
Two mass sequencing projects reported over 52,00 piRNAs associated with MIWI and over 1,000 piRNAs associated with MILI, respectively (Aravin et al. 2006; Girard et al. 2006). In these studies as well, piRNAs were found to be asymmetrically distributed about the genome; over 96% are clustered at a few hundred sites. These clusters exhibit interesting characteristics (Figure 3): some are unidirectional, in that the piRNAs appear to be encoded by one strand or the other; whereas many other clusters are bidirectional, with a central piRNA-poor region flanked by piRNA arms encoded by the opposite strand. The clusters tend to be located in intergenic or gene-poor regions of the genome. piRNAs in mice were found not to be overly enriched for transposon-encoding sequences; only 17% were found to map to repeated elements in the mouse genome. MIWI-associated piRNA sequences possessed a strong bias at the first nucleotide for uracil (94.2% of the piRNAs found) (Girard et al. 2006)MILI-associated piRNAs also show a strong bias for 5′U (88%) (Aravin et al. 2006). MIWI-associated piRNAs are often 30-31 nt long; whereas MILI-associated piRNAs are l26-28nt long, both much longer than siRNAs and miRNAs. It was further found that while piRNA sequences themselves are not conserved in mammals per se, cloning of piRNA sized small ncRNAs from humans and rats showed that many of the larger piRNA clusters are syntenic (Girard et al. 2006). Isolation of rat piRNAs was carried out in the Kingston group (Lau et al. 2006). Rat piRNAs display similar characteristics to mouse piRNAs. Interestingly, humans and rat piRNAs also display a strong preference for U at their 5′ end (Lau et al. 2006). Finally although these RNAs were not isolated in association with PIWI proteins, piRNA-like RNAs have been identified in platypus and marsupials (Devor et al. 2008; Murchison et al. 2008). Consistent with other mammalian piRNAs there is a preference for a 5′U, they cluster to discrete genomic sites and are longer in length compared to siRNAs and miRNAs.
Figure 3.
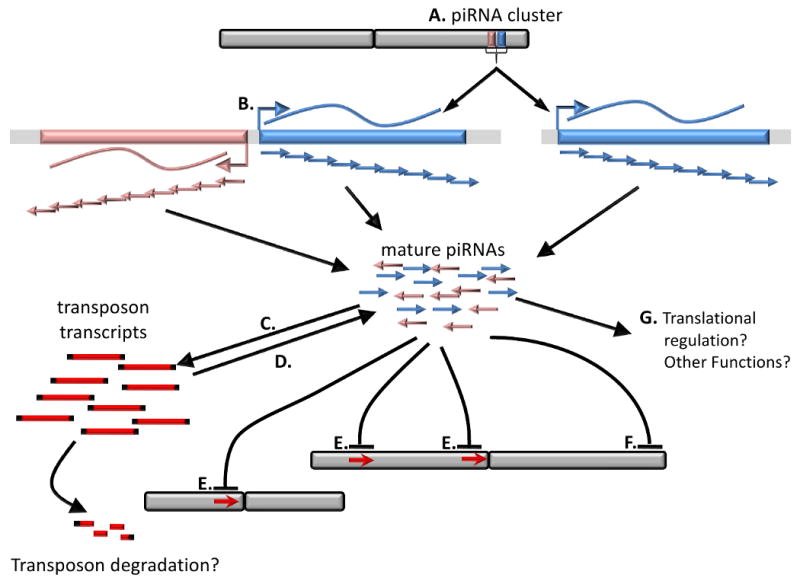
The Biogenesis and function of piRNAs. piRNAs are processed from long precursors encoded by long primary transcripts. A. Often piRNAs cluster to arrays that appear to bi-directionally transcribed, while less often are primary transcripts derived from one strand. B. The clusters are transcribed as long transcripts that go through primary processing to give rise to mature piRNAs, the mechanism of primary processing is not understood. C. piRNAs are believed to be vectors for transposon mRNA regulation, which leads potentially to degradation. D. At the same time, post-transcriptional amplification leads to more mature piRNAs (see figure 4 and text). E. several lines of evidence show that PIWI proteins, presumably through piRNAs lead to epigenetic repression of transposon encoding regions (red arrow). F. Other epigenetic functions of PIWI-proteins may exist, at least in Drosophila more than one line of evidence indicate that PIWI-proteins have a role in telomere length maintenance and epigenetic control. The epigenetic control appears to involve piRNAs as sequence recognition molecules. G. Evidence for a role of piRNAs in translational control and other functions exists but remain to be stringently tested.
B. piRNAs in insects
In a sense, piRNAs was first cloned in Drosophila without knowing they are piRNAs (Aravin et al. 2003). Unbeknownst to the Tuschl group in their cloning for small RNAs between 20 and 30 nucleotides, they isolated 4,000 small RNAs between 20 and 30 from Drosophila and identified a subset that corresponded to repeated elements in the genome, specifically transposable elements. This group of small RNAs, typically larger (24-26 nts) than miRNAs (21-22 nts), was named repeat-associated small interfering RNAs (rasiRNAs).
The purposeful isolation of piRNAs from Drosophila followed a similar path to that in the discovery of mammalian piRNAs. As mentioned above, rasiRNAs had been discovered prior to systematic screens carried out in Drosophila. The first evidence that piRNAs and rasiRNAs were of the same population of small RNAs became apparent when the Zamore group showed rasiRNA levels were not affected by mutations in siRNA machinery (Ago2 and DCR2) (Vagin et al. 2006). In addition, the su(ste) locus had long been known to be affected by AUB. Furthermore, rasiRNAs were known to associate with Piwi proteins but not with Ago1. Collectively, this data suggested there was a distinct pathway for biogenesis and function of rasiRNAs. The Siomi group would find similar evidence independently, in which they isolated several novel rasiRNAs by cloning the small RNAs associated with PIWI (Saito et al. 2006). This would later be followed up by cloning the piRNAs associated with AGO3 (Gunawardane et al. 2007). Although the number of piRNAs cloned by this point was only in the hundreds, it was observed in Gunawardane et al 2007, that there is 10nt complementarity between the piRNAs associating with AGO3 and AUB, the implications of which were further expanded in following piRNA isolations and explained in greater detail later in this review.
These were followed by two systematic screens for Drosophila piRNAs, which led to identification of two populations of PIWI-associated piRNAs of 12,903 and 13,904 piRNAs with only 10% (1,362) overlap (Brennecke et al. 2007; Yin and Lin 2007). In addition, approximately 20,000 AGO3-associated piRNAs and 10,000 AUB-associated piRNAs were identified (Brennecke et al. 2007). As in mammals, piRNAs were shown to cluster in the Drosophila genome at specific loci. However, unlike mammals these clusters mostly correspond to transposon sequences. One extremely large cluster on the X-chromosome had been genetically identified as the flamenco region, a known regulator of transposons that is itself comprised of many degenerated transposon sequences. PIWI and AUB show a preference for a 5′ U, while Ago3-associated piRNAs were observed to contain an A at position 10. There was considerable complementarity in the sequences of piRNAs associated with AUB and PIWI in comparison to Ago3 (more so between Ago3 and AUB). There is considerable strand bias of Ago3 to the sense strand of a transposon, while AUB and PIWI show a preference for the antisense strand of a transposon. This has ramifications for a potential amplification of piRNAs to be discussed later in this review.
Putative piRNAs have been reported in at least one other insect, the silkworm Bombyx mori (Kawaoka et al. 2008). Over 38 000 putative piRNAs were isolated from B. mori ovaries. Approximately 1/3 of these piRNAs are rasiRNAs, and, like Drosophila, the antisense piRNAs have a strong 5′U preference and sense piRNAs have a strong 10A preference. This suggests that the biogenesis of piRNAs is similar among insect species.
C. piRNAs in zebrafish
In zebrafish, Ziwi- and Zili-associated piRNAs have been identified by deep sequencing projects (Houwing et al. 2007; Houwing et al. 2008). These piRNAs were found to be 26-28nt long. Unlike mice and rats, but like Drosophila, piRNAs in zebrafish are present in both the male and female germline. Ziwi piRNAs have a 5′ U bias while Zili piRNAs have a preference for A at position 10. This again suggests a similar mechanism of piRNA biogenesis among insect species.
V. Biogenesis of piRNAs
A. The Precursor
Several lines of evidence point to a large single stranded RNA or a pair of divergently transcribed large single stranded RNAs as precursors for piRNAs derived from the same cluster. First, piRNAs were found to map to clusters in the genome. piRNAs line up end-to-end or overlap slightly in a strand specific manner (Figure 3). RT-PCR using primers derived from the same strand of a cluster showed that transcripts much longer than mature piRNAs were being generated from at least two rat piRNA clusters (Lau et al. 2006).
A strong line of evidence for long piRNA precursors can be found in a study that cloned total small RNAs from mice testis (Ro et al. 2007). This study found that the most predominant species of small RNA are piRNA-like. Although this study was not isolating small RNAs associated with PIWI proteins, these isolated sequences share many of the characteristics of piRNAs. A close examination of a cluster found a significant number of the isolated piRNAs mapped to one strand, often in an end-to-end manner, suggesting that there were two long precursors transcribed from a shared bidirectional promoter. One arm of this cluster was predicted to encode a 11.7kb precursor; this prediction was tested using reverse transcriptase PCR, which yielded 5 large ∼2.3kb fragments from the predicted 11.7kb precursor. The products were found to be encoded by the piRNA cluster, with no intermingled, non-transcribed sequence. The primers used in this study were overlapping, suggesting the fragments originated from the same precursor. Northern analysis using a probe against the 5′ end of the predicted precursor showed an 11.7kb band expressed specifically in testis. Although it remains to be determined if these sequences are true piRNAs, this work shows very convincing evidence that piRNA-like sequences are derived from long mRNA precursors.
B. Post-transcriptional Processing
It is not very clear how the putative precursors are processed into mature piRNAs. Since PIWI proteins do have the RNase H fold and some PIWI-type proteins have been tested and show slicer activity, it is quite possible that PIWI proteins chop up the precursor into mature piRNAs. In support of this, mutations of miwi and mili show a significant decrease in mature piRNAs, although it remains to be seen if the actual precursor is made in wild type levels in these mutants.
C. Post Transcriptional Amplification--Ping Pong Model
The distinct sequence features of piRNAs associated with different PIWI proteins has led to the hypothesis that there is a post-transcriptional amplification of piRNA cleavage process. Many isolated piRNAs associated with Ago3 are often complementary to piRNAs associated with AUB and PIWI. Furthermore, the piRNAs found in association with PIWI/AUB are often anti-sense to transposon sequences, with a strong preference for a uridine at the first nucleotide (1U); whereas AGO3-associated piRNAs have a strong preference for adenine at position 10 (10A) (Brennecke et al. 2007). It has been suggested that clusters of piRNAs arise (at least in Drosophila) from genomic arrays of defective transposons, transcribed in an anti-sense direction. These defective transposons serve as a pool from which transposon-complementary piRNAs are made. According to this model, these precursors are cleaved into piRNAs, either directly by AUB and PIWI, or are loaded indirectly into PIWI/AUB RNPs. The piRNAs that have complementarity to transposons lead to a targeting of the PIWI/AUB RNPs to actively transcribed transposons. These transposons are cleaved, resulting in sense piRNAs being created and loaded into directly or indirectly into AGO3 RNPs. The resulting sense piRNAs then target the large antisense precursors, leading to cleavage of the precursor and the formation of more antisense piRNAs to associate with AUB and PIWI. This results in a feedback system amplifying the initial pool of piRNAs; this proposed system has been termed the “ping-pong model” (Figure 4).
Figure 4.
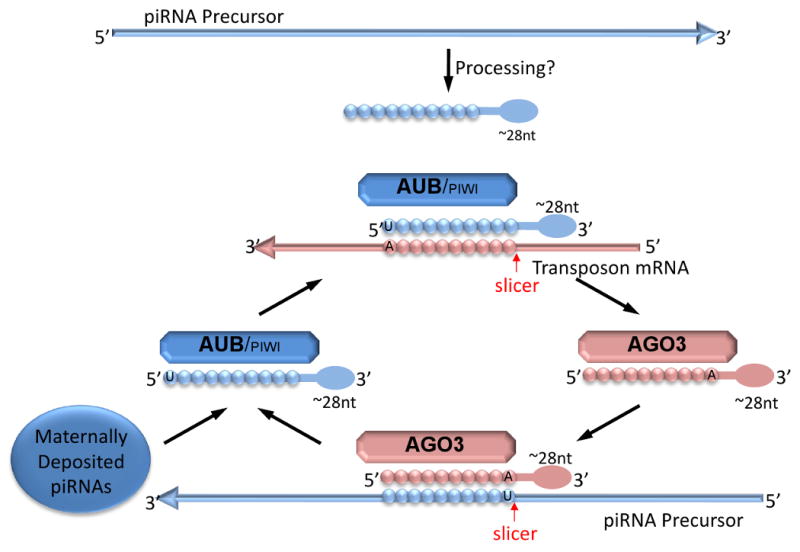
The Ping-Pong model for piRNA amplification in Drosophila. Mammalian post-transcriptional amplification appears to be a similar process. See text for detaied description of the model.
One prediction of the Ping-pong model is that there should be a relative increase in abundance of complementary piRNAs due to the amplification. Indeed, Brennecke et al. reported an abundance of piRNAs complementary to each other, specifically with piRNAs associated with AGO3 and AUB (Brennecke et al. 2007). There has not been such a reported increase of piRNAs complementary to AGO3 and PIWI. Although piRNA cloning from Drosophila has not been saturated, these data suggest that either PIWI functions outside of the ping-pong model of biogenesis, or that its function is not needed directly for either the formation of piRNAs from defective transposons-encoding sequences or through transposon degradation. The potential predominance of AUB and AGO3 in the ping-pong model suggests that the ping pong mechanism is cytoplasmic. Therefore one would not expect to see PIWI-associated piRNAs complementary to AGO3, unless there is transport of piRNAs to and from the nucleus. It will be interesting to determine then, if there are different subsets of piRNAs are found in the nucleus versus the cytoplasm.
Evidence for post-transcriptional amplification of piRNAs has been found in vertebrates as well. It had been reported that there a significant portion of piRNAs in mice often have an A at the 10th nt. Interestingly, the subset of piRNAs with A at position 10 rarely map uniquely to the genome, suggesting they may not be derived from a primary genomic sequence (as the majority of mammalian piRNAs are). Additional evidence for a mammalian ping-pong biogenesis mechanism arrived with the cloning of prenatal MILI and MIWI2 piRNAs (Aravin et al. 2008). Prenatal piRNAs display complementarity between MILI- and MIWI2-associated sequences. MILI piRNAs had already been reported to have a 1U bias, but it was found that MIWI2 had the signature 10A seen in AGO3 sequences. Additionally, MILI-associated piRNAs often match the sense strand of piRNA clusters and/or transposons, suggesting they arise from cleavage of a genomically encoded piRNA cluster. MIWI2 has a strong bias for the sequences corresponding to sense transposon sequences, and has been suggested to have a similar function to that of AGO3 in flies. Several examples of sequences with complementarity at the first 10 nt of 1U and 10A piRNAs have been reported between MILI and MIWI.
At present, Ping-pong model is an attractive hypothesis mostly based on bioinformatic analysis but needs experimental support. Several questions still remain unanswered with regard to the ping-pong model. Is the ping-pong mechanism active predominantly in the cytoplasm? Do PIWI proteins have slicer activity in vivo? What co-factors, if any, are needed for the slicer activity? During Drosophila oogenesis, the function of two putative nucleases, Zucchini and Squash are needed to repress retrotransposons, both these nucleases interact with AUB and are needed for rasiRNAs production. Suggesting some piRNA pathway components other than the PIWI proteins have a role in slicing precursor or transposons into rasi/piRNAs (Pane et al. 2007). Which also begs the question “are transposons ever degraded into sequences smaller than a piRNA?” Furthermore, how are the piRNAs transported from one RNP to another, do AGO3 and AUB strictly co-localize in the cytoplasm? Are the resulting piRNAs transported back into the nucleus, at least the subset that is associated with PIWI or MIWI2? Answers to these questions will greatly advance our understanding the piRNA biogenesis mechanism.
VI. Regulatory Functions of piRNAs and PIWI proteins
A. PIWI proteins as epigenetic regulators
The first evidence that PIWI proteins have a role in epigenetic function was found in Drosophila, where AUB and PIWI were found to act as regulators of position-effect variegation (PEV) (Pal-Bhadra et al. 2004). PEV is the phenomenon observed in Drosophila where the expression of a euchromatic gene is silenced when it is localized in proximity to a heterochromatic region, as in the case of chromosomal re-arrangements and tandem repeats of transgenes (Dorer and Henikoff 1994; Dorer and Henikoff 1997; Martin-Morris et al. 1997; Fanti et al. 1998). For example, repeated array of transgenes recruit heterochromatin-inducing machinery, leading to transcriptional silencing of the transgenes. This is an example of how repeated elements in the genome, such as the ste locus and transposons, are kept under repressive regulation. Artificial PEV is observed in a repeated transgene that contains the white gene that is responsible for red eye color. Arrangement of white+ in tandem repeats leads to suppression of red eye color. There is a de-repression leading to red eyes by mutations in aub and piwi, among other genes (Pal-Bhadra et al. 2004). Consistent with this, Heterochromatin Protein 1a (HP1a) is mis-localized from the regions containing the white+ tandem repeats in piwi and aub mutants. HP1a, as the name implies, participates in the formation of heterochromatin. This is accomplished through recruitment of the SU(VAR)3-9 histone methylase, which methylates Histone 3 at Lys 9 (H3K9me) (Pal-Bhadra et al. 2004). Compellingly, HP1a and PIWI physically bind to each other in vivo and in vitro (Brower-Toland et al. 2007). In flies lacking this interaction, there is a loss of heterochromatin - specifically, PEV is not as effective, leading to loss of repression of the white transgene. As further evidence of a potential epigenetic function, PIWI is present in the nucleus and directly associates with chromosomes (Brower-Toland et al. 2007).
PIWI can also function as an epigenetic activator (Yin and Lin 2007), suggesting local factors and protein-protein interactions may influence whether PIWI sets up a repressive or permissive chromatin environment. In a screen for suppression of the piwi mutant phenotype, a specific P-element (P{A4-4}) inserted in the sub-telomeric region (a.ak.a., telomere associated sequence, or TAS) in the right arm of the 3rd chromosome (3R-TAS) was found to rescue the PIWI mutant phenotype (Smulders-Srinivasan and Lin 2003). Specifically, the P{A4-4}) insertion rescues the loss of germline maintenance seen in piwi mutants. Isolation of piRNAs associated with PIWI showed that a single unique piRNA, 3R-TAS1 piRNA, is derived from the 3R-TAS region (Yin and Lin 2007). In piwi mutants, the 3R-TAS region was found to be heterochromatic, while in the wild type flies the region is semi-euchromatic. In the piwi mutant background, the insertion of P{A4-4} can restore the euchromatic signature of 3R-TAS. The presence of the 3R-TAS1-encoding sequence contained in the 3R-TAS has led to the intriguing suggestion that PIWI-piRNA complex may recruit epigenetic factors to specific genomic loci to organize chromatin status. Finally, many of the epigenetic effects described here were observed in somatic cells - although PIWI and most of its sibling proteins are enriched in the germline. PIWI is expressed in somatic cells, most noticeably on the polytene chromosomes of Drosophila salivary glands (Brower-Toland et al. 2007).
The epigenetic functions of PIWI proteins are conserved in vertebrates. As discussed in the next section, transposon-coding regions are not methylated in the absence of MILI and MIWI2; this loss of DNA methylation is consistent with a loss of epigenetic control (Aravin et al. 2008). The DNA methylation of transposons is dependent on DNMT3L, an isoform of Dnmt3a and b which lacks methylation activity. piRNAs are normally produced in dnmt3l mutant mice, suggesting that, if piRNAs or involved with PIWI proteins in DNA methylation, they both serve as upstream mediators of epigenetic control. Additionally, the isolation of RIWI (Rat PIWI) complexes identified rRecQ1 as a potential RIWI protein complex component (Lau et al. 2006); the homologous neurospora QDE-3 has a role in gene silencing in combination with an AGO protein (Catalanotto et al. 2002), further suggesting PIWI proteins are intimately involved in transcriptional gene silencing.
A model for how PIWI could accomplish this function comes from Schizosaccaromyces Pombe. In this fission yeast, siRNAs can induce heterochromatin formation at centromeres. The siRNA machinery is recruited by siRNA complementary to nascent transcripts derived from the centromere, in turn recruiting the HP1a homologue SWI6. This then leads to heterochromatin formation (Volpe et al. 2002; Verdel et al. 2004; Buhler et al. 2006). It is tempting to propose an analogous model of piRNA-mediated silencing, and determination of whether PIWI proteins affect epigenetic control in a similar fashion or through direct targeting of PIWI complexes to genomic sites by piRNA-DNA interaction (or other, unpredicted mechanisms) is of tremendous interest.
B. Aubergine and the Stellate Locus
The role of AUB in regulating the status of the Stellate locus represents an interesting case of the function of PIWI proteins in epigenetic regulation. Drosophila males which lack the Y chromosome (XO) are sterile and form crystalline structures in spermatocytes (Hardy et al. 1984). In addition, these males also are defective in meiosis (Palumbo et al. 1994). The formation of crystals in XO is suppressed in flies containing a deletion of a small region of the X chromosome referred to as the stellate (ste) locus (Livak 1984). In addition, it was found that a deletion of a heterchromatic region of the Y chromosome also leads to male sterility. This locus is referred to as suppressor of stellate su(ste) (Hardy et al. 1984) because in flies lacking the su(ste) region or in XO males, there is an increase of transcripts from the ste locus (Livak 1990). The ste protein has been identified as having homology with β-subunit of Casein Kinase 2 (CK2) (Bozzetti et al. 1995). The STE protein is a major constituent of the crystals that form in spermatocytes (Bozzetti et al. 1995). CK2 has a role in the phosphorylation of many nuclear targets, from these roles it is postulated that a misregulation of nuclear targets in su(ste) mutants are responsible for the meiotic defects (Tritto et al. 2003).
The mechanism by which the repression of the euchromatic ste locus by the heterochromatic region su(ste), occurs has been of intense interest. The sequencing of the su(ste) locus showed that there is considerable homology between the ste and su(ste) loci (Kalmykova et al. 1998). Later it was found that both strands of the su(ste) locus were transcribed. The injection of dsRNA derived from the specific sequence in su(ste) that shares homology to the ste locus is sufficient for the repression of the ste locus (Aravin et al. 2001). The spindle-E, a Drosophila gene with roles in oogenesis, and dsRNA metabolism, when mutant also shows defects in suppression of the ste locus (Aravin et al. 2001). The repression of the ste was also defective in mutations of aub (Schmidt et al. 1999). This leads to the exciting possibility that there was endogenous gene regulation by dsRNA-like mechanisms related to PIWI proteins.
C. PIWI Proteins in DNA Elimination
Consistent with the above epigenetic model is work from Tetrahymena. As previously mentioned, TW1P is needed for DNA elimination. Work has shown that DNA sequences to be eliminated show the heterochromatin marker H3K9 (Liu et al. 2004). Mutations that decrease H3K9 methylation show a decreased efficiency in DNA elimination. A small noncoding RNA known as scanning RNAs (scnRNAs) is dependent on TWI1P for formation (Aronica et al. 2008). Mutations affecting scnRNA accumulation decrease H3K9 accumulation on DNA sequences to be eliminated. In a matter very similar to Pombe scanning RNAs bind to nascent RNA transcribed from DNA sequences to be eliminated (Aronica et al. 2008). This shows that in Tetrahymena at least a PIWI protein can affect epigenetic control through a non-coding RNA. It will be interesting to see if this mechanism is conserved through higher eukaryotes.
D. piRNAs and PIWI proteins as regulators of transposon activity
Several recent papers and reviews have presented the considerable, albeit largely correlative, evidence that piRNAs and PIWI-type proteins regulate transposon activity. We will briefly summarize this data here. First, as previously mentioned, a considerable portion of piRNAs isolated to date map to transposon-encoding regions (although this is highly variable from species to species)(Girard and Hannon 2008). Second the flamenco region, a heterochromatin region on the X chromosome required to repress global transposon activity in a PIWI dependent manner (Sarot et al. 2004), encodes one of the largest piRNA clusters in flies (Brennecke et al. 2007). Disruption of the flamenco region leads to a decrease in piRNA expression and a corresponding increase in transposon activity (Brennecke et al. 2007). The flamenco locus is considered a junkyard for defective transposons, the transcription of which acts a large transcript for piRNA biogenesis. This has lead to a master control locus hypothesis, which large piRNA clusters act as regulators of transposon activity. Considerable evidence shows that mutations of aub likewise show increased transposon activity (Vagin et al. 2004; Savitsky et al. 2006; Chen et al. 2007b; Pelisson et al. 2007; Shpiz et al. 2007). However, there is no evidence that this increase in transposon activity contributes to piwi or aub mutant phenotypes.
Similar transposon-related functions of vertebrate PIWI-type proteins have been reported. Originally, only ∼20% of piRNAs isolated from mice from mammals correspond to transposon encoding regions. Further piRNA cloning led to the characterization of prenatal mouse piRNAs which have a much larger representation of transposon sequences, although the biological significance of the prenatal piRNAs is not clear (Aravin et al. 2008). Consistent with PIWI interacting with HP1a and having a role in heterochromatin formation, MILI and MIWI2 are both needed for the methylation of transposon encoding genomic regions (Aravin et al. 2007; Kuramochi-Miyagawa et al. 2008). Without MILI and MIWI2 methylation, there is an increase in transposon activity (Aravin et al. 2007; Kuramochi-Miyagawa et al. 2008). piRNAs themselves may have a role in transposon repression, as the deletion of a small piRNA cluster in mice leads to increased transposon activity, consistent of a role for piRNAs in transposon regulation (Xu et al. 2008). The evidence for the posttranscriptional amplification of piRNAs in mammals also supports a conserved role in transposon regulation in metazoans.
Recent work shows that in Drosophila maternally deposited piRNAs are able to inoculate the next generation against transposon activity (Brennecke et al. 2008).These maternal piRNAs may serve as a primer for the first round of piRNA generation. Consistent with role in epigenetic control, this piRNAs inoculate the flies for the whole next generation, suggesting the piRNAs have a role in epigenetic control. Without piRNAs being loaded into the early germline, there would not be protection against transposons. How these piRNAs are loaded into the early embryo is not known, however, one vehicle that might be used to carry piRNAs from one generation to the next at least in the germline are polar granules. Polar granules traffic many proteins and RNAs necessary for germ cell formation. Similar structures are seen in the germline of many metazoan species, including the mammalian testis in which similar structures known as chromatoid bodies are found. As presented earlier in this chapter, AUB is a known polar granule component and strong evidence shows that PIWI is also a polar granule component (Harris and Macdonald 2001; Megosh et al. 2006; Thomson et al. 2008). Additionally MIWI is a known chromatoid body component. Besides passing on genetic information from one generation to the next, metazoan germlines share additional similarities. One of these is transcriptional quiescence; the lack of piRNA precursor transcription necessitates the maternal loading of piRNAs, potentially to suppress transposons during germ cell formation. This function may have special importance in Drosophila, where early embryos are largely transcriptionally quiescent and thus would depend on maternally-deposited piRNAs for a long period of embryogenesis. The presence of maternally loaded piRNAs would serve as an inoculation against transposons until zygotic transcription of piRNA precursors can ward off transposons.
E. PIWI proteins and DNA integrity
One common feature of the absence of PIWI proteins is DNA damage as measured by γ-H2Av foci; γ-H2Av is a H2AX variant presents at sites of double stranded DNA (dsDNA) damage (Klattenhoff et al. 2007). It has been proposed that such dsDNA breaks are a result of over active transposons, however there is significant evidence that dsDNA breaks are a cause of transposon activity not necessarily a result of it (Klattenhoff et al. 2007). There is still an increase in γ-H2Av foci in double mutants of aub and ovaries deficient in ATR or CHK2. Leaving open the question of whether the increased transposon activity is responsible for the dsDNA breaks. The role of PIWI proteins in dsDNA break repair are the increase of γ-H2Av foci in MILI and MIWI2 mutants (Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007). Consistent with a role for mammalian PIWI-type proteins in dsDNA repair is the presence of RecQ1 in RIWI complexes (Lau et al. 2006). A highly conserved role of RecQ enzymes are in dsDNA break repair through recombination (Hunter 2008). Evidence has been recovered that transposition of transposons to sites of dsDNA damage is needed for or at least contributes to repair of this damage (Morrish et al. 2002; Zingler et al. 2005; Morrish et al. 2007). Interestingly one study shows that in mammals the repair of the DNA damage through a transposon mRNA sequence, at the telomeres (Morrish et al. 2007). This suggests that transposon have been adapted to repairing DNA damage, and may explain the need for tight control of transposon activity but not an entire elimination of transposons from the genome.
F. Cytoplasmic function of PIWI proteins and piRNAs
As previously mentioned, MIWI and piRNAs co-fractionate with polysomes, indicating a potential role for MIWI and piRNAs in translation control (Grivna et al. 2006a). Further work shows that MIWI associates with mRNAs, in polysomes and RNP fractions (Grivna et al. 2006b). Isolation of small RNAs from polysome fractions containing MIWI show that piRNAs are in a complex with MIWI in polysomes. Strongly suggesting that any role MIWI may in translation control is in association with piRNAs. Consistent with a role in translational control, MIWI is associated with eIF4E, the mRNA cap binding protein with central roles in translational control. Additionally, MIWI is in a complex with DICER. Although some miRNAs are still produced in miwi-/- testis, some are severely depleted or reduced. MIWI may thus be involved in miRNA-mediated translational control. Furthermore, MIWI target mRNAs are severely down-regulated in miwi-/- mutants, suggesting that MIWI is required for mRNA stability (Deng and Lin 2001).
Similarly, MILI appears to be involved in translational control (Unhavaithaya et al. 2008). MILI forms a stable RNA-independent complex with eIF3a, and associates with the eIF4E and eIF4G containing m7G cap-binding complex. In isolated 7-dpp testicular seminiferous tubules that contain only stem cells in the germline, the mili mutation has no effect on the cellular mRNA level yet significantly reduced the rate of protein synthesis. These observations indicate that MILI may positively regulate translation and that such regulation is required for germline stem cell self-renewal.
VII. Concluding Remarks
The discovery of PIWI proteins and especially piRNAs has revealed a new dimension of gene regulation. It is evident that PIWI proteins are involved in germline development of many metazoan species. It is also evident that PIWI proteins associate with and are needed for the biogenesis of piRNAs. However, it is not clear exactly what role PIWI proteins play in the biogenesis of piRNAs and what is the function of piRNAs. Certainly one of the immediate focuses of PIWI/piRNAs research will be to determine every step involved in creating individual piRNAs. It is also imperative to investigate how PIWI proteins achieve epigenetic and translational regulation, how these mechanisms are related to biological functions such as transposon silencing, germline determination, germline stem cell maintenance, meiosis, and other gametogenic events. Moreover, it is important to explore how piRNAs contribute to the functions of PIWI proteins and whether piRNAs have PIWI-independent functions. Answers to these questions will significantly advance our understanding of gene regulation.
Acknowledgments
We thank Jonathan Saxe for valuable comments on the manuscript. We apologize to those whose works are not cited here due to space limitation. The stem cell work performed in the Lin laboratory is supported by National Institutes of Health Grants HD33760, HD37760S1, and HD42042, the Connecticut Stem Cell Research Fund, the G. Harold and Leila Mathers Foundation, and the Stem Cell Research Foundation.
Bibliography
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5(2):337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11(13):1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316(5825):744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22(16):2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D, Jr, Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31(1):67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Beyret E, Lin H. Piwi-interacting RNAs (piRNAs) In: Appasani K, editor. MicroRNAs From Basic Science to Disease Biology. Cambridge University Press; Cambridge: 2008. pp. 489–496. [Google Scholar]
- Bozzetti MP, Massari S, Finelli P, Meggio F, Pinna LA, Boldyreff B, Issinger OG, Palumbo G, Ciriaco C, Bonaccorsi S, et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc Natl Acad Sci U S A. 1995;92(13):6067–6071. doi: 10.1073/pnas.92.13.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322(5906):1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21(18):2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125(5):873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16(7):790–795. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16(19):2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Shen R, Ye Y, Pu XA, Liu X, Duan W, Wen J, Zimmerer J, Wang Y, Liu Y, Lasky LC, Heerema NA, Perrotti D, Ozato K, Kuramochi-Miyagawa S, Nakano T, Yates AJ, Carson WE, 3rd, Lin H, Barsky SH, Gao JX. Precancerous stem cells have the potential for both benign and malignant differentiation. PLoS ONE. 2007a;2(3):e293. doi: 10.1371/journal.pone.0000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007b;17(7):637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. Asymmetric germ cell division and oocyte determination during Drosophila oogenesis. Int Rev Cytol. 2001;203:93–138. doi: 10.1016/s0074-7696(01)03005-4. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2(6):819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Devor EJ, Huang L, Samollow PB. piRNA-like RNAs in the marsupial Monodelphis domestica identify transcription clusters and likely marsupial transposon targets. Mamm Genome. 2008;19(78):581–586. doi: 10.1007/s00335-008-9109-x. [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77(7):993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147(3):1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66(1):37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Fanti L, Dorer DR, Berloco M, Henikoff S, Pimpinelli S. Heterochromatin protein 1 binds transgene arrays. Chromosoma. 1998;107(5):286–292. doi: 10.1007/s004120050310. [DOI] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18(3):136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006a;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006b;103(36):13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hardy RW, Lindsley DL, Livak KJ, Lewis B, Siversten AL, Joslyn GL, Edwards J, Bonaccorsi S. Cytogenetic analysis of a segment of the Y chromosome of Drosophila melanogaster. Genetics. 1984;107(4):591–610. doi: 10.1093/genetics/107.4.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128(14):2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55(4):577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 1990;109(2):425–433. doi: 10.1242/dev.109.2.425. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27(20):2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129(1):69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Hunter N. The RecQ DNA helicases: Jacks-of-all-trades or master-tradesmen? Cell Res. 2008;18(3):328–330. doi: 10.1038/cr.2008.33. [DOI] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci U S A. 1974;71(4):1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Dobritsa AA, Gvozdev VA. Su(Ste) diverged tandem repeats in a Y chromosome of Drosophila melanogaster are transcribed and variously processed. Genetics. 1998;148(1):243–249. doi: 10.1093/genetics/148.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119(2):447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Katsuma S, Kishino H, Kohara Y, Mita K, Shimada T. Bombyx small RNAs: Genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008 doi: 10.1016/j.ibmb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12(1):45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335(6191):611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990;4(6):905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, Schweyer S, Engel W, Nayernia K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15(2):201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- Leedham SJ, Wright NA. Expansion of a mutated clone: from stem cell to tumour. J Clin Pathol. 2008;61(2):164–171. doi: 10.1136/jcp.2006.044610. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104(16):6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124(12):2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426(6965):465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Liu X, Sun Y, Guo J, Ma H, Li J, Dong B, Jin G, Zhang J, Wu J, Meng L, Shou C. Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer. 2006;118(8):1922–1929. doi: 10.1002/ijc.21575. [DOI] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci U S A. 2004;101(6):1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107(4):611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics. 1990;124(2):303–316. doi: 10.1093/genetics/124.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP. Assembly of the Drosophila germ plasm. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- Martin-Morris LE, Csink AK, Dorer DR, Talbert PB, Henikoff S. Heterochromatic trans-inactivation of Drosophila white transgenes. Genetics. 1997;147(2):671–677. doi: 10.1093/genetics/147.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16(19):1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110(6):689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446(7132):208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Kheradpour P, Sachidanandam R, Smith C, Hodges E, Xuan Z, Kellis M, Grutzner F, Stark A, Hannon GJ. Conservation of small RNA pathways in platypus. Genome Res. 2008;18(6):995–1004. doi: 10.1101/gr.073056.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9(2):315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14(6):1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo G, Bonaccorsi S, Robbins LG, Pimpinelli S. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics. 1994;138(4):1181–1197. doi: 10.1093/genetics/138.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12(6):851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81(4):1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21(25):3988–3999. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310(5752):1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Song R, Nguyen D, Jin J, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA. 2007;13(10):1693–1702. doi: 10.1261/rna.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman EE, Lasko P. Germline development in vertebrates and invertebrates. Cell Mol Life Sci. 1999;55(89):1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20(16):2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166(3):1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20(3):345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, Schafer U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics. 1999;151(2):749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129(4):1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Uneva A, Kim M, Klenov M, Rozovsky Y, Georgiev P, Savitsky M, Kalmykova A. Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol Biol Evol. 2007;24(11):2535–2545. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- Smulders-Srinivasan TK, Lin H. Screens for piwi suppressors in Drosophila identify dosage-dependent regulators of germline stem cell division. Genetics. 2003;165(4):1971–1991. doi: 10.1093/genetics/165.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10(12):1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305(5689):1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Szakmary A, Cox DN, Wang Z, Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15(2):171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14(7):841–853. [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev. 2008;125(910):865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritto P, Specchia V, Fanti L, Berloco M, D'Alessandro R, Pimpinelli S, Palumbo G, Bozzetti MP. Structure, regulation and evolution of the crystal-Stellate system of Drosophila. Genetica. 2003;117(23):247–257. doi: 10.1023/a:1022960632306. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a piRNA binding protein, is required for germline stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2008 doi: 10.1074/jbc.M809104200. Published online, December 29, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1(1):54–58. [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18(12):861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456(7224):921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20(13):1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122(5):1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, Hecht NB. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol Reprod. 2008;79(1):51–57. doi: 10.1095/biolreprod.108.068072. [DOI] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426(6965):468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450(7167):304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, Hanschmann KM, Morrish TA, Lower J, Schumann GG. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15(6):780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]


