Abstract
The DEAD-box RNA helicases p68 (DDX5) and p72 (DDX17) have been shown to act as transcriptional co-activators for a diverse range of transcription factors, including estrogen receptor α (ERα). Here, we show that, although both proteins interact with and co-activate ERα in reporter gene assays, siRNA-mediated knockdown of p72, but not p68, results in a significant inhibition of estrogen-dependent transcription of endogenous ERα-responsive genes and estrogen-dependent growth of MCF-7 and ZR75-1 breast cancer cells. Furthermore, immunohistochemical staining of ERα-positive primary breast cancers for p68 and p72 indicate that p72 expression is associated with an increased period of relapse-free and overall survival (p=0.006 and p=0.016 respectively), as well as being inversely associated with Her2 expression (p=0.008). Conversely, p68 shows no association with relapse-free period, or overall, survival but it is associated with an increased expression of Her2 (p=0.001), AIB-1 (p<0.001) and higher tumour grade (p=0.044). Our data thus highlight a crucial role for p72 in ERα co-activation and estrogen-dependent cell growth and provide evidence in support of distinct but important roles for both p68 and p72 in regulating ERα activity in breast cancer.
Keywords: p68 RNA helicase, p72 RNA helicase, Estrogen receptor α, gene regulation, breast cancer, tamoxifen
Introduction
The DEAD box subfamily of RNA helicases, originally so named on the basis of the presence of a conserved motif having the sequence Asp-Glu-Ala-Asp, have been implicated in cellular processes involving the regulation of RNA structure, including pre-mRNA processing, RNA export, RNA degradation, ribosome assembly, translation and miRNA maturation (Fuller-Pace, 2006; Linder et al., 1989; Tanner and Linder, 2001). The p68 RNA helicase (DDX5) and the closely related p72 RNA helicase (DDX17) are two key members of the DEAD box RNA helicases that are essential for viability; p68-/- mice die in utero and p72-/- mice die shortly after birth, with mice for both knockouts exhibiting severe developmental defects (Fukuda et al., 2007).
Recent findings have shown that p68 and p72 act as transcriptional co-regulators required for the action of diverse transcription factors. Evidence for this was first forthcoming through the demonstration that p68 associates with estrogen receptor α (ERα), with preferential interaction being observed for ERα phosphorylated at serine 118 (Endoh et al., 1999). p72 has also been shown to be an ERα co-activator (Watanabe et al., 2001). p68 also acts as a transcriptional co-activator of the ERα-related androgen receptor (Clark et al., 2008), the tumour suppressor p53 (Bates et al., 2005) and Runx2, a regulator of osteoblast differentiation (Jensen et al., 2008). Both p68 and p72 have also been shown to co-activate MyoD, acting as regulators of muscle differentiation (Caretti et al., 2006). Although the mechanisms of p68 and p72 action as transcriptional co-activators remain to be defined, their interaction with other co-activators, including the histone acetyltransferases CBP/p300 and P/CAF (Rossow and Janknecht, 2003; Shin and Janknecht, 2007) and steroid receptor co-activator (SRC) family of nuclear receptor co-activators (Watanabe et al., 2001), which themselves interact with CBP/p300 and P/CAF (Goodman and Smolik, 2000), is suggestive of roles for p68 and p72 in the regulation of histone modification by histone acetyltransferases. A potential role for the RNA binding/helicase activity of p68/p72 has been suggested by the finding that they co-immunoprecipitate with ERα, SRC and the RNA coactivator SRA (Watanabe et al., 2001), although RNA helicase activity per se does not appear to be required for co-activation in reporter assays (Endoh et al., 1999). Finally, p68 and p72 also have potential as co-repressors through association with the histone deacetylase HDAC1 (Wilson et al., 2004), the association being promoted by sumoylation of p68 (Jacobs et al., 2007).
Expression of ERα is one of the key features of breast cancer, the most frequently observed cancer in women in industrialised nations, with ERα expression being found in approximately 70% of tumours. Its presence is both prognostic and predictive of response to endocrine therapies. Determination of ER status of tumours is therefore crucial in the management of ER-positive breast cancers by treatment with endocrine agents, such as tamoxifen, that inhibit ERα activity (Ali and Coombes, 2002), or aromatase inhibitors that reduce the levels of circulating estrogens. However, following an initial response to tamoxifen, a significant proportion of patients relapse. These tumours often remain ERα positive and may respond to an alternative endocrine agent, demonstrating ERα-dependence for the continued growth of these tumours (Buzdar and Howell, 2001; Morris and Wakeling, 2002), and indicating altered ERα function as a possible mechanism underlying treatment failure.
ERα is a member of the nuclear receptor superfamily of ligand-activated transcription factors (Mangelsdorf et al., 1995). Activation of gene expression by ERα is mediated by two transcription activation domains, AF1 and AF2, which act in a promoter- and cell-specific manner. AF1, located N-terminal to the DNA binding domain (DBD), functions in a ligand-independent manner and its activity can be stimulated through growth factor-regulated signalling cascades in the absence of estrogen (Bunone et al., 1996). AF1 activity is enhanced by activating phosphorylation of serines 104, 106 and 118 via the mitogen-activated protein kinase (MAPK) pathway (Ali et al., 1993; Le Goff et al., 1994; Thomas et al., 2008), and by cyclin-dependent protein kinases CDK2 (Ser-104/Ser106) (Trowbridge et al., 1997) and CDK7 (Ser-118) (Chen et al., 2000). Additionally, these phosphorylation sites are substrates for GSK-3ß (Medunjanin et al., 2005; Grisouard et al., 2007). AF2 is integral to the ligand-binding domain (LBD) and its activity requires estrogen binding by the LBD, which results in a conformational change facilitating co-regulator recruitment (Glass, 1994).
Transcriptional regulation by ERα requires the action of a plethora of transcriptional co-regulators at estrogen-responsive gene promoters, which mediate chromatin remodelling [reviewed in (Klinge, 2000)]. Extensive chromatin immunoprecipitation (ChIP) analysis of model estrogen-responsive genes, in particular the pS2 gene, has shown that these co-regulators are recruited and dissociated in an ordered, cyclical manner, where the promoter is initially activated to produce one round of transcribed mRNA, then is reset to an inactive state before the next cycle of transcription begins (Metivier et al., 2003; Shang et al., 2000). In the cycles of co-regulator recruitment, p68 is one of the earliest ERα co-activators to be recruited, suggestive of a particularly important role for p68 in the regulation of estrogen-responsive gene expression.
Here we have characterised further the importance of p68 and p72 in gene regulation by ERα, in particular using RNA interference-mediated downregulation of p68 and p72 in the MCF-7 breast cancer cell line. Furthermore, we have investigated the expression of both proteins in a cohort of human breast cancers and correlated their expression with a number of prognostic markers. These studies have identified p72 as a key factor in the regulation of estrogen-dependent cell growth and have identified associations with improved patient survival, indicating that it plays an important role in breast cancer pathogenesis.
Results
p68 and p72 co-activate ERα in synergy with SRC-1
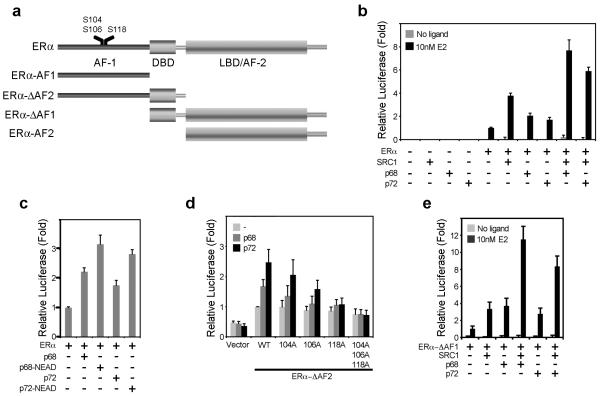
In order to examine p68/p72 co-activator function we transfected COS-1 cells with an estrogen-responsive firefly luciferase reporter plasmid and combinations of full-length ERα or mutants lacking either AF1 or AF2 (Figure 1a), together with p68, p72 and the well-characterised steroid receptor co-activator SRC-1. In agreement with previous reports (Endoh et al., 1999; Watanabe et al., 2001), ERα activity was stimulated by co-transfection with p68 and p72 (Figure 1b); this co-activation was synergistically enhanced by SRC-1. As has also previously been described (Endoh et al., 1999), the RNA helicase activity of p68 is dispensable since co-transfection with a helicase-inactive p68 mutant did not prevent stimulation of ERα activity by p68 (Figure 1c), nor was co-activation of ERα blocked by a similar mutation in p72.
Figure 1.
p68 and p72 co-activate ERα AF1 and AF2 in synergy with SRC-1 but independently of RNA helicase activity.
a) A schematic representation of ERα, with positions of the functional domains, is shown. Positions of serines 104, 106 and 118 are highlighted. Also depicted are the ERα deletion constructs used in this study. b) COS-1 cells were transfected with an estrogen-responsive firefly luciferase reporter gene, the renilla luciferase control plasmid (RLTK), together with p68, p72 and SRC-1, as shown. Reporter gene activities were calculated relative to the renilla luciferase activity, to control for transfection efficiency, and are shown relative to the reporter gene activity for ERα in the presence of 10 nM 17β-estradiol (E2). c) COS-1 cells were transfected as above, with p68 and p72 mutants in which the DEAD-box motif had been mutated to a helicase-inactive “NEAD” sequence.. d) Mutation of serines 104, 106 and 118 reduces co-activation of the ERα AF1 region by p68 and p72. e) p68 and p72 co-activate the ERα LBD/AF2 in the presence of 10nM E2. (b-d) The mean reporter activities of at least three independent experiments are shown, with the error bars representing the standard errors of the mean (SEM).
p68 was originally identified as an ERα co-activator following in vitro purification of proteins that interacted preferentially with ERα phosphorylated at ser-118 by MAPK (Endoh et al., 1999). In reporter assays using truncated ERα lacking the LBD/AF2, p68 and p72 stimulated AF1 activity (Fig. 1d). Whilst mutation of Ser-104 or Ser-106 did not prevent co-activation by p68 or by p72, mutation of Ser-118 gave little or no stimulation of ERα activity by p68 and p72, with mutation of all three residues completely preventing co-activation of AF1 by p68 and p72 altogether. Interestingly, however, both p68 and p72 stimulated the activity of an ERα deletion mutant lacking AF1 (Fig. 1e), suggesting that co-activation of ERα by p68/p72 involves the ERα DBD and/or AF2, in addition to AF1. Again this co-activation was synergistically enhanced by SRC-1.
p68 and p72 interact with ERα in-vitro and in vivo in an estrogen-independent manner
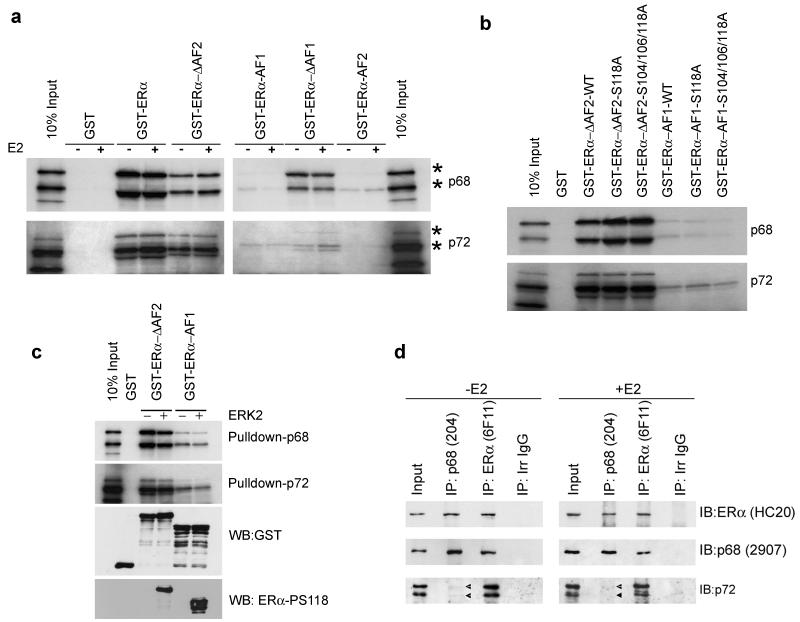
Using GST-pulldowns, an in-vitro interaction between ERα and p68 was observed, the interaction being estrogen-independent (Fig. 2a). Interestingly, deletion mutants lacking AF1 or the AF2 interacted with p68, whilst only weak interactions were observed for AF1 or AF2, indicative of interaction of p68 and p72 with the ERα DBD. Similar results were obtained for p72, although interaction with the mutant lacking AF1 was weaker than for p68, indicating that the interaction of p68 and p72 with ERα is similar, at least in-vitro. The lack of interaction between ERα-AF1 and p68/p72 could be due to a requirement for AF1 phosphorylation. However, interaction of AF1-DBD (ERα-ΔAF2) and the AF1 region with p68 and p72 was unaffected by mutation of S118 or of S104/S106/S118 (Fig. 2b). Nor was interaction between these ERα mutants and p68 or p72 stimulated by in-vitro phosphorylation with ERK2 MAPK (Fig. 2c). Taken together, these findings show that regions in addition to the AF1 are important for the interaction between ERα and p68/p72 and indicate that the ERα DBD is likely to be important for interaction of ERα with p68 and p72.
Figure 2.
p68 and p72 interact with ERα in a ligand-indepdent manner in vitro and in vivo.
(a-b) GST-pulldowns were carried out using 35S-labelled, in vitro translated p68 and p72 and GST-fusions of full-length ERα or regions of ERα, as shown. Double bands are observed for p68 and p72 (marked *); these are the result of internal translation initiation. Full-length (upper band) and N-terminal truncated forms of p68 and p72 are indicated. c) GST-ERα-ΔAF2 and GST-ERa-AF1 were phosphorylated in-vitro with ERK2 following pulldown using glutathione beads. In vitro transcribed/translated p68 or p72 were then added and pulldowns completed as above. Western blotting with an ERα-phospho-S118-specific antibody demonstrated phosphorylation of the GST-tagged proteins. d) Co-immunoprecipitation of p68 and p72 with ERα. Nuclear extracts were prepared from MCF-7 cells grown for 3 days in estrogen-free medium then treated with either 10nM 17β-estradiol (E2) or a vehicle control for 6 hours. Lysates were immunoprecipitated with antibodies to either ERα (6F11) or p68 (PAb204) and analysed by western blotting for ERα (HC20), p68 (2907) and p72 (DRD K12). The doublet observed in the p72 western blot shows the p72 and p82 isoforms of p72, which have been reported previously (Uhlmann-Schiffler et al., 2002; Ogilvie et al., 2003).
Co-immunoprecipitation of nuclear extracts prepared from MCF-7 cells treated with 10nM estrogen for 6 hours showed that p68 and p72 co-immunoprecipitated with ERα in an estrogen-independent manner (Figure 2d). Furthermore, we found that while ERα co-immunoprecipitated both p68 and p72, p68 co-immunoprecipitated ERα efficiently, but only co-immunoprecipitated a very small amount of p72, implying that ERα-p68 and ERα-p72 complexes may be distinct within the cell.
RNAi-mediated knockdown of p68 and p72 indicates that p72 is required for expression of estrogen-regulated genes
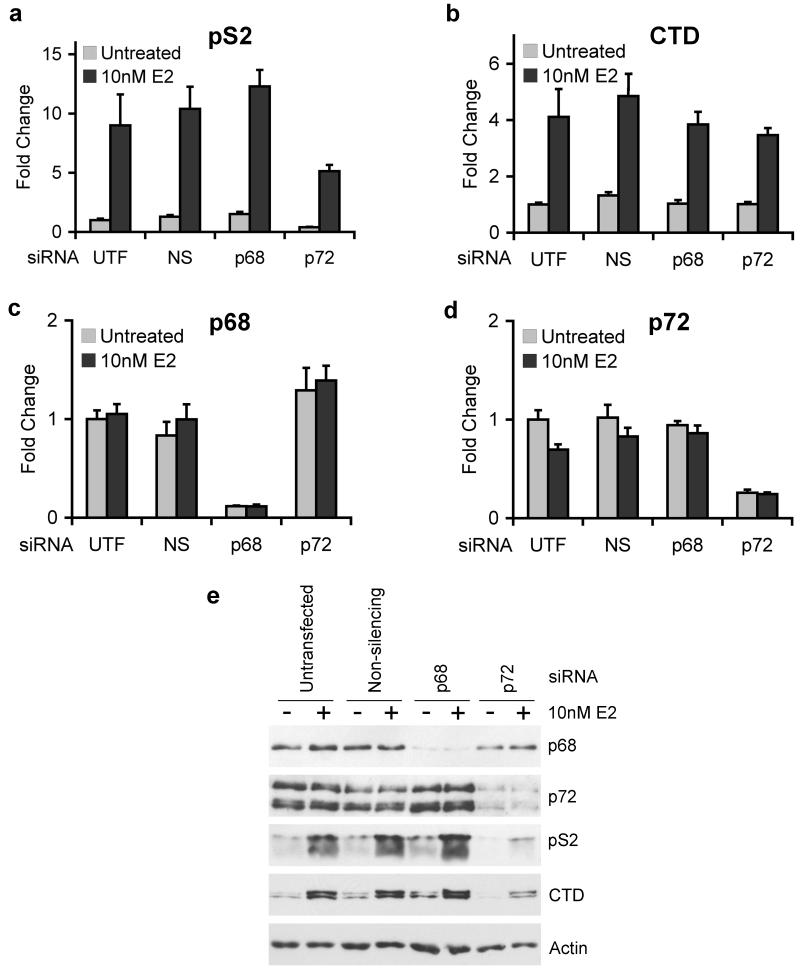
To determine whether p68 and p72 are required for expression of endogenous estrogen-responsive genes, we knocked down expression of these proteins by transfection of specific siRNA molecules, which have previously been described (Bates et al., 2005). Knockdown of p72 resulted in significantly reduced pS2 expression at both the mRNA and protein levels (Figures 3a, e). For Cathepsin D (CTD), knockdown of p72 led to a significant decrease in protein expression although there was only a small, but reproducible, decrease in mRNA levels (Figures 3b, e). Surprisingly, p68 knockdown did not affect pS2 or CTD expression (Figures 3a, b, e). Knockdown of p68 and p72 was confirmed by quantitative RT-PCR and western blotting (Figures 3c, d). Interestingly, we still observed estrogen stimulation of pS2 and CTD expression following p72 knockdown, although the overall level is markedly reduced, suggesting that p68 or other ERα co-regulators can compensate for p72, at least in part.
Figure 3.
siRNA-mediated knockdown of p68 and p72 indicates that p72 is required for expression of estrogen-regulated genes.
MCF-7 cells were grown in estrogen-free medium for 72 hours and then transfected using siRNAs against p68 or p72, or a non-silencing control (NS). 56 hours after transfection, the cells were treated with 10nM 17β-estradiol (E2) or an equivalent volume of ethanol for 16 hours and then harvested for RNA or protein. cDNA prepared from these cells was used to carry out Taqman quantitative RT-PCR to examine the expression of the endogenous estrogen-responsive genes pS2 (a) and Cathepsin D (b), and western blotting was used to examine expression of these genes at the protein level, with actin as a loading control (e). All graphs (a-d) are the means of three independent experiments −/+ S.E.M.
RNAi depletion of p72 inhibits the estrogen-dependent growth of cells
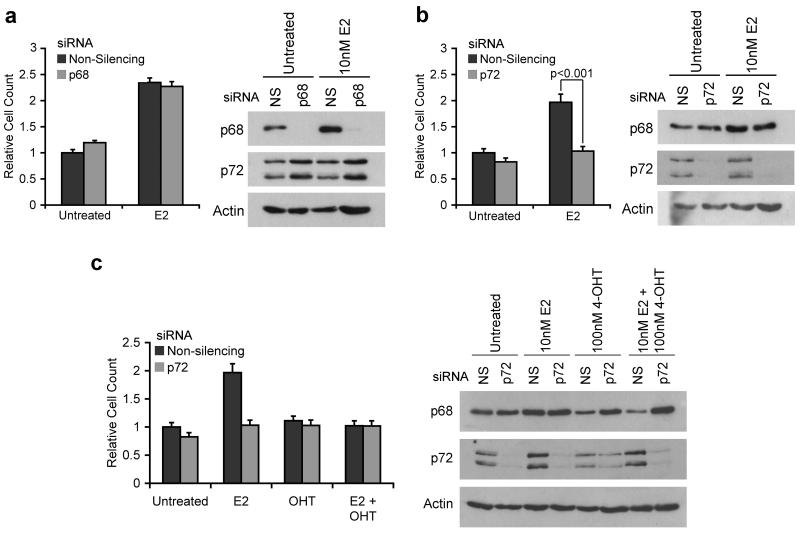
Since p72 appears to be important for stimulation of estrogen-regulated gene expression we determined the effect of p68 or p72 knockdown on estrogen-stimulated growth of MCF-7 cells. Whilst knockdown of p68 had no significant effect on cell growth compared to a non-silencing siRNA (Figure 4a), knockdown of p72 (Figure 4b) significantly reduced cell growth. To test whether the effects of p72 were estrogen-specific, we treated these cells with 4-hydroxytamoxifen (4-OHT) alone or in combination with estrogen. We found that tamoxifen treatment does not further slow the growth of the cells, suggesting that the inhibition of MCF-7 cell growth following p72 knockdown is due to inhibition of ERα activity (Figure 4c). For all experiments, knockdown of p68 and p72 was confirmed by western blotting. Knockdown of p72 in ZR75-1 cells similarly inhibited estrogen-stimulated growth, indicating that p72 is also required for ER function in these cells (Supplementary Figure 1).
Figure 4.
siRNA mediated knockdown of p72, but not p68, results in a slowdown of estrogen-dependent growth of breast cancer cells.
MCF-7 cells were grown for 3 days in estrogen-free medium then transfected with siRNAs against p68 or p72, or a non-silencing control (NS), in the presence or absence of 10nM 17β-estradiol (E2) and/or 100nM 4-hydroxytamoxifen (4-OHT). Cells were trypsinised and counted 4 days after transfection to assess cell growth. a) Effect of p68 knockdown; b) Effect of p72 knockdown; c) Effect of p72 knockdown on growth in the presence of E2, OHT, or a combination of both ligands. Western blots showing knockdown of p68 and p72 at the time of counting accompany each graph. Cell counts are shown as relative to the untreated non-silencing control. Each count was carried out in triplicate within each experiment, and results are shown as the mean −/+ S.E.M. of at least 3 independent experiments. In all western blots actin was used as a loading control.
p72 expression is associated with better prognosis in breast cancer
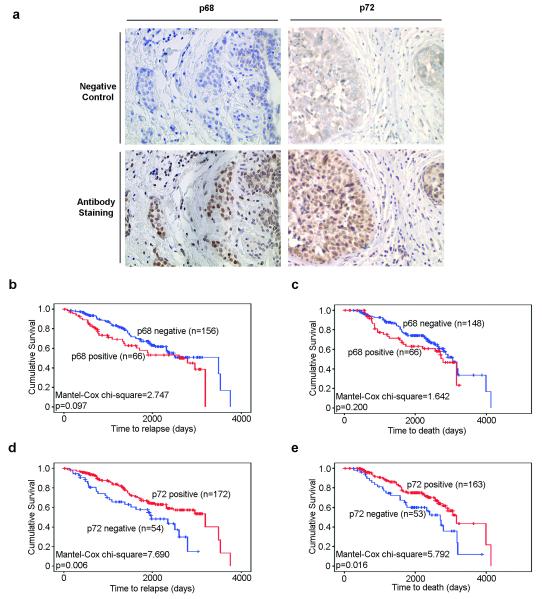
In order to assess their significance in breast cancer pathogenesis, immunohistochemical staining of 233 ERα positive tumours was performed for p68 and p72 (Figure 5a). p68 and p72 staining data were obtained for 226 and 229 cases respectively, of which 67 were p68 positive, and 173 were p72 positive. These tumours had previously been immunostained for ERα phosphorylated at S118, as well as a series of other markers including Her2, PR (Sarwar et al., 2006) and AIB-1. We correlated expression of p68, p72 and these markers with other pathological and clinical data for each tumour, including histological grade, lymph node involvement, time to relapse and death (Table 1). There was no statistically significant association between p68 and p72 expression in this group of patients (p=0.397). p68 expression was associated with Her2 and AIB-1 positivity (p=0.001 and p<0.001, respectively), as well as with increasing tumour grade (p=0.044), all of which are markers of poor prognosis (Osborne et al., 2003). However, no association was identified between p68 expression and either relapse-free or overall survival (p=0.097 and p=0.200 respectively) in our patient cohort (Figure 5b, c). Conversely, p72 expression was associated with a favourable prognosis. Most strikingly, p72 positive tumours were associated with Her2 negativity (p=0.008), progesterone receptor positivity (p=0.037) and reduced likelihood of relapse (p=0.025) and cancer death (p=0.014). In agreement with this, Kaplan-Meier analysis demonstrated that p72 positive tumours are associated with a longer relapse-free period and overall survival (p=0.006 and p=0.016 respectively) (Figure 5d, e). However, multivariate analysis showed that p72 was not a significant predictor of relapse-free or overall survival (Table 2). Lymph node positivity, as well as Her2 and AIB1 expression were significant predictors of relapse-free survival (p<0.001, p=0.003 and p=0.024 respectively), whilst lymph node positivity, Her2 positivity and tumour grade predicted overall survival (p=0.001, p=0.017 and p=0.008 respectively).
Figure 5.
Immunohistochemical staining of breast cancer biopsies for p68 and p72 RNA helicases. a) Representative examples of p68 and p72 negative and positive human breast cancer sections (magnification: X20) are shown. Kaplan-Meier survival curves showed no association of p68 with disease-free (b) or overall (c) survival, whereas p72 positivity was associated with increased disease-free (d) and overall (e) survival in breast cancer patients.
Table 1.
Relationships between p68 and p72 expression and clinical features.
| p68 Negative (%)b,c |
p68 Positivea,c (%) |
Chi- squaredd |
P- valuee |
p72 Negative (%)b,c |
p72 Positivea,c (%) |
Chi- squaredd |
P- valuee |
|
|---|---|---|---|---|---|---|---|---|
| p68 status | ||||||||
| Negative | - | - | - | - | 42 (75) | 116 (69) | 0.716 | 0.397 |
| Positive | - | - | 14 (25) | 52 (31) | ||||
| NDf | - | - | 0 | 5 | ||||
|
| ||||||||
| p72 status | ||||||||
| Negative | 42 (27) | 14 (21) | 0.716 | 0.397 | - | - | - | - |
| Positive | 116 (73) | 52 (79) | - | - | ||||
| ND | 1 | 1 | - | - | ||||
|
| ||||||||
| Relapse | ||||||||
| No | 101 (65) | 37 (56) | 1.487 | 0.223 | 27 (50) | 115 (67) | 5.003 | 0.025 |
| Yes | 55 (35) | 29 (44) | 27 (50) | 57 (33) | ||||
| Unknown | 3 | 1 | 2 | 1 | ||||
|
| ||||||||
| Death | ||||||||
| No | 101 (68) | 40 (61) | 1.185 | 0.276 | 28 (53) | 116 (71) | 6.051 | 0.014 |
| Yes | 47 (32) | 26 (39) | 25 (47) | 47 (29) | ||||
| Unknown | 11 | 1 | 3 | 10 | ||||
|
| ||||||||
| AIB-1 | ||||||||
| Negative | 95 (60) | 20 (30) | 17.259 | <0.001 | 31 (55) | 84 (50) | 0.482 | 0.487 |
| Positive | 63 (40) | 47 (70) | 25 (45) | 84 (50) | ||||
| ND | 1 | 0 | 0 | 5 | ||||
|
| ||||||||
| Her2 | ||||||||
| Negative | 135 (86) | 44 (66) | 12.073 | 0.001 | 38 (68) | 144 (84) | 7.098 | 0.008 |
| Positive | 22 (14) | 23 (34) | 18 (32) | 27 (16) | ||||
| ND | 2 | 0 | 0 | 2 | ||||
|
| ||||||||
| Lymph Node | ||||||||
| Negative | 49 (37) | 21 (36) | 0.025 | 0.875 | 13 (28) | 60 (41) | 2.835 | 0.092 |
| Positive | 82 (63) | 37 (64) | 34 (72) | 85 (59) | ||||
| Unknown | 28 | 9 | 9 | 28 | ||||
|
| ||||||||
| Grade | ||||||||
| I | 27 (18) | 6 (9) | 7 (13) | 28 (17) | ||||
| II | 99 (64) | 39 (59) | 6.251 | 0.044 | 31 (57) | 109 (64) | 2.574 | 0.276 |
| III | 28 (18) | 21 (22) | 16 (30) | 33 (19) | ||||
| Unknown | 5 | 1 | 2 | 3 | ||||
|
| ||||||||
| PR | ||||||||
| Negative | 47 (30) | 20 (30) | 0.002 | 0.965 | 22 (39) | 43 (25) | 4.334 | 0.037 |
| Positive | 112 (70) | 47 (70) | 34 (61) | 130 (75) | ||||
p68 or p72 positive score, >10% nuclei positive.
Percentage of cases in each category.
Data for p68 were available from 226/233 tumours, data for p72 were available from 229/233 tumours
Pearson’s chi-squared test. The analysis was carried out using known samples only.
A value of P<0.05 denotes statistical significance
Not determined
Table 2.
Odds ratios for significant predictive factors for i) period of relapse-free survival and ii) period of overall survival.
| i) Period of disease-free survival | ||||
|---|---|---|---|---|
| Factor | Odds Ratio | 95% Confidence Intervals | p-value | |
| Lymph node metastasis |
6.109 | 2.628 | 14.202 | <0.001 |
| Her2 expression |
2.219 | 1.311 | 3.756 | 0.003 |
| AIB-1 expression |
1.805 | 1.079 | 3.019 | 0.024 |
| ii) Period of overall survival | ||||
|---|---|---|---|---|
| Factor | Odds Ratio | 95% Confidence Intervals | p-value | |
| Lymph node metastasis |
4.391 | 1.877 | 10.271 | 0.001 |
| Tumour grade | 1.957 | 1.188 | 3.226 | 0.008 |
| Her2 expression |
1.967 | 1.130 | 3.427 | 0.017 |
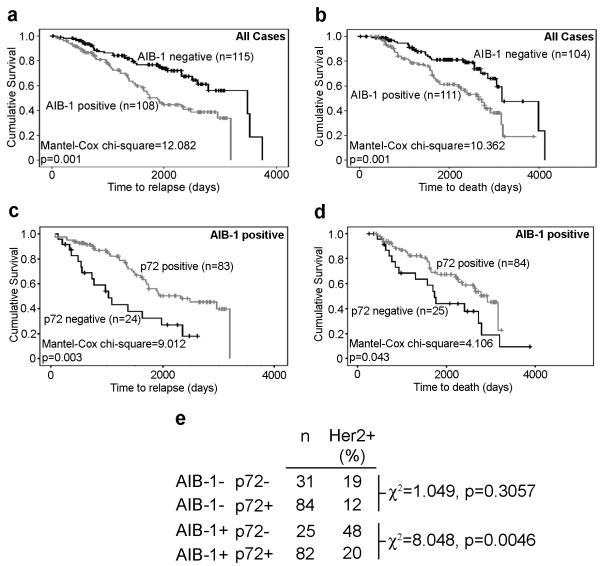
Dysregulation of ERα-mediated inhibition of Her2 expression is one mechanism through which breast tumours may become resistant to anti-estrogen treatment. AIB-1 is thought to play a crucial role in this dysregulation by competing for ERα binding with Pax2 and reversing ERα-dependent repression of Her2 (Hurtado et al., 2008). We examined the effects of expression of p72 in AIB-1 positive tumours and its relation to Her2 expression. As has been previously described in a number of studies (Osborne et al., 2003), AIB-1 positivity in the tumours was associated with poorer prognosis (Figure 6a, b). While expression of p72 had little effect on the prognosis of patients with AIB-1 negative tumours, p72 expression in AIB-1 positive tumours was associated with an increased period of relapse-free and overall survival (Figure 6c, d, p=0.003 and p=0.043 respectively). Furthermore, the proportion of Her2 positive tumours was significantly lower in AIB-1 positive tumours that were also positive for p72 (p=0.005, Figure 6e). Expression of p68 did not show any association with prognosis in AIB-1 positive tumours, although the proportion of Her2 positive tumours was significantly increased in AIB-1 positive tumours expressing p68 (p=0.005), nor did p72 expression influence the prognosis of Her2 positive tumours (Supplementary Figures 2, 3).
Figure 6.
Expression of p72 in AIB-1 positive tumours is associated with Her2 negativity and improved patient prognosis.
AIB-1 expression is associated with shorter relapse-free (a) and overall survival (b) in our patient cohort. However, in AIB-1 positive patients, p72 positivity is associated with a longer relapse-free period (c) and overall survival (d). The proportion of Her2 positive tumours is significantly less in AIB-1 positive tumours expressing p72 than in those that were p72 negative (e).
Discussion
Recent studies have shown that the related p68 and p72 RNA helicases regulate gene expression by acting as transcriptional co-regulators for diverse transcription factors (Fuller-Pace, 2006). The first evidence for a role in gene regulation for p68 and p72 was reported for ERα, where p68 was identified on the basis of a preferential association with ERα phosphorylated at S118 in the transcription activation function AF1 (Endoh et al., 1999). Subsequently p72 was also shown to co-activate ERα (Watanabe et al., 2001). Here we have undertaken a study of the importance of p68 and p72 in estrogen-regulated gene expression in the ERα-positive and estrogen-dependent MCF-7 cells. Our findings indicate that, while both proteins can act as ERα co-activators, p72 appears to play a more critical role in estrogen signalling than p68.
In agreement with previous reports, both p68 and p72 act as ERα co-activators in a reporter gene assay, in a manner that is independent of the p68 and p72 ATPase and helicase activity. This implies that the N- and C-terminal regions of these proteins are important for co-activation and is consistent with our previous work, which also showed that these regions are critical for transcriptional regulation (Wilson et al., 2004). Furthermore, although ser-118 is important for co-activation by p68 and p72 when assessing ERα lacking the LBD/AF2, another region, most likely the ERα DBD, is also important for interaction with p68/p72.
Surprisingly, siRNA-mediated knockdown of p68 and p72 showed that whilst p72 is important for the regulation of estrogen-responsive genes and for the estrogen-stimulated growth of cells, p68 does not appear to be required. On the other hand, immunohistochemical staining showed that p68 positivity is associated with markers of poor prognosis in ERα-positive breast cancer, suggesting that p68 and p72 may in fact have distinct roles in the regulation of ERα activity in breast cancer cells.
Investigation of the expression of p68 and p72 in a panel of breast cancers revealed that increased p72 expression is associated with a favourable prognosis. Patients whose cancers were p72 positive had a significantly increased period of disease-free and overall survival. However, p72 was not itself an independent predictor of patient outcome. The implication from our data is that p72 is required for estrogen-dependent cell growth. Therefore, tumours expressing p72 are likely to be estrogen sensitive, and so treatment with endocrine agents may be more effective in p72-positive patients. Since the tumours in our cohort were all ERα positive, we were unable to ascertain whether a relationship between ERα and p72 expression exists. However, meta-analysis of publicly available datasets in Oncomine (Rhodes et al., 2007) showed a positive association between ERα and p72 mRNA in 10 independent studies (Supplementary Table 1), which supports the hypothesis that p72 is required for estrogen signalling.
A significantly greater proportion of p72 negative tumours were positive for Her2, which is associated with poor response to endocrine treatment in ERα-positive tumours (Osborne et al., 2003). Interestingly, AIB-1, another marker of endocrine resistance and poor prognosis (Osborne et al., 2003), did not correlate with p72 expression. Overexpression of AIB-1, usually through amplification of 20q13 (Anzick et al., 1997), is thought to override ERα-mediated repression of Her2 by competing out Pax2 (Hurtado et al., 2008). As would be expected, AIB-1 and Her2 expression are strongly correlated in our dataset (p=0.002, data not shown). However, we found that in AIB-1 positive tumours the presence of p72 is associated with lower Her2 positivity compared to the p72-negative AIB1-positive tumours, which is suggestive of a role for p72 in the ERα-mediated repression of Her2 expression. The role of p68 in breast cancer is less clear. Although p68 positivity appears to have no overall effect on survival, there is a strong association between the presence of p68 and markers of poor prognosis, in particular Her2 and AIB-1.
In summary, our data provide evidence for distinct roles for p68 and p72 in regulating ERα activity. Furthermore, we have shown that p72 is important for transcriptional regulation by ERα and estrogen-dependent cell growth in breast cancer cells. Finally, immunohistochemical staining indicates that p72 is a marker of good prognosis in breast cancer.
Materials and Methods
Cell maintenance
MCF-7, ZR75-1 and COS-1 cells were routinely maintained in Dulbecco’s Modified Eagles Medium (DMEM) supplemented with 10% foetal calf serum (FCS), 2mM L-glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin (all supplied by Invitrogen, Paisley, UK) in 5% CO2 at 37°C. Prior to transfection, cells were placed for 72 hours in DMEM without phenol red (Invitrogen) supplemented with 5% double dextran-charcoal stripped FCS (First Link, Birmingham, UK), 2mM glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin.
Plasmids
Plasmids expressing full length and deletion mutants of ERα have previously been described (Tora et al., 1989), as have the GST-ERα fusion proteins and the estrogen-responsive luciferase reporter gene (Lopez-Garcia et al., 2006). p68, p72 and the DEAD box mutants have also previously been described (Bates et al., 2005). The SRC-1 expression plasmid was a kind gift of Dr M. Parker. pERE3-TATA-luc has also previosuly been described (Thomas et al. 2008). A thymidine kinase promoter Renilla luciferase reporter plasmid (RLTK; Promega, UK) was used to control for transfection efficiency.
siRNA Transfection
siRNA transfections were carried out using Lipofectamine RNAiMax (Invitrogen) by reverse transfection. siRNA oligos against p68, p72 and a non-silencing control have been previously described (Bates et al., 2005). 17ß-estradiol (Sigma, UK) was added to a final concentration of 10 nM, 56 hours following transfection, and cells were harvested after a further 16 hours for RNA or protein preparation. An equal volume of ethanol (vehicle) was added to the ‘no ligand’ controls.
Reporter gene assays
COS-1 cells were transiently transfected with ERα or ERα deletion constructs, together with p68, p72, SRC1, together with pERE3-TAT-luc and pRL-TK reporter genes, as previously described (Thomas et al., 2008). Luciferase activities were determined using Dual-Glo reagents (Promega). Firefly luciferase levels were corrected for transfection efficiency using corresponding renilla luciferase levels. All experiments were independently repeated at least three times, and the data graphed as mean values, with error bars representing the standard errors of the mean.
GST-pulldown assay
GST-tagged proteins were expressed in Rosetta BL21 E. coli cells and purified on glutathione beads (GE Healthcare) as previously described (Bates et al., 2005). p68 and p72 were translated and 35S-labelled using the TNT in-vitro transcription/translation kit (Promega). The GST pull-down of in-vitro-translated protein was carried out as described by (Hsieh et al., 1999). Where GST-tagged proteins were phosphorylated, the following modifications were made to the protocol. Following binding of GST lysates, the beads were washed and resuspended in 2.5 bed volumes of 1xMAPK buffer (New England Biolabs), containing 200μM ATP. 0.5μl per 30μl of buffer of activated ERK2 (New England Biolabs) was added to the beads, which were then incubated at 30°C for 30 minutes. Following this, the beads were washed and the protocol proceeded with incubation with 35S-labelled proteins as previously described (Hsieh et al., 1999).
Nuclear extract preparation and co-immunoprecipitation
Nuclear extracts were prepared as described previously (Bates et al., 2005) from MCF-7 cells grown in estrogen-depleted media (DMEM containing 5% dextran-coated charcoal-stripped FCS) for 72 hours with the addition of 10nM 17β-estradiol or an equivalent volume of ethanol for 6 hours, prior to extract preparation. Immunoprecipitations were carried out using antibodies against ERα (6F11; Novocastra, Newcastle-upon-Tyne, UK) and p68 (PAb204; Millipore, USA) as described previously (Bates et al., 2005).
Western blot analysis
Western blot analysis was carried out as described previously (Bates et al., 2005) using antibodies against p68 (PAb204 and 2907 (rabbit polyclonal raised against the C-terminal 15 amino acids of p68)); p72 (DRD-K12 (rabbit polyclonal raised against amino acids 12-26 of p72)); ERα (HC20; Santa Cruz Biotechnologies, CA, USA); ERα-phospho-S118 (Cell Signalling Technology, MA, USA); pS2 (FL-84, Santa Cruz Biotechnologies); Cathepsin D (CTD-19; Abcam, Cambridge, UK); and actin (A2066; Sigma-Aldrich, Poole, UK). Appropriate HRP-conjugated anti-mouse and anti-rabbit secondary antibodies for immunoblotting were purchased from DAKO (Ely, UK).
Cell growth assays
Cells were transfected in 6-well plates with siRNA, as described above, except with the addition of 10nM 17ß-estradiol (E2), 100nM 4-hydroxytamoxifen (4-OHT, Sigma-Aldrich), a combination of both ligands, or an equivalent volume of ethanol at the time of transfection. Transfections were carried out in triplicate. 96 hours after transfection, cells were trypsinised and counted using a haemocytometer. Following counting, cells were washed twice in PBS and lysed in RIPA buffer. Lysates were sonicated and cleared by centrifugation at 14,000 rpm for 15 mins. Protein concentrations were measured by Bradford assay (Sigma-Aldrich), equilibrated and analysed by Western blotting.
Quantitative RT-PCR
Total RNA was extracted from cells using the RNeasy kit (Qiagen) according to the manufacturer’s protocol. 1μg of RNA was treated with RQ1 RNase-free DNase (Promega) and reverse-transcribed using M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Quantitative (Taqman) RT-PCR was carried out using the Stratagene MX3005-P instrument (Agilent). Taqman gene expression assays (Applied Biosystems) were purchased for each gene (Supplementary Table 2). β-actin was used as a control and fold changes and differences between samples were calculated using the ΔΔCt method in Microsoft Excel.
Immunohistochemistry
Two hundred and thirty three patients with primary invasive breast cancer, who had undergone surgery at Charing Cross Hospital between 1981 and 2003 were selected on the basis of the availability of clinical details at presentation and follow-up, including time to relapse, time to death, ER and PR status (Sarwar et al., 2006). ER and PR status was confirmed using ER (VP-E613; Vector Laboratories), PR (MU328-UC; Biogenex, UK) antibodies, and Her2 status was determined, as described (Jiang et al., 2007). The clinico-pathological characteristics of the patient cohort are shown in Table 1. Immunohistochemical staining and scoring was carried out as previously described (Sarwar et al., 2006). Immunostaining and scoring for AIB1 was carried out as described (Hurtado et al., 2008). Staining for p68 and p72 was carried out using the antibodies PAb204 (Stevenson et al., 1998) and DRD-K12 respectively. p68 and p72 were scored positive if greater than 10% of tumour cell nuclei were stained. This study fulfilled the Institutional Ethics Review Board’s guidelines for the use of stored tissues samples.
Statistical analyses
All statistical analyses were carried out using SPSS v14.0. Comparisons of cell counts were carried out by t-test. Immunohistochemical scores for p68 and p72 were compared with clinico-pathological features using the Pearson chi-squared test. Survival analyses were carried out using the Kaplan-Meier survival function and the Mantel-Cox Log-rank test. Multivariate analysis was carried out using the Cox Proportional Hazards model.
Supplementary Material
Acknowledgements
This work was supported by grants from the Breast Cancer Campaign, Association for International Cancer Research, Cancer Research UK and the Breast Cancer Research Trust. We are grateful for support from the NIHR Biomedical Research Centre funding scheme, and thank David Meek, Malcolm Parker and lab members for helpful discussions.
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- Ali S, Metzger D, Bornert JM, Chambon P. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993;12:1153–60. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, et al. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005;24:543–53. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunone G, Briand PA, Miksicek RJ, Picard D. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–83. [PMC free article] [PubMed] [Google Scholar]
- Buzdar A, Howell A. Advances in aromatase inhibition: clinical efficacy and tolerability in the treatment of breast cancer. Clin Cancer Res. 2001;7:2620–35. [PubMed] [Google Scholar]
- Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11:547–60. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Chen D, Riedl T, Washbrook E, Pace PE, Coombes RC, Egly JM, et al. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell. 2000;6:127–37. [PubMed] [Google Scholar]
- Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–46. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, et al. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–72. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–15. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–77. [PubMed] [Google Scholar]
- Grisouard J, Medunjanin S, Hermani A, Shukla A, Mayer D. Glycogen synthase kinase-3 protects estrogen receptor alpha from proteasomal degradation and is required for full transcriptional activity of the receptor. Mol Endocrinol. 2007;21:2427–39. doi: 10.1210/me.2007-0129. [DOI] [PubMed] [Google Scholar]
- Hsieh JK, Chan FS, O’Connor DJ, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–93. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–6. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AM-F, Nicol SM, Hislop RG, Jaffray EG, Hay RT, Fuller-Pace FV. SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene. 2007;26:5866–5876. doi: 10.1038/sj.onc.1210387. [DOI] [PubMed] [Google Scholar]
- Jensen ED, Niu L, Caretti G, Nicol SM, Teplyuk N, Stein GS, et al. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. J Cell Biochem. 2008;103:1438–51. doi: 10.1002/jcb.21526. [DOI] [PubMed] [Google Scholar]
- Jiang J, Sarwar N, Peston D, Kulinskaya E, Shousha S, Coombes RC, et al. Phosphorylation of estrogen receptor-alpha at Ser167 is indicative of longer disease-free and overall survival in breast cancer patients. Clin Cancer Res. 2007;13:5769–76. doi: 10.1158/1078-0432.CCR-07-0822. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with co-activators and co-repressors. Steroids. 2000;65:227–51. doi: 10.1016/s0039-128x(99)00107-5. [DOI] [PubMed] [Google Scholar]
- Le Goff P, Montano MM, Schodin DJ, Katzenellenbogen BS. Phosphorylation of the human estrogen receptor. Identification of hormone-regulated sites and examination of their influence on transcriptional activity. J Biol Chem. 1994;269:4458–66. [PubMed] [Google Scholar]
- Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, et al. Birth of the D-E-A-D box. Nature. 1989;337:121–2. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia J, Periyasamy M, Thomas RS, Christian M, Leao M, Jat P, et al. ZNF366 is an estrogen receptor corepressor that acts through CtBP and histone deacetylases. Nucleic Acids Res. 2006;34:6126–36. doi: 10.1093/nar/gkl875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medunjanin S, Hermani A, De Servi B, Grisouard J, Rincke G, Mayer D. Glycogen synthase kinase-3 interacts with and phosphorylates estrogen receptor alpha and is involved in the regulation of receptor activity. J Biol Chem. 2005;280:33006–14. doi: 10.1074/jbc.M506758200. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Morris C, Wakeling A. Fulvestrant (‘Faslodex’)--a new treatment option for patients progressing on prior endocrine therapy. Endocr Relat Cancer. 2002;9:267–76. doi: 10.1677/erc.0.0090267. [DOI] [PubMed] [Google Scholar]
- Ogilvie VC, Wilson BJ, Nicol SM, Morrice NA, Saunders LR, Barber GN, et al. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 2003;31:1470–80. doi: 10.1093/nar/gkg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–61. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossow KL, Janknecht R. Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene. 2003;22:151–6. doi: 10.1038/sj.onc.1206067. [DOI] [PubMed] [Google Scholar]
- Sarwar N, Kim JS, Jiang J, Peston D, Sinnett HD, Madden P, et al. Phosphorylation of ERalpha at serine 118 in primary breast cancer and in tamoxifen-resistant tumours is indicative of a complex role for ERalpha phosphorylation in breast cancer progression. Endocr Relat Cancer. 2006;13:851–61. doi: 10.1677/erc.1.01123. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shin S, Janknecht R. Concerted activation of the Mdm2 promoter by p72 RNA helicase and the coactivators p300 and P/CAF. J Cell Biochem. 2007;101:1252–65. doi: 10.1002/jcb.21250. [DOI] [PubMed] [Google Scholar]
- Stevenson RJ, Hamilton SJ, MacCallum DE, Hall PA, Fuller Pace FV. Expression of the ‘DEAD box’ RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J Pathol. 1998;184:351–9. doi: 10.1002/(SICI)1096-9896(199804)184:4<351::AID-PATH1235>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–62. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Thomas RS, Sarwar N, Phoenix F, Coombes RC, Ali S. Phosphorylation at serines 104 and 106 by Erk1/2 MAPK is important for estrogen receptor-alpha activity. J Mol Endocrinol. 2008;40:173–84. doi: 10.1677/JME-07-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–6. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge JM, Rogatsky I, Garabedian MJ. Regulation of estrogen receptor transcriptional enhancement by the cyclin A/Cdk2 complex. Proc Natl Acad Sci U S A. 1997;94:10132–7. doi: 10.1073/pnas.94.19.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann-Schiffler H, Rossler OG, Stahl H. The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J Biol Chem. 2002;277:1066–75. doi: 10.1074/jbc.M107535200. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF- 1) with an RNA coactivator, SRA. EMBO J. 2001;20:1341–52. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.