Abstract
Polymerization of actin filaments directed by the Arp2/3 complex supports many types of cellular movements1. However, questions remain regarding the relative contributions of Arp2/3 complex versus other mechanisms of actin filament nucleation to processes such as path finding by neuronal growth cones owing to the lack of simple methods to inhibit Arp2/3 complex reversibly in living cells. Here we describe two classes of small molecules that bind to different sites on Arp2/3 complex and inhibit its ability to nucleate actin filaments. CK-636 binds between Arp2 and Arp3 where it appears to block movement of Arp2 and Arp3 into their active conformation. CK-548 inserts into the hydrophobic core of Arp3 and alters its conformation. Both classes of compounds inhibit formation of actin filament comet tails by Listeria and podosomes by monocytes. Two inhibitors with different mechanisms of action provide a powerful approach for studying Arp2/3 complex in living cells.
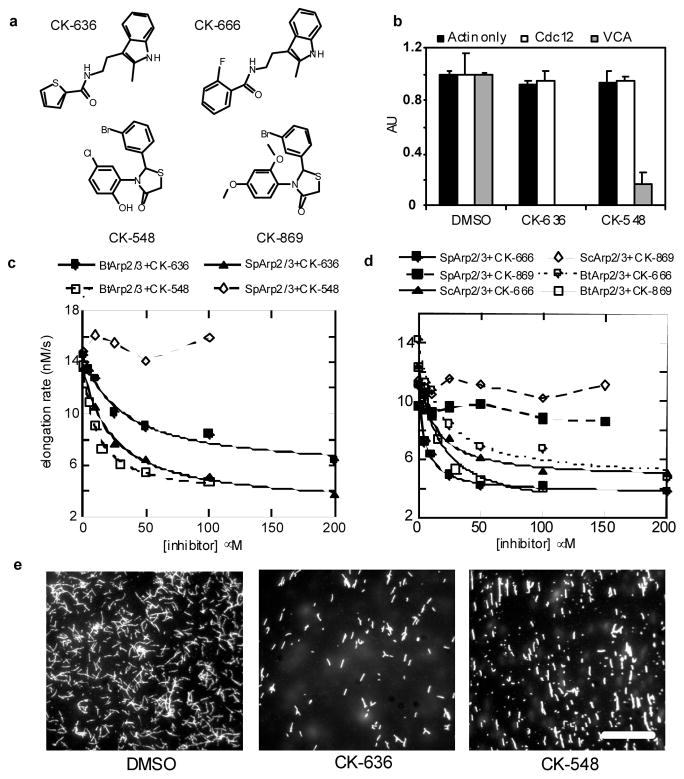
We used fluorescence assays to screen a library of >400,000 small molecules for inhibitors of actin polymerization stimulated by human (Hs) Arp2/3 complex and either full-length HsWASp with acrylodan-actin or HsWASp residues 105–502 with pyrenyl-actin. These screens each identified two inhibitors of human and bovine Arp2/3 complex, CK-0944636 (abbreviated CK-636) and CK-0993548 (abbreviated CK-548) (Fig. 1a). These compounds inhibited bovine (Bt) Arp2/3 complex with IC50 values of 32 μM for CK-636 and 11 μM for CK-548 (Fig. 1c). CK-636 inhibited actin polymerization stimulated by fission yeast Arp2/3 complex (SpArp2/3 complex, IC50 = 24 μM), but 100 μM CK-548 did not (Fig. 1c, Table S1). Fluorescence microscopy of the products of these reactions stained with Alexa 488 phalloidin showed branched actin filaments in controls (Fig. 1e, left panel). Samples with 100 μM CK-636 contained fewer branched filaments (Fig. 1e, center panel), while samples with 100 μM CK-548 contained only unbranched filaments (Fig. 1e, right panel). We tested a number of compounds structurally related to CK-548 or CK-636 that had no effect on actin polymerization at concentrations up to 200 μM and are useful as controls for experiments with cells (Fig. 2g). Table S2 lists one inactive compound from each class.
Figure 1.
Two classes of small molecules inhibit nucleation of actin filaments by Arp2/3 complex. a, Structures of CK-636, CK-548, CK-666 and CK-869. b, Inhibition of HsArp2/3 complex by CK-636 and CK-548. The time course of actin polymerization was monitored by the fluorescence increase of pyrenyl-actin. Conditions: 20 μM compound or DMSO, 2.5 μM 15% pyrenyl-actin alone or with 100 nM Cdc12(FH2) or 6 nM HsArp2/3 complex and 300 nM WASp-VCAThe maximum polymerization rate is expressed in arbitrary units. Error bars, s.d., n=4) c, Inhibition of Bos taurus (Bt) and Schizosaccharomyces pombe (Sp) Arp2/3 complexes by CK-636 and CK-548. The time course of polymerization was measured as in (1b). Conditions: 4 μM 15% pyrenyl-actin, 5 nM SpArp2/3 complex, and 1 μM N-WASp-VCA, or 3 μM 30% pyrenyl-actin, 5 nM BtArp2/3 complex and 1 μM N-WASp-VCA. CK548 was insoluble at 200 μM under the conditions used for this assay. The maximum polymerization rate of actin alone under these conditions was 4.6 nM/s. d, Effect of CK-666 and CK-869 on the polymerization of actin with bovine and yeast Arp2/3 complexes. Conditions as in 1(c) with either 3 μM 30% pyrenyl-actin and 5 nM BtArp2/3 complex or 4 μM 20% pyrenyl-actin plus 20 nM SpArp2/3 complex or 5 nM ScArp2/3 complex. Both compounds reduced the maximum polymerization rate of samples with BtArp2/3 complex to the basal rate without Arp2/3 complex but CK869 did not inhibit either yeast Arp2/3 complex. e, Fluorescence micrographs of the products of actin polymerization assays stained with Alexa 488-phalloidin. Actin (3.6 μM) was polymerized with 6 nM HsArp2/3 complex, 100 nM WASp-105-502, 300 nM Cdc42 and 100 μM CK-636 Scale bar = 20 μm.
Figure 2.
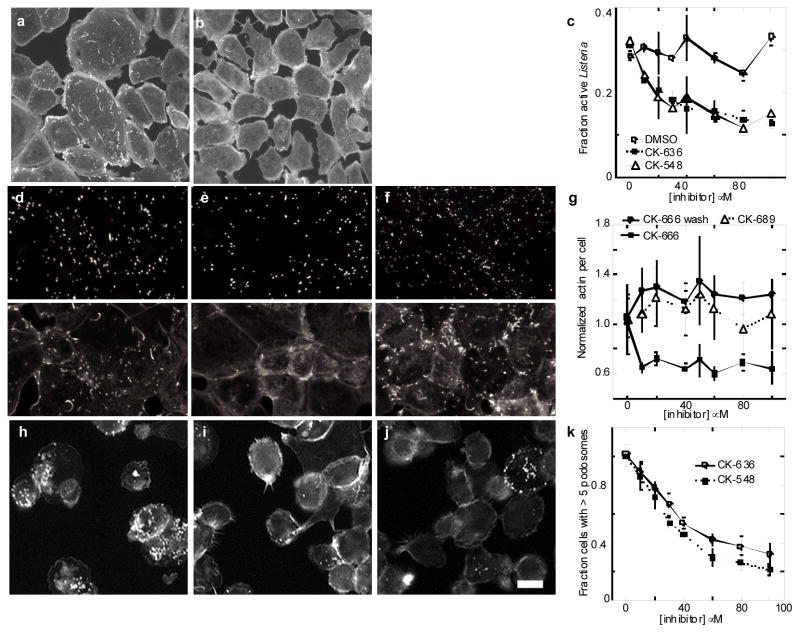
Inhibition of actin assembly in live cells by CK-548, CK-636 and CK-666. a–g, Formation of actin filament comet tails by Listeria infecting SKOV3 cells. a–b, Fluorescence micrographs of fixed cells stained with rhodamine phalloidin. a, Cells incubated at 37°C for 90 min with 0.1% DMSO had Listeria comet tails. b, Cells incubated at 37°C for 90 min with 100 μM CK548 in 0.1% DMSO had no actin comet tails. c, Dependence of the fraction of Listeria with comet tails on the concentrations of CK-636 and CK-548. Error bars, s.d., n=3. d–g, Effects of CK-666 on actin fluorescence around Listeria in SKOV3 cells. Infected cells were treated with 40 μM CK-666 for 60 min followed by a 60 min washout. The pairs of fluorescence micrographs show anti-Listeria fluorescence (top) and Alexa 488 phalloidin fluorescence (bottom). d, Control without CK-666 for 60 minutes. e, CK-666 for 60 minutes. f, CK-666 for 60 minutes followed by 60 minutes washout. g, Dependence of the mean actin fluorescence around each Listeria cell on the concentrations of CK-666 (active) and CK-689 (inactive) for 60 minutes. Fluorescence recovered when CK-666 was washed out for 60 minutes. Error bars are standard deviations of the mean fluorescence values from four separate experiments each with around 5000 Listeria per condition. A fluorescence value of 0.6 corresponds to background actin fluorescence. h–k, Effect of compounds on podosome formation by THP-1 derived monocyte cells. Adhered cells were treated with DMSO or compounds for 15 minutes, then fixed and stained with Alexa 568 phalloidin. h, 0.1 % DMSO control. i, 100 μM CK-636 and j, 100 μM CK-548 reduce formation of podosomes. k, Fraction of cells with >5 podosomes versus inhibitor concentration, normalized to DMSO control. Error bars, s.d., n=3. Scale bar = 20 μm.
The following control experiments showed that both compounds interact with Arp2/3 complex rather than actin or WASp. At 20 μM, neither compound inhibited polymerization of actin alone or polymerization stimulated by formin Cdc12p (Fig. 1b), so neither compound interacts directly with actin. Since actin can polymerize on its own, neither compound eliminated actin polymerization in mixtures with Arp2/3 complex and WASp or N-WASP-VCA (Fig. 1c-d). Both compounds inhibited polymerization stimulated by HsArp2/3 complex and ActA, an Arp2/3 complex activating factor from Listeria monocytogenes2, suggesting that they do not act on WASp (Supplementary Fig. 1). A fluorescence anisotropy assay showed that neither compound had a large effect on Arp2/3 complex binding N-WASP-VCA (Supplementary Fig. 2). The affinity of rhodamine-N-WASP-VCA for BtArp2/3 complex was the same with 50 μM CK-636 (Kd = 470 ± 50 nM) as controls (Kd = 510 ± 30 nM), but 50 μM CK548 decreased the affinity approximately two-fold (Kd = 1.0 ± 0.2 μM).
We used a Listeria motility assay to determine whether CK-636 and CK-548 can inhibit actin polymerization mediated by Arp2/3 complex in live cells. Both compounds reduced the formation of actin filament comet tails by Listeria in infected SKOV3 cells (Fig. 2a–c). The concentration dependence of this inhibition gave IC50 values of 22 μM for CK-636 and 31 μM for CK-548. We used the actin polymerization and Listeria comet tail assays to search for more potent inhibitors related to CK-636 and CK-548.
Compound CK-0944666 (abbreviated CK-666) has a fluorobenzene rather than the thiophene ring of CK-636 (Fig. 1a). CK-666 was a better inhibitor of actin polymerization with either BtArp2/3 complex (IC50 = 32 μM for CK-636 verses 17 μM for CK-666) or SpArp2/3 complex (IC50 = 24 μM for CK-636 versus 5 μM for CK-666) (Fig. 1d, Table S1). Both CK-636 and CK-666 had IC50 values of 4 μM for inhibiting HsArp2/3 complex. CK-666 reduced actin polymerization around intracellular Listeria to background levels (Fig. 2e and g) at lower concentrations than CK-636 (Fig. 2c). Actin filament halos and comet tails reformed when the compounds were removed after one hour of treatment (Fig. 2f–g). Inactive control compound CK-689 had no effect on comet tail formation (Fig. 2g).
CK-0157869 (abbreviated CK-869) has methoxy groups in the ortho and para positions replacing the ortho-hydroxy and meta-chlorine substituents of CK-548 (Fig. 1a). CK-869 inhibited actin polymerization with BtArp2/3 complex similar to CK-548 (IC50 = 11 μM for both compounds), but CK869 inhibited comet tail formation by Listeria more effectively (IC50 = 7 μM for CK-869 vs. 31 μM for CK-548). Like CK-548, CK-869 did not inhibit either yeast Arp2/3 complex (Fig. 1d).
Both classes of compounds also inhibited the formation of podosomes by the THP-1 monocyte cell line (Fig. 2h–k). Podosomes are adhesive structures that depend on Arp2/3 complex, WASp and actin polymerization3. Podosomes also depend on microtubules4, but neither compound at 100 μM disrupted the microtubule network in THP-1 cells (Supplementary Fig. 3). Neither class of compound shows irreversible or off target morphological effects on cells, such as apoptosis, at concentrations up to 100 μM over 24 hours. Our most potent compound, CK-666, had no effect on the mitotic index of SKOV cells at concentrations up to 80 μM, while inhibiting actin assembly around Listeria completely at 10 μM. Fifty micromolar CK-666 or CK-869 each altered the morphology and slightly slowed but did not stop the gliding motility of fish keratocytes (data not shown), as expected, since ~10% of Arp2/3 complex (~500 nM) would still be active. Thus Listeria comet tails and monocyte podosomes are more sensitive than lamellar motility to inhibition of Arp2/3 complex.
We determined x-ray crystal structures of BtArp2/3 complex with either CK-636 or CK-548 bound using molecular replacement with the apo-form BtArp2/3 complex as the search model (1K8K.pdb)5. Structures of Arp2/3 complex co-crystallized with the compounds and with compounds soaked into the crystals were nearly identical, but the resolution of soaked crystals was higher (2.70 Å for CK-636 and 2.85 Å for CK-548, Table S3). As in previous crystal structures of BtArp2/3 complex5–7, none of these structures had appreciable electron density for subdomains 1 and 2 of Arp2, which appears only after chemical crosslinking7.
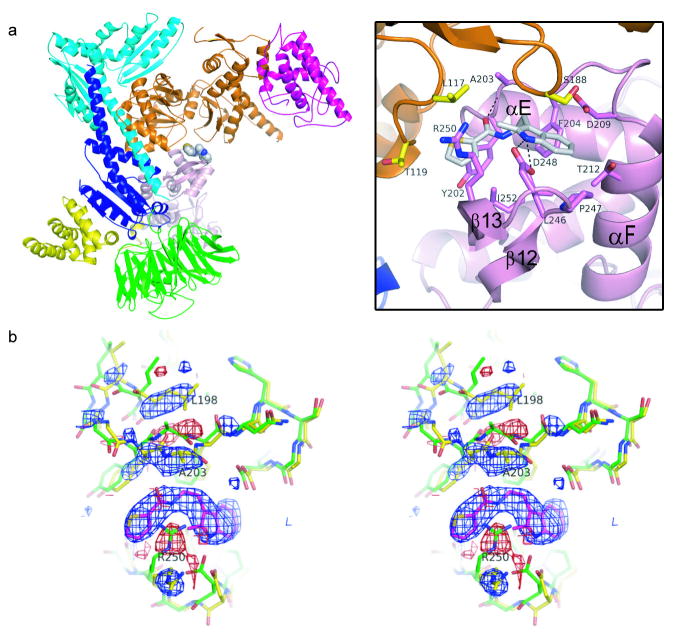
After one round of rigid body refinement, strong Fo-Fc electron density was present at the interface between Arp2 and Arp3 in maps calculated with data from crystals soaked in 1 mM CK-636 (Fig. 3). This density improved upon further refinement (Supplementary Fig. 4a) and CK-636 was modeled into the density when the Rf was 29.2 %. CK-636 binds in a pocket between subdomain 4 of Arp2 and subdomain 1 of Arp3. The contact surface is mainly hydrophobic and buries 161 Å2 of the accessible surface of Arp2 and 79 Å2 of Arp3. CK-636 forms hydrogen bonds with two residues in the pocket. The side chain of Asp248 forms a hydrogen bond with the nitrogen in the indole ring of the inhibitor and the backbone amide of Ala203 forms a hydrogen bond with the carbonyl oxygen on the linker between the two ring systems. Both trans- and cis- conformations of the amide functional group of CK-636 fit into the electron density with the hydrogen bonds described above preserved in both conformations. Because trans-amides are generally 1–5 kcal/mol more stable than cis-amides in solution 8, and the cis-conformation is rarely observed in small molecules, we modeled CK-636 in the trans-conformation.
Figure 3.
Crystal structure of BtArp2/3 complex with bound CK-636. Color code: Arp3, orange; Arp2, pink; ARPC1, green; ARPC2, cyan; ARPC3, magenta, ARPC4, blue; ARPC5, yellow. a, Ribbon diagram with a detail of binding pocket. b, Stereo diagram of an Fo-Fc electron density map showing changes caused by CK-636. Positive (blue) and negative (red) difference densities indicate the position of CK-636 and small conformational changes of the protein. The apo-structure (1K8K pdb) is shown in green stick representation and the final structure in yellow. The density is contoured at 3.5 σ and was generated using structure factors calculated after one round of rigid body refinement and the data for the CK-636-soaked crystal.
A difference electron density map shows that CK-636 causes minor conformational changes in the complex (Fig. 3b). Arp2 Arg250 from β strand 13 rotates out of the pocket and adopts an extended conformation. Arp2 Ala203 from the loop (residues 199-208) connecting αE to αF moves toward the pocket, causing the side chain of Leu198 to adopt a new rotamer. These changes and small changes in the backbone of residues surrounding the pocket result in an overall RMSD of 0.63 Å for an overlay of subdomains 3 and 4 (191 Cα atoms) of Arp2 with and without inhibitor bound.
We modeled CK-666 into the CK-636 binding pocket with the aromatic fluorine pointed toward a concave surface in the back wall of the pocket formed by Tyr202 in Arp2 and residues 118 to 120 in Arp3 (Supplementary Fig. 6). One hundred steps of conjugate gradient minimization in CNS9 showed that CK-666 is stable in this conformation and does not clash with residues in the pocket. The fluorine atom provides additional van der Waals interactions with the back wall of the pocket, and the benzene ring completely fills the hydrophobic pocket created by Ile252, Tyr202 and the aliphatic portion of Thr119. These interactions may allow CK-666 to bind more tightly than CK-636.
During formation of a branch Arp2 moves 31 Å relative to Arp3 from its position in inactive Arp2/3 complex10, so the location of the CK-636 binding pocket between Arp2 and Arp3 suggests that CK-636 and CK-666 lock Arp2/3 complex in an inactive conformation. CK-636 at a concentration of 100 μM did not interfere with N-WASp-VCA binding to Arp2/3 complex (Supplementary Figure 2), but it prevented WASp-VCA binding from increasing the fluorescence of Arp2/3 complex loaded with etheno-ATP (Supplementary Figure 5). Thus CK-636 inhibits a conformational change caused by activator binding and supports the hypothesis that CK-636 locks the complex in an inactive conformation.
The residues contributed by Arp2 and Arp3 to form the CK-636 binding pocket are conserved across a broad range of species (Table S4), so we expect CK-636 and CK-666 will be useful for inhibiting Arp2/3 complex from diverse species. These residues are also conserved in actin, but CK-636 does not inhibit actin polymerization at concentrations up to 200 μM. Only half of the binding site is available on an actin monomer, and the two half-sites are not juxtaposed in actin filaments11.
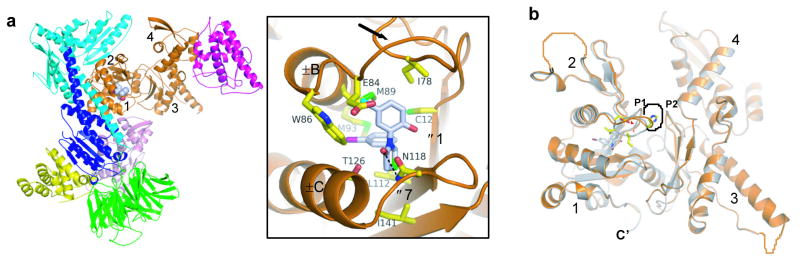
CK-548 has a single stereo center and was used as a racemic mixture in our experiments. The 2S enantiomer binds in a hydrophobic cavity in the core of the Arp3 subunit (Fig. 4a and Supplementary Fig. 4b). The isolated 2R enantiomer, which does not fit in the binding pocket, should provide a useful control for in vivo experiments. Binding of CK548 requires a substantial conformational change: the loop (residues 76-85) connecting β6 (73-75) and αB (86-98) in subdomain 1 of Arp3 flips upward 7.2 Å, exposing the binding site in the hydrophobic core of Arp3 (Fig. 4b). CK-548 contacts 239 Å2 of the inner surface of this cavity, which includes residues Cys12, Trp86, Met89, Met93, Leu112 and Ile78. A hydrogen bond between the side chain amide of Asn118 and the carbonyl oxygen of CK548 anchors the inhibitor to the loop between β7 (110-114) and αC (120-132), which forms the bottom lip of the cavity. The ability of CK-548 to bind crystallized Arp2/3 complex suggests that the β6/αB loop can open the hydrophobic binding pocket both in crystals and in solution, even in the absence of inhibitor. The average B-factor for all residues in the loop is 63 Å2 in the closed conformation and 82 Å2 in the open conformation, suggesting that the loop is more flexible when open.
Figure 4.
Crystal structure of BtArp2/3 complex with bound CK548. The color coding is the same as in Figure 3. a, Ribbon diagram with detail of binding pocket. Black arrow marks the loop (residues 76–85) connecting β6 (73–75) and αB (86–98) in subdomain 1 of Arp3, which flips up to accommodate inhibitor binding. b, Ribbon diagrams of Arp3 with bound CK-548 (orange) overlaid onto the apo-BtArp2/3 structure (1K8K.pdb, grey). Black dotted line indicates the binding pocket for ATP or ADP. Orange dotted lines indicate disordered regions of the structure. Small red arrowheads indicate alternative conformations of the sensor loop. CK-548 is shown in stick representation with grey carbon atoms and select residues in the sensor loop from the 1K8K structure are shown as sticks with yellow carbon atoms.
When modeled into the CK-548 binding site, the para-methoxy group of CK-869 projects toward the upper lip of the pocket and is sandwiched by the side chains of Ile78, Glu84 and Met89 (Supplemetary Fig. 7). The ortho-methoxy group points toward a crease formed at the ends of strands β1 and β7, which could be exploited to design higher affinity inhibitors.
Both CK-548 and CK-869 inhibited human and bovine Arp2/3 complexes, but neither inhibited budding or fission yeast Arp2/3 complexes (Fig. 1c, d). The residues that contact CK548 are conserved in Arp2/3 complexes of most mammals, but tryptophan replaces Met93 at the back of the CK-548 binding pocket in many other species, including budding and fission yeast (Table S5). The bent side chain of Met93 allows CK-548 to bind, but tryptophan at this position blocks binding to non-mammalian Arp3 and actin.
The mechanism of action of CK-548/869 is less apparent than that of CK-636/666. CK-548 decreased the affinity of SpWsp1-Rho-VCA for Arp2/3 complex two-fold, but this should be inconsequential under the conditions of our assay. The conformation induced by CK-548/869 must interfere with one or more of the reactions along the pathway to branch formation, such as binding of the pointed end of Arp3 to a mother filament10, nucleotide binding to Arp3, or conformational changes that activate branch formation.
METHODS SUMMARY
Compounds were purchased from Chemdiv, San Diego CA: CK-0944636 (Catalog number 8012-5103), CK-0993548 (Catalog number K205-1650), CK-0944666 (Catalog number 8012-5153) and CK-0157869 (Catalog number K205-0942). We purified native Arp2/3 complex from human platelets12, bovine thymus6, Schizosaccharomyces pombe13 and Saccharomyces cerevisiae (Supplemental methods), actin from chicken skeletal muscle14 and recombinant HsWASp, WASp105-502, WASp-VCA and Cdc4212, N-WASp-VCA 428-505 (Supplemental methods), GST-ActA 36-170 (Supplemental methods) and S. pombe Cdc12p(FH2)-His 973-139015 from E. coli. We used standard assays to measure polymerization of pyrenyl-actin16 and to visualize actin filaments by fluorescence microscopy17. Binding of etheno-ATP to Arp2/3 complex was performed as described previously with slight modifications18. We crystallized BtArp2/3 complex7 with either 0.5 mM CK-548 or 1 mM CK-636 in DMSO or soaked these compounds into crystals for 24 hours before freezing in liquid nitrogen. Diffraction data were collected at beamline X29A at Brookhaven National Laboratories. SKOV3 cells were infected with Listeria monocytogenes and fixed with 2% formaldehyde, permeabilized with 0.1% Triton-X in PBS, stained with Listeria antibody (US Biologics, Cleveland, Ohio) and Alexa Fluor 568 phalloidin (Molecular Probes, Eugene, OR), and imaged by fluorescence microscopy. We used an Isodata threshold on background-subtracted images of Listeria to isolate individual bacterium and measure the ratio of colocalized actin to Listeria fluorescence. Monocyte THP-1 cells were differentiated in 50 nM phorbol myristate acetate (Sigma-Aldrich-Fluka) to form podosomes before treatment with compounds. Black molly keratocytes19 were observed by time-lapse phase contrast microscopy.
METHODS
Protein Purification
Human, bovine and S. pombe Arp2/3 complexes were purified as previously described11,12,20. S. cerevisiae Arp2/3 complex was purified from strain BN020 (MAT a or α, ura3-52, his3-200, leu2-3, lys2-801, trp1-1, Δarp2::TRP1, Δarp3::HIS, [pRS315-Arp3], [ pRS317-ARP2]). Cultures were grown overnight at 30° C to OD600 = 1 and pelleted by centrifuging at 5000g for 8 min. Cells were washed in lysis buffer (20 mM Tris pH 8.0, 50 mM NaCl, 1 mM EDTA) and pelleted. The pellet was frozen in liquid nitrogen and stored at −80° C. Cells were thawed and 1 mL lysis buffer was added per g wet cell pellet. Protease inhibitor tablets (Sigma) were added (1 tablet per 100 mL cell suspension) and cells were lysed by five passes at 30,000 psi on a model 110EH Microfluidizer (Microfluidics). The lysate was spun at 30K g for 20 min and the supernatant was collected and centrifuged for 1 hr 15 min at 125 Kg. The supernatant was filtered through cheesecloth and proteins were precipitated with 50% (NH4)2SO4. Pelleted proteins were resuspended in 50 mL of PKME (25 mM PIPES pH 7.0, 50 mM KCl, 1mM EGTA, 3mM MgCl2, 1 mM EGTA, 1 mM DTT and 0.1 mM ATP) and dialyzed against the same buffer overnight. The dialysate was purified on a GST-N-WASp-VCA affinity column and Arp2/3 complex-containing fractions were pooled and concentrated to 1 mL before loading onto a Superdex200 gel filtration column (GE Healthcare) equilibrated in 20mM Tris pH 8.0 and 100 mM NaCl. Peak fractions were pooled and concentrated to approximately 10– 20 μM, flash frozen in liquid nitrogen, and stored at −80°C. Bovine N-WASp-VCA (residues 428–505) was amplified from into pGEX-2T-NWASp-VCA (a gift of Henry Higgs) and cloned onto pGV67 using BamHI and EcoRI restriction sites. This vector was used to transform E. coli strain BL21(DE3). N-WASp-VCA was expressed and purified as previously described11. Chicken skeletal muscle was purified and labeled with pyrene iodoacetamide21. Recombinant full length human WASp was purified from 293 cells and recombinant human WASp (residues 105–502) was purified from E. coli for the initial screens with HsArp2/3 complex. Recombinant Listeria ActA residues 36–170 were expressed as an N-terminal GST fusion and purified from E. coli. S. pombe formin Cdc12 FH2 domain (residues 973–1390) was purified from E. coli.
Actin Polymerization assay
We measured the nucleation activity of Arp2/3 complex from the time course of actin polymerization. Polymerization reactions of 100 μL were assembled as previously described12 and fluorescence measurements were made at 8 s intervals in a 96 well plate using a Gemini XPS spectrofluorimeter (Molecular Devices) with an excitation wavelength of 365 nm and an emission wavelength of 407 nM. The rate of polymerization at each time point was determined by calculating the slope in 5 point intervals and multiplying by [total polymer]/Δ RFU, where [total polymer] is the total actin minus the critical concentration (0.1 μM) and ΔRFU is RFUmax - RFUmin. We plotted the maximum elongation rate verses inhibitor concentration and fit the curve using the equation:
The products of polymerization reactions were diluted into Alexa 488-phalloidin and observed by fluorescence microscopy16.
Fluorescence anisotropy
The endogenous cysteine (Cys431) near the end of the N-terminus in purified N-WASp-VCA was labeled with rhodamine maleimide12. Fixed concentrations of Rho-N-WASp-VCA were titrated with BtArp2/3 complex and the fluorescence anisotropy measured12. Binding constants were determined by fitting the anisotropy curves to the following equation:
where rf is the signal of the free receptor (R), rb is the signal of the bound receptor, and [L] is the total concentration of the ligand (species titrated). rb and Kd were fit using Kaleidagraph (Synergy Software).
Etheno-ATP fluorescent assay
Binding of etheno-ATP to Arp2/3 complex was performed as described previously with slight modifications17,18. Human Arp2/3 complex (0.7μM) was incubated with 2.5 μM etheno-ATP, 0.2 M acylamide (used as a etheno-ATP fluorescence quencher) in 2mM Trs-HCl pH 8.0, 50 mM KCl, 0.05 mM EGTA 0.8 mM MgCl2, 0 and 0.5 mM DTT. Ethano-ATP emission spectrums were acquired upon excitation at 340 nm using spectrofluorometer FluoroMax (Horiba JY). Inhibition of Arp2/3 complex by 100 μM CK-636 was performed in the presence or absence of 2.8 μM WASp-VCA.
Listeria comet, motility and podosome assays
SKOV3, THP-1 and Listeria monocytogenes (ATCC 984) cells were from ATCC. SKOV3 cells were cultured in RPMI with 5% FBS without antibiotic at 37°C and 5% CO2. Listeria were grown in shaking culture in brain heart infusion (Difco Inc, DF0037-15) at 37°C. SKOV3 cells were infected with Listeria monocytogenes, incubated for 90 min and then treated for 60 minutes with either compounds dissolved in DMSO or a DMSO control before fixing with 2% formaldehyde. Fixed cells were permeabilized with 0.1% Triton-X in PBS at RT for 15 min and reacted with a 1:200 dilution of Listeria antibody (US Biologics, Cleveland, Ohio) in PBS for 60 min at RT. Antibody was removed and cells were stained with a 1:100 dilution of Alexa Fluor 568 phalloidin and a 1:400 dilution Alexa Fluor 488 goat anti-rabbit secondary antibody (both from Molecular Probes, Eugene, OR) for 60 min at RT. After washing, cells were imaged at 20x with an Axon Instruments automated fluorescence microscope system (Molecular Devices). The THP-1 monocyte cell line was grown in RPMI with 10% FBS/2mM L-glutamine, differentiated in 50 nM phorbol myristate acetate (Sigma-Aldrich-Fluka) for 48 hrs at 37°C, trypsinized and plated at 5×105 cells/ml in black Nunc glass-bottom 96-well plates. After incubation at 37°C for 2 hr, cells were treated with compounds for 15 min before fixing and staining with Alexa Fluor 568 phalloidin and DAPI (Sigma-Aldrich-Fluka) as above. Supplemental Figure 2 details the metrics used for podosome quantification. Black molly keratocytes were isolated19, treated briefly with trypsin and observed by time lapse phase contrast microscopy on plastic Petri dishes at room temperature.
X-ray Crystallography
Crystals of BtArp2/3 complex were grown at 4°C using the hanging drop vapor diffusion method as previously described7. Crystals were soaked for 24 hrs in 18 % polyethylene glycol 8000, 50 mM HEPES pH 7.5, 100 mM potassium thiocyanate, 20 % glycerol and either 0.5 mM CK548 or 1mM CK636 in DMSO before freezing in liquid nitrogen. Data was collected at beamline X29A at Brookhaven National laboratories and processed with HKL200022. To generate initial models, protein atoms from 1K8K were refined against the data using 30 cycles of minimization in which each subunit was allowed to move as a separate rigid body. CK548 structure was refined in Refmac23 with topology files generated in ccp4i sketcher. The CK636 structure was refined in CNS8 using topology files generated with the prodrg server24.
Supplementary Material
Acknowledgments
This work was supported by Cytokinetics, Inc., NIH research grant GM-066311 to TDP, NSF Graduate Research Fellowship to CDM and a Ruth Kirschstein postdoctoral fellowship GM074374-02 to BJN. The authors thank Lisa Belmont, Zarak Khurshid, Obidimma Ezizika, Juliet Lee, Stephanie Leuenroth and Hongli Chen for help with the project.
Footnotes
AUTHOR CONTRIBUTIONS
BJN, NT, AR, DWP, ZJ and JH designed and carried out experiments; CDM analyzed data; RS and TDP supervised research; BJN, NT, AR, JH and TDP wrote the paper.
AUTHOR INFORMATION
Structural data have been deposited in Protein Data Bank with accession codes 3DXK (CK-0944636) and 3DXM (CK-0993548).
References
- 1.Goley ED, Welch MD. The Arp2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 2.Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 3.Linder S, et al. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocation of the Arp2/3 complex. J Immunol. 2000;165:221–225. doi: 10.4049/jimmunol.165.1.221. [DOI] [PubMed] [Google Scholar]
- 4.Gimona M, Buccione R, Courtneidge SA, Linder S. Curr Opin Cell Biol. 2008;20:235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 6.Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci USA. 2004;101:15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jabs A, Weiss MS, Hilgenfeld R. Non-proline cis peptide bonds in proteins. J Mol Biol. 1999;286:291–305. doi: 10.1006/jmbi.1998.2459. [DOI] [PubMed] [Google Scholar]
- 9.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 10.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 12.Tomasevic N, et al. Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry. 2007;46:3494–3502. doi: 10.1021/bi062152y. [DOI] [PubMed] [Google Scholar]
- 13.Nolen BJ, Pollard TD. Structure and biochemical properties of fission yeast Arp2/3 complex lacking the Arp2 subunit. J Biol Chem. 2008;283:26490–26498. doi: 10.1074/jbc.M802607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacLean-Fletcher S, Pollard TD. Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem Biophys Res Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- 15.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 17.Blanchoin L, et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 18.Dayel MJ, Holleran EA, Mullins RD. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc Natl Acad Sci USA. 2001;98:14871–14876. doi: 10.1073/pnas.261419298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Ishihara A, Theriot JA, Jacobson K. Principles of locomotion for simple-shaped cells. Nature. 1993;362:167–171. doi: 10.1038/362167a0. [DOI] [PubMed] [Google Scholar]
- 20.Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–22. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 21.Pollard TD. Polymerization of ADP-actin. J Cell Biol. 1984;99:769–77. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data dollected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Dodson EJ, Winn M, Ralph A. Collaborative Computational Project, number 4: providing programs for protein crystallography. Methods Enzymol. 1997;277:620–33. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 24.Schuettelkopf AW, van Aalten DMF. PRODRG- a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;D60 doi: 10.1107/S0907444904011679. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.