Abstract
Anaplastic lymphoma kinase (ALK), a receptor tyrosine kinase in the insulin receptor superfamily, was initially identified in constitutively activated oncogenic fusion forms – the most common being nucleophosmin-ALK – in anaplastic large-cell lymphomas, and subsequent studies have identified ALK fusions in diffuse large B-cell lymphomas, systemic histiocytosis, inflammatory myofibroblastic tumors, esophageal squamous cell carcinomas and non-small-cell lung carcinomas. More recently, genomic DNA amplification and protein overexpression, as well as activating point mutations, of ALK have been described in neuroblastomas. In addition to those cancers for which a causative role for aberrant ALK activity is well validated, more circumstantial links implicate the full-length, normal ALK receptor in the genesis of other malignancies – including glioblastoma and breast cancer – via a mechanism of receptor activation involving autocrine and/or paracrine growth loops with the reported ALK ligands, pleiotrophin and midkine. This review summarizes normal ALK biology, the confirmed and putative roles of ALK in the development of human cancers and efforts to target ALK using small-molecule kinase inhibitors.
Keywords: anaplastic large-cell lymphoma, anaplastic lymphoma kinase, esophageal squamous cell carcinoma, glioblastoma, inflammatory myofibroblastic tumor, midkine, neuroblastoma, non-small-cell lung carcinoma, pleiotrophin, targeted cancer therapy, tyrosine kinase inhibitor
Anaplastic lymphoma kinase receptor tyrosine kinase
Normal anaplastic lymphoma kinase gene & protein
As well as playing vital roles in the transmission of extracellular cues that control diverse normal functions, including proliferation, survival and differentiation, a number of receptor tyrosine kinases (RTKs) have also been implicated in oncogenesis [1-4]. Anaplastic lymphoma kinase (ALK) – one of the insulin receptor (IR) superfamily of RTKs, members of which also include the IGF-1 receptor, the RTK neurotrophin receptors and hepatocyte growth factor/scatter factor receptor (MET) – is an example of one such RTK with roles in both normal development and oncogenesis. ALK is encoded by a genomic locus found at the chromosomal band 2p23 in the human [5,6] and on the distal mouse chromosome 17 [7]. ALK (which is also known by the cluster designation CD246) was originally identified as a result of the cloning of the nucleolar protein nucleophosmin (NPM)–ALK fusion gene in anaplastic large-cell lymphomas (ALCLs) [5,8]. An association between ALCL and the t(2;5)(p23;q35) chromosomal rearrangement was reported in the late 1980s [9-13], and the genes involved in this translocation were identified in 1994 as those encoding NPM at 5q35 and the novel ALK RTK at 2p23 [5]. ALCLs account for approximately 2.5–5% of all human non-Hodgkin’s lymphomas (NHLs), and are most common in young patients, comprising 30–40% of pediatric large-cell lymphomas [14,15]. The genes encoding the full-length ALK protein in mouse and man (human ALK sequences, Genbank accession numbers: U62540 and U66559; mouse Alk cDNA accession number D83002) were cloned in 1997 [16,17]. The presence of an ALK counterpart in Drosophila (DAlk) has also been confirmed by Loren et al. (GenBank accession number AAF36990) [18].
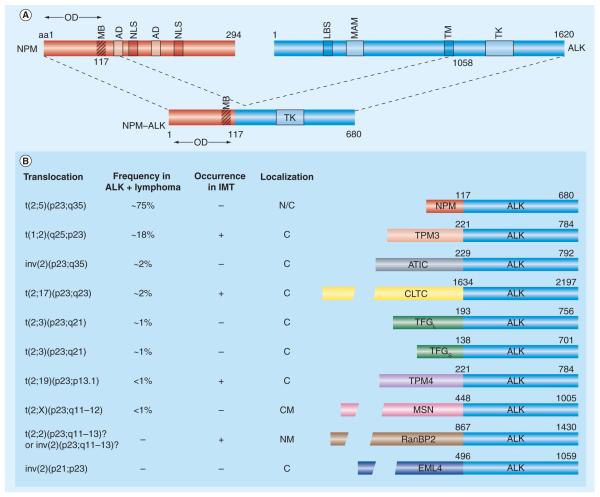
As with other RTKs, ALK possesses an extracellular ligand-binding region, a transmembrane-spanning domain and a cytoplasmic kinase catalytic region (Figure 1A). ALK shows significant homology to leucocyte tyrosine kinase (LTK), another RTK in the IR superfamily that is, to date, of uncertain normal or pathologic function [6,16,17]. Despite the high degree of similarity shared by ALK and LTK, which would suggest possible redundant physiologic functions, simultaneous deletion of the two genes in mice is not associated with obvious deleterious effects (Xue L, Morris SW, Unpublished Data). The 6226-bp ALK cDNA encodes for a 177-kDa polypeptide; post-translational modifications, such as N-glycosylation, generate a mature ALK RTK of approximately 200–220 kDa [17,19,20]. ALK is a single-chain transmembrane protein of 1620 amino acids (aa) in the human and 1621 or 1701 aa in the mouse and fruit fly, respectively [16-18]. The 1030-aa extracellular domain of human ALK contains several motifs (Figure 1A), including a 26-aa N-terminal signal peptide sequence, as well as the binding site (located at residues 391–401) for the endogenous ligands reported to activate ALK, pleiotrophin (PTN) and midkine (MK) [21,22]. The 28-aa transmembrane domain is followed by a 64-aa juxtamembrane segment that contains a binding site (aa 1093–1096) for phosphotyrosine-dependent interaction with the IR substrate-1. The kinase domain (KD) of ALK includes a three-tyrosine-containing motif (tyrosines 1278, 1282 and 1283) within its activation loop. As with other IR superfamily RTKs, these tyrosine residues represent major autophosphorylation sites that regulate the activation loop confirmation, occluding access of ATP to the ATP-binding pocket in its nonphosphorylated state and swinging outward and away from the binding pocket to allow unimpeded entry of ATP during the kinase-activation process following phosphorylation of the triplet tyrosines [23]. The 244-aa ALK C-terminus contains a phosphotyrosine-dependent binding site (aa 1504–1507) for the substrate protein Src homolgy 2 domain containing (SHC), as well as an interaction site for the phosphotyrosine-dependent binding of phospholipase C-γ (ALK aa 1603–1606) [24-26].
Figure 1.
(A) ALK receptor tyrosine kinase and the NPM–ALK fusion protein created by t(2;5). Fusion of the chromosome 5 gene encoding NPM to the chromosome 2 gene encoding ALK generates the chimeric tyrosine kinase, NPM–ALK. NPM contains an OD (residues 1–117) a putative MB (residues 104–115), two ADs (Asp/Glu-rich acidic domain; residues 120–132 and 161–188) that function as acceptor regions for nucleolar targeting signals and two NLS (residues 152–157 and 191–197). ALK contains a single MAM domain, a region of approximately 170 aa homologous to the extracellular portions of a number of functionally diverse proteins that may have an adhesive function (residues 480–635). The LBS for pleiotrophin and midkine (ALK residues 391–401) is indicated. Note that the entire intracytoplasmic portion of ALK, exclusive of the TM, is incorporated into the NPM–ALK and all other ALK chimeric proteins.
(B) Representative ALK fusion proteins, the chromosomalrearrangements that generate them, their occurrence in ALK-positivelymphomas and IMTs and their subcellular localizations. A partiallisting of the more common oncogenic ALK fusions is shown; a total of15 different ALK fusions have now been described (those not shown are ALO17–ALK, CARS–ALK, MYH9–ALK, SEC31L1– ALK and TFGXL–ALK). The exact frequency of the various ALK fusions expressed in IMT has not yet been determined. To date, six ALK fusions (CARS–ALK, CLTC–ALK, RANBP2–ALK, SEC31L1–ALK, TPM3–ALK and TPM4–ALK) have been identified in IMT. The TPM3–ALK, TPM4–ALK and CLTC–ALK fusions have been detected in both classical null or T-cell anaplastic large-cell lymphomas and IMT, whereas CARS-ALK, RANBP2–ALK and SEC31L1–ALK occur in IMT but have not yet been described in anaplastic large-cell lymphoma. CLTC–ALK and, to a lesser extent, NPM–ALK, also occur in rare B-cell plasmablastic/immunoblastic non-Hodgkin’s lymphomas. Two independent reports have also recently described the occurrence of the TPM4–ALK fusion in squamous cell carcinomas of the esophagus [177,178], and several studies have identified the presence of the novel ALK fusion, EML4–ALK, in a subset of non-small-cell lung cancers [180-187]. aa: Amino acid; AD: Acidic amino acid domain; ALK: Anaplastic lymphoma kinase; ATIC: 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; C: Cytosolic; CLTC: Clathrin heavy chain; CM: Cell membrane; EML: Echinoderm microtubule-associated protein-like; LBS: Ligand-binding site; IMT: Inflammatory myofibroblastic tumor; MAM: Meprin/A5/protein tyrosine phosphatase Mu; MB: Metal-binding domain; MSN: Moesin; N: Nuclear; NLS: Nuclear localization signal; NPM: Nucleophosmin; NM: Nuclear membrane; OD: Oligomerization domain; RanBP2: Ran-binding protein 2; TFG: TRK-fused gene; TK: Tyrosine kinase catalytic domain; TM: Transmembrane domain; TPM3: Non-muscle tropomyosi.
Normal ALK function
Initial studies of human ALK mRNA expression demonstrated the presence of 6.5- and 8.0-kb transcripts in rhabdomyo sarcoma tumors and in normal tissues, mainly in the central and peripheral nervous systems, with no or very minimal expression in other tissues [5]. Subsequent studies confirmed the expression of Alk transcripts in murine brain and spinal cord [16,17], and in situ hybridization studies showed Alk expression to be restricted mainly to specific regions of the developing mouse brain and peripheral nervous system – the thalamus, hypothalamus, midbrain, olfactory bulb and selected cranial, as well as dorsal root, ganglia and the myoenteric plexus of embryonic mice, beginning at day 11 of embryogenesis. The levels of Alk mRNA decrease near the end of gestation and are detected at only very low quantities in neonates; immunoblotting studies have also shown the levels of Alk protein to decrease substantially after birth [16]. Vernersson and colleagues recently reported additional in situ hybridization and immunostaining expression ana lysis of Alk in the mouse, corroborating and expanding upon earlier studies; this study identified additional Alk expression in the retinal neural and pigment layers, the lens and optic nerve and in portions of the tongue, testis and ovary [27]. The restricted normal tissue distribution of the ALK protein in adult human tissues was confirmed by anti-ALK immunocytochemical studies, in which only rare scattered neural cells, pericytes and endothelial cells in the brain were shown to be immunoreactive [19,28]. DAlk mRNA and protein expression in the fruit fly is also highly regulated, with expression mainly in the brain and ventral nerve cord. However, the DAlk protein has also been detected at stage 11 in the developing fly mesoderm [18], and more recent immunostaining studies using DAlk-mutant Drosophila strains have confirmed mRNA and protein expression in the digestive tract musculature throughout embryonic development [29-31].
The nervous system-predominant expression pattern of Alk suggests that the RTK could play an important role in the physiological development and function of this tissue [16-19]. Intriguingly, however, Alk function is not required for the viability of knockout mice, which possess a full life span and have no readily obvious abnormalities (Xue L, Morris SW, Unpublished Data). This is the case despite the observation in Caenorhabditis elegans that the ALK ortholog is implicated in the inhibition of presynaptic neural differentiation, with downregulation of ceAlk by a specific Skp, Cullin, F-box-like ubiquitin ligase complex required for the maturation of neural synapses in the worm [32]. Furthermore, recently published work from Bazigou et al. has demonstrated DAlk to function in the development of the neuronal circuit assembly in the fly visual system [33]. In these studies, DAlk was shown to be expressed and required in target neurons present in the optic lobe, while the DAlk ligand, Jelly Belly (Jeb; vide infra), is produced by photoreceptor axons and functions in the eye to control target selection by axons in the lamina and medulla. In addition to this critical nervous system-related function in the fly, earlier studies of DAlk had clearly shown that the receptor plays an essential role in the development of the gut musculature of Drosophila, with its absence being associated with deficits in the specification of muscle-founder cells responsible for visceral gut formation [29-31,34].
Some degree of clarity regarding Alk function in mammals has been provided by Bilsland and colleagues, who reported the characterization of independently generated Alk knockout mice [35]. The investigators identified no apparent developmental, anatomical or locomotor deficits in these knockout animals. Interestingly, however, the mice were reported to display an ‘antidepressant profile’ – an age-dependent increase in basal hippocampal progenitor proliferation, similar to that observed with chronic antidepressant treatment or neurotransmitter modulation. Such hippocampal neurogenesis has been correlated with the regulation of mood and is hypothesized to be required for antidepressant efficacy based on studies in murine models; furthermore, hippocampal neurogenesis has also been postulated to be required for several aspects of learning and memory [36-38]. Consistent with their increased hippocampal progenitor proliferation, Alk knockout mice demonstrated enhanced performance in novel object recognition/location tests, as well as increased struggle time in tail suspension and Porsolt swim tests. Neurochemical ana lysis of the knockout mice revealed increased basal dopaminergic signaling tone selectively within the prefrontal cortex. Collectively, these data suggest antagonism of ALK as a potential new approach for the therapeutic management of cognitive and/or mood disorders. Within the context of these findings, it is also interesting to note that certain polymorphisms within the ALK gene locus have been reported to segregate with the development of schizophrenia in some families [39].
ALK ligands
The expression of ALK predominately in the nervous system together with the observation that the distribution of ALK mRNA and protein partially overlaps with that reported for members of the TRK neurotrophin RTKs both suggest that ALK could serve as a receptor for a neurotrophic factor(s) [40]. Recently, the related proteins PTN and MK have been reported as being ligands for ALK in mouse and man [21,22], while the Jeb protein was described as a ligand for DAlk in the fruit fly [29,30]. The 18-kDa heparin-binding growth factor PTN induces neurite outgrowth, but also has mitogenic activity for a variety of cell lineages [41-44]. The expression of PTN is similar to that of ALK, with high levels in the nervous system during fetal development and decreased expression following birth [45]. The other putative mammalian ALK ligand, MK, was originally described as a retinoic acid-inducible, developmentally regulated, heparin-binding neurotrophic factor that shows 45% identity to PTN [46]. Functions attributed to MK are similar to those of PTN, including the direction of neurite connections, effects upon neuronal migration and a possible role in angiogenesis [46-58]. MK is expressed in the brain as well as other organs, with highest levels generally at midgestation and, similar to PTN, downregulation at birth. The DAlk ligand, Jeb, is structurally dissimilar to PTN and MK, and is produced and secreted in the ventral somatic mesoderm of the fruit fly. Englund and colleagues and Lee et al. observed a similar phenotype in Jeb and DAlk mutant flies [29,30], further implicating the two proteins as playing a role in the specification of visceral mesoderm precursors into muscle-founder cells and fusion-competent cells [59]. Immunohistochemical studies demonstrated that, while Jeb and DAlk are expressed in adjacent cells, the two proteins are colocalised at areas of contact and both are essential for the activation of the RAS/MAPK pathway, with high-affinity binding of Jeb to DAlk [30] and uptake of Jeb in vivo dependent on the presence of DAlk [29]. To date, a mammalian counterpart to Jeb has not been identified, nor does evidence exist to support a role for ALK in muscle specification in mammals. Likewise, although the Drosophila counterparts to PTN and MK – the so-called miple-1 and -2 genes, respectively – have been identified, their exact functions in the fly and whether they may also bind and activate DAlk remain uncertain [60].
Pleiotrophin, MK and ALK appear to play roles in the regulation of cell survival. PTN and MK themselves were implicated in the regulation of apoptosis some time ago [45] and more recent studies by Bowden et al. suggested that this activity is regulated, at least in part, by ALK [61]. One mechanism by which ALK transduces survival signals probably involves PTN-stimulated phosphorylation and activation of the serine/threonine kinases PI3-kinase and protein kinase B (PKB/Akt), which are anti-apoptotic proteins [62]. A role for PTN (and MK) in angiogenesis has also been reported [45,49,58,63-68], and Stoica et al. postulated that ALK may mediate this process, citing ALK mRNA and protein expression in umbilical endothelial cells [21,22].
It should be mentioned that controversy remains regarding the identity of PTN and MK as bona fide ALK receptor ligands. For example, although certain experimental results have supported such a biological connection [69], several investigators have had difficulty reproducing experimental data showing the binding and activation of ALK by PTN [70-72]. In addition, receptors for PTN other than ALK have been reported – the receptor protein tyrosine phosphatase ζ1 (PTPRZ1, also known as RPTPβ/ζ) [73,74] and the heparan sulphate proteoglycan N-syndecan (syndecan 3) [75-79]; likewise, MK receptors other than ALK have been described – neuroglycan C [48], PTPRZ1 (which, like ALK, it shares with PTN) [50], the low-density lipoprotein receptor-related protein [53] and α4β1- and αβ1-integrins [54]. Thus, a role for PTN- and MK-mediated activation of ALK in the regulation of normal physiologic cell responses, such as angiogenesis and neurogenesis, remains to be unequivocally established, and at least some of the effects of PTN and MK may be transduced by other receptors. Partial clarity regarding this issue with respect to PTN appeared to have been provided by the 2005 studies of Lu et al., in which the two naturally occurring forms of PTN (15- and 18-kDa polypeptides, which differ by the processing of 12 C-terminal aa residues), were shown to differentially bind and activate ALK (PTN15) or PTPRZ1 (PTN18) [80]. In studies performed with glioblastoma cell lines, PTN15 promoted cell proliferation in an ALK-dependent fashion, while PTN18 mediated cell migration in a PTPRZ1-dependent manner. Thus, these data suggested that some of the confusion in the literature regarding the binding of PTN and ALK might be due to the differential binding of the two PTN isoforms to different receptors; however, a follow-up study published in 2007 from another group using neuroblastoma and glioblastoma cells failed to show evidence for binding of ALK by either PTN isoform [72].
A different mechanism for ALK activation by PTN has also recently been suggested by Perez-Pinera and colleagues [81]. These investigators reported that phosphorylation of ALK in PTN-stimulated cells is mediated indirectly through the PTN/ PTPRZ1 signaling pathway, without actual interactions between PTN and ALK. This model posits that, in cells not stimulated by PTN, PTPRZ1 dephosphorylates ALK at the sites that undergo autophosphorylation during activation of the kinase; by contrast, when PTN binds PTPRZ1 (which results in inactivation of the membrane-spanning receptor phosphatase) in PTN-stimulated cells, the autophosphorylation sites in ALK are no longer dephosphorylated and tyrosine phosphorylation/activation of ALK occurs. This unique mechanism of ALK activation was only recently described (in 2007), and follow-up studies further examining it have, thus, not yet appeared in the literature. A substantial body of data exists linking PTN and MK to the proliferation, survival and metastasis of various tumors, and to tumor-associated angiogenesis [45,47,68,82]; thus, the elucidation of the exact role played by ALK as opposed to other receptors in the binding and transmission of PTN- and MK-mediated signals is of critical importance and requires substantial further investigation.
Role of ALK in cancer pathogenesis
Full-length ALK receptor in tumorigenesis
Several tumor types are known to express the full-length ALK RTK. For example, full-length ALK receptor protein has been detected in cell lines and/or primary specimens representing a variety of tumors including neuroblastomas, neuroectodermal tumors [20,22,83-85], glioblastomas [62,80,86,87] and melanoma [70]. Furthermore, the full-length ALK cDNA was originally cloned from an Rh30 rhabdomyosarcoma cell line cDNA library [5,17,19], and expression of the full-length protein has subsequently been reported to occur in a subset of rhabdomyosarcoma tumors [88-91]. In addition, anti-ALK immunoreactivity has been observed in tissue immunostaining studies of other malignancies, such as breast carcinoma, malignant peripheral nerve-sheath tumors, and lipogenic tumors [70,89,91-93]; in these tumors, however, whether the full-length or rather fusion forms of ALK might be expressed in immuno positive cases has not yet been unequivocally determined (indeed, it has been noted that at least some breast carcinomas appear to exhibit an ALK immunostaining pattern more characteristic of certain ALK fusion proteins than the full-length receptor) [92]. With the exception of neuroblastoma (in which a pathogenic role for full-length ALK expression seems clear, vide infra), the pathogenic significance – if any – of ALK receptor expression in each of the aforementioned tumor types remains uncertain at this time. It has been postulated that autocrine and/or paracrine growth loops involving PTN and/or MK may be driving the growth of tumors that express the full-length normal ALK receptor, and data suggestive of such a mechanism have been generated in studies of glioblastoma (the tumor, thus far, best-characterized in this regard [62,80,87]) but this has not yet been unequivocally proven for any tumor type.
Neuroblastoma: overexpression & activating point mutations of full-length ALK
Neuroblastoma is the most common extracranial solid tumor of childhood and is an embryonal tumor derived from the developing neural crest [94]. With current treatment protocols for high-risk neuroblastoma involving myeloablative chemotherapy with peripheral stem cell rescue, improved event-free survival is observed; however, long-term survival remains poor [95].
By single nucleotide polymorphism (SNP) array ana lysis, it was recently reported that ALK amplification occurs both in cell lines and primary neuroblastomas [96]; these data corroborated earlier reports that had also found ALK expression and DNA amplification in neuroblastoma [20,83-85]. As previously described, Alk is normally expressed in the nervous system and its deregulation, therefore, might be expected to drive abnormal development and growth of neural lineage tumors, such as neuroblastoma [5,16,17,27,96,97]. Full-length ALK protein expression was described in 92% (22 out of 24 specimens) of primary neuroblastomas by Lamant and colleagues [20], although no clear association with clinical or prognostic features was identified in this small number of cases. Other translational research reports had suggested links associating the putative ALK ligands PTN and MK with neuroblastoma clinically [98-101], although the exact pathophysiologic importance of these links and their relevance to ALK remains uncertain. For example, Nakagawara et al. found high levels of PTN mRNA expression in neuroblastomas to correlate with a favorable prognosis (whereas MK expression was not associated with disease stage) [98], and Calvet et al. found low levels of PTN to be associated with the acquisition by neuroblastoma of resistance to the chemotherapeutic agent irinotecan in vivo [101]. These findings would seem to be inconsistent with a role for PTN or MK in promoting the autocrine or paracrine ALK-mediated growth of neuroblastomas.
Anaplastic lymphoma kinase had also been found to be constitutively activated due to high-level overexpression as a result of gene amplification in neuroblastoma cell lines [84]; in these studies, activated ALK was shown to form a stable complex with hyperphosphorylated ShcC, a SHC adaptor molecule, and to modify the responsiveness of the MAPK pathway to growth factors. Mice lacking ShcB and ShcC exhibit significant loss of sympathetic neurons, suggesting that these two kinase signaling substrate adaptors act to support sympathetic development and survival [102]. Suppression of activated ALK in neuroblastoma cells was found to reduce the phosphorylation of ShcC and downstream targets of ALK, such as MAPKs p42 and p44, as well as Akt, and to induce apoptotic changes in the cells [84]. Collectively, these aforementioned data supported a potential role for deregulated ALK signaling in neuroblastoma genesis and/or progression.
In order to determine whether ALK might be activated in neuro blastoma by mechanisms other than amplification, George et al. recently sequenced the ALK gene in a cohort of 93 primary neuroblastoma specimens [103]. This sequence ana lysis of the entire open reading frame of ALK identified five novel mis-sense mutations; sequence variants were identified in conserved positions in the ALK tyrosine kinase (TK) domain, four being singletons and one found to be recurrent in four of the 93 samples. Sequence ana lysis of matched normal control DNA from this set of patients revealed that in the eight patients, two mutations were germline and six were somatic. The most common mutation was a recurrent cytosine-to-adenine change in exon 23 that results in a phenylalanine-to-leucine substitution at codon 1174 (F1174L) within the ALK KD; this somatic mutation was identified in 4% (four out of 93) of primary tumors. None of these mutations appeared to be frequently occurring SNPs (none were found in dbSNP or the Catalogue of Somatic Mutations in Cancer [two SNP databases]), and these alterations were not detected by genotyping of 270 samples collected by the International Hap Map Consortium [104].
The F1174L, as well as an R1275Q, ALK mutation rendered IL-3-dependent murine lymphoid BaF3 cells cytokine-independent for their growth, indicating that both mutations are kinase-activating [103]; however, expression of neither wild-type ALK nor the sequence variants T1151M or A1234T led to IL-3-independent BaF3 growth. Moreover, when expressed in BaF3 cells, the F1174L mutation induced constitutive activation and autophosphorylation of ALK; the R1275Q mutation was also associated with constitutive ALK phosphorylation, but to a lesser extent. By contrast, neither the T1151M nor A1234T alterations caused ALK activation, suggesting that these sequence variations may actually be uncommon SNPs that have yet to be reported.
Anaplastic lymphoma kinase was found to be mutated in six out of 30 neuroblastoma cell lines as well, including three different cell lines (Kelly, SH-SY5Y and LAN-1), with the F1174L mutation also shown to be present in four of the 93 primary tumor samples examined [103]. The R1275Q mutation was found in the neuroblastoma cell line SMS–KCNR, while a mutation that had not been observed in the primary tumor specimen, F1245V, was identified in the CHLA-90 cell line. To test the sensitivity of the tumor-derived mutant ALK proteins to ALK inhibition, neuroblastoma cell lines harboring specific ALK mutants were treated with increasing concentrations of a recently described ALK small-molecule inhibitor, NVP-TAE684 (vide infra [103]. All of the constitutively activated point mutants of ALK were inhibited by NVP-TAE684, the most sensitive to the small molecule being the F1174L mutation; for example, the IC50 values for inhibition of the proliferation of two neuroblastoma cell lines harboring the F1174L mutation, SH-SY5Y and Kelly, were 258 nM and 416 nM, respectively. In agreement with the findings of George et al. [103], these two neuroblastoma cell lines had also recently been reported by an independent group of investigators to be responsive to NVP-TAE684, but the basis for the sensitivity was not elucidated [103,105]. Furthermore, in experiments performed in parallel by George et al. to corroborate their results with NVP-TAE684, a second ALK small-molecule inhibitor – a pyridone compound ATP-competitive ALK small-molecule inhibitor, CRL151104A (codeveloped by ChemBridge Research Laboratories Inc. [San Diego, CA, USA] and investigators from the Steve Morris laboratory at St Jude Children’s Research Hospital [Memphis, TN, USA]; vide infra, Section 3E) was tested against neuroblastoma cell lines containing activated ALK mutant proteins; dose-dependent growth inhibition of the Kelly (F1174L), SH-SY5Y (F1174L) and CHLA-90 (F1245V) neuroblastoma cell lines was observed in the presence of CRL151104A, with IC50 values of 610, 500 and 370 nM, respectively (George RE, Look AT, Morris SW; Unpublished Data).
Of note, the SMS–KCNR cell line expressing the ALK R1275Q mutation was resistant to both NVP-TAE684 and CRL151104A, with IC50 values of 4.9 and 3.8 μM, respectively (George RE, Look AT, Morris SW; Unpublished Data) [103]. The basis for the resistance of this cell line that expresses the R1275Q-activating allele is not clear; however, Ba/F3 cells expressing this mutation became IL-3 independent and were sensitive to both NVP-TAE684 and CRL151104A (IC50 values of 328 and 370 nM, respectively). The SMS–KCNR cell line may, therefore, have acquired additional molecular abnormalities during its adaptation to tissue culture that render it independent of the activity of the mutated ALK kinase. Other neuroblastoma cell lines that expressed ALK but lacked mutations were also resistant to the growth-inhibitory properties of NVP-TAE684 and CRL151104A (with IC50 values for the compounds ranging from 1.67 to 4.33 μM); statistical ana lysis performed to compare the IC50 values of the sensitive neuroblastoma cell lines Kelly, SH-SY5Y and CHLA-90 with the IC50 values of nine lines expressing wild-type ALK showed the difference to be highly significant (p = 0.0015).
Treatment with either of the ALK inhibitors (NVP-TAE684, 200 nM; CRL151104A, 1 μM) resulted in 3–12-fold increases in the percentage of apoptotic cells in the inhibitor-sensitive cell lines Kelly (F1174L) and SH-SY5Y (F1174L) after only 24 h, in contrast to the resistant SMS–KCNR (R1275Q) and IMR-5 (wild-type ALK) cell lines, which lacked any increase in terminal deoxynucleotidyl transferase mediated dUTP nick-end labeling activity (George RE, Look AT, Morris SW; Unpublished Data) [103]. Cytotoxicity was also associated with G1-phase arrest and substantial reductions in S-phase cell fractions as measured by flow cytometry [103]. The inhibitor-sensitive cell lines, such as Kelly (F1174L), demonstrated reduced phosphorylation of ALK and of the downstream effectors ERK 1/2, STAT3 and Akt upon treatment. By contrast, there was no apparent effect on the phosphorylation of ALK or its downstream targets in the resistant cell line IMR-5. Moreover, shRNA knockdown of ALK in the Kelly and SH-SY5Y cell lines that harbor the F1174L mutation was associated with a reduction in cell growth and viability by comparison with cells transduced with a control shRNA [103].
Concurrent with the findings of George and colleagues, three other groups of investigators independently described mutations of ALK in neuroblastomas as well [106-108]. In one of these studies, Mosse et al. studied the rare subset of neuroblastomas that are inherited as an autosomal dominant trait with incomplete penetrance [106]. A genome-wide scan for linkage in 20 neuroblastoma pedigrees was performed at approximately 6000 SNPs. A single region consistent with linkage was discovered at 2p23–24 delimited by SNPs rs1862110 and rs2008535 and containing 104 genes, including the candidate oncogenes MYCN and ALK. No sequence variations were discovered during complete resequencing of the MYCN coding region and 18 kb of surrounding genomic DNA. However, eight of the 20 pedigrees studied showed single base substitutions in highly conserved nucleotides within the ALK TK domain that segregated with the disease. To determine if ALK alterations might also be somatically acquired, Mosse et al. examined a representative set of 491 diagnostic neuroblastoma primary tumors from sporadically occurring cases for copy number alterations on a 550K SNP array, identifying a focal unbalanced gain of a large genomic region at 2p, including the ALK locus, in 112 cases (22.8%: partial trisomy) and an additional 16 cases (3.3%) of high-level focal ALK amplification. Aberrant ALK copy number status (either gain or amplification) was associated with an aggressive clinical phenotype, such as metastasis at diagnosis (p < 0.0001) and death from disease (p = 0.0003). Sequencing of ALK in a subset of 167 neuroblastomas from high-risk patients and 27 human neuroblastoma cell lines (all established from high-risk patients) revealed 14 out of 167 (8.4%) primary specimens and ten out of 27 (35.7%) cell lines to possess single-base point mutations. In total, in their sequencing of both familial and sporadic neuroblastoma cases, Mosse et al. identified mutations at eight different codons (G1128A, M1166R, I1171N, F1174I, F1174L, R1192P, F1245C, F1245V, I1250T and R1275Q) within the ALK KD; each of the mutations identified were shown experimentally and/or predicted to be kinase activating [106]. Neuroblastoma cell lines expressing ALK mutants or containing amplified ALK demonstrated marked inhibition of proliferation in response to knockdown of expression by ALK-directed siRNAs. These data revealed that heritable mutations in the ALK KD are the major genetic determinant of familial neuroblastoma and independently corroborated the observation that somatically acquired alterations in ALK also frequently occur in sporadic neuroblastomas.
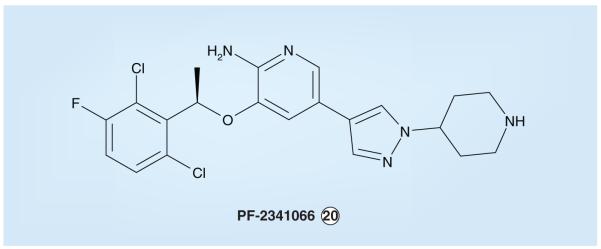
Two additional reports regarding ALK abnormalities in neuroblastoma described findings similar to George et al. and Mosse and colleagues [103,106-108]. Specifically, Chen et al. identified mis-sense mutations in 13 out of 215 fresh tumors (6.1%) and eight out of 24 neuroblastoma cell lines (33%) [108]; ALK mutation was found to be highly correlated with MYCN amplification, and 12 out of 13 of the mutations in the primary samples were found in patients with high-risk stage III or IV disease. In addition to F1174 and R1275 mutations, mutations of T1087 (T1087I) and K1062 (K1062M) were identified. Together with cases shown to have high-grade amplification of the ALK gene, abnormal ALK signaling was implicated in 16 out of 151 (11%) of the advanced neuroblastomas examined by these investigators. Janoueix-Lerosey et al. identified both germline and somatic ALK mutations and, in addition to a predominance of F1174 and R1275 abnormalities, found mutations at R1192 (R1192P) and Y1278 (Y1278S); mutations were identified in nine out of 28 (32.1%) neuroblastoma cell lines and seven out of 115 (6.1%) frozen tumor samples [107]. Interestingly, one of the cell lines (CLB-BAR) in which no activating point mutations of ALK were identified, expressed a truncated ALK-immunoreactive protein with high kinase activity, suggesting an alternative mechanism of activation, such as an ALK fusion. These investigators also found that 26 out of 592 neuroblastomas (4.4%) demonstrated higher than twofold copy number increases of the ALK genomic locus and that 135 out of 592 (22.8%) showed additional cases of lower level gains, indicating that more than 25% of the neuroblastomas contain copy number increases of ALK. Janoueix-Lerosey and colleagues also found the dual ALK/MET small-molecule inhibitor PF-2341066 (Pfizer, vide infra) to inhibit the growth of neuroblastoma cell lines expressing ALK-activating point mutations selectively compared with a neuroblastoma line that expresses neither ALK nor MET. Collectively, these four recent reports indicate that ALK is a promising and novel therapeutic target in neuroblastoma and support the performance of clinical trials to test small-molecule ALK inhibitors for the treatment of this pediatric cancer [103,106-108].
Full-length ALK in diffuse large B-cell lymphoma
In 1997, Delsol and colleagues reported full-length ALK protein expression in a rare subtype of poor-prognosis B-cell NHL that probably comprises less than 0.5–1% of all B-cell lymphomas [109]. However, in 2003, this group and others described the presence of certain ALK fusions – coexpressed with full-length ALK in at least some of the cases – in these NHLs (see the later section entitled ‘ALK fusion proteins in hematopoietic tumors: ALK-positive diffuse large B-cell lymphoma’). The oncogenic importance of full-length ALK expression in the genesis of these B-cell NHLs is not clear. It is more likely that their development and progression depend upon the expression of the constitutively activated ALK fusion proteins, rather than the full-length, normal ALK receptor, which would be ligand dependent for its activation.
ALK fusion proteins
A number of RTKs have been implicated in oncogenesis due to genetic abnormalities ranging from point mutations, gene amplification, or fusion to heterologous genes; mechanisms that result in the activation of their kinase catalytic domains. This kinase activation induces growth factor-independent proliferation, cellular transformation, anti-apoptotic signaling and resistance to drugs and gamma irradiation employed during therapy [2-4,110]. As described above, activating point mutations of ALK have recently been identified for the first time in cancer, in neuro blastoma; furthermore, genomic DNA amplification of the ALK gene locus at human chromosome 2p23 and resultant overexpression of normal ALK at sufficiently high levels to result in its activation occurs in a subset of neuroblastomas [20,83-85,103,106-108]. However, the most common mechanism of constitutive ALK activation currently known to occur in cancer involves chromosomal rearrangements (also known as ‘translocations’) that interrupt the ALK gene at 2p23 and fuse it with another gene, resulting in the creation of oncogenic ALK fusion genes and proteins. The first ALK fusion protein identified (and the most common ALK chimera found in ALCLs), NPM–ALK, as well as several of the so-called ‘variant’ ALK fusion proteins (i.e., ALK fusions other than NPM–ALK), are shown in Figure 1B
Except for MSN–ALK and nonmuscle myosin heavy chain–ALK (which differ only slightly from all other ALK fusions with respect to the portion of ALK incorporated into them), all chimeric ALK proteins contain the same region of the protein – aa 1058–1620 of the full-length normal receptor – that comprises the complete intracellular segment, including the TK catalytic domain. All ALK fusion proteins share two critical features:
An N-terminal partner protein that is widely expressed in normal cells, the gene promoter of which controls the aberrant transcription of the ALK chimeric mRNA and the expression of its encoded fusion protein;
The presence of an oligomerization domain of some type in the sequence of the ALK fusion partner, which mediates constitutive self association of the ALK fusion and, in turn, causes constant ALK KD activation [111,112].
Self association of ALK fusion proteins via these domains mimics the ligand-mediated aggregation and activation of the full-length wild-type ALK receptor; the mechanism by which ALK kinase activity is normally controlled. However, because ALK chimeric proteins are always homo-oligomerized, they experience constitutive activation and, therefore, continuously transmit growth-promoting cellular signals [111,113-119]. The constitutive activation of the ALK KD in these fusion proteins leads to an additional shared feature, the phosphorylation and aberrant activation of multiple downstream substrates and signaling cascades involved in the neoplastic transformation of cells. The description and details of these pathways are beyond the scope of this review; rather, we refer interested readers to other recent reviews concerning ALK as well as selected reports that describe primary data concerning novel ALK substrates and signaling mechanisms [24,120-133].
ALK fusion proteins in hematopoietic tumors
Anaplastic large-cell lymphoma
To date, within the various types of hematopoietic malignancies, chimeric ALK proteins have been reported only in lymphomas, and not in preleukemias, acute or chronic leukemias or myeloproliferative disorders. The cloning of the NPM–ALK fusion generated by the t(2;5)(p23;q35) chromosomal rearrangement permitted the facile diagnosis of NPM–ALK-expressing ALCLs using methodologies such as FISH and reverse transcriptase (RT)-PCR assays; furthermore, given that normal hematopoietic cells do not express detectable levels of the normal full-length ALK protein, the use of ALK-specific antibodies has helped to revolutionize the diagnosis of ALCL (because of the often bizarre and heterogeneous morphology of ALCLs, these lymphomas had previously often been quite difficult to diagnose unequivocally) [120,134]. Using immunostaining and other methods, 60–80% of ALCLs have been found to be ALK-positive. ALK-positive lymphoma cells express the cell surface-marker protein CD30 and exhibit a cytotoxic T-cell or null (i.e., lacking expression of both T- and B-cell markers) phenotype [135-140].
The detection of ALK in NHLs has resulted in the designation of the entity ‘ALK-positive lymphoma’ [135]; often informally referred to as ‘ALKomas’, this entity is now officially classified as ALK-positive ALCL in the WHO’s classification of NHL [141]. These tumors most frequently occur in children and young adults and have a slight male predominance. ALK-positive ALCLs are highly aggressive tumors but tend to have a generally good prognosis following treatment with cyclophosphamide, doxorubicin, vincristine and prednisone (CHOP)-based combination chemotherapy regimens [142-145].
The most common ALK fusion protein in NHL is NPM–ALK, being found in approximately 75–80% of all ALK-positive ALCLs [120]. Since the NPM–ALK protein has a characteristic nuclear and cytoplasmic subcellular localization, the absence of nuclear ALK labeling in an ALK-positive ALCL indicates the expression of one of the variant ALK fusions in the tumor cells [134,136,137]. ALK fusion proteins not only self-associate, they also hetero-associate with the normal counterpart of their partner proteins; consequently, the localization of ALK fusions in tumor cells is determined largely by the normal subcellular distribution of their partner proteins. For example, CLTC–ALK (which is found in ALCL, diffuse large B-cell lymphoma (DLBCL) and inflammatory myofibroblastic tumors, vide infra) exhibits a granular cytoplasmic staining due to its hetero-association with the normal clathrin protein present in the coated vesicles; while Ran binding protein 2-ALK (which occurs in inflammatory myofibroblastic tumors) demonstrates nuclear membrane-associated staining due to its oligomerization with Ran-binding protein 2 (also known as NUP358), a nuclear pore protein. All ALK-positive ALCLs – whether positive for NPM–ALK or for one of the variant ALK fusions – have similar good treatment outcomes (especially in comparison to ALK-negative ALCL), with approximately 80% of children and 50–60% of adults with ALK-positive ALCL experiencing cures following therapy with CHOP-based chemotherapy [139,142-145].
Anaplastic lymphoma kinase-negative ALCLs possessing either a null or T-cell phenotype exist and are subdivided into primary cutaneous ALCL and primary systemic ALK-negative ALCL. Approximately 20% of the primary systemic ALCLs that occur in children and adolescents, and 50–60% of these cases in adults, are ALK negative. Primary systemic ALK-negative lymphomas – the molecular genetic cause(s) of which remain unknown – are histologically indistinguishable from primary systemic ALK-positive ALCLs, with ALK expression being the sole identifier capable of discriminating between them. The correct diagnosis of primary cutaneous ALCL is extremely important, given that such tumors, although ALK-negative, are responsive to treatment and have a markedly better outcome compared with primary systemic ALK-negative ALCL [145-148]. Likewise, it is critical that the diagnosis of primary systemic ALK-negative ALCL is made without equivocation; CHOP-based therapies that cure 60–80% of primary systemic ALK-positive ALCL patients result in the cure of only approximately 40% of primary systemic ALK-negative ALCL patients [138,145]. A hope for the future is that primary systemic ALK-positive ALCL patients will be treated with improved efficacy, using new, less toxic (compared with CHOP) therapies that incorporate an ALK inhibitor(s) and, in addition, that novel therapies, more effective than CHOP, can be identified for patients with primary systemic ALK-negative ALCL to improve the prognosis for these cases.
ALK-positive diffuse large B-cell lymphoma
The expression of ALK fusion proteins in lymphoma had been thought to be restricted to ALK-positive ALCL; however, in 2003, five groups of investigators independently described the expression of either CLTC–ALK or NPM–ALK in a rare form of B-cell NHL [149-153]. This subset of B-cell lymphoma is comprised of poor-prognosis tumors, with an inferior clinical outcome following therapy compared with typical ALK-positive ALCLs. Although epithelial membrane antigen-positive, as with most ALK-positive ALCLs, these B-cell NHLs lack CD30 (by contrast, typical ALK-positive ALCLs invariably express CD30) and, consistent with their B-cell immunophenotype, they express intracytoplasmic IgA as well as the plasma cell-associated marker p63. The tumor cells in these aggressive lymphomas share the same plasmablastic morphology (monomorphic large immunoblasts) and immunophenotype as the B-cell lymphomas that had been previously described by Delsol et al. to express the full-length ALK receptor [109]. In fact, one of the cases included in the 2003 study from Gascoyne et al. [149] was also described in the original paper by Delsol et al. in 1997 [109]. FISH and RT-PCR studies revealed the presence of t(2;17)(p23;q23) and CLTC–ALK mRNA in the neoplastic cells of this case, while immunocytochemical labeling studies confirmed a granular ALK-staining pattern in the tumor cells that was essentially identical to that observed in cases of CLTC–ALK-positive ALCL. In contrast to Gascoyne et al., De Paepe and colleagues found no evidence of mRNA encoding for the ALK extracellular domain in the tumors they examined [151]. Although no results regarding the activation status of the CLTC–ALK fusion protein in these B-cell tumors were provided in these studies, the presence of a self-association motif in the CLTC portion of CLTC–ALK predicts the constitutive oligomerization and activation of the fusion kinase, thus driving lymphomagenesis. In another study, Onciu et al. reported the t(2;5)(p23:q35) and NPM–ALK mRNA in cases of B-cell lymphoma that were more or less phenotypically identical to the CLTC–ALK-positive B-cell NHLs described by others [152]. Additional ALK-positive DLBCLs have also been subsequently reported, and the reports taken as a whole suggest that these cases comprise less than 0.5–1% of DLBCL patients [154]. Regardless of the relative contributions of full-length normal ALK (when expressed in these cases) versus the CLTC–ALK or NPM–ALK fusions in the development of these lymphomas, ALK-targeted therapy is likely to benefit ALK-positive DLBCL patients, who unfortunately tend to have poor outcomes following management with currently available therapies.
ALK-positive systemic histiocytosis
Also, in 2008, ALK fusions were described in another hematopoietic neoplasm, systemic histiocytosis [155]. Three cases of this previously uncharacterized form of histiocytosis, which presents in early infancy, exhibited ALK immunoreactivity and one case characterized expressed TPM3–ALK. These infants present with massive hepatosplenomegaly, anemia and thrombocytopenia and, in some cases, life-threatening complications during the acute disease phase. Two of the three patients were treated with dexamethasone and etoposide; all three experienced slow disease resolution over months (including the patient who received no specific therapy) and were alive at follow-up. The ALK-transformed cell of origin is not clear but appears to be of either macrophage or dendritic cell lineage. It is not clear at this juncture whether ALK-positive systemic histiocytosis is truly a bona fide malignancy or rather represents an incompletely transformed, nonmalignant hyperproliferation of macrophages or dendritic cells, driven by activated ALK. Regardless, ALK small-molecule inhibitors would be predicted to benefit patients with this disease.
ALK fusion proteins in nonhematopoietic tumors
Inflammatory myofibroblastic tumor
In 1999, Griffin et al. identified ALK gene rearrangement and ALK protein expression in three cases of inflammatory myofibroblastic tumor (IMT) [156]. IMTs are rare, with best estimates suggesting approximately 150–200 new cases diagnosed in the USA annually. As spindle cell proliferations that involve malignant myofibroblasts and infiltrating nonmalignant inflammatory cells in a collagenous matrix, these neoplasms most often occur in the soft tissue and viscera of children and young adults [157,158]. IMTs are, not infrequently, difficult to distinguish from other benign or malignant spindle cell neoplasms or from a variety of inflammatory conditions; for example, entities with which IMTs can be confused include nodular fasciitis, gastrointestinal stromal tumors (GISTs) and fibromatosis [158]. Usually indolent and curable by complete surgical resection when possible, IMTs can, nonetheless, cause clinical management problems due to local recurrences following incomplete resection. Furthermore, approximately 10–15% of cases develop a more aggressive phenotype, including the occurrence of metastases. Therapies other than surgical resection (i.e., chemotherapy and radiotherapy) are minimally effective against IMTs.
Frequencies of ALK expression in IMT vary in the literature from 36 to 61.8% of cases [118,156-173]. The TPM3–ALK, TPM4–ALK, CLTC–ALK and ATIC–ALK fusion proteins – all of which have been reported in lymphoma – are now known to also occur in IMTs. Three other ALK fusions, CARS–ALK, RANBP2–ALK and SEC31L1–ALK, have been recently described in IMT but not, as yet, in NHL [118,120,160-163]. As with ALK-positive ALCL, ALK-positive IMTs tend to occur in younger patients. Whether the presence of ALK fusion proteins will prove to be a means of identifying prognostically-meaningful subtypes of IMT has not yet been fully examined; however, there is some suggestion that ALK expression may identify IMTs that have superior outcomes compared with ALK-negative IMT cases. For example, Chan et al. reported that patients with ALK-positive IMT were alive and well at follow-up, which was up to 17 years after diagnosis [166]. Coffin and colleagues described ALK reactivity to be associated with local recurrence but not distant metastasis, which was confined to ALK-negative lesions; furthermore, the lack of ALK expression was associated with a higher age overall and death from disease or distant metastases [171].
The detection of ALK expression is also becoming an approach to help pathologists discriminate IMT from some of its mesenchymal tumor and other mimics [89,158,159,170-173]. Small-molecule inhibitor therapy will be useful in those cases of ALK-positive IMTs in which local recurrences and/or distant metastases occur. In addition, short-term therapy with an ALK inhibitor could be employed to shrink unresectable IMTs, allowing their complete removal and surgical cure.
Esophageal squamous cell carcinoma
In late 2006, a report describing the expression of the TPM4–ALK fusion protein in squamous cell carcinomas (SCCs) of the esophagus in a population of Iranian patients appeared in the literature [174]. This manuscript described a proteomics approach that was used to identify proteins either under- or over-represented in esophageal SCCs; ALK – in the form of TPM4–ALK – was among those proteins over-represented. A total of 45 esophageal SCCs were examined in this study, but the number that were TPM4–ALK-positive was not noted. A second, independent proteomics-based profiling study of esophageal SCCs – in this case, from Chinese patients – that also reported TPM4–ALK expression in these cancers was published in 2007 [175]. In neither of these reports was the expression of TPM4–ALK independently confirmed using other methodologies (e.g., anti-ALK immunostaining, RT-PCR and/or FISH to identify the t(2;19)(p23;p13) chromosomal translocation that generates this ALK fusion gene), nor was the incidence of TPM4–ALK-expressing tumors carefully examined; additional studies will be required to address these issues. Survival rates at 5 years for esophageal SCC patients following best available therapies are only approximately 5–15%, with median survival being less than 1 year [176]. The constitutive kinase activity of the TPM4–ALK fusion would be expected to drive SCC growth and, conversely, inhibition of this activity could potentially be associated with substantial anti-tumor responses, perhaps similar to the robust responses observed for ALK fusion-expressing lymphomas in preclinical studies [120]. Therefore, the expression of activated forms of ALK could provide a compelling novel therapeutic target for esophageal SCC, an otherwise very treatment-refractory and extremely poor-prognosis tumor.
Non-small-cell lung carcinoma
The role of ALK fusions in nonhematopoietic tumors expanded yet further in 2007 with the description by Soda and colleagues of the expression of the novel EML4–ALK chimeric protein in five out of 75 (6.7%) Japanese patients with non-small-cell lung carcinoma (NSCLC) [177]; notably, the patient population harboring EML4–ALK was found to occur exclusive of those cases in this cohort that harbored activating mutations of either the EGF receptor (EGFR) or KRAS. Shortly thereafter, the existence of ALK fusions in lung cancer was independently corroborated in another 2007 manuscript from a different group of investigators who found six out of 137 (4.4%) Chinese lung cancer patients to express ALK fusions (EML4–ALK, three patients; TFG–ALK, one patient; X–ALK [unidentified ALK fusion partners], two patients) [178]. Follow-up studies in 2008 have confirmed the expression of ALK fusions in lung carcinomas. In a report of 221 Japanese lung cancer patients, five cases (2.3%) were found to be EML4–ALK-positive; interestingly, all five of these patients were from the cohort of 149 adenocarcinomas of the lung examined in this study (3.4% incidence) and none out of 72 lung cancers of other histologies (squamous cell, small-cell or neuroendocrine) were ALK-positive [179]. A study of 603 patients from two population-based NSCLC cohorts from the USA and Switzerland (containing 75% adenocarcinomas and 25% squamous cell carcinomas) revealed 16 (2.7%) to be ALK-positive; the ALK locus was also found to undergo genomic DNA amplification in three additional cases [180]. Shinmura et al. examined 77 NSCLCs(50 of which were adenocarcinomas) from Japanese patients for EML4–, NPM–, TPM3–, CLTC–, ATIC– and TFG–ALK fusion transcripts by RT-PCR and subsequent sequencing ana lysis, and found two adenocarcinomas (2.6% overall incidence; 4% incidence in adenocarcinomas) to express EML4–ALK fusions (none of the other ALK fusion transcripts were identified); no somatic mutations were detected in the mutation cluster regions of the EGFR, KRAS and PIK3CA genes in the two EML4–ALK-positive carcinomas [181]. Another 2008 study examined 104 lung carcinomas from Japanese patients for the EML4–ALK transcript using an RT-PCR method, and identified a single positive adenocarcinoma; 555 gastrointestinal (96 color-ectal, 96 gastric, 112 esophageal, 232 hepatic, 11 cholangiocellular and eight pancreatic) and 90 breast cancers were also examined specifically for expression of EML4–ALK in this report and none were found to be positive [182]. In addition, Koivunen et al. recently characterized 305 primary NSCLCs (138 from the USA and 167 from Korea), as well as 83 NSCLC cell lines, for the EML4–ALK fusion; eight of the 305 (2.6%) tumors and three of the 83 (3.6%) cell lines were found to be positive [183]. This study, as well as a separate report [184], also described the expression of novel isoforms of EML4–ALK in NSCLC that are subtly different from those originally described [177].
Collectively, at least two common themes seem to have emerged thus far from these studies of ALK pathogenesis in lung cancer:
ALK fusions appear to be restricted to patients with adenocarcinoma, mostly in patients with minimal or no smoking history;
ALK abnormalities seem to occur exclusive of other commonly observed genetic abnormalities such as EGFR and KRAS mutations.
In total, at the time of the writing of this review, 43 out of 1522 (2.8%) lung cancer patients (of all histologies) had been reported to be ALK-positive. An issue that has not yet been addressed in lung cancer (nor in any cancer other than neuroblastoma) concerns the possibility that ALK might also be aberrantly activated by mechanisms other than the generation of fusion forms of the kinase (e.g., by point mutations). Regardless of the exact mechanism of ALK activation, the fact that the signaling pathways by which ALK mediates oncogenic transformation are shared in large part with other TKs, such as the EGFR, suggests the likelihood for robust anti-tumor responses in many lung cancer patients treated with an ALK small-molecule inhibitor – for example, similar to the responses that have been observed in the clinic for lung cancer patients treated with EGFR small-molecule inhibitors, especially in patients whose tumors harbor activating EGFR mutations [185]. Preclinical data using an ALK small-molecule inhibitor in a recently described mouse model of EML4–ALK-expressing lung tumorigenesis, in which rapid disappearance of tumors and prolongation of survival were observed with inhibitor administration [186], also bode well for the likelihood of excellent anti-tumor responses in patients with ALK-positive lung cancers.
ALK small-molecule inhibitor development
TK inhibitors currently approved for cancer therapy
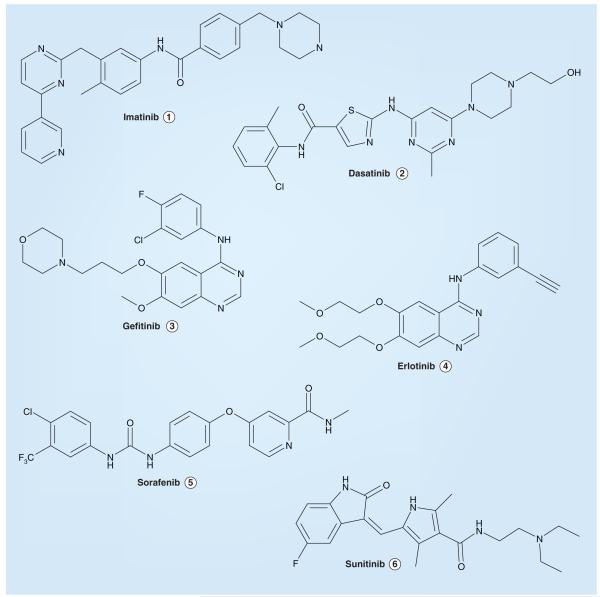
There are presently no small-molecule inhibitors of ALK approved for clinical use. However, substantial precedent exists for the development and clinical efficacy of small-molecule inhibitors of other TKs that are likewise involved in the genesis of cancer. Gleevec® (imatinib, Figure 2, STI571, Novartis) was the first member of the now ever-expanding group of clinically approved TK inhibitors. Imatinib binds tightly to the inactive form of the KD of ABL (Abelson proto-oncogene), and inhibits this kinase (in the form of the chimeric breakpoint cluster region (BCR)–ABL kinase in chronic myeloid leukemia [CML]), as well as c-KIT and the PDGF receptor [187-189]. Imatinib is used clinically in the treatment of CML, GIST and selected additional malignancies [190-192]. Continual and prolonged exposure to imatinib is necessary for optimal therapeutic responses in CML and, as a consequence, ABL mutations and other mechanisms arise that lead to imatinib resistance in a substantial portion of patients [193,194].
Figure 2.
Selected tyrosine kinase inhibitors approved for clinical anticancer indications.
To address the issue of imatinib resistance and intolerance, second-generation BCR–ABL inhibitors such as Sprycel® (dasatinib, BMS-354825, Bristol–Myers Squibb; Figure 2) have been recently developed and approved for clinical use [195]. Dasatinib inhibits both the inactive and active forms of BCR–ABL, as well as SRC family TKs [196,197]. Other novel BCR–ABL inhibitors are also in various stages of development; for example, in addition to dasatinib, the phenylaminopyrimidine derivative Tasigna® (nilotinib, not shown, AMN107, Novartis) was recently approved for the therapy of imatinib-resistant CML [198,199].
Both Iressa® (gefitinib, ZD1839, AstraZeneca; Figure 2) and Tarceva® (erlotinib, OSI-774, OSI Pharmaceuticals/Genentech/Roche; Figure 2) are ATP-competitive inhibitors that target the EGFR TK [200-202], which is overexpressed or mutated in a variety of human cancers. Both of these EGFR small-molecule inhibitors lead to substantial clinical anti-tumor responses in certain patients; for example, approximately 10–30% of NSCLC patients demonstrate very good responses to these agents – the frequency depending on the exact patient population examined and reflecting for the most part the variable incidence of inhibitor-sensitive activating point mutations of EGFR that drive tumor growth and tend to occur more commonly in East Asians, women, nonsmokers and patients with adenocarcinoma histology [203]. However, based on the lack of a survival benefit of gefitinib therapy compared with placebo in a pivotal Phase III trial of advanced recurrent NSCLC, in the USA, the drug is now accessible only to patients who had shown benefit from it prior to the restriction of its availability. As a result, erlotinib, which has demonstrated evidence of survival benefit in advanced NSCLC, has largely supplanted gefitinib in the clinic for this indication; erlotinib is also approved in combination with gemcitabine for the treatment of locally advanced, unresectable, or metastatic pancreatic cancer [204,205].
Nexavar® (sorafenib, BAY43–9006, Bayer/Onyx; Figure 2) and Sutent® (sunitinib, SU11248, Pfizer; Figure 2) are both multi-targeted kinase inhibitors. While sorafenib inhibits the RAF serine/threonine kinase, as well as the PDGF receptor, c-KIT and VEGF receptor (VEGFR)-2 and -3 TKs, sunitinib blocks all of the VEGFRs (VEGFR-1, -2 and -3) and the c-KIT, FLT3, CSF1R and RET TKs [206-208]. Both drugs have been approved by the US FDA as treatment options for advanced renal cell carcinoma; sorafenib is also approved for the treatment of advanced hepatocellular carcinoma, while sunitinib is approved also for the treatment of GIST, especially tumors that are imatinib resistant [207-209].
Given the proven success of kinase inhibitor drug development to date and the involvement of mutant forms of ALK in a variety of cancer types, targeting ALK in these malignancies is a logical therapeutic approach for which substantial and compelling target validation now exists [120]. One recently reported example of this validation is represented by the work of McDermott et al., who tested various kinase inhibitors against a panel of 602 cell lines derived from a variety of human cancer types and found that a selective ALK inhibitor (NVP-TAE684, vide infra) suppressed the growth of a subset of the tumor lines examined, including cell lines derived from anaplastic large-cell lymphomas, NSCLCs and neuroblastomas – cancers all known to harbor genomic ALK alterations, as noted herein [105,210].
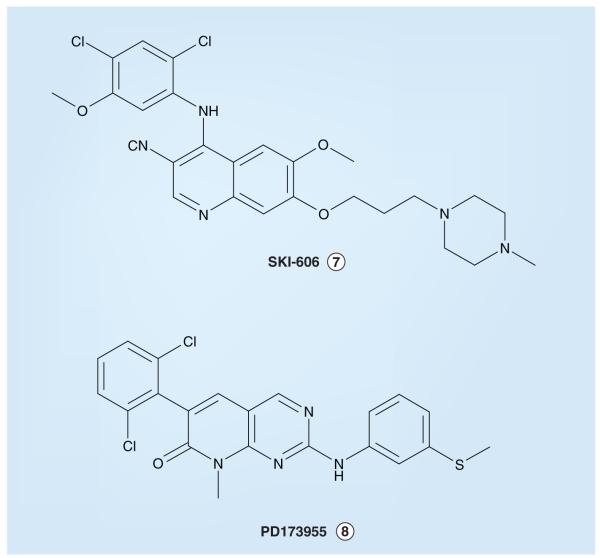
The development of ALK small-molecule inhibitors has been somewhat hampered since an X-ray crystal structure of the catalytic domain of the kinase has yet to be reported. Nevertheless, some useful data with respect to the ALK KD have been communicated. For example, Gunby et al. reported characterization of a leucine gatekeeper residue (L1196 of the full-length normal ALK receptor, which is equivalent to L256 in the NPM–ALK oncogenic fusion) that sterically impedes the entrance of small-molecule inhibitors into the ATP-binding site of the activated KD [211]. This observation was confirmed via site-specific mutation studies; while wild-type NPM–ALK was insensitive to several ABL inhibitors, mutated NPM–ALK proteins (e.g., an L256T-NPM–ALK mutant protein in which a smaller threonine residue was substituted for the larger leucine at position 256) were relatively sensitive to SKI-606 and PD173955 (Figure 3) at lower concentrations (IC50 values of 150 nM and 2.5 μM, respectively) but remained insensitive to imatinib (IC50 > 100 μM).
Figure 3.
Small molecules shown experimentally to inhibit a ‘gatekeeper residue-mutant’ form of anaplastic lymphoma kinase (ALK; L256T-NPM–ALK) but not wild-type ALK.
Naturally occurring inhibitors of ALK & their derivatives
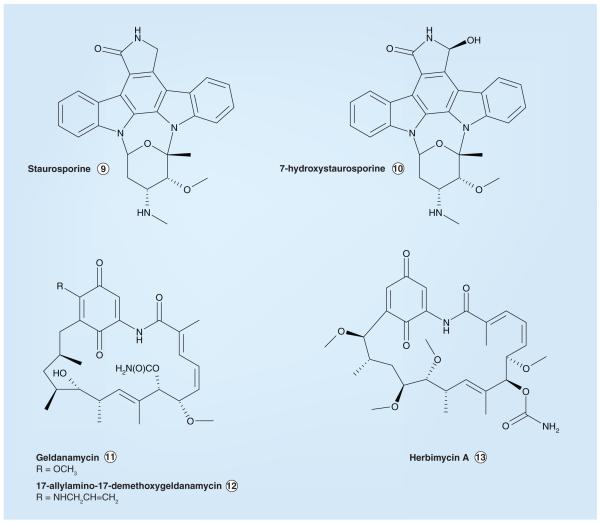
There are no naturally occurring selective inhibitors of ALK. However, two structurally related natural product compounds have been reported to inhibit ALK (as well as other kinases); staurosporine and 7-hydroxystaurosporine (Figure 4) possess IC50 values of 150 nM and 5 μM, respectively, in the presence of 30 μM ATP in an ELISA-based ALK assay [212]. Both staurosporine and 7-hydroxystaurosporine inhibit a number of the kinases within the human kinome [213,214]. Three additional naturally occurring or derivative compounds that affect ALK activity are geldanamycin, 17-allylamino-17-demethoxygeldanamycin and herbimycin A (Figure 4). However, unlike the other small-molecule ALK inhibitors described in this review (which compete with ATP for binding to the ALK KD), these three structurally related compounds inhibit ALK differently – by increasing the proteasome-mediated degradation of the ALK protein [215-217]. Cephalon (Frazer, PA) has synthesized staurosporine-based compounds that exhibit enzymatic ALK IC50 values less than 5 nM and cell-based ALK-inhibitory activity with IC50s <30 nM (CEP-14083 and CEP-14513, Figure 5); however, these compounds have unfavorable physical properties that preclude their use in vivo [218,219,401].
Figure 4.
Naturally occurring inhibitors of anaplastic lymphoma kinase and selected derivative compounds.
Figure 5. Staurosporine-based anaplastic lymphoma kinase (ALK) inhibitors from Cephalon.
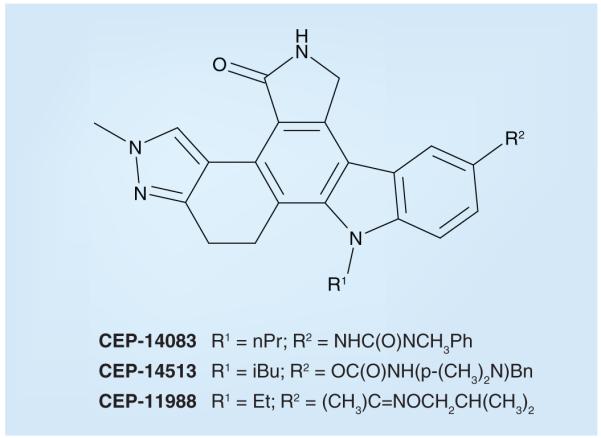
In contrast to CEP-14083 and CEP-14513, both of which are potent ALK inhibitors (enzymatic IC50 2–4 nM; cellular IC50 10–30 nM for both), the structurally related CEP-11988 displays only weak ALK inhibition (enzymatic IC50 > 20 μM; IC50 for inhibition of cell-based ALK tyrosine phosphorylation > 30 μM). From [221].
Synthetic inhibitors of ALK inspired by homology modeling
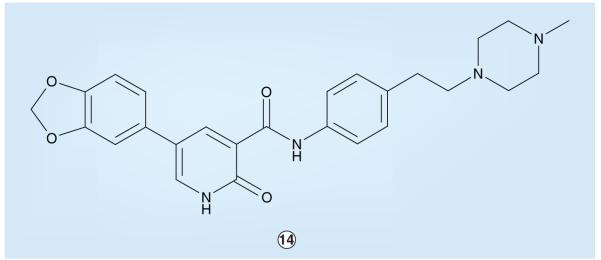
Work between ChemBridge Research Laboratories and St Jude Children’s Research Hospital established that pyridone-based compounds can serve as potent inhibitors of ALK [220,402,403]. For example, based on the screening of a computationally focused small-molecule library and initial chemistry-based optimization, pyridone 14 (Figure 6) was found to inhibit ALK with an enzymatic IC50 of 380 nM. Pyridone 14 displayed selective inhibition within the IR superfamily; the enzymatic IC50 value for the IRK was 3.6 μM and the IGF1R was inhibited to an even lesser extent [220]. In cellular assays, compound 14 possessed nonselective anti proliferative properties with IC50 values in the 4.5–18 μM range. Further optimization and experiments with this particular pyridone series have not yet been reported.
Figure 6.
ChemBridge–St Jude pyridone-based anaplastic lymphoma kinase inhibitor.
Another ChemBridge–St Jude compound that displays ALK-inhibitory activity is pyridone 15 (Figure 7). Compound 15 was discovered while targeting various oncogenic TKs including the ALK-homologous IGF1R [301]. Pyridone 15 demonstrates reasonably potent ALK inhibition, with an IC50 of less than 0.5 μM in enzyme assays. However, this pyridone was nonselective and exhibited only slightly less potent inhibition of other kinases of the IR superfamily (e.g., the IR and IGF1R); furthermore, although it displayed cellular antiproliferative activity, pyridone 15-associated cytotoxicity was also nonselective, the activity against nonmalignant cells (e.g., BaF3) being of similar magnitude to cancer cell lines transformed by NPM–ALK (e.g., Karpas-299) or other activated kinases such as BCR–ABL (e.g., K562), with IC50 values between 0.5 and 5.0 μM for all cell lines tested. Despite these liabilities, pyridone 15 could represent a chemotype amenable to further optimization for ALK-inhibitory properties.
Figure 7.
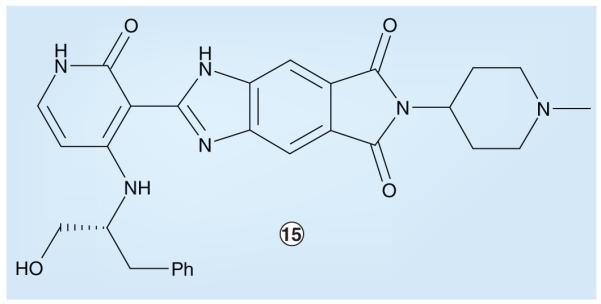
ChemBridge–St Jude IGF1 receptor-targeted inhibitor with anaplastic lymphoma kinase-inhibitory activity.
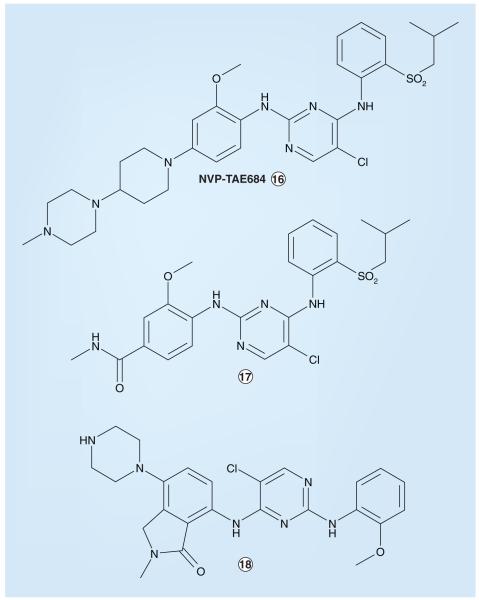
Investigators affiliated with the Genomics Institute of the Novartis Research Foundation (San Diego, CA, USA) identified an ALK inhibitor, NVP-TAE684 (Figure 8), using a cellular screen of a kinase-directed small-molecule library to search for compounds that were selectively cytotoxic to cells oncogenically transformed by NPM–ALK [221,404]. While the identification of this inhibitor did not involve homology modeling per se, these investigators were able to provide a possible explanation regarding the selectivity of NVP-TAE684 for ALK relative to other kinases by the virtual docking of the small molecule into an ALK homology model based on the published crystal structure of the IR KD in an ‘active’ (‘DFG-in’) confirmation. This analysis revealed the orthomethoxy group attached to the 2-aniline substituent to project into a small groove located between the side chains of residues L258 and M259 of the NPM–ALK KD; molecular modeling showed that bulkier amino acids at the L258 position (as found in most other kinases at the analogous position in their KDs) would lead to steric clash with NVP-TAE684, suggesting that this leucine residue in ALK is a major determinant of selectivity for this particular inhibitor [221]. NVP-TAE684 has a cellular IC50 value of 3 nM and displays selective cytotoxicity against BaF3 cells engineered to express NPM–ALK; no effects on the survival of parental BaF3 cells were observed at concentrations up to 1 μM [221]. In other cellular cytotoxicity studies, the IC50 against Karpas-299 and SU-DHL-1 human anaplastic large-cell lymphoma cells was in the range 2–5 nM, while growth inhibition associated with NVP-TAE684 in non-ALK-dependent cell lines was substantially higher, ranging from 0.5 to 3 μM [221]. In addition to anaplastic large-cell lymphoma, NVP-TAE684 has been independently reported to impair the growth of other malignancies that harbor activated forms of ALK, including NSCLC and neuro blastoma [105]. Additional compounds from Novartis-associated investigators that have the same amino-pyrimidine core as NVP-TAE684 and somewhat less potent ALK-inhibitory activities (IC50 values between 0.01 and 1.0 μM) have also been reported in the patent literature (Figure 8) [302]. NVP-TAE684 is not currently being tested as a clinical agent [105]; the further preclinical/clinical development status of compounds 17, 18 or other ALK inhibitors of this chemotype has not been reported.
Figure 8.
Amino-pyrimidine anaplastic lymphoma kinase inhibitors from Novartis.
Synthetic inhibitors of ALK inspired by high-throughput screening
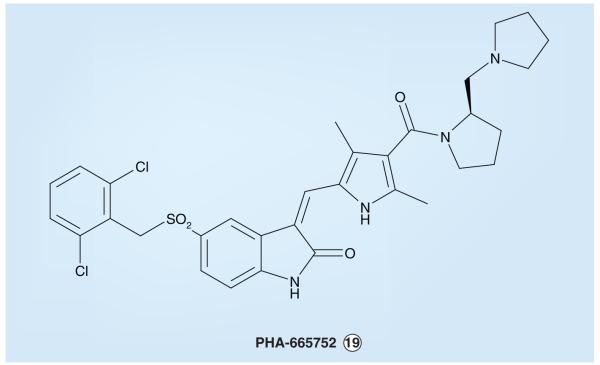
As part of its kinase inhibitor programs, Pfizer studied the small molecule PHA-665752 (Figure 9) as an inhibitor of MET, a member of the IR superfamily of TKs, as is ALK, that is also deregulated and thought to contribute to the genesis of various human cancers [222-225]. PHA-665752 has an IC50 of 9 nM in MET enzyme assays and displays a more than 50-fold selective MET inhibition compared with a large panel of other tyrosine, as well as serine/threonine kinases [222]. In vivo xenograft studies with PHA-665752 showed the inhibitor to reduce NCI-H69 (small-cell lung cancer) and NCI-H441 (NSCLC) tumor size in mice by 99 and 75%, respectively [223].
Figure 9.
PHA-665752, a MET (hepatocyte growth factor/scatter factor receptor)-selective inhibitor that has provided insight for anaplastic lymphoma kinase inhibitor design.
During the process of generating yet further optimized kinaseinhibitors, the dichloro compound PF-2341066 (Figure10) was synthesized by Pfizer scientists [226-228]. The 2,6-dihalogenated aryl lipophilic region, as seen in PHA-665752, is maintained in PF-2341066 but within the context of a compound possessing a lower overall molecular weight and lesser conformational strain. The hinge binder is changed from an oxindole in PHA-665752 to an amino-pyridine in PF-2341066. PF-2341066 exhibits good oral bioavail-ability and is a potent inhibitor not only of MET, but also ALK, with mean cellular IC50 values for inhibition of tyrosine phosphorylation of these kinases of 11 and 24 nM, respectively [226-228]. This dual ALK/MET inhibitor blocked the proliferation of NPM–ALK-positive ALCL cells (Karpas-299 and SU-DHL-1) with IC50 values of 32 and 43 nM, respectively, and showed a desirable selectivity profile; as an example, the IC50 observed for U-937, an acute myeloid leukemia cell line that expresses neither activated ALK nor MET proteins, was 257 nM [228]. As expected, PHA-665752, which has minimal affinity for the ALK KD (ALK IC50 > 10 μM) [228], failed to exhibit cytotoxicity similar to PF-2341066 in these NPM–ALK-positive ALCL cell lines. PF-2341066 has also been demonstrated to cause complete tumor regression when administered at an optimal dose in mice bearing Karpas-299 xenografts (Figure 11) [228]. PF-2341066 is currently nearing the completion of an initial Phase I clinical trial study.
Figure 10.
The Pfizer dual anaplastic lymphoma kinase/MET (hepatocyte growth factor/scatter factor receptor) inhibitor PF-2341066.
Figure 11. Dose-dependent anticancer activity of PF-2341066 in a Karpas-299 xenograft model.
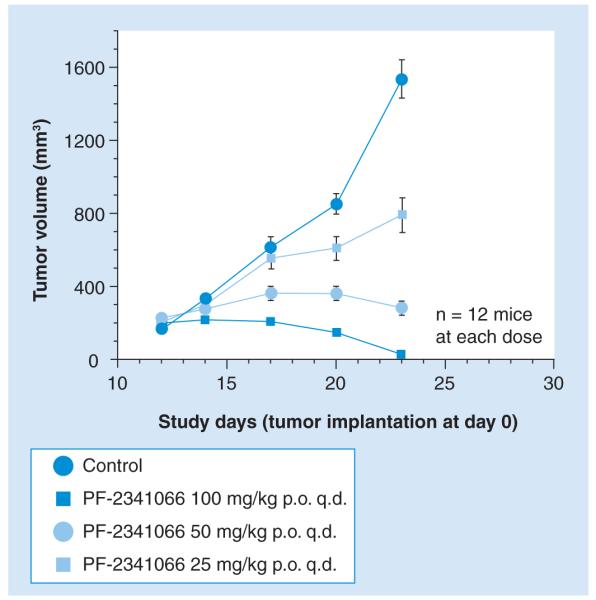
Immunocompromized mice bearing established (200-mm3) subcutaneous tumors of the human nucleolar protein nucleophosmin–anaplastic lymphoma kinase (ALK)-positive anaplastic large-cell lymphoma cell line Karpas-299 were treated with the Pfizer dual ALK/MET (hepatocyte growth factor/scatter factor receptor) inhibitor PF-2341066 at the indicated dosing regimens or with vehicle only. Treatments were administered from day 11 to day 23 except for the 100 mg/kg group, which received treatment through day 28. The mean tumor volumes at the time of the final measurements were significantly less in each of the three treatment groups compared with the control cohort (p < 0.001, one-way ANOVA). A subset of the mice treated at the 100 mg/kg/day dose of PF-2341066 (three mice total) experienced tumor regrowth after prolonged observation following the cessation of therapy; the tumors in these mice underwent complete regression again following a 13-day course of PF-2341066 100 mg/kg/day (not shown). p.o.: Orally; q.d.: Once daily. Modified with permission from [231].
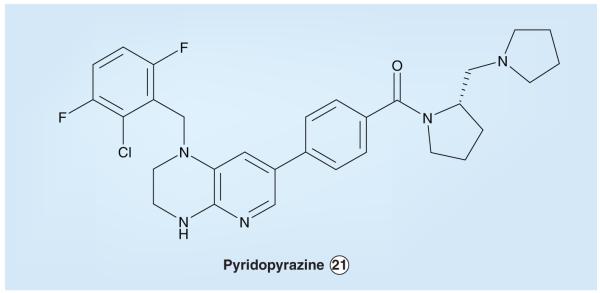
Scientists at Cephalon noted the promising activity reported for PF-2341066 and have developed a set of α-amino pyridinyl-based ALK inhibitors with one major difference – the Cephalon compounds have an additional constraint in the amino aryl region. The compound reported to have the best ALK-inhibitory activity is pyridopyrazine 21 (Figure 12), with an ALK IC50 value of 3 nM. Compound 21 also had activity against MET, the IC50 value being 650 nM [303]. Structure-activity relationship studies with this set of inhibitors revealed that ALK prefers halogenated aromatic substituents (even more preferably at the 2,6-aryl positions), and substituents at the benzyl position are tolerated without major loss of activity. However, activity is severely hampered when the aforementioned hydrocarbon linkage is replaced with a carbonyl linkage. In addition, substituents about the pyrazine ring and alkylation of pyrazine nitrogen results in reduced potency [229].
Figure 12.
Dual anaplastic lymphoma kinase/MET (hepatocyte growth factor/scatter factor receptor) inhibitor from Cephalon.
Synthetic inhibitors of ALK for which only partial characterization is currently available
There are two small-molecule ALK inhibitors of interest – CRL151104A and WZ-5-126 – for which only partial characterization (and no structural information) have been publically reported to date.
CRL151104A was developed by investigators from ChemBridge Research Laboratories and St Jude Children’s Research Hospital and is a third-generation pyridone compound ATP-competitive ALK inhibitor. Although the structure of this inhibitor has not been publically disclosed, a patent that describes the inhibitor chemotype was recently published [304]. CRL151104A has a distinct pattern of inhibition focused on ALK, with an enzymatic IC50value of 9.75 nM when measured at 100 μM ATP. In addition, when tested at 50 nM, CRL151104A showed little or no inhibition of ten structurally diverse serine/threonine kinases selected to represent the major branches of the entire serine/threonine kinome. Cellular cytotoxicity assays examining five target NPM–ALK-dependent human ALCL cell lines (Karpas-299, SU-DHL1, JB6, SUP-M2, UConnL2) and 12 unrelated control cell lines (HEK293, CHO-K1, MCF7, HepG2, H292, H520, H1581, H596, H522, CHAGO-K1, UMC-11 and SK-MES1) demonstrated very potent inhibition of all five NPM–ALK-expressing lymphoma lines (IC50s < 100 nM); by contrast, the lowest IC50s observed for the 12 non-ALK-dependent cell lines was approximately 2.5 μM, indicating a minimum 25-fold differential cytotoxicity. Aberrant forms of ALK harboring activating point mutations, such as F1174L and R1275Q that have been described in neuroblastoma [103,106-108], are sensitive to this ALK inhibitor. Western blot ana lysis of the tyrosine phosphorylation status of NPM–ALK immuno precipitated from various ALCL cell lines treated with CRL151104A at submicromolar concentrations has been described to result in complete inhibition of the kinase activity of ALK. As noted, the chemical synthesis and complete characterization of the biological responses to CRL151104A are yet to be reported in the biomedical research literature.
Another new ALK inhibitor, WZ-5-126, was recently noted by McDermott et al. to potently inhibit the growth of two NSCLC cell lines containing ALK gene rearrangements, NCI-H2228 and NCI-H3122 [105]. WZ-5-126 possesses an IC50 value against ALK of 3.4 nM, based on in vitro kinase assay profiling; this small-molecule also potently inhibits several other tyrosine and serine/threonine kinases in enzymatic assays, for example LTK (5.8 nM), ROS1 (8.2 nM), PTK2/FAK1 (9.2 nM), TNK1/CD38-negative kinase (14 nM), TNK2/ACK1 (18 nM), ULK1 (32 nM), DCAMKL1 (33 nM) and PTK2B/FAK2 (33 nM). WZ-5-126 demonstrates approximately 23-fold selectivity for ALK compared with the homologous IR kinase (enzymatic IR – 77 nM) [105]. To date, no data characterizing WZ-5-126 in in vivo tumor models have been reported; similarly, neither the structure nor synthesis of the compound have yet been described.
Conclusion & expert commentary
Various mutant forms of ALK are now well validated as the proximate cause of several human malignancies including both relatively uncommon cancers, such as ALCL and inflammatory myofibroblastic tumor, as well as subsets of common neoplasms, such as lung cancer. Small-molecule ALK inhibitor development is still at an early stage, with only a single agent (PF-2341066) having entered clinical trials to date. Industry interest in ALK as an anticancer target has, heretofore, been diminished due to the relatively small market that had been represented by those malignancies unequivocally known to be driven by abnormal ALK signaling; it is anticipated that the increasing number and types of cancers now being shown to be associated with aberrations of ALK will foster greater interest in inhibitor discovery and development.
As with ABL and EGFR small-molecule kinase inhibitors, it is quite likely that resistance to ALK inhibitors will also emerge in a portion of patients treated with these agents; such a likelihood should provide further impetus to develop more than one ALK inhibitor for clinical use – perhaps both additional ATP-competitive inhibitors as well as non-ATP-competitive, allosteric inhibitors – to fully optimize the management of responsive cancers. The availability of several inhibitors would also permit the testing of possible combination therapy as a means to potentially help prevent the development of resistance [230]. Based on the preceding experience with other kinase inhibitors, it is expected that patients whose tumors harbor genetic abnormalities that cause ALK activation (e.g., the truncation and creation of fused forms, activating point mutations, genomic DNA amplification and resultant overexpression) will be the most highly responsive to small-molecule inhibitor therapy; thus, the molecular definition of the ALK status of tumors should be considered as a criterion for evaluation and selection of patients for clinical trials that seek to assess the efficacy of these compounds.
Five-year view
The recognition during the past year that tumorigenic ALK fusions occur in common human cancers, such as esophageal and lung carcinomas, together with the recent discovery that ALK can also be oncogenically activated in the pediatric malignancy neuro blastoma by point mutations, will prompt investigators to perform comprehensive surveys of various cancers to assess the possibility that ALK could play a pathogenic role in them. While the results of such surveys cannot be predicted, it is likely that causative links to subsets of still more malignancies will be made with genetic abnormalities of ALK and/or that alternative mechanisms of ALK activation will be discovered in cancers already known to express mutant forms of ALK (e.g., activating point mutations of ALK may be identified in lung cancers, in addition to the recently described ALK fusions).
Interest in ALK small-molecule inhibitor development has only recently heightened, due largely to the expanding number of malignancies in which aberrant ALK activity is being implicated. It is expected that an increasing number of inhibitors will be at various stages of development during the next 5 years. Given the very marked anti-tumor responses and minimal toxicities of the ALK inhibitors characterized to date in preclinical and clinical studies, it is also highly likely that the clinical development path for such agents will be quite rapid; as such, at least one ALK small-molecule inhibitor should be approved for clinical use within the coming 5-year timeline.
As has been the case with ATP-competitive inhibitors targeting other oncogenic kinases, resistance mutations of ALK will almost certainly emerge following continuous clinical exposure to any one inhibitor (prolonged inhibitor administration will likely be required to optimally treat solid tumors such as ALK-positive esophageal and lung carcinomas, for example), necessitating the clinical development of second and subsequent generation inhibitors. It is also quite probable that small-molecule inhibitors will vary in their ability to abrogate ALK activation due to different oncogenic point mutations within the ALK KD, thus providing additional incentive for the clinical development of more than one inhibitor.
Key issues.
The anaplastic lymphoma kinase (ALK) receptor tyrosine kinase is a single-chain member of the insulin receptor kinase superfamily that is expressed normally in the central and peripheral nervous sytems.
Among other possible neural functions, ALK may play a physiologic role in aspects of normal behavior and cognition. As such, ALK could potentially be a therapeutic target of utility for the management of certain neurobehavioral and psychiatric disorders.
Oncogenic fusion forms of ALK – two of the most common being nucleolar protein nucleophosmin (NPM)–ALK (which occurs mainly in anaplastic large-cell lymphomas) and EML4–ALK (in lung cancers) – are of pathogenic importance in subsets of anaplastic large-cell lymphomas, diffuse large B-cell lymphomas, systemic histiocytosis, inflammatory myofibroblastic tumors, non-small-cell lung carcinomas and, possibly, esophageal squamous cell cancers.
Oncogenic point mutations as well as genomic DNA amplification and overexpression of ALK have recently been discovered in the pediatric malignancy neuroblastoma, raising the possibility that still more human tumors may harbor such pathogenic cancer-associated ALK genetic abnormalities.
Although yet to be fully validated, ALK may also contribute to the genesis of other human tumors (e.g., glioblastoma) via autocrine and/or paracrine growth-promoting loops involving the putative ALK ligands pleiotrophin and midkine.
Preclinical and early clinical experience with ALK small-molecule inhibitor compounds has demonstrated extremely robust anti-tumor efficacy together with little or no target-associated toxicity.
Efforts to develop ALK small-molecule inhibitors are currently still at a relatively early stage, with only one such inhibitor (PF-2341066) in the clinic and nearing the completion of a Phase I trial.
ALK represents a highly attractive and tractable target for small-molecule inhibitor-based anticancer drug development.
Acknowledgments
Financial & competing interests disclosure Thomas R Webb is a coinventor on patents that describe the synthesis and characterization of anaplastic lymphoma kinase (ALK) small-molecule inhibitors designed by ChemBridge Research Laboratories. A Thomas Look and Stephan W Morris are coinventors on patents concerned with the discovery of ALK, the use of methods to detect ALK for diagnostic purposes, and the targeting of ALK for therapeutic indications. Studies cited in this review performed in the authors’ laboratories were supported in part by NIH grants R01 CA69129 (Stephan Morris) and Cancer Center CORE grant CA21765, and by the American Lebanese Syrian Associated Charities, St Jude Children’s Research Hospital.
Footnotes
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Thomas R Webb, Department of Chemical Biology and Therapeutics, St Jude Children’s Research Hospital, 332 North Lauderdale Street, Mail Stop 1000, Memphis, TN 38105, USA.
Jake Slavish, Department of Chemical Biology and Therapeutics, St Jude Children’s Research Hospital, 332 North Lauderdale Street, Mail Stop 1000, Memphis, TN 38105, USA.
Rani E George, Department of Pediatric Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Dana 640E, 44 Binney Street, Boston, MA 02115, USA.
A Thomas Look, Department of Pediatric Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Dana 640E, 44 Binney Street, Boston, MA 02115, USA.
Liquan Xue, Departments of Pathology and Oncology, St Jude Children’s Research Hospital, 332 North Lauderdale Street, Mail Stop 343, Memphis, TN 38105, UA.
Qin Jiang, Departments of Pathology and Oncology, St Jude Children’s Research Hospital, 332 North Lauderdale Street, Mail Stop 343, Memphis, TN 38105, USA.
Xiaoli Cui, Departments of Pathology and Oncology, St Jude Children’s Research Hospital, 332 North Lauderdale Street, Mail Stop 343, Memphis, TN 38105, USA.
Walter B Rentrop, Departments of Pathology and Oncology, St Jude Children’s Research Hospital, 332 North Lauderdale Street, Mail Stop 343, Memphis, TN 38105, USA.
Stephan W Morris, Departments of Pathology and Oncology, St Jude Children’s Research Hospital, Thomas Tower – Room 4026, Mail Stop 343, 262 Danny Thomas Place, Memphis, TN 38105-3678, USA.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Schlessinger J. Cell signalling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Kolibaba KS, Druker BJ. Protein kinases and cancer. Biochim. Biophys. Acta. 1997;1333:F217–F248. doi: 10.1016/s0304-419x(97)00022-x. [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 4.Scheijen B, Griffin JD. Tyrosine kinase oncogenes in normal hematopoiesis and hematological disease. Oncogene. 2002;21:3314–3333. doi: 10.1038/sj.onc.1205317. [DOI] [PubMed] [Google Scholar]
- 5.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112.Describes the initial cloning and characterization of nucleolar protein nucleophosmin (NPM)–anaplastic lymphoma kinase (ALK).
- 6.Shiota M, Fujimoto J, Semba T, Satoh H, Yamamoto T, Mori S. Hyperphosphorylation of a novel 80 kDa protein tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574.Describes the initial cloning and characterization of NPM–ALK.
- 7.Mathew P, Morris SW, Kane JR, et al. Localization of the murine homolog of the anaplastic lymphoma kinase (Alk) gene on mouse chromosome 17. Cytogenet. Cell. Genet. 1995;70:143–144. doi: 10.1159/000134080. [DOI] [PubMed] [Google Scholar]
- 8.Pulford K, Mason DY. CD246 Antigen. Leucocyte Typing VII. In: Mason DY, et al., editors. White Cell Differentiation Antigens. Oxford University Press; Oxford, UK: 2002. [Google Scholar]
- 9.Kaneko Y, Frizzera G, Edamura S, et al. A novel translocation, t(2;5)(p23;q35), in childhood phagocytic large T-cell lymphoma mimicking malignant histiocytosis. Blood. 1989;73:806–813. [PubMed] [Google Scholar]
- 10.Le Beau MM, Bitter MA, Larson RA, et al. The t(2;5)(p23;q35): a recurring chromoso mal abnormality in Ki-1-positive anaplastic large cell lymphoma. Leukemia. 1989;3:866–870. [PubMed] [Google Scholar]
- 11.Rimokh R, Magaud JP, Berger F, et al. A translocation involving a specific breakpoint (q35) on chromosome 5 is characteristic of anaplastic large cell lymphoma (‘Ki-1 lymphoma’) Br. J. Haematol. 1989;71:31–36. doi: 10.1111/j.1365-2141.1989.tb06270.x. [DOI] [PubMed] [Google Scholar]
- 12.Bitter MA, Franklin WA, Larson RA, et al. Morphology in Ki-1(CD30)-positive non-Hodgkin’s lymphoma is correlated with clinical features and the presence of a unique chromosomal abnormality, t(2;5) (p23;q35) Am. J. Surg. Pathol. 1990;14:305–316. doi: 10.1097/00000478-199004000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Mason DY, Bastard C, Rimokh R, et al. CD30-positive large cell lymphomas (‘Ki-1 lymphoma’) are associated with a chromosomal translocation involving 5q35. Br. J. Haematol. 1990;74:161–168. doi: 10.1111/j.1365-2141.1990.tb02560.x. [DOI] [PubMed] [Google Scholar]
- 14.Sandlund JT, Pui CH, Sandlund JT, et al. Clinicopathlogic features and treatment outcome of chidren with large cell lymphoma and the t(2;5)(p23;q35) Blood. 1994;84:2467–2471. [PubMed] [Google Scholar]
- 15.Kadin ME, Morris SW. The t(2;5) in human lymphomas. Leuk. Lymphoma. 1998;29:249–256. doi: 10.3109/10428199809068562. [DOI] [PubMed] [Google Scholar]
- 16.Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–449. doi: 10.1038/sj.onc.1200849.Describes the initial cloning and characterization of the full-length ALK receptor tyrosine kinase (RTK).
- 17.Morris SW, Naeve C, Mathew P, et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062.Describes the initial cloning and characterization of the full-length ALK RTK.
- 18.Loren CE, Scully A, Grabbe C, et al. Identification and characterization of DAlk: a novel Drosophila melanogaster RTK which drives ERK activation in vivo. Genes Cells. 2001;6:531–544. doi: 10.1046/j.1365-2443.2001.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulford K, Lamant L, Morris SW, et al. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)–ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. 1997;89:1394–1404. [PubMed] [Google Scholar]
- 20.Lamant L, Pulford K, Bischof D, et al. Expression of the ALK tyrosine kinase gene in neuroblastoma. Am. J. Pathol. 2000;156:1711–1721. doi: 10.1016/S0002-9440(10)65042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoica GE, Kuo A, Aigner A, et al. Identification of anaplastic lymphoma kinase as a receptor for the growth factor pleiotrophin. J. Biol. Chem. 2001;276:16772–16779. doi: 10.1074/jbc.M010660200.Describes pleiotrophin as an ALK ligand.
- 22.Stoica GE, Kuo A, Powers C, et al. Midkine binds to anaplastic lymphoma kinase (ALK) and acts as a growth factor for different cell types. J. Biol. Chem. 2002;277:35990–35999. doi: 10.1074/jbc.M205749200.Describes midkine as an ALK ligand.
- 23.Tartari CJ, Gunby RH, Coluccia AM, et al. Characterization of some molecular mechanisms governing autoactivation of the catalytic domain of the anaplastic lymhoma kinase. J. Biol. Chem. 2008;283:3743–3750. doi: 10.1074/jbc.M706067200. [DOI] [PubMed] [Google Scholar]
- 24.Degoutin J, Vigny M, Gouzi JY. ALK activation induces Shc and FRS2 recruitment: signaling and phenotypic outcomes in PC12 cells differentiation. FEBS Lett. 2007;581:727–734. doi: 10.1016/j.febslet.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 25.Turner SD, Yeung D, Hadfield K, Cook SJ, Alexander DR. The NPM–ALK tyrosine kinase mimics TCR signaling pathways, inducing NFAT and AP-1 by RAS-dependent mechanisms. Cell. Signal. 2007;19:740–747. doi: 10.1016/j.cellsig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Bai RY, Dieter P, Peschel C, Morris SW, Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-γ to mediate its mitogenicity. Mol. Cell. Biol. 1998;18:6951–6961. doi: 10.1128/mcb.18.12.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernersson E, Khoo NKS, Henriksson ML, Roos G, Palmer RH, Hallberg B. Characterization of the expression of the ALK receptor tyrosine kinase in mice. Gene Exp. Patterns. 2006;6:448–461. doi: 10.1016/j.modgep.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Falini B, Pileri S, Zinzani PL, et al. ALK+ lymphoma: clinico-pathological findings and outcome. Blood. 1999;93:2697–2706. [PubMed] [Google Scholar]
- 29.Englund C, Loren CE, Grabbe C, Deleuil F, Varshney GK, Palmer RH. Jeb signals via the DAlk receptor tyrosine kinase to drive visceral muscle fusion. Nature. 2003;425:512–516. doi: 10.1038/nature01950. [DOI] [PubMed] [Google Scholar]
- 30.Lee H-H, Norris A, Weiss JB, Frasch M. Drosophila jelly belly signals through the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature. 2003;425:507–512. doi: 10.1038/nature01916.The two preceding studies identify the Drosophila Alk ligand, jelly belly, and describe a role for the RTK in muscle development in the fly.
- 31.Loren CE, Englund C, Grabbe C, Hallberg B, Hunter T, Palmer RH. A crucial role for the anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO J. 2003;4:1–6. doi: 10.1038/sj.embor.embor897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- 33.Bazigou E, Apitz H, Johansson J, et al. Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell. 2007;128:961–975. doi: 10.1016/j.cell.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 34.Varshney GK, Palmer RH. The bHLH transcription factor Hand is regulated by Alk in the Drosophila embryonic gut. Biochem. Biophys. Res. Commun. 2006;351:839–846. doi: 10.1016/j.bbrc.2006.10.117. [DOI] [PubMed] [Google Scholar]
- 35.Bilsland JG, Wheeldon A, Mead A, et al. Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacology. 2008;33:685–700. doi: 10.1038/sj.npp.1301446. [DOI] [PubMed] [Google Scholar]
- 36.Monje ML, Toda H, Palmer TD. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 2003;302:1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- 37.Santarelli L, Saxe M, Gross C, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 38.Bruel-Jungerman E, Laroche S, Rampon C. New neurons in the dentate gyrus are involved in the expression of enhanced long-term memory following environmental enrichment. Eur. J. Neurosci. 2005;21:513–521. doi: 10.1111/j.1460-9568.2005.03875.x. [DOI] [PubMed] [Google Scholar]
- 39.Kunugi H, Hashimoto R, Okada T, et al. Possible association between nonsynonymous polymorphisms of the anaplastic lymphoma kinase (ALK) gene and schizophrenia in a Japanese population. J. Neural. Transm. 2006;113:1569–1573. doi: 10.1007/s00702-006-0436-3. [DOI] [PubMed] [Google Scholar]
- 40.Barbacid M. Structural and functional properties of the TRK family of neurotrophin receptors. Ann. NY Acad. Sci. 1995;766:442–458. doi: 10.1111/j.1749-6632.1995.tb26693.x. [DOI] [PubMed] [Google Scholar]
- 41.Li YS, Milner PG, Chauhan AK, et al. Cloning and expression of a developmentally regulated protein that induces mitogenic and neurite outgrowth activity. Science. 1990;250:1690–1694. doi: 10.1126/science.2270483. [DOI] [PubMed] [Google Scholar]
- 42.Fang W, Hartmann N, Chow DT, Riegel AT, Wellstein A. Pleiotrophin stimulates fibroblasts and endothelial and epithelial cells and is expressed in human cancer. J. Biol. Chem. 1992;267:25889–25897. [PubMed] [Google Scholar]
- 43.Wellstein A, Fang WJ, Khatri A, et al. A heparin-binding growth factor secreted from breast cancer cells homologous to a developmentally regulated cytokine. J. Biol. Chem. 1992;267:2582–2587. [PubMed] [Google Scholar]
- 44.Chauhan AK, Li YS, Deuel TF. Pleiotrophin transforms NIH-3T3 cells and induces tumors in nude mice. Proc. Natl Acad. Sci. USA. 1993;90:679–682. doi: 10.1073/pnas.90.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulte AM, Wellstein A. Pleiotrophin and related molecules. In: Bicknell R, Lewis CM, Ferrara N, editors. Tumour Angiogenesis. Oxford University Press; NY, USA: 1997. [Google Scholar]
- 46.Kadomatsu K, Huang RP, Suganuma T, Murata F, Muramatsu T. A retinoic acid responsive gene MK found in the teratocarcinoma system is expressed in a spatially and temporally controlled manner during mouse embryogenesis. J. Cell. Biol. 1990;110:607–616. doi: 10.1083/jcb.110.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadomatsu K, Muramatsu T. Midkine and pleiotrophin in neural development and cancer. Cancer Lett. 2004;204:127–143. doi: 10.1016/S0304-3835(03)00450-6. [DOI] [PubMed] [Google Scholar]
- 48.Ichihara-Tanaka K, Oohira A, Rumsby M, Muramatsu T. Neuroglycan C is a novel midkine receptor involved in process elongation of oligodendroglial precursor-like cells. J. Biol. Chem. 2006;281:30857–30864. doi: 10.1074/jbc.M602228200. [DOI] [PubMed] [Google Scholar]
- 49.Kojima S, Inui T, Muramatsu H, et al. Dimerization of midkine by tissue transglutaminase and its functional implication. J. Biol. Chem. 1997;272:9410–9416. doi: 10.1074/jbc.272.14.9410. [DOI] [PubMed] [Google Scholar]
- 50.Maeda N, Ichihara-Tanaka K, Kimura T, Kadomatsu K, Muramatsu T, Noda M. A receptor-like protein-tyrosine phosphatase PTPζ/RPTPβ binds a heparin-binding growth factor midkine. Involvement of arginine 78 or midkine in the high affinity binding to PTP-ζ. J. Biol. Chem. 1999;274:12474–12479. doi: 10.1074/jbc.274.18.12474. [DOI] [PubMed] [Google Scholar]
- 51.Matsubara S, Take M, Pedraza C, Muramatsu T. Mapping and characterization of a retinoic acid-responsive enhancer of midkine, a novel heparin-binding growth/differentiation factor with neurotrophic activity. J. Biochem. (Tokyo) 1994;115:1088–1096. doi: 10.1093/oxfordjournals.jbchem.a124462. [DOI] [PubMed] [Google Scholar]
- 52.Muramatsu T. Midkine and pleiotrophin: two related proteins involved in development, survival, inflammation and tumourigenesis. J. Biochem. (Tokyo) 2002;132:359–371. doi: 10.1093/oxfordjournals.jbchem.a003231. [DOI] [PubMed] [Google Scholar]
- 53.Muramatsu H, Zou K, Sakaguchi N, Ikematsu S, Sakuma S, Muramatsu T. LDL receptor-related protein as a component of the midkine receptor. Biochem. Biophys. Res. Commun. 2000;270:936–941. doi: 10.1006/bbrc.2000.2549. [DOI] [PubMed] [Google Scholar]
- 54.Muramatsu H, Zou P, Suzuki H, et al. α4β1- and αβ1-integrins are functional receptors for midkine, a heparin-binding growth factor. J. Cell. Sci. 2004;117:5404–5415. doi: 10.1242/jcs.01423. [DOI] [PubMed] [Google Scholar]
- 55.Obata Y, Kikuchi S, Lin Y, Yagyu K, Muramatsu T, Kumai H. Tokyo Research Group on Prevention of Gastric Cancer. Serum midkine concentrations and gastric cancer. Cancer Sci. 2005;96:54–56. doi: 10.1111/j.1349-7006.2005.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salama RH, Muramatsu H, Zou P, Okayama M, Muramatsu T. Midkine, a heparin-binding growth factor, produced by the host enhances metastasis of Lewis lung carcinoma. Cancer Lett. 2006;233:16–20. doi: 10.1016/j.canlet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 57.Zou P, Muramatsu H, Miyata T, Muramatsu T. Midkine, a heparin-binding growth factor, is expressed in neural precursor cells and promotes their growth. J. Neurochem. 2006;99:1470–1479. doi: 10.1111/j.1471-4159.2006.04138.x. [DOI] [PubMed] [Google Scholar]
- 58.O’Brien T, Cranston D, Fuggle S, Bicknell R, Harris AL. The angiogenic factor midkine is expressed in bladder cancer, and overexpression correlates with a poor outcome in patients with invasive cancers. Cancer Res. 1996;56:2515–2518. [PubMed] [Google Scholar]
- 59.Weiss JB, Suyama KL, Lee HH, Scott MP. Jelly belly: a Drosophila LDL receptor repeat-containing signal required for mesoderm migration and differentiation. Cell. 2001;107:387–398. doi: 10.1016/s0092-8674(01)00540-2. [DOI] [PubMed] [Google Scholar]
- 60.Englund C, Birve A, Falileeva L, Grabbe C, Palmer RH. Miple1 and miple2 encode a family of MK/PTN homologues in Drosophila melanogaster. Dev. Genes Evol. 2006;216:10–18. doi: 10.1007/s00427-005-0025-8. [DOI] [PubMed] [Google Scholar]
- 61.Bowden ET, Stoica GE, Wellstein A. Anti-apoptotic signaling of pleiotrophin through its receptor, anaplastic lymphoma kinase. J. Biol. Chem. 2002;277:35862–35868. doi: 10.1074/jbc.M203963200. [DOI] [PubMed] [Google Scholar]
- 62.Powers C, Aigner A, Stoica GE, McDonnell K, Wellstein A. Pleiotrophin signaling through anaplastic lymphoma kinase is rate-limiting for glioblastoma growth. J. Biol. Chem. 2002;277:14153–14158. doi: 10.1074/jbc.M112354200. [DOI] [PubMed] [Google Scholar]
- 63.Czubayko F, Schulte AM, Berchem GJ, Wellstein A. Melanoma angiogenesis and metastasis modulated by ribozyme targeting of the secreted growth factor pleiotrophin. Proc. Natl Acad. Sci. USA. 1996;93:14753–14758. doi: 10.1073/pnas.93.25.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schulte AM, Lai S, Kurtz A, Czubayko F, Riegel AT, Wellstein A. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl Acad. Sci. USA. 1996;93:14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choudhuri R, Zhang HT, Donnini S, Ziche M, Bicknell R. An angiogenic role for the neurokines midkine and pleiotrophin in tumorigenesis. Cancer Res. 1997;57:1814–1819. [PubMed] [Google Scholar]
- 66.Beecken WD, Kramer W, Joans D. New molecular mediators in tumor angiogenesis. J. Cell. Mol. Med. 2000;4:262–269. doi: 10.1111/j.1582-4934.2000.tb00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Christman KL, Fang O, Kim AJ, et al. Pleiotrophin induces formation of functional neovasculature in vivo. Biochem. Biophys. Res. Commun. 2005;332:1146–1152. doi: 10.1016/j.bbrc.2005.04.174. [DOI] [PubMed] [Google Scholar]
- 68.Mikelis C, Koutsioumpa M, Papadimitriou E. Pleiotrophin as a possible new target for angiogenesis-related diseases and cancer. Recent Patents Anticancer Drug Discov. 2007;2:175–186. doi: 10.2174/157489207780832405. [DOI] [PubMed] [Google Scholar]
- 69.Bernard-Pierrot I, Delbe J, Rouet V, et al. Dominant negative effectors of heparin affin regulatory peptide (HARP) angiogenic and transforming activities. J. Biol. Chem. 2002;277:32071–32077. doi: 10.1074/jbc.M202747200. [DOI] [PubMed] [Google Scholar]
- 70.Dirks WG, Fahnrich S, Lis Y, Becker E, MacLeod RA, Drexler HG. Expression and functional analysis of the anaplastic lymphoma kinase (ALK) gene in tumor cell lines. Int. J. Cancer. 2002;100:49–56. doi: 10.1002/ijc.10435. [DOI] [PubMed] [Google Scholar]
- 71.Moog-Lutz C, Degoutin J, Gouzi JY, et al. Activation and inhibiton of anaplastic lymphoma kinase receptor tyrosine kinase activity by monoclonal antibodies and absence of agonist activity of pleiotrophin. J. Biol. Chem. 2005;280:26039–26048. doi: 10.1074/jbc.M501972200. [DOI] [PubMed] [Google Scholar]
- 72.Mathivet T, Mazot P, Vigny M. In contrast to agonist monoclonal antibodies, both C-terminal truncated form and full length form of pleiotrophin failed to activate vertebrate ALK (anaplastic lymphoma kinase) Cell Signal. 2007;19:2434–2443. doi: 10.1016/j.cellsig.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 73.Meng K, Rodriguez-Pena A, Dimitrov T, et al. Pleiotrophin signals increased tyrosine phosphorylation of β-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase β/ζ. Proc. Natl Acad. Sci. USA. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maeda N, Nishiwaki T, Shintani T, Hamanaka H, Noda M. 6B4 proteoglycan/phosphacan, an extracellular variant of receptor-like protein-tyrosine phosphatase ζ/RPTPβ, binds pleiotrophin/heparin-binding growth-associated molecule (HB-GAM) J. Biol. Chem. 1996;271:21446–21452. doi: 10.1074/jbc.271.35.21446. [DOI] [PubMed] [Google Scholar]
- 75.Raulo E, Julkunen I, Merenmies J, Pihlaskari R, Rauvala H. Secretion and biological activities of heparin-binding growth-associated molecule. Neurite outgrowth-promoting and mitogenic actions of the recombinant and tissue-derived protein. J. Biol. Chem. 1992;267:11408–11416. [PubMed] [Google Scholar]
- 76.Raulo E, Chernousov MA, Carey DJ, Nolo R, Rauvala H. Isolation of a neuronal cell surface receptor of heparin binding growth-associated molecule (HB-GAM). Identification as N-syndecan (syndecan-3) J. Biol. Chem. 1994;269:12999–13004. [PubMed] [Google Scholar]
- 77.Hida H, Jung CG, Wu CZ, et al. Pleiotrophin exhibits a trophic effect on survival of dopaminergic neurons in vitro. Eur. J. Neurosci. 2003;17:2127–2134. doi: 10.1046/j.1460-9568.2003.02661.x. [DOI] [PubMed] [Google Scholar]
- 78.Furuta M, Shiraishi T, Okamoto H, Mineta T, Tabuchi K, Shiwa M. Identification of pleiotrophin in conditioned medium secreted from neural stem cells by SELDI-TOF and SELDI-tandem mass spectrometry. Brain Res. Dev. Brain Res. 2004;152:189–197. doi: 10.1016/j.devbrainres.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 79.Landgraf P, Wahle P, Pape HC, Gundelfinger ED, Kreutz MR. The survival-promoting peptide Y-P30 enhances binding of pleiotrophin to syndecan-2 and -3 and supports its neuritogenic activity. J. Biol. Chem. 2008;283:25036–25045. doi: 10.1074/jbc.M800963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu KV, Jong KA, Kim GY, et al. Differential induction of glioblastoma migration and growth by two forms of pleiotrophin. J. Biol. Chem. 2005;280:26953–26964. doi: 10.1074/jbc.M502614200. [DOI] [PubMed] [Google Scholar]
- 81.Perez-Pinera P, Zhang W, Chang Y, Vega JA, Deuel TF. Anaplastic lymphoma kinase is activated through the pleiotrophin/receptor protein-tyrosine phosphatase β/ζ signaling pathway: an alternative mechanism of receptor tyrosine kinase activation. J. Biol. Chem. 2007;282:28683–28690. doi: 10.1074/jbc.M704505200. [DOI] [PubMed] [Google Scholar]
- 82.Perez-Pinera P, Berenson JR, Deuel TF. Pleiotrophin, a multifunctional angiogenic factor: mechanisms and pathways in normal and pathological angiogenesis. Curr. Opin. Hematol. 2008;15:210–214. doi: 10.1097/MOH.0b013e3282fdc69e. [DOI] [PubMed] [Google Scholar]
- 83.Miyake I, Hakomori Y, Shinohara A, et al. Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene. 2002;21:5823–5834. doi: 10.1038/sj.onc.1205735. [DOI] [PubMed] [Google Scholar]
- 84.Osajima-Hakomori Y, Miyake I, Ohira M, Nakagawara A, Nakagawa A, Sakai R. Biological role of anaplastic lymphoma kinase in neuroblastoma. Am. J. Pathol. 2005;167:213–222. doi: 10.1016/S0002-9440(10)62966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyake I, Hakomori Y, Misu Y, et al. Domain-specific function of ShcC docking protein in neuroblastoma cells. Oncogene. 2005;24:3206–3215. doi: 10.1038/sj.onc.1208523. [DOI] [PubMed] [Google Scholar]
- 86.Shao CK, Su ZL, Feng ZY, Rao HL, Tang LY. Significance of ALK gene expression in neoplasms and normal tissues. Ai Zheng. 2002;21:58–62. [PubMed] [Google Scholar]
- 87.Grzelinski M, Bader N, Czubayko F, Aigner A. Ribozyme-targeting reveals the rate-limiting role of pleiotrophin in glioblastoma. Int. J. Cancer. 2005;117:942–951. doi: 10.1002/ijc.21276. [DOI] [PubMed] [Google Scholar]
- 88.Falini B, Bigerna B, Fizzotti M, et al. ALK expression defines a distinct group of T/null lymphomas with a wide morphological spectrum. Am. J. Pathol. 1998;153:875–886. doi: 10.1016/S0002-9440(10)65629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cessna MH, Zhou H, Sanger WG, et al. Expression of ALK1 and p80 in inflammatory myofibroblastic tumor and its mesenchymal mimics: a study of 135 cases. Mod. Pathol. 2002;15:931–938. doi: 10.1097/01.MP.0000026615.04130.1F. [DOI] [PubMed] [Google Scholar]
- 90.Pillay K, Govender D, Chetty R. ALK protein expression in rhabdomyosarcomas. Histopathology. 2002;41:461–467. doi: 10.1046/j.1365-2559.2002.01534.x. [DOI] [PubMed] [Google Scholar]
- 91.Li X-Q, Hisaoka M, Shi D-R, Zhu X-Z, Hashimoto H. Expression of anaplastic lymphoma kinase in soft tissue tumors: an immunohistochemical and molecular study of 249 cases. Hum. Pathol. 2004;35:711–721. doi: 10.1016/j.humpath.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 92.Perez-Pinera P, Chang Y, Astudillo A, Mortimer J, Deuel TF. Anaplastic lymphoma kinase in expressed in different subtypes of human breast cancer. Biochem. Biophys. Res. Commun. 2007;358:399–403. doi: 10.1016/j.bbrc.2007.04.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Perez-Pinera P, Garcia-Suarez O, Menendez-Rodriguez P, et al. The receptor protein tyrosine phosphatase (RPTP)β/ζ is expressed in differerent subtypes of human breast cancer. Biochem. Biophys. Res. Commun. 2007;362:5–10. doi: 10.1016/j.bbrc.2007.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park JR, Eggert A, Caron H. Neuroblastoma: biology, prognosis, and treatment. Pediatr. Clin. North Am. 2008;55:97–120. doi: 10.1016/j.pcl.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 95.Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N. Engl. J. Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 96.George RE, Attiyeh EF, Li S, et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS ONE. 2007;2:e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hurley SP, Clary DO, Copie V, Lefcort F. Anaplastic lymphoma kinase is dynamically expressed on subsets of motor neurons and in the peripheral nervous system. J. Comp. Neurol. 2006;495:202–212. doi: 10.1002/cne.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakagawara A, Milbrandt J, Muramatsu T, et al. Differential expression of pleiotrophin and midkine in advanced neuroblastomas. Cancer Res. 1995;55:1792–1797. [PubMed] [Google Scholar]
- 99.Ikematsu S, Nakagawara A, Nakamura Y, et al. Correlation of elevated level of blood midkine with poor prognostic factors of human neuroblastomas. Br. J. Cancer. 2003;88:1522–1526. doi: 10.1038/sj.bjc.6600938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barthlen W, Flaadt D, Girgert R, et al. Significance of heparin-binding growth factor expression on cells of solid pediatric tumors. J. Pediatr. Surg. 2003;38:1296–1304. doi: 10.1016/s0022-3468(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 101.Calvet L, Geoerger B, Regairaz M, et al. Pleiotrophin, a candidate gene for poor tumor vasculature and in vivo neuroblastoma sensitivity to irinotecan. Oncogene. 2006;25:3150–3159. doi: 10.1038/sj.onc.1209348. [DOI] [PubMed] [Google Scholar]
- 102.Sakai R, Henderson JT, O’Bryan JP, Elia AJ, Saxton TM, Pawson T. The mammalian ShcB and ShcC phosphotyrosine docking proteins function in the maturation of sensory and sympathetic neurons. Neuron. 2000;28:819–833. doi: 10.1016/s0896-6273(00)00156-2. [DOI] [PubMed] [Google Scholar]
- 103.George RE, Sanda T, Hanna M, et al. Activating mutations in the ALK tyrosine kinase provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397.One of the first descriptions of the occurrence of activating point mutations of ALK in human cancer.
- 104.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 105.McDermott U, Iafrate AJ, Gray NS, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186.Description of the sensitivity of 602 cell lines, representing a wide variety of human malignancies, to ALK small-molecule inhibitors.
- 106.Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261.One of the first descriptions of the occurrence of activating point mutations of ALK in human cancer.
- 107.Janoueix-Lerosey I, Lequin D, Brugieres L, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398.One of the first descriptions of the occurrence of activating point mutations of ALK in human cancer.
- 108.Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399.One of the first descriptions of the occurrence of activating point mutations of ALK in human cancer.
- 109.Delsol G, Lamant L, Mariame B, et al. A new subtype of large B-cell lymphoma expressing the ALK kinase and lacking the 2;5 translocation. Blood. 1997;89:1483–1490. [PubMed] [Google Scholar]
- 110.Hannah AL. Kinases as drug discovery targets in hematologic malignancies. Curr. Mol. Med. 2005;5:625–642. doi: 10.2174/156652405774641106. [DOI] [PubMed] [Google Scholar]
- 111.Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin’s lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol. Cell. Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mason DY, Pulford KA, Bischof D, et al. Nucleolar localization of the nucleophosmin-anaplastic lymphoma kinase is not required for malignant transformation. Cancer Res. 1998;58:1057–1062. [PubMed] [Google Scholar]
- 113.Fujimoto J, Shiota M, Iwahara T, et al. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5) Proc. Natl Acad. Sci. USA. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rosenwald A, Ott G, Pulford K, et al. t(1;2) (q21;p23) and t(2;3)(p23;q21): two novel variant translocations of the t(2;5) (p23;q35) in anaplastic large cell lymphoma. Blood. 1999;94:362–364. [PubMed] [Google Scholar]
- 115.Pulford K, Falini B, Cordell J, et al. Biochemical detection of novel anaplastic lymphoma kinase proteins in tissue sections of anaplastic large cell lymphoma. Am. J. Pathol. 1999;154:1657–1663. doi: 10.1016/S0002-9440(10)65421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ma Z, Cools J, Marynen P, et al. Inv(2) (p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood. 2000;95:2144–2149. [PubMed] [Google Scholar]
- 117.Ma Z, Hill DA, Collins MH, et al. Fusion of ALK to the Ran-binding protein 2 (RANBP2) in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;37:98–105. doi: 10.1002/gcc.10177. [DOI] [PubMed] [Google Scholar]
- 118.Tort F, Pinyol M, Pulford K, et al. Molecular characterization of a new ALK translocation involving moesin (MSN-ALK) in anaplastic large cell lymphoma. Lab. Invest. 2001;81:419–426. doi: 10.1038/labinvest.3780249. [DOI] [PubMed] [Google Scholar]
- 119.Trinei M, Lanfrancone L, Campo E, et al. A new variant anaplastic lymphoma kinase (ALK)-fusion protein (ATIC–ALK) in a case of ALK-positive anaplastic large cell lymphoma. Cancer Res. 2000;60:793–798. [PubMed] [Google Scholar]
- 120.Li R, Morris SW. Development of anaplastic lymphoma kinase (ALK) small-molecule inhibitors for cancer therapy. Med. Res. Rev. 2008;28:372–412. doi: 10.1002/med.20109.Provides a recent comprehensive description of the normal and pathogenic functions of ALK.
- 121.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat. Rev. Cancer. 2008;8:11–23. doi: 10.1038/nrc2291.Provides a recent comprehensive description of the normal and pathogenic functions of ALK.
- 122.Marzec M, Kasprzychka M, Liu X, et al. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–5614. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- 123.Leventaki V, Drakos E, Medeiros LJ, et al. NPM–ALK oncogenic kinase promotes cell-cycle progression through activation of JNK/cJUN signaling in anaplastic large-cell lymphoma. Blood. 2007;110:1621–1630. doi: 10.1182/blood-2006-11-059451. [DOI] [PubMed] [Google Scholar]
- 124.Voena C, Conte C, Ambrogio C, et al. The tyrosine phosphatase Shp2 interacts with NPM–ALK and regulates anaplastic lymphoma cell growth and migration. Cancer Res. 2007;67:4278–4286. doi: 10.1158/0008-5472.CAN-06-4350. [DOI] [PubMed] [Google Scholar]
- 125.Galietta A, Bunby RH, Redaelli S, et al. NPM/ALK binds and phosphorylates the RNA/DNA-binding protein PSF in anaplastic large-cell lymphoma. Blood. 2007;110:2600–2609. doi: 10.1182/blood-2006-01-028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sjostrom C, Seiler C, Crockett DK, Tripp SR, Johnson KS Elenitoba, Lim MS. Global proteome profiling of NPM/ALK-positive anaplastic large cell lymphoma. Exp. Hematol. 2007;35:1240–1248. doi: 10.1016/j.exphem.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 127.Staber PB, Vesely P, Haq N, et al. The oncoprotein NPM–ALK of anaplastic large-cell lymphoma induces JUNB transcription via ERK1/2 and JunB translation via mTOR signaling. Blood. 2007;110:3374–3383. doi: 10.1182/blood-2007-02-071258. [DOI] [PubMed] [Google Scholar]
- 128.Zhang Q, Wang HY, Liu X, Wasik MA. STAT5A is epigenetically silenced by the tyrosine kinase NPM1–ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1–ALK expression. Nat. Med. 2007;13:1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- 129.Bohling SD, Jenson SD, Crockett DK, Schumacher JA, Elenitoba-Johnson KS, Lim MS. Analysis of gene expression profile of TPM3–ALK positive anaplastic large cell lymphoma reveals overlapping and unique patterns with that of NPM–ALK positive anaplastic large cell lymphoma. Leuk. Res. 2008;32:383–393. doi: 10.1016/j.leukres.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 130.Cho-Vega JH, Vega F, Medeiros LJ. An attractive therapeutic target, mTOR pathway, in ALK+ anaplastic large cell lymphoma. Adv. Anat. Pathol. 2008;15:105–112. doi: 10.1097/PAP.0b013e318166139f. [DOI] [PubMed] [Google Scholar]
- 131.Colomba A, Courilleau D, Ramel D, et al. Activation of Rac1 and the exchange factor Vav3 are involved in NPM–ALK signaling in anaplastic large cell lymphomas. Oncogene. 2008;27:2728–2736. doi: 10.1038/sj.onc.1210921. [DOI] [PubMed] [Google Scholar]
- 132.Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Coronas S, Lagarrigue F, Ramel D, et al. Elevated levels of PtdIns5P in NPM–ALK–transformed cells: implication of PIKfyve. Biochem. Biophys. Res. Commun. 2008;372:351–355. doi: 10.1016/j.bbrc.2008.05.062. [DOI] [PubMed] [Google Scholar]
- 134.Pulford K, Roberton HM, Jones M. Antibody techiques used in the study of anaplastic lymphoma kinase-positive ALCL. Methods Mol. Med. 2005;115:271–294. doi: 10.1385/1-59259-936-2:271. [DOI] [PubMed] [Google Scholar]
- 135.Benharroch D, Meguerian-Bedoyan Z, Lamant L, et al. ALK-positive lymphoma: a single disease with a broad spectrum of morphology. Blood. 1998;91:2076–2084. [PubMed] [Google Scholar]
- 136.Kinney MC, Kadin ME. The pathologic and clinical spectrum of anaplastic large cell lymphoma and correlation with ALK gene dysregulation. Am. J. Clin. Pathol. 1999;111:S56–S67. [PubMed] [Google Scholar]
- 137.Morris SW, Xue L, Ma Z, Kinney MC. Alk+ CD30+ lymphomas: a distinct molecular genetic subtype of non-Hodgkin’s lymphoma. Br. J. Haematol. 2001;113:275–295. doi: 10.1046/j.1365-2141.2001.02574.x. [DOI] [PubMed] [Google Scholar]
- 138.Gascoyne RD, Aoun P, Wu D, et al. Prognostic significance of anaplastic lymphoma kinase (ALK) protein expression in adults with anaplastic large cell lymphoma. Blood. 1999;93:3913–3921. [PubMed] [Google Scholar]
- 139.Falini B, Pulford K, Pucciarini A, et al. Lymphomas expressing ALK fusion protein(s) other than NPM–ALK. Blood. 1999;94:3509–3515. [PubMed] [Google Scholar]
- 140.Haralambieva E, Pulford KA, Lamant L, et al. Anaplastic large-cell lymphomas of B-cell phenotype are anaplastic lymphoma kinase (ALK) negative and belong to the spectrum of diffuse large B-cell lymphomas. Br. J. Haematol. 2000;109:584–591. doi: 10.1046/j.1365-2141.2000.02045.x. [DOI] [PubMed] [Google Scholar]
- 141.Delsol G, Ralfkaier E, Stein H, Wright D, Jaffe ES. Anaplastic large cell lymphoma. Pathology and genetics. In: Jaffe E, Harris NL, Stein H, et al., editors. World Health Organisation of Tumours. Tumours of Haematopoietic and Lymphoid Tissues. IACR; Lyon, France: 2001. [Google Scholar]
- 142.Brugieres L, Deley MC, Pacquement H, et al. CD30+ anaplastic large-cell lymphoma in children: analysis of 82 patients enrolled in two consecutive studies of the French Society of Pediatric Oncology. Blood. 1998;92:3591–3598. [PubMed] [Google Scholar]
- 143.Brugieres L, Quartier P, Le Deley MC, et al. Relapses of childhood anaplastic large-cell lymphoma: treatment results in a series of 41 children – a report from the French Society of Pediatric Oncology. Ann. Oncol. 2000;11:53–58. doi: 10.1023/a:1008352726155. [DOI] [PubMed] [Google Scholar]
- 144.Williams DM, Hobson R, Imeson J, Gerrard M, McCarthy K, Pinkerton CR. Anaplastic large cell lymphoma in childhood: analysis of 72 patients treated on The United Kingdom Children’s Cancer Study Group chemotherapy regimens. Br. J. Haematol. 2002;117:812–820. doi: 10.1046/j.1365-2141.2002.03482.x. [DOI] [PubMed] [Google Scholar]
- 145.Savage KJ, Harris NL, Vose JM, et al. International Peripheral T-Cell Lymphoma Project ALK-anaplastic large-cell lymphoma is clinically and immunophentoypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-cell Lymphoma Project. Blood. 2008;111:5496–5504. doi: 10.1182/blood-2008-01-134270. [DOI] [PubMed] [Google Scholar]
- 146.Droc C, Cualing HD, Kadin ME. Need for an improved molecular/genetic classification for CD30+ lymphomas involving the skin. Cancer Control. 2007;14:124–132. doi: 10.1177/107327480701400205. [DOI] [PubMed] [Google Scholar]
- 147.Kinnney MC, Jones D. Cutaneous T-cell and NK-cell lymphomas: the WHO-EORTC classification and the increasing recognition of specialized tumor types. Am. J. Clin. Pathol. 2007;127:670–686. doi: 10.1309/MTTM86UT1XFQL7RV. [DOI] [PubMed] [Google Scholar]
- 148.Querfeld C, Kuzel TM, Guitart J, Rosen ST. Primary cutaneous CD30+ lymphoproliferative disorders: new insights into biology and therapy. Oncology. 2007;21:689–696. [PubMed] [Google Scholar]
- 149.Gascoyne RD, Lamant L, Martin-Subero JI, et al. ALK-positive diffuse large B-cell lymphoma is associated with clathrin-ALK rearrangements: report of six cases. Blood. 2003;102:2568–2571. doi: 10.1182/blood-2003-03-0786. [DOI] [PubMed] [Google Scholar]
- 150.Chikatsu N, Kojima H, Suzukawa K, et al. ALK+, CD30−, CD20− large B-cell lymphoma containing anaplastic lymphoma kinase (ALK) fused to clathrin heavy chain gene (CLTC) Mod. Pathol. 2003;16:828–832. doi: 10.1097/01.MP.0000081729.40230.1F. [DOI] [PubMed] [Google Scholar]
- 151.De Paepe P, Baens M, van Krieken H, et al. ALK activation by the CTLC-ALK fusion is a recurrent event in B-cell lymphoma. Blood. 2003;102:2638–2641. doi: 10.1182/blood-2003-04-1050. [DOI] [PubMed] [Google Scholar]
- 152.Onciu M, Behm FG, Downing JR, et al. ALK-positive plasmablastic B-cell lymphoma with expression of the NPM–ALK fusion transcript: report of two cases. Blood. 2003;102:2642–2644. doi: 10.1182/blood-2003-04-1095. [DOI] [PubMed] [Google Scholar]
- 153.Reichard KK, McKenna RW, Kroft SH. ALK-positive B-cell lymphoma: a report of three cases. Mod. Pathol. 2003;16:250A. doi: 10.1038/modpathol.3800742. [DOI] [PubMed] [Google Scholar]
- 154.Reichard KK, McKenna RW, Kroft SH. ALK-positive diffuse large B-cell lymphoma: report of four cases and review of the literature. Mod. Pathol. 2007;20:310–319. doi: 10.1038/modpathol.3800742. [DOI] [PubMed] [Google Scholar]
- 155.Shinmura K, Kageyama S, Tao H, et al. ALK+ histiocytosis: a novel type of systemic histiocytic proliferative disorder of early infancy. Blood. 2008;112:2965–2968. doi: 10.1182/blood-2008-03-147017. [DOI] [PubMed] [Google Scholar]
- 156.Griffin CA, Hawkins AL, Dvorak C, Henkle C, Ellingham T, Perlman EJ. Recurrent involvement of 2p23 in inflammatory myofibroblastic tumors. Cancer Res. 1999;59:2776–2780.Initial report implicating a role for ALK in the genesis of inflammatory myofibroblastic tumors.
- 157.Coffin CM, Patel A, Perkins S, Elenitoba–Johnson KS, Perlman E, Griffin CA. ALK1 and p80 expression and chromosomal rearrangements involving 2p23 in inflammatory myofibroblastic tumor. Mod. Pathol. 2001;14:569–576. doi: 10.1038/modpathol.3880352. [DOI] [PubMed] [Google Scholar]
- 158.Gleason BC, Hornick JL. Inflammatory myofibroblastic tumours: where are we now? J. Clin. Pathol. 2008;61:428–437. doi: 10.1136/jcp.2007.049387. [DOI] [PubMed] [Google Scholar]
- 159.Cook JR, Dehner LP, Collins MH, et al. Anaplastic lymphoma kinase (ALK) expression in the inflammatory myofibroblastic tumor: a comparative immunohistochemical study. Am. J. Surg. Pathol. 2001;25:1364–1371. doi: 10.1097/00000478-200111000-00003. [DOI] [PubMed] [Google Scholar]
- 160.Lawrence B, Perez-Atayde A, Hibbard MK, et al. TPM3–ALK and TPM4–ALK oncogenes in inflammatory myofibroblastic tumors. Am. J. Pathol. 2000;157:377–384. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bridge JA, Kanamori M, Ma Z, et al. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am. J. Pathol. 2001;159:411–415. doi: 10.1016/S0002-9440(10)61711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cools J, Wlodarska I, Somers R, et al. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- 163.Panagopoulos I, Nilsson T, Domanski HA, et al. Fusion of the SEC31L1 and ALK genes in an inflammatory myofibroblastic tumor. Int. J. Cancer. 2006;118:1181–1186. doi: 10.1002/ijc.21490. [DOI] [PubMed] [Google Scholar]
- 164.Yousem SA, Shaw H, Cieply K. Involvement of 2p23 in pulmonary inflammatory pseudotumors. Hum. Pathol. 2001;32:428–433. doi: 10.1053/hupa.2001.23523. [DOI] [PubMed] [Google Scholar]
- 165.Sirvent N, Hawkins AL, Moeglin D, et al. ALK probe rearrangement in a t(2;11;2) (p23;p15;q31) translocation found in a prenatal myofibroblastic fibrous lesion: toward a molecular definition of an inflammatory myofibroblastic tumor family? Genes Chromosomes Cancer. 2001;31:85–90. doi: 10.1002/gcc.1121. [DOI] [PubMed] [Google Scholar]
- 166.Chan JK, Cheuk W, Shimizu M. Anaplastic lymphoma kinase expression in inflammatory pseudotumors. Am. J. Surg. Pathol. 2001;25:761–768. doi: 10.1097/00000478-200106000-00007. [DOI] [PubMed] [Google Scholar]
- 167.Sigel JE, Smith TA, Reith JD, Goldblum JR. Immunohistochemical analysis of anaplastic lymphoma kinase expression in deep soft tissue calcifying fibrous pseudotumor: evidence of a late sclerosing stage of inflammatory myofibroblastic tumor? Ann. Diagn. Pathol. 2001;5:10–14. doi: 10.1053/adpa.2001.21474. [DOI] [PubMed] [Google Scholar]
- 168.Chun YS, Wang L, Nascimento AG, Moir CR, Rodeberg DA. Pediatric inflammatory myofibroblastic tumor: anaplastic lymphoma kinase (ALK) expression and prognosis. Pediatr. Blood Cancer. 2005;45:796–801. doi: 10.1002/pbc.20294. [DOI] [PubMed] [Google Scholar]
- 169.Mergan F, Jaubert F, Sauvat F, et al. Inflammatory myofibroblastic tumor in children: clinical review with anaplastic lymphoma kinase, Epstein–Barr virus, and human herpesvirus 8 detection analysis. J. Ped. Surg. 2005;40:1581–1586. doi: 10.1016/j.jpedsurg.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 170.Montgomery EA, Shuster DD, Burkart AL, et al. Inflammatory myofibroblastic tumors of the urinary tract: a clinicopathologic study of 46 cases, including a malignant example of inflammatory fibrosarcoma and a subset associated with high-grade urothelial carcinoma. Am. J. Surg. Pathol. 2006;30:1502–1512. doi: 10.1097/01.pas.0000213280.35413.1b. [DOI] [PubMed] [Google Scholar]
- 171.Coffin CM, Hornick JL, Fletcher CD. Inflammatory myofibroblastic tumor: comparison of clinicopathologic, histologic, and immunohistochemical features including ALK expression in atypical and aggressive cases. Am. J. Surg. Pathol. 2007;31:509–520. doi: 10.1097/01.pas.0000213393.57322.c7. [DOI] [PubMed] [Google Scholar]
- 172.Swain RS, Tihan T, Horvai AE. Inflammatory myofibroblastic tumor of the central nervous system and its relationship to inflammatory pseudotumor. Hum. Pathol. 2008;39:410–419. doi: 10.1016/j.humpath.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 173.Qui X, Montgomery E, Sun B. Inflammatory myofibroblastic tumor and low-grade myofibroblastic sarcoma: a comparative study of clinicopathologic features and further observations on the immunohistochemical profile of myofibroblasts. Hum. Pathol. 2008;39:846–856. doi: 10.1016/j.humpath.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 174.Jazii FR, Najafi Z, Malekzadeh R, et al. Identification of squamous cell carcinoma associated proteins by proteomics and loss of β tropomyosin expression in esophageal cancer. World J. Gastroenterol. 2006;12:7104–7112. doi: 10.3748/wjg.v12.i44.7104.Initial report of ALK involvement in esophageal squamous cell carcinoma.
- 175.Du X-L, Hu H, Lin D-C, et al. Proteomic profiling of proteins dysregulated in Chinese esophageal squamous cell carcinoma. J. Mol. Med. 2007;85:863–875. doi: 10.1007/s00109-007-0159-4.Initial report of ALK involvement in esophageal squamous cell carcinoma.
- 176.Aklilu M, Ilson DH. Targeted agents and esophageal cancer – the next step? Semin. Radiat. Oncol. 2007;17:62–69. doi: 10.1016/j.semradonc.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 177.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945.Provides an initial description of a pathogenic role for ALK in the genesis of human non-small-cell lung carcinoma.
- 178.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025.Provides an initial description of a pathogenic role for ALK in the genesis of human non-small-cell lung carcinoma.
- 179.Inamura K, Takeuchi K, Togashi Y, et al. EML4–ALK fusion is linked to histological characteristics in a subset of lung cancers. J. Thorac. Oncol. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- 180.Perner S, Wagner PL, Demichelis F, et al. EML4–ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shinmura K, Kageyama S, Tao H, et al. EML4–ALK fusion transcripts, but no NPM–, TPM3–, CLTC–, ATIC–, or TFG–ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer. 2008;61(2):163–169. doi: 10.1016/j.lungcan.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 182.Fukuyoshi Y, Inoue H, Kita Y, Utsunomiya T, Ishida T, Mori M. EML4–ALK fusion transcript is not found in gastrointestinal and breast cancers. Br. J. Cancer. 2008;98:1536–1539. doi: 10.1038/sj.bjc.6604341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4–ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168.One of a number of studies confirming the role of aberrant ALK activity in the causation of a subset of human lung cancer; also touches upon mechanisms that may confer resistance to ALK inhibitor therapy (e.g., concomitant activation in tumors of other tyrosine kinases such as the EGF receptor and ERBB2).
- 184.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4–ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 185.Wheatley-Price P, Shepherd FA. Epidermal growth factor receptor inhibitors in the treatment of lung cancer: reality and hopes. Curr. Opin. Oncol. 2008;20:162–175. doi: 10.1097/CCO.0b013e3282f335a3. [DOI] [PubMed] [Google Scholar]
- 186.Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4–ALK-positive lung cancer. Proc. Natl Acad. Sci. USA. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Buchdunger E, Zimmermann J, Mett H, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56(1):100–104. [PubMed] [Google Scholar]
- 188.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr–Abl positive cells. Nat. Med. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 189.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 190.Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N. Engl. J. Med. 2002;346(9):645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 191.Talpaz M, Silver RT, Druker BJ, et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a Phase 2 study. Blood. 2002;99(6):1928–1937. doi: 10.1182/blood.v99.6.1928. [DOI] [PubMed] [Google Scholar]
- 192.van Oosterom AT, Judson I, Verweij J, et al. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumors: a Phase I study. Lancet. 2001;358(9291):1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- 193.Roumiantsev S, Shah NP, Gorre ME, et al. Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of Tyr-253 in the abl kinase domain P-loop. Proc. Natl Acad. Sci. USA. 2000;99(16):10700–10705. doi: 10.1073/pnas.162140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.de Silva MV Chandu, Reid R. Gastrointestinal stromal tumors (GIST): C-kit mutations, CD117 expression, differential diagnosis and targeted cancer therapy with imatinib. Path. Oncol. Res. 2003;9(1):13–19. doi: 10.1007/BF03033708. [DOI] [PubMed] [Google Scholar]
- 195.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305(5682):399–402. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 196.Lombardo LJ, Lee FY, Chen P, et al. Discovery of N-(2-chloro-6-methylphenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent anti-tumor activity in preclinical assays. J. Med. Chem. 2004;47(27):6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 197.Abdelhalim A, Barcos M, Block AW, et al. Remission of Philadelphia chromosome-positive central nervous system leukemia after dasatinib therapy. Leuk. Lymphoma. 2007;48(5):1053–1056. doi: 10.1080/10428190701258370. [DOI] [PubMed] [Google Scholar]
- 198.Weisberg E, Manley PW, Cowan-Jacob SW, Hochhaus A, Griffin JD. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukemia. Nat. Rev. Cancer. 2007;7(5):345–356. doi: 10.1038/nrc2126. [DOI] [PubMed] [Google Scholar]
- 199.Quintas-Cardama A, Kantarjian H, Cortes J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nat. Rev. Drug Discov. 2007;6(10):834–848. doi: 10.1038/nrd2324. [DOI] [PubMed] [Google Scholar]
- 200.Meric JB, Faivre S, Monnerat C, et al. Zd 1839 “Iressa”. Bull. Cancer. 2000;87(12):873–876. [PubMed] [Google Scholar]
- 201.Hightower M. Erlotinib (OSI-774, Tarceva), a selective epidermal growth factor receptor tyrosine kinase inhibitor, in combination with chemotherapy for advanced non-small-cell lung cancer. Clin. Lung Cancer. 2003;4(6):336–338. doi: 10.1016/s1525-7304(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 202.Baselga J, Albanell J. Targeting epidermal growth factor receptor in lung cancer. Curr. Oncol. Rep. 2002;4(4):317–324. doi: 10.1007/s11912-002-0007-1. [DOI] [PubMed] [Google Scholar]
- 203.Kumar A, Petri ET, Halmos B, Boggon TJ. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J. Clin. Oncol. 2008;26:1742–1751. doi: 10.1200/JCO.2007.12.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Perez-Soler R. Phase II clinical trial data with the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib (OSI-774) in non-small-cell lung cancer. Clin. Lung Cancer. 2004;6(Suppl 1):S20–S23. doi: 10.3816/clc.2004.s.010. [DOI] [PubMed] [Google Scholar]
- 205.Senderowicz AM, Johnson JR, Sridhara R, Zimmerman P, Justice R, Pazdur R. Erlotinib/gemcitabine for first-line treatment of locally advanced or metastatic adenocarcinoma of the pancreas. Oncology. 2007;21(14):1696–1706. [PubMed] [Google Scholar]
- 206.Ho QT, Kuo CJ. Vascular endothelial growth factor: biology and therapeutic applications. Int. J. Biochem. Cell Biol. 2007;39(7–8):1349–1357. doi: 10.1016/j.biocel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Larkin JM, Eisen T. Kinase inhibitors in the treatment of renal cell carcinoma. Crit. Rev. Oncol. Hematol. 2006;60(3):216–226. doi: 10.1016/j.critrevonc.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 208.Oudard S, George D, Medioni J, Motzer R. Treatment options in renal cell carcinoma: past, present and future. Ann. Oncol. 2007;18(Suppl 10):25–31. doi: 10.1093/annonc/mdm411. [DOI] [PubMed] [Google Scholar]
- 209.Joensuu H. Sunitinib for imatinib-resistant GIST. Lancet. 2006;368(9544):1303–1304. doi: 10.1016/S0140-6736(06)69489-0. [DOI] [PubMed] [Google Scholar]
- 210.McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc. Natl Acad. Sciences USA. 2007;104(50):19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gunby RH, Ahmed S, Sottocornola R, et al. Structural insights into the ATP binding pocket of the anaplastic lymphoma kinase by site-directed mutagenesis, inhibitor binding analysis, and homology modeling. J. Med. Chem. 2006;49(19):5759–5768. doi: 10.1021/jm060380k. [DOI] [PubMed] [Google Scholar]
- 212.Gunby RH, Tartari CJ, Porchia F, Donella-Deana A, Scapozza L, Gambacorti-Passerini C. An enzyme-linked immunosorbent assay to screen for inhibitors of the oncogenic anaplastic lymphoma kinase. Haematologica. 2005;90:988–990. [PubMed] [Google Scholar]
- 213.Karaman MW, Herrgard S, Treiber DK, et al. A quanti-tative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 214.Fuse E, Kuwabara T, Sparreboom A, Sausville EA, Figg WD. Review of UCN-01 development: a lesson in the importance of clinical pharmacology. J. Clin. Pharmacol. 2005;45:394–403. doi: 10.1177/0091270005274549. [DOI] [PubMed] [Google Scholar]
- 215.Bonvini P, Rosa HD, Vignes N, Rosolen A. Ubiquitination and proteasomal degradation of nucleophosmin-anaplastic lymphoma kinase induced by 17-allylamino-demethoxygeldanamycin: role of the co-chaperone carboxyl heat shock protein 70-interacting protein. Cancer Res. 2004;64:3256–3264. doi: 10.1158/0008-5472.can-03-3531. [DOI] [PubMed] [Google Scholar]
- 216.Georgakis GV, Li Y, Rassidakis GZ, Medeiros LJ, Younes A. The HSP90 inhibitor 17-AAG synergizes with doxorubicin and U0126 in anaplastic large cell lymphoma irrespective of ALK expression. Exp. Hematol. 2006;34:1670–1679. doi: 10.1016/j.exphem.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 217.Turturro F, Arnold MD, Frist AY, Pulford K. Model of inhibition of the NPM–ALK kinase activity by herbimycin A. Clin. Cancer Res. 2002;8:240–245. [PubMed] [Google Scholar]
- 218.Wan W, Albom MS, Lu L, et al. Anaplastic lymphoma kinase activity is essential for the proliferation and survival of anaplastic large-cell lymphoma cells. Blood. 2006;107:1617–1623. doi: 10.1182/blood-2005-08-3254.Describes the ALK small-molecule inhibitor tool compounds and their use to providing validation of ALK as a cancer target.
- 219.Piva R, Pellegrino E, Mattioli M, et al. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J. Clin. Invest. 2006;116:3171–3182. doi: 10.1172/JCI29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li R, Xue L, Zhu T, et al. Design and synthesis of 5-aryl-pyridone-carboxamides as inhibitors of anaplastic lymphoma kinase. J. Med. Chem. 2006;49(3):1006–1015. doi: 10.1021/jm050824x. [DOI] [PubMed] [Google Scholar]
- 221.Galkin AV, Melnick JS, Kim S, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM–ALK. Proc. Natl. Acad. Sci. USA. 2007;104(1):270–275. doi: 10.1073/pnas.0609412103.Describes ALK small-molecule inhibitor tool compounds and their use to provide further validation of ALK as a cancer target.
- 222.Christensen JG, Schreck R, Burrows J, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive anti-tumor activity in vivo. Cancer Res. 2003;63(21):7345–7355. [PubMed] [Google Scholar]
- 223.Puri N, Khramtsov A, Ahmed S, et al. A selective small molecule inhibitor of c-Met, PHA665752, inhibits tumorigenicity and angiogenesis in mouse lung cancer xenografts. Cancer Res. 2007;67:3529–3534. doi: 10.1158/0008-5472.CAN-06-4416. [DOI] [PubMed] [Google Scholar]
- 224.Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 225.Knudsen BS, vande Woude G. Showering c-Met-dependent cancers with drugs. Curr. Opin. Genet. Dev. 2008;18:87–96. doi: 10.1016/j.gde.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 226.Cui JJ, Botrous I, Shen H, et al. Structure based drug design for the discovery of clinical candidate PF-2341066 as potent and highly selective c-Met inhibitor. Presented at: 235th ACS National Meeting; New Orleans, LA, USA. 6–10 April 2008. [Google Scholar]
- 227.Zou HY, Li Q, Lee JH, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive anti-tumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Research. 2007;67(9):4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 228.Christensen JG, Zou HY, Arango EA, et al. Cytoreductive anti-tumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol. Cancer Ther. 2007;6(12 Pt 1):3314–3322. doi: 10.1158/1535-7163.MCT-07-0365.Report of preclinical studies that confirm the efficacy of the dual ALK/MET (hepatocyte growth factor/scatter factor receptor) small-molecule inhibitor PF-2341066 (which is currently in late-stage Phase I clinical trial) in ALK-driven tumors, such as anaplastic large-cell lymphoma.
- 229.Weinberg LR, Albom MS, Angeles TS, et al. Synthesis and SAR of 1,2,3,4-tetrahydro-pyrido[2,3-b]pyrazines as c-Met and ALK inhibitors. Presented at: 235th ACS National Meeting; New Orleans, LA, USA. 6–10 April 2008. [Google Scholar]
- 230.Wodarz D, Komarova NL. Emergence and prevention of resistance against small molecule inhibitors. Semin. Cancer Biol. 2005;15:506–614. doi: 10.1016/j.semcancer.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 301.Gregor VE, Liu Y, Anikin A, et al. Tricyclic compound derivatives useful in the treatment of neoplastic diseases, inflammatory disorders and immunomodulatory disorders. WO 2008021369 A2 CHEMBRIDGE RESEARCH LABORATORIES INC. 2008
- 302.Kawahara ET, Miyake T, Roesel J. Preparation of pyrimidine compounds as FAK and/or ALK inhibitors. WO 2006021457 A2 NOVARTIS A.-G. Switzerland; NOVARTIS PHARMA GMBH. 2006
- 303.Ahmed G, Bohnstedt A, Breslin HJ, et al. Fused bicyclic derivatives of 2,4-diaminopyrimidine as alk and c-Met inhibitors. WO 2008051547 A1 CEPHALON INC. USA; PHARMACOPEIA DRUG DISCOVERY, INC. 2008
- 304.Gregor VE, Liu Y, Anikin A, et al. Tricyclic compound derivatives useful in the treatment of neoplastic diseases, inflammatory disorders and immunomodulatory disorders. US0171769 A1 CHEMBRIDGE RESEARCH LABORATORIES INC. USA. 2008
- 401.Cephalon www.cephalon.com.
- 402.Chembridge Corporation www.chembridge.com.
- 403.St Jude Children’s Research Hospital www.stjude.org.
- 404.Genomics Institute of the Novartis Research Foundation www.gnf.org.