Abstract
We previously discovered a germ cell-specific spermatogenesis and oogenesis basic helix-loop-helix transcription factor, Sohlh2. We generated Sohlh2-deficient mice to understand physiologic consequences of Sohlh2 deletion. We discovered that Sohlh2-knockout adult female mice are infertile due to lack of ovarian follicles. Sohlh2-deficient ovaries can form primordial follicles and, despite limited oocyte growth, do not differentiate surrounding granulosa cells into cuboidal and multilayered structures. Oocytes are rapidly lost in Sohlh2-deficient ovaries, and few are present by 14 days of postnatal life. However, the primordial oocytes are abnormal at the molecular level because they misexpress numerous germ cell- and oocyte-specific genes, including Sohlh1, Nobox, Figla, Gdf9, Pou5f1, Zp1, Zp3, Kit, Oosp1, Nlrp14, H1foo, and Stra8. Our findings show that Sohlh2 is a critical factor for maintenance and differentiation of the oocyte during early oogenesis.
Keywords: oocyte development, oogenesis, primordial follicle, Sohlh2, transcription factor
Sohlh2 is an essential transcriptional regulator of mammalian oogenesis and mediates its action in part by disrupting oocyte-specific pathways and Kit expression.
INTRODUCTION
The follicle formation in the mouse ovary begins shortly after birth with the breakdown of germ cell clusters. Germ cell clusters break down, and most oocytes become enveloped by a layer of flat pregranulosa somatic cells [1, 2]. Some primordial follicles are recruited to grow into primary and larger follicles [3]. The formation of primordial follicles and their transition to primary follicles are critical events associated with oocyte loss, transition from prenatal to postnatal life, significant hormonal changes, and initiation of the transcription of numerous genes, including genes preferentially expressed in oocytes such as Zp1 (zona pellucida 1), Zp2, and Zp3, as well as Gdf9 (growth differentiation factor 9) and Pou5f1 (also known as Oct4) [4–9]. The molecular mechanisms involved in initiating early folliculogenesis and gene transcription have begun to unravel.
Several transcription factors are known to affect the regulation of oocyte-specific genes during early folliculogenesis. Figla (factor in the germline alpha) is a well-known oocyte-specific basic helix-loop-helix (bHLH) transcription factor. Figla regulates expression of many genes in the ovary, including Zp2, Pou5f1, Nlrp14, Nlrp4f, and Nlrp4b [10, 11]. Nobox (newborn ovary homeobox gene) is also necessary for expression of several key oocyte-specific genes, including Gdf9 and Pou5f1 [9, 12, 13]. Sohlh1 (spermatogenesis- and oogenesis-specific bHLH transcription factor 1) is another germ cell-specific gene that encodes a bHLH protein [14]. Sohlh1 lies upstream of Lhx8, Zp1, and Zp3 and is preferentially expressed in primordial oocytes, a pattern distinct from that of Figla and Nobox [14].
We recently discovered a novel bHLH transcription factor, SOHLH2, which is preferentially expressed in germ cells of the embryonic ovary and in oocytes of primordial and primary follicles [15]. The Sohlh2 pattern of expression mimics that of Sohlh1, and the 2 proteins share 47% identity in their bHLH domain. In the present study, we demonstrate that Sohlh2 deficiency accelerates postnatal oocyte loss in the ovary and causes infertility in female mice. In addition, Sohlh2 deficiency affects the expression of numerous oocyte-specific genes in the ovary. These findings show that Sohlh2 is critical in early follicle formation and oocyte differentiation and that Sohlh1 and Sohlh2 are independently required for successful oocyte differentiation.
MATERIALS AND METHODS
Targeting Construct, Genotyping, and Colony Generation
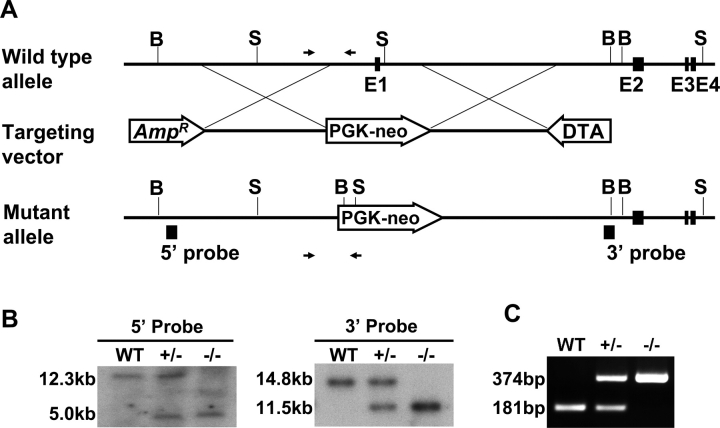
A targeting construct was prepared as previously described [16]. Exon 1 of the mouse Sohlh2 gene was replaced with a neomycin-resistance gene flanked by 5.0 kb (5′) and 5.0 kb (3′) of homologous DNA sequences (Fig. 1A). The vector was linearized and electroporated into embryonic stem cells (AB2.1), kindly provided by Dr. Martin M. Matzuk (Baylor College of Medicine). Targeted clones were screened by Southern blot analysis using 5′ and 3′ probes located outside of the homologous vector arms (Fig. 1B). The embryonic stem cells, identified as heterozygous for the targeted Sohlh2 mutation, were microinjected into the C57BL/6 blastocysts to produce chimeric mice that carried the mutation into the germline. These mice were mated with C57BL/6 wild-type (WT) mice to generate Sohlh2 heterozygous animals that were subsequently crossed to produce offspring from the second generation for analysis. Mice were genotyped by Southern blot analysis and PCR analysis. The PCR genotyping was performed using primer sets WT-L (5′-ACT TCA CCC TTG TCC TGC AT-3′) and WT-R (5′-GGG GAC CTG AAA TCA ATC CT-3′) to amplify a 181-nucleotide WT band and using primer sets WT-L (5′-ACT TCA CCC TTG TCC TGC AT-3′) and KO-R (5′-GCC AGA GGC CAC TTG TGT AG-3′) to yield a 374-nucleotide mutant band. Hot Start Taq polymerase (Qiagen, Valencia, CA) was activated at 95°C for 15 min, and PCR was performed for 30 cycles at 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, with a final extension at 72°C for 10 min.
FIG. 1.
Sohlh2-knockout strategy. A) The targeting vector was designed to delete exon 1 encoding the ATG site and nucleotides upstream of exon 1. For positive and negative selections, PGK-neo (PL-452) and pDTA.3 (DTA) cassettes were used. Genomic regions amplified by PCR for genotyping are indicated by arrows. B) Southern blot analysis of tail DNA extracted from WT, Sohlh2+/–, and Sohlh2–/– mice. We used two external probes to distinguish the Sohlh2 WT alleles (12.3 kb for the 5′ probe and 14.8 kb for the 3′ probe) and null alleles (5.0 kb for the 5′ probe and 11.5 kb for the 3′ probe). Genomic DNA was isolated from mouse tails, digested with BmtI (for the 5′ probe) and SpeI (for the 3′ probe), and then hybridized to 32P-labeled 5′ and 3′ DNA probes. C) The PCR analysis for genotyping of WT, Sohlh2+/–, and Sohlh2–/– mice. The sizes of predicted WT and mutant allele amplification products are 181 bp and 374 bp, respectively.
Animal Breeding and Experiments
All murine experiments were performed on animals of C57BL/6/129S6/SvEv hybrid background. All experimental and surgical procedures complied with the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Agricultural Animal Care and Use Committee of Baylor College of Medicine. Litters were weaned at 3 wk, and breeding pairs were set up at age 6–7 wk. One mating pair was placed per cage and was inspected daily for the presence of litters. Both WT and Sohlh2-deficient mice were euthanized and examined (n = 5/day) in every experiment.
Histology and Histomorphometric Analysis
Ovaries collected from newborn (P0) mice and from mice at Postnatal Days 7, 14, and 21 (n = 5) were fixed in 10% buffered formalin. Fixed tissues were serially sectioned (5 μm) and were stained with hematoxylin-eosin or with periodic acid-Schiff. At least 5 pairs of ovaries of each genotype were subjected to gross and microscopic analysis for each time point.
Immunohistochemistry
For immunohistochemistry, we used antibodies against SOHLH1 and LHX8 proteins. Polyclonal rabbit antibodies against SOHLH1 and polyclonal guinea pig antibodies against LHX8 were generated by Cocalico Biologicals (Reamstown, PA) [9, 14]. Immunohistochemistry was performed as described previously [17]. In brief, sections (5-μm thick) were cut and mounted onto polylysine-coated microscope slides (Fisher Scientific, Pittsburgh, PA). After blocking, diluted primary antibodies were applied to each section. Visualization of positive signal were performed using Vectastain kit (Vector Laboratories, Inc., Burlingame, CA) according to the manufacturer's instructions. Preimmune serum was used in control sections.
RNA Isolation, RT-PCR, and Real-Time PCR
The newborn ovaries isolated from the WT, Sohlh2+/– , and Sohlh2–/– mice were stored in RNAlater solution (Ambion, Austin, TX). Total RNA was extracted using the RNeasy mini kit (Qiagen). Two micrograms of tRNA was reverse transcribed using the Superscript system (Invitrogen, Rockville, MD). The PCR was performed as previously described [9, 18]. The sequences of primers for RT-PCR are available on request from the correspondence author. Mouse beta-actin was used as a control for equivalent RNA levels. Real-time PCR was performed as previously described [12]. Primer sequences used in the real-time PCR analysis are given in Supplemental Table 1 (available at www.biolreprod.org). Mouse Gapdh was used for the endogenous control because Gapdh transcript levels were unaffected by Sohlh2 deficiency. Each real-time PCR experiment was performed on at least 3 independent pools of WT and Sohlh2–/– newborn ovaries. Each real-time experiment was repeated in triplicate for each individual pool of ovaries. The relative amount of transcripts was calculated by the ΔΔCT method. The mean (SE) was calculated for the triplicate measurements, and the relative amount of target gene transcripts was plotted. Significance was determined using Student t-test.
RESULTS
Sohlh2 Deficiency Affects Ovarian Development
Sohlh2 encodes a bHLH transcriptional regulator with homologues in humans and other mammals [15]. In females, Sohlh2 transcripts are detected in the embryonic ovary as early as Embryonic Day 12.5 [15] before oocyte entry into meiosis I. To examine the physiologic role of Sohlh2 in the ovary during oogenesis, we generated a targeted deletion of the Sohlh2 first exon and 500 bp upstream of the first exon, which presumably contains a portion of the Sohlh2 promoter region (Fig. 1A). The mutant allele does not transcribe Sohlh2 mRNA (Fig. 1C). The genotype was confirmed by Southern blot analysis (Fig. 1B) and by PCR analysis (Fig. 1C).
Fertility was tested by mating heterozygous or homozygous mutant Sohlh2 female mice with WT male mice during a 6-month period. Like WT mice, heterozygous (Sohlh2+/–) female mice were fertile, and Sohlh2+/– reproductive organs were grossly normal compared with those of WT mice (Fig. 2, A, B, and D). In contrast, all Sohlh2–/– females tested (n = 5) were infertile, and their ovaries showed atrophy at age 3 wk (Fig. 2, A, C, and E). The 3-wk-old Sohlh2–/– ovaries contained few oocytes or follicles (Fig. 2E).
FIG. 2.
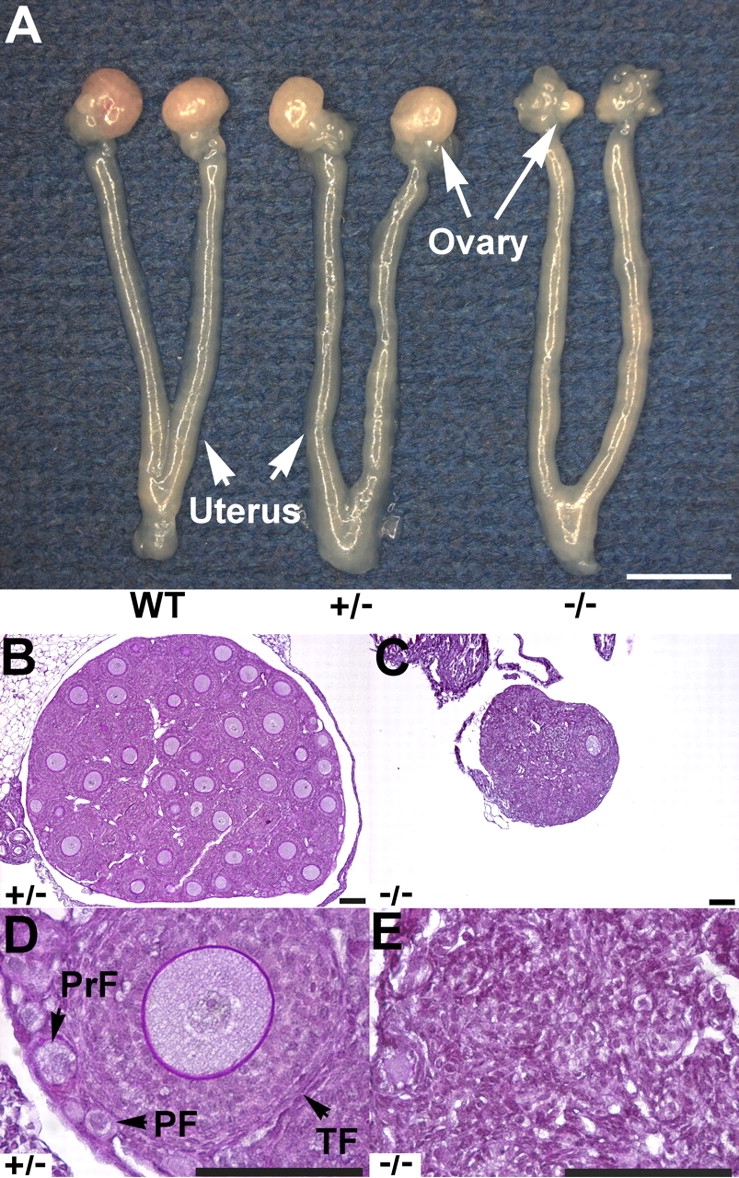
Sohlh2-knockout anatomy and histology. A) Female reproductive tracts were dissected from 3-wk-old WT, Sohlh2+/–, and Sohlh2–/– mice. Note atrophic ovaries and uterine horns in the Sohlh2–/– mice. Bar = 3 mm. B–E) Histology of 3-wk-old Sohlh2+/–and Sohlh2–/– ovaries. Sohlh2–/– ovaries lack follicles, while a magnified view of the Sohlh2+/– ovary shows well-defined primordial follicle (PF), primary follicle (PrF), and growing follicle (TF). At least five pairs of ovaries were examined, and representative sections are shown. Bar = 100 μm.
Sohlh2 Is Essential in Postnatal Oocyte Differentiation and Survival
Because Sohlh2−/− ovaries at the time of sexual maturity (6 wk) were depleted of oocytes, we examined the onset of oocyte loss in postnatal Sohlh2-deficient ovaries by studying the ovarian histology at Day 0 (Postnatal Day 0), Day 7, and Day 14 (Fig. 3). Wild-type and Sohlh2–/– newborn ovaries did not show gross differences in morphology or histology (Fig. 3, A and B). Germ cell clusters and primordial follicles (defined as oocytes ≤20 μm surrounded by flat granulosa cells) were present in WT and Sohlh2–/– newborn ovaries. At 7 days postpartum, Sohlh2–/– ovaries contained primordial follicles and follicles containing oocytes that were larger than 20 μm, but these oocytes were enveloped by a single layer of flat granulosa cells and did not meet the strict criteria for primary follicles (defined as oocytes >20 μm surrounded by cuboidal granulosa cells) (Fig. 3D). By histologic examination, there are normal-appearing primordial follicles at Postnatal Day 7 but few “normal” primary follicles. Beyond primordial follicles, large oocytes and flat granulosa cells characterize the remaining follicles. However, the premature activation is not complete because rare follicles can escape early death (Fig. 2C) and become multilayered follicles. We hypothesize that such escape may be due to the incomplete suppression of other germ cell-specific transcriptional regulators such as Figla and Nobox, which may partially compensate for Sohlh2 deficiency.
FIG. 3.
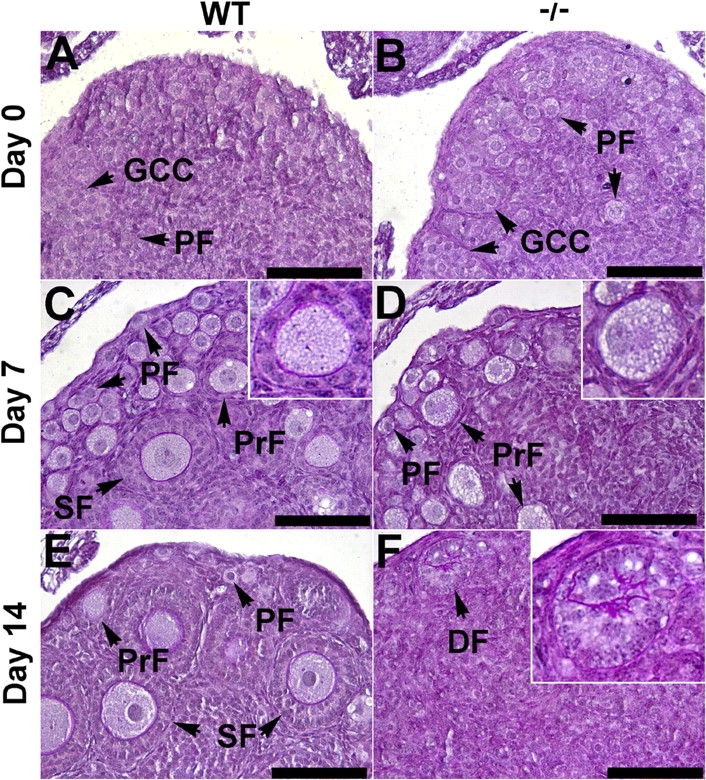
Histology of WT and Sohlh2–/–ovaries at Postnatal Day 0 (Day 0), Day 7, and Day 14. A–F) Periodic acid-Schiff staining of ovaries dissected from WT and Sohlh2–/– mice. A, C, and E) Day 0, Day 7, and Day 14 WT ovaries displaying germ cell clusters (GCC), primordial-like follicles (PF), primary follicles (PrF), and secondary follicles (SF). B) At Day 0, Sohlh2–/– newborn ovaries contain GCC and PF. D) At Day 7, periodic acid-Schiff staining of 7-day-old ovaries shows fewer PF and PrF in Sohlh2–/– ovaries. Note that the granulosa cells that surround PrF in Sohlh2–/– ovaries do not differentiate from flattened to cuboidal shape, as seen in WT ovaries. F) At Day 14, Sohlh2–/– ovaries contain few follicles and a degenerated follicle (DF). Bar = 100 μm.
At Postnatal Day 14, Sohlh2–/– ovaries contained few oocytes, and most follicles were in the state of degeneration (Fig. 3F). Occasional multilayered follicles were visible at 3 wk of postnatal life, but why few follicles escaped early death is unclear. Sohlh2-deficient ovaries were atrophic and depleted of follicles by 3 wk of postnatal life. These observations implicate that Sohlh2 is required for survival and differentiation of oocytes after birth.
Expression of Transcription Factors in Sohlh2–/– Ovaries
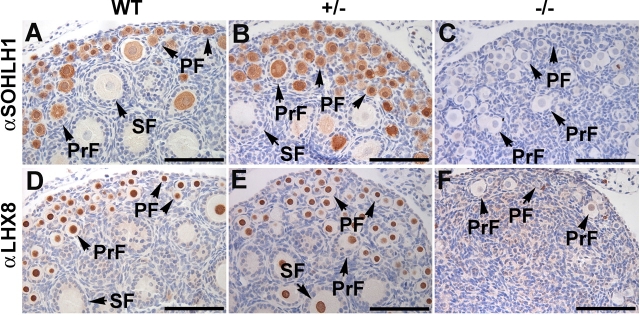
We examined expression of germ cell-specific transcriptional regulators Sohlh1 and Lhx8 in Sohlh2–/–ovaries (Fig. 4). Sohlh1, like Sohlh2, is a bHLH transcription factor preferentially expressed in oocytes of primordial follicles [14]. Sohlh1 is necessary for the expression of several key oocyte-specific genes, including Gdf9, Pou5f1, Nobox, and Lhx8 [14]. SOHLH1 protein is detectable in the oocytes of primordial and primary follicles of 7-day-old WT and Sohlh2+/– ovaries. However, SOHLH1 protein is not detectable by immunohistochemistry in the oocytes of Sohlh2–/–ovaries (Fig. 4, A–C). We also examined expression of LHX8 protein in Sohlh2-deficient ovaries. Lhx8 is a member of the LIM-homeobox transcription factor family and is preferentially expressed in germ cells and growing follicles in the mouse ovary [14]. Lhx8 regulates oocyte-specific genes such as Gdf9, Pou5f1, and Kit [19] and lies downstream of Sohlh1. Immunohistochemistry using anti-LHX8 antibodies shows that LHX8 protein is detectable in the nuclei of oocytes in the follicles of WT and Sohlh2+/– ovaries but not in Sohlh2–/–ovaries. Therefore, Sohlh2 deficiency is associated with lower expression of SOHLH1 and LHX8 proteins.
FIG. 4.
Expression of SOHLH1 and LHX8 proteins in 7-day-old ovaries. A–C) Anti-SOHLH1 antibodies (αSOHLH1) detected germ cell-specific transcription factor SOHLH1 in Postnatal Day 7 WT and Sohlh2+/– ovaries, as shown by brownish cytoplasmic staining of primordial follicles (PF), primary follicles (PrF), and secondary follicles (SF). SOHLH1 protein was not significantly expressed in Sohlh2–/– ovaries. D–F) Antibodies against LHX8 (αLHX8) localize LHX8 protein within the oocyte nuclei of PF, PrF, and SF in 7-day-old WT and Sohlh2+/– ovaries but not in Sohlh2–/– ovaries. Bar = 100 μm.
Molecular Changes in Sohlh2−/− Ovaries
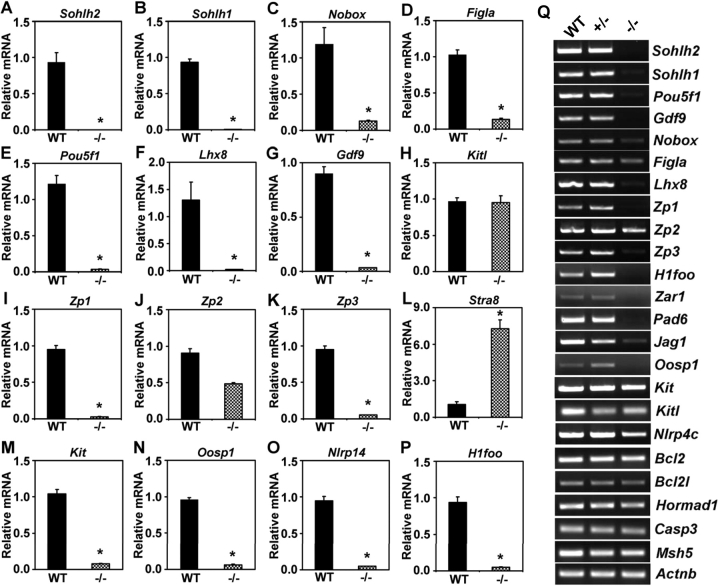
Sohlh2 is a germ cell-specific transcriptional regulator with an expression pattern that mimics that of Sohlh1. Histology of Sohlh2-deficient ovaries shows remarkable similarity to that of Sohlh1-deficient ovaries. We previously described how Sohlh1 deficiency causes misexpression of numerous genes preferentially expressed in the germ cells [14]. Therefore, we examined whether genes preferentially expressed in the oocyte are also misexpressed in Sohlh2–/– ovaries (Fig. 5). Transcript levels of many genes, including Sohlh1, Pou5f1, Nobox, Figla, Gdf9, Zp1, Zp3, and Lhx8, are reduced or undetectable in Sohlh2–/– ovaries compared with WT ovaries. Lower transcript levels of Sohlh1 in Sohlh2-deficient ovaries are consistent with the immunohistochemistry showing diminished levels of SOHLH1 protein in Sohlh2–/– ovaries (Fig. 4C). We previously described how Sohlh1 deficiency leads to decreased levels of Lhx8, Zp1, and Zp3 transcripts [14]. The Lhx8, Zp1, and Zp3 mRNAs are also significantly reduced in Sohlh2–/– ovaries (Fig. 5, F, I, and K). In addition to Lhx8, Zp1, and Zp3, the transcript levels of Pou5f1 and Gdf9 are drastically down-regulated in Sohlh2–/– ovaries (Fig. 5, E and G). We have previously shown that Pou5f1 and Gdf9 lie downstream of Nobox [13]. Nobox transcript levels are also reduced approximately 9-fold in Sohlh2–/– ovaries (Fig. 5C). The reduction of the level of Pou5f1 and Gdf9 transcripts in Sohlh2-deficient ovaries may be due to the decrease of Nobox transcripts in Sohlh2–/– ovaries. Figla transcript levels are also decreased approximately 8-fold in Sohlh2–/– ovaries (Fig. 5D). Figla is also known as a regulator of numerous oocyte-specific genes, including the zona pellucida genes [10, 11].
FIG. 5.
Misexpression of oocyte-specific genes in Sohlh2–/– newborn ovaries. A–P) Expression of select genes in WT and Sohlh2–/– newborn ovaries was assessed by real-time PCR: Sohlh2 (A), Sohlh1 (B), Nobox (C), Figla (D), Pou5f1 (E), Lhx8 (F), Gdf9 (G), Kitl (H), Zp1 (I), Zp2 (J), Zp3 (K), Stra8 (L), Kit (M), Oosp1 (N), Nlrp14 (O), and H1foo (P). Three independent pools of ovaries were used for each experiment, and each experiment was repeated in triplicate. Data are normalized to Gapdh expression and are given as the mean relative quantity (compared with WT), with error bars representing the SEM. Student t-test was used to calculate P values. *, P < 0.001. Q) RNA expression of selected genes in WT, Sohlh2+/–, and Sohlh2–/– newborn ovaries using semiquantitative RT-PCR.
We also examined the expression of genes implicated in germ cell survival such as Bax, Bcl2, Casp3, Kitl, and Kit. Bax, Bcl2, and Casp3 transcripts were not significantly misexpressed in Sohlh2–/– ovaries (Supplemental Fig. 1 [available at www.biolreprod.org]). However, Kit receptor is down-regulated in Sohlh2–/– ovaries (Fig. 5M), whereas its ligand, Kitl, is not misexpressed (Fig. 5H). The deficiency of Kit may explain the initial block in primordial to primary ovarian follicle transition. Although Sohlh2-deficient ovaries form primordial-like follicles, the oocytes are molecularly immature, as shown by the continual expression of Stra8 (stimulated by retinoic acid gene 8) transcripts in Sohlh2–/– newborn ovaries (Fig. 5L). Stra8 is highly expressed in the germ cells of the Embryonic Day 14 female gonad, but its expression declines drastically after Embryonic Day 15, so that no transcripts are detectable in the postnatal ovary [20]. Stra8 expression in Sohlh2–/– ovaries indicates that Sohlh2 may indirectly have a role in suppressing embryonic Stra8 expression.
We also discovered that several genes preferentially expressed in the oocytes, but with undefined function in ovarian development, are significantly misexpressed in Sohlh2–/– newborn ovaries. NLRP14 is a leucine-rich repeat protein and a member of the NACHT nucleoside triphosphatase family and is down-regulated almost 10-fold in Sohlh2–/– ovaries (Fig. 5O). In addition to NLRP14, OOSP1 (oocyte-secreted protein 1) and H1FOO (oocyte-specific H1 variant) mRNA levels are down-regulated in Sohlh2–/– ovaries (Fig. 5, N and P). OOSP1 is exclusively expressed in the oocytes [21]. OOSP1 is predicted to be secreted, but its role in the ovary remains unknown. H1FOO is a predominant linker protein in the mouse oocyte and remains expressed beyond fertilization [22]. It has been suggested that H1FOO is important for chromatin remodeling during development [22].
Misexpression of Key Genes Is Shared by Sohlh2 and Sohlh1 Mouse Mutants
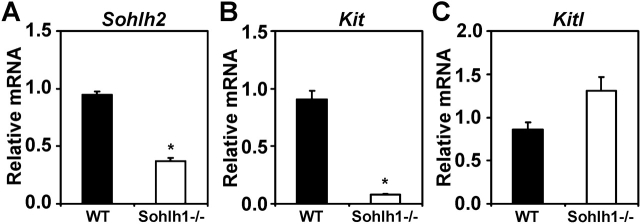
Sohlh1 and Sohlh2 share similar patterns of expression in the developing gonad, with protein and RNA expression uniquely and preferentially confined to the primordial oocytes. We have shown that Sohlh2 deficiency causes misexpression of transcriptional regulators Nobox, Figla, Lhx8, and Pou5f1, just as we previously observed in Sohlh1-deficient ovaries [14]. We also examined the expression of prosurvivor factors Kit and Kitl in Sohlh1-deficient ovaries. Kit is significantly down-regulated in Sohlh1–/– ovaries, as we discovered in Sohlh2–/– ovaries. Kitl transcripts are not misexpressed in Sohlh1 or Sohlh2 mutants (Fig. 6, B and C).
FIG. 6.
Sohlh2, Kit, and Kitl expression in Sohlh1-deficient ovaries. A–C) Results of real-time PCR are shown for relative mRNA expression of Sohlh2 (A), Kit (B), and Kitl (C) in WT and Sohlh1–/– ovaries. Data are normalized to Gapdh expression and are given as the mean relative quantity (compared with WT), with error bars representing the SEM. Student t-test was used to calculate P values. *, P < 0.001.
We also examined the expression of Sohlh2 and Sohlh1 relative to each other's deficiency. Sohlh2 transcript expression is reduced by almost 60% in Sohlh1-deficient newborn ovaries, while Sohlh1 expression is reduced by more than 90% in Sohlh2-deficient newborn ovaries (Fig. 6A).
DISCUSSION
We previously discovered Sohlh2 by using the BLAST program of the National Center for Biotechnology Information (Bethesda, MD) to discover bHLH domains that share homology to Sohlh1. SOHLH1 and SOHLH2 share a 47% amino acid identity in the bHLH domain but little homology outside of the bHLH domain. Tissue-specific bHLH proteins have critical roles in the differentiation of many tissues, as exemplified by MyoD and MyoD-like genes during muscle differentiation [23, 24] and typically bind to a consensus sequence called an E-box (CANNTG) [25].
Several germ cell-specific bHLH transcription factors have been shown to be critical in early differentiation of oocytes. For example, Figla and Sohlh1 are preferentially expressed in the gonads. FIGLA is a well-known bHLH transcription factor expressed in the embryonic ovary and throughout folliculogenesis [26]. Despite its expression in the embryonic ovary, Figla deficiency causes a block in the formation of primordial follicles [11]. Gene expression profiling in Figla-deficient embryonic and newborn ovaries, compared with that in WT ovaries, shows that significant RNA changes are detectable after birth but that few RNA changes are observed before birth [10]. SOHLH1 is another bHLH transcription factor that is preferentially expressed in germ cells, but (unlike Figla) Sohlh1 expression is confined to primordial oocytes [27]. Sohlh1 deficiency causes rapid postnatal loss of oocytes so that by 2 wk after birth most germ cells are lost [14, 27]. Primordial-like structures form in Sohlh1-deficient ovaries. Sohlh2 (like Sohlh1) is preferentially expressed after birth in primordial and primary ovarian follicles [15]. Sohlh2-deficient mice phenocopy Sohlh1 remarkably well. Sohlh2 deficiency caused accelerated postnatal loss of oocytes, and early histology showed oocyte growth but absent proliferation and differentiation of surrounding somatic cells. Few advanced follicular types were visualized in Sohlh2–/– ovaries, with complete loss of oocytes and follicles. The phenotype in Sohlh2 knockouts is remarkably similar to that of Sohlh1, and our results show that the role of Sohlh2 is not redundant in the ovary.
Similar to Sohlh1–/– mice, Sohlh2 deficiency causes fast elimination of oocytes from the ovary. Extensive loss of oocytes occurs during normal fetal ovarian development, but the molecular explanation of this process is yet to be understood. Apoptosis (programmed cell death) is critical in the regulation of ovarian function and development. Factors such as Bax, Bcl2, and Casp3 are considered primary executioners and regulators of ovarian follicle death or survival. Bcl2 is a prosurvival factor expressed in the developing follicles [28]. Bcl2 deficiency causes reduction in the number of primordial oocytes; however, Bcl2-null mice are fertile [29]. Sohlh2–/– ovaries express Bcl2, although its transcription levels are reduced in the mutant (Supplemental Fig. 1B). Bax is known as a pro-apoptotic member of the Bcl2 family, and Bax-deficient ovaries contain increased number of oocytes per ovary [30]. Bax transcripts were not misexpressed in Sohlh2–/– ovaries (Supplemental Fig. 1A). Caspases mediate the process of ovarian apoptosis. Casp3 is expressed in the granulosa cells of atretic follicles [31]. Casp3 knockouts have little effect on early folliculogenesis [32], and Casp3 transcripts levels are not affected in Sohlh2–/– ovaries (Supplemental Fig. 1C). Sohlh2–/– mice phenotypically do not phenocopy any of the known apoptosis gene deficiencies. Further studies are required to investigate the contribution of the apoptosis pathway to the postnatal loss of oocytes in the Sohlh2 mutants. In addition to apoptotic genes, Kit and Kitl are known to have a critical role for the survival and differentiation of germ cells [33–35]. Kit is significantly reduced in Sohlh1–/– and Sohlh2–/– ovaries (Fig. 5M), whereas Kitl is not misexpressed in the mutants. It is unclear whether Kit is directly regulated by Sohlh1 or Sohlh2. However, the reduction of Kit transcript in Sohlh1–/– and Sohlh2–/– ovaries may account for the accelerated loss of oocytes after birth.
Sohlh2–/– ovaries misexpress two bHLH transcription factors, Sohlh1 and Figla, which are known as transcriptional regulators and express numerous oocyte-specific genes in the ovaries [10, 14]. We have previously shown that SOHLH1 can bind Lhx8, Zp1, and Zp3 promoters via E-box [14]. Lhx8, Zp1, and Zp3 are significantly down-regulated in Sohlh2–/– ovaries (Fig. 5). FIGLA regulates the expression of numerous oocyte-specific genes [10] that are also misexpressed in Sohlh1–/– and Sohlh2–/– ovaries. Although FIGLA, SOHLH1, and SOHLH2 share a common genetic pathway, none of the factors are redundant.
The phenotype observed in Sohlh2–/– ovaries may be in part due to the diminished expression of Sohlh1 and Figla. It is also likely that these transcription factors cross-regulate each other, directly or indirectly, because Sohlh2 expression is also diminished in Sohlh1-deficient ovaries.
Stra8 is up-regulated in Sohlh2–/– ovaries (Fig. 5L). Stra8 is normally expressed in germ cells at approximately Embryonic Day 14.5, a time when germ cells enter meiosis I, and thereafter disappears, with no detectable postnatal expression [20]. Germ cells in Stra8-null ovaries fail to undergo premeiotic DNA replication, meiotic chromosome condensation, cohesion, synapsis, and recombination [36]. The presence of Stra8 in Sohlh2–/– ovaries suggests that Sohlh2 may have a role in suppressing Stra8 expression in the postnatal ovary. These results raise a question of whether Stra8 suppression is critical for completion of meiosis I before birth. Meiotic genes such as Msh5, Rec8, and Spo11 [37–40] are not misexpressed in Sohlh2–/– newborn ovaries (Supplemental Fig. 1). However, we cannot rule out the absence of novel yet unidentified germ cell-specific components of meiosis.
Sohlh2 is down-regulated in Sohlh1–/– ovaries (Fig. 6A), and Sohlh1 is down-regulated in Sohlh2–/– ovaries (Fig. 5B). Based on real-time PCR, it seems that Sohlh1 expression is more severely down-regulated in Sohlh2–/– ovaries. As already described, genes down-regulated in Sohlh2–/– ovaries are also down-regulated in Sohlh1–/– ovaries (Fig. 5). These results indicate that Sohlh1 and Sohlh2 share similar pathways and potentially cross-regulate each other's transcription directly or indirectly. We do not know whether SOHLH1 and SOHLH2 can form heterodimers to effect oocyte differentiation or whether synergism between the two pathways exists. Clearly, neither Sohlh1 nor Sohlh2 is redundant because both knockouts display severe phenotypes. Future studies are necessary to determine whether synergism between these two pathways exists.
Supplementary Material
Footnotes
1Supported by the National Institute of Health Grant HD44858, and March of Dimes grant #6-FY08-313.
REFERENCES
- Pepling ME.From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis 2006; 44: 622–632.. [DOI] [PubMed] [Google Scholar]
- Pepling ME, Spradling AC.Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol 2001; 234: 339–351.. [DOI] [PubMed] [Google Scholar]
- Kezele P, Skinner MK.Regulation of ovarian primordial follicle assembly and development by estrogen and progesterone: endocrine model of follicle assembly. Endocrinology 2003; 144: 3329–3337.. [DOI] [PubMed] [Google Scholar]
- Dean J.Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J Reprod Immunol 2002; 53: 171–180.. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM.Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 1996; 383: 531–535.. [DOI] [PubMed] [Google Scholar]
- Dube JL, Wang P, Elvin J, Lyons KM, Celeste AJ, Matzuk MM.The bone morphogenetic protein 15 gene is X-linked and expressed in oocytes. Mol Endocrinol 1998; 12: 1809–1817.. [DOI] [PubMed] [Google Scholar]
- Elvin JA, Yan C, Matzuk MM.Growth differentiation factor-9 stimulates progesterone synthesis in granulosa cells via a prostaglandin E2/EP2 receptor pathway. Proc Natl Acad Sci U S A 2000; 97: 10288–10293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M, Wang X, Wolgemuth DJ, Scholer H.Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev 1998; 71: 89–98.. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM.NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 2004; 305: 1157–1159.. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davies H, Sims LP, Levy SE, Dean J.Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol 2007; 7: e67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyal SM, Amleh A, Dean J.FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development 2000; 127: 4645–4654.. [DOI] [PubMed] [Google Scholar]
- Choi Y, Qin Y, Berger MF, Ballow DJ, Bulyk ML, Rajkovic A.Microarray analyses of newborn mouse ovaries lacking Nobox. Biol Reprod 2007; 77: 312–319.. [DOI] [PubMed] [Google Scholar]
- Choi Y, Rajkovic A.Characterization of NOBOX DNA binding specificity and its regulation of Gdf9 and Pou5f1 promoters. J Biol Chem 2006; 281: 35747–35756.. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, Rajkovic A.Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A 2006; 103: 8090–8095.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballow DJ, Xin Y, Choi Y, Pangas SA, Rajkovic A.Sohlh2 is a germ cell-specific bHLH transcription factor. Gene Expr Patterns 2006; 6: 1014–1018.. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG.Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 2005; 33: e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Rajkovic A, Viveiros MM, Burns KH, Eppig JJ, Matzuk MM.Identification of Gasz, an evolutionarily conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile-alpha motif, and a basic leucine zipper. Mol Endocrinol 2002; 16: 1168–1184.. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Yan MSC, Klysik M, Matzuk M.Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil Steril 2001; 76: 550–554.. [DOI] [PubMed] [Google Scholar]
- Choi Y, Ballow DJ, Xin Y, Rajkovic A.Lim homeobox gene, Lhx8, is essential for mouse oocyte differentiation and survival. Biol Reprod 2008; 79(3):442–449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke DB, Koubova J, Page DC.Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol 2003; 262: 303–312.. [DOI] [PubMed] [Google Scholar]
- Yan C, Pendola FL, Jacob R, Lau AL, Eppig JJ, Matzuk MM.Oosp1 encodes a novel mouse oocyte-secreted protein. Genesis 2001; 31: 105–110.. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Hennebold JD, Macfarlane J, Adashi EY.A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 2001; 128: 655–664.. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R.MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 1993; 75: 1351–1359.. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD.Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A 1989; 86: 5434–5438.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, Weintraubf H, Baltimoreb D.Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 1989; 58: 537–544.. [DOI] [PubMed] [Google Scholar]
- Liang L, Soyal SM, Dean J.FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 1997; 124: 4939–4947.. [DOI] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A.Sohlh1 is essential for spermatogonial differentiation. Dev Biol 2006; 294: 161–167.. [DOI] [PubMed] [Google Scholar]
- Van Nassauw L, Tao L, Harrisson F.Distribution of apoptosis-related proteins in the quail ovary during folliculogenesis: BCL-2, BAX and CPP32. Acta Histochem 1999; 101: 103–112.. [DOI] [PubMed] [Google Scholar]
- Ratts VS, Flaws JA, Kolp R, Sorenson CM, Tilly JL.Ablation of bcl-2 gene expression decreases the numbers of oocytes and primordial follicles established in the post-natal female mouse gonad. Endocrinology 1995; 136: 3665–3668.. [DOI] [PubMed] [Google Scholar]
- Perez GI, Robles R, Knudson CM, Flaws JA, Korsmeyer SJ, Tilly JL.Prolongation of ovarian lifespan into advanced chronological age by Bax-deficiency. Nat Genet 1999; 21: 200–203.. [DOI] [PubMed] [Google Scholar]
- Fenwick MA, Hurst PR.Immunohistochemical localization of active caspase-3 in the mouse ovary: growth and atresia of small follicles. Reproduction 2002; 124: 659–665.. [DOI] [PubMed] [Google Scholar]
- Takai Y, Canning J, Perez GI, Pru JK, Schlezinger JJ, Sherr DH, Kolesnick RN, Yuan J, Flavell RA, Korsmeyer SJ, Tilly JL.Bax, caspase-2, and caspase-3 are required for ovarian follicle loss caused by 4-vinylcyclohexene diepoxide exposure of female mice in vivo. Endocrinology 2003; 144: 69–74.. [DOI] [PubMed] [Google Scholar]
- Dolci S, Williams DE, Ernst MK, Resnick JL, Brannan CI, Lock LF, Lyman SD, Boswell HS, Donovan PJ.Requirement for mast cell growth factor for primordial germ cell survival in culture. Nature 1991; 352: 809–811.. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Toksoz D, Nishikawa S, Nishikawa S, Williams D, Zsebo K, Hogan BL.Effect of Steel factor and leukaemia inhibitory factor on murine primordial germ cells in culture. Nature 1991; 353: 750–752.. [DOI] [PubMed] [Google Scholar]
- Sakata S, Sakamaki K, Watanabe K, Nakamura N, Toyokuni S, Nishimune Y, Mori C, Yonehara S.Involvement of death receptor Fas in germ cell degeneration in gonads of Kit-deficient Wv/Wv mutant mice. Cell Death Differ 2003; 10: 676–686.. [DOI] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC.In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 2006; 38: 1430–1434.. [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M.Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci U S A 2005; 102: 737–742.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, Kolodner R, Pollard JW, Kucherlapati R.Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet 1999; 21: 123–127.. [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD.The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000; 6: 975–987.. [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ.Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 2005; 8: 949–961.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.