Abstract
Mesenchymal stem cells (MSCs) are considered to be one of the most promising therapeutic cell sources as they encompass a plasticity of multiple cell lineages. The challenge in using these cells lies in developing well-defined protocols for directing cellular differentiation to generate a desired lineage. In this study, we investigated the effect of 5-azacytidine, a DNA demethylating agent, on osteogenic differentiation of MSCs. The cells were exposed to 5-azacytidine in culture medium for 24 h prior to osteogenic induction. Osteogenic differentiation was determined by several the appearance of a number of osteogenesis characteristics, including gene expression, ALP activity, and calcium mineralization. Pretreatment of MSCs with 5-azacytidine significantly facilitated osteogenic differentiation and was accompanied by hypomethylation of genomic DNA and increased osteogenic gene expression. Taking dlx5 as a representative, methylation alterations of the “CpG island shore” in the promoter caused by 5-azacytidine appeared to contribute to osteogenic differentiation.
Keywords: 5-Azacytidine, Mesenchymal stem cells, Osteogenic differentiation, DNA methylation, Epigenetic
Introduction
Bone tissue engineering and reconstructive surgery have become a major focus in the newly emerging field of regenerative medicine (Meijer et al. 2007). In this respect, stem cell-based transplantation therapy appears to hold particular promise. Mesenchymal stem cells (MSCs) are considered to be one of the most useful cell sources for clinical application in tissue regeneration, including bone repair, because they can produce multiple tissues, such as bone, cartilage, fat, tendon, muscle, liver and neurons (Koc and Lazarus 2001; Anjos-Afonso et al. 2004). Compared with embryonic stem (ES) cells, the therapeutic applications of MSCs present several advantages; for example, MSCs are readily accessible from bone marrow, they possess lower risks of immunorejection or of teratoma formation and they are not subject to the same ethical controversies (Chen et al. 2008).
The essential prerequisite for the clinical application of MSCs is to develop well-defined protocols for directing cellular differentiation into a distinct lineage, followed by in vitro selective isolation and proliferation. This necessitates reducing the likelihood of spontaneous differentiation of MSC into divergent lineages, which could in turn reduce the efficacy of cell transplantation therapy. There have been several protocols available for osteogenic differentiation from MSCs; for example, the protocol based on a chemical cocktail comprising dexamethasone, l-ascorbic acid (vitamin C), and beta-glycerol phosphate (Jaiswal et al. 2000). However, a highly reproducible method to acquire abundant functional osteoblasts still remains elusive.
In recent years chemicals such as histone deacetylase inhibitors (HDACi), which participate in epigenetic modification by histone acetylation of chromatins, have emerged as a new class of chemotherapeutic drugs for cancer clinical therapy. Their main virtue lies in their ability to regulate the expression of specific genes involved in proliferation, differentiation, and apoptosis (Chen et al. 2006). Because of their biological similarities to cancer cells, stem cells have also been tested for responses to HDACi, especially in terms of stem cell specialization. Thus, differentiation events such as cardiomyocyte differentiation from embryonic stem cells (Hosseinkhani et al. 2007), neuronal differentiation from neural stem cells (Hsieh et al. 2004) and osteogenic differentiation from MSCs (Chen et al. 2007) have been investigated. These studies in turn have initiated new investigations into the relationship between epigenetic modifications of chromatin and stem cell differentiation.
DNA methylation is one of the most important epigenetic mechanisms, with known involvement in diverse genetic events such as gene expression, chromatin modification, X chromosome inactivation, genomic imprinting, and endogenic gene silencing (Sulewska et al. 2007). In addition, alterations in DNA methylation are closely related to the presentation of many diseases, including cancer. DNA methylation is also crucial for maintaining pluripotency and self-renewal of stem cells.
Genes that maintain pluripotency are usually activated when hypomethylated, while genes associated with differentiation are repressed by hypermethylation (Fouse et al. 2008). Currently, much attention is being paid to the effects of DNA methylation on stem cell differentiation. Nevertheless, relatively little documentation exists regarding the effects of DNA demethylation on stem cell differentiation. In the present study, we used 5-azacytidine, an analog of cytidine and a useful demethylating agent for epigenetic research (Christman 2002), to examine osteogenic differentiation in MSCs and its underlying mechanism. Pretreatment of MSCs with 5-azacytidine considerably reduced methylation of promoters, resulting in increased osteogenic gene expression and cellular differentiation. The results presented here provide new insight into the role of DNA methylation in stem cell specialization, and introduce a well-defined and efficient strategy for promoting osteogenesis in MSCs.
Materials and methods
Experimental animals
A total of 6–8-week-old male ICR mice obtained from the Laboratory Animal Unit of Zhejiang Academy of Medical Sciences (Hangzhou, People’s Republic of China) were used in the experiments.
Isolation and culture of MSCs from mouse bone marrow
Mice were sacrificed by cervical dislocation, bone marrow cells were collected by flushing femurs with Iscove’s modified Dulbecco’s medium (IMDM, Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone, Rockville, MD), 100 U/mL penicillin (Gibco BRL, Rockville, MD), and 100 mg/mL streptomycin (Gibco BRL) (Medium A). Cells were seeded on T-175 flasks (Greiner Bio-One GmbH, Frickenhausen, Germany) at concentration of 5 × 106/mL cells. After 24 h, non-adherent cells and debris were removed. Cells were harvested by 0.25% trypsin-EDTA (Sigma) when they had grown to 90% confluence. For most of the experiments, we used MSCs at the 3rd–5th passage. Media were changed twice a week during culture.
Identification of isolated MSCs
The isolated MSCs were identified by their antigen expression profiles. Briefly, cells were harvested by trypsinization and incubated with the following rat to mouse monoclonal antibodies: FITC-conjugated CD11b and CD45, PE-conjugated CD44, CD73, CD90 and SCA-1 (all from Caltag Laboratories, San Diego, CA, USA). The cell fluorescence signals were determined using a FACScan flow cytometer (Becton Dickinson, San Jose, CA) equipped with an argon laser, with emission at 488 nm. At least 10,000 events were collected. Data were analyzed with Cell Quest Software (Becton Dickinson).
Effect of 5-azacytidine on MSC viability
To evaluate appropriate dosage for subsequent experiments, the cytotoxic effects of 5-azacytidine on MSCs was examined. Cells were cultured in 96-well plates at a concentration of 2 × 103 cells/well in Medium A, and various concentrations (0, 2.5, 5, 10, 20, 40, 80, 120 or 160 μM) of 5-azacytidine were added when cells had reached 50% confluence. After 24 h, cellular viability was assessed with the MTT assay (Freimoser et al. 1999). Cell numbers were also counted by hemocytometer. Measurements were performed in quadruplicate, and percent viability was calculated relative to the untreated samples.
Osteogenic induction in MSCs
To test for osteogenic induction, MSCs at 50% confluence were pretreated with different concentrations (0, 10, 20 or 40 μM) of 5-azacytidine for 24 h. The cells were then incubated in a differentiation medium for 2 weeks, with medium changes every 3 days. The differentiation medium was composed of IMDM supplemented with 10% FBS, 10−8 M dexamethasone (Calbiochem, Darmstadt, Germany), 50 μM ascorbic acid 2-phosphate (Fluka Chemie GmbH, Buchs, Switzerland), and 10 mM β-glycerol phosphate (Sigma).
Alkaline phosphatase assays
Alkaline phosphatase (ALP) activity was detected by biochemical assay and histochemical staining of cells after osteogenic induction for 7 or 14 days. In the biochemical assay, cells were washed three times in PBS and resuspended in lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate). After sonication, the lysates were spun at 3,000 rpm for 15 min at 4 °C. Enzyme activity in the supernatants was determined using an ALP Detection Kit (Sigma) according to the manufacturer’s instructions. For histochemical staining of ALP, a modified Gomori staining was performed as previously reported (Imre and Fekete 1983). The percentage of cells staining positive for ALP was determined by counting cell numbers in 10 contiguous fields after random starts.
Determination of calcium mineralization
Cells were fixed in 4% paraformaldehyde after 14 days of culture, and stained with 0.1% alizarin red S (Sigma) for 20 min. Mineralization was quantified by measurement of the average photodensity of calcium mineralization with a pathology image analysis system (Mike Audi Image Analysis Inc. China). The data from 5-azacytidine pretreated samples were presented as a percentage of control. Measurements were carried out in duplicate and each experiment was repeated at least three times.
Osteogenic gene expression by real-time PCR analysis
After osteogenic induction for 7, 11 or 14 days, total RNA was isolated using TRIZOL (Invitrogen) according to the manufacturer’s instructions and quantified by UV spectroscopy. One microgram of total RNA was reverse-transcribed to complementary DNA using Super Script III first-strand synthesis system (Invitrogen) with oligo (dT) and random hexamer primers. Real-time PCR was performed with Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) according to the manufacturer’s instructions. The PCR profile was as follows: 1 cycle at 94 °C for 2 min, 40 cycles at 94 °C for 20 s, 65 °C for 20 s, and 72 °C for 20 s. Signals were detected with a Realplex5 Real-Time PCR System instrument (Eppendorf, Germany). Gene-specific primers for GAPDH, dlx5, runx2, col1a1, osterix, and osteocalcin are listed in Table 1. All transcript levels were normalized to that of GAPDH.
Table 1.
Primers and annealing temperatures used for RT-PCR and bisulfite sequencing
| Gene | Sequence (5′ → 3′) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|
| RT-PCR | |||
| β-actin-F | ACACCTTCTACAATGAGCTG | 816 | 56 |
| β-actin-R | CTGCTTGCTGATCCACATCT | ||
| Runx2-F | CCTGAACTCTGCACCAAGTC | 234 | 55 |
| Runx2-R | GAGGTGGCAGTGTCATCATC | ||
| Col1a1-F | TGGACGCCATCAAGGTCTACTGC | 455 | 56 |
| Col1a1-R | GGAGGTCTTGGTGGTTTTGTATTCG | ||
| Osterix-F | CCTCTGCGGGACTCAACAAC | 355 | 56 |
| Osterix-R | TGCCTGGACCTGGTGAGATG | ||
| Osteocalcin-F | CAGACAAGTCCCACACAGCAGC | 165 | 56 |
| Osteocalcin-R | TGTTCACTACCTTATTGCCCTCC | ||
| Bisulfite sequencing | |||
| Dlx5 region1-F | TTAGTAGGTAGGAAATAATGGG | 707 | 52 |
| Dlx5 region1-R | CGATTCTTAATACTCTTTTCTTACT | ||
| Dlx5 region1-F | TGGAGTGGATGTAGGTAATG (nested) | 446 | 55 |
| Dlx5 region1-R | TATAAACTAAAAAACAATTAAACAC (nested) | ||
| Dlx5 region2-F | GAGTTATGATAGGAGTGTTTGAT | 365 | 50 |
| Dlx5 region2-R | CCTACCTAACTAACTAATAACACT | ||
| Dlx5 region2-F | TGTTTGATAGAAGAGTTTTAAGTATT (nested) | 183 | 56 |
| Dlx5 region2-R | AAACAAAAATAAAAAAACAATAACC (nested) | ||
Collagen type I immunofluorescence staining
MSCs were fixed in 4% paraformaldehyde after 14 days of culture. After washing with PBS, they were blocked with 5% normal goat serum and then incubated with Rat Anti-Mouse monoclonal antibodies against collagen type I (1:100, Abcam, USA). Sequentially, the samples were incubated with secondary FITC-labeled goat anti-rat antibodies (1:200 Caltag Laboratories) to detect the primary antibodies. Images were collected and analyzed with a Zeiss LSM 510 laser scanning confocal microscope (Germany).
Methylation analysis of genomic DNA
Genome-wide methylation levels in 5-azacytidine pretreated MSCs were analyzed by a combination of methylation-insensitive digestion method that cuts CCGG sites regardless of methylation status (Msp I, TaKaRa) and methylation-sensitive digestion (Hpa II, TaKaRa), that cuts only if the internal site C is unmethylated. Total genomic DNA was isolated using a DNeasy Tissue Kit (Qiagen, Hilden, Germany) as per the manufacturer’s instructions. After treatment with Rnase A for 30 min at 37 °C, 2 μg of genomic DNA was digested with Hpa II and Msp I (20 U in 40 μL reaction volume) for 16 h at 37 °C. Digestion products (20 μL) were visualized by electrophoresis in an ethidium bromide stained 0.8% agarose gel.
Bisulfite sequencing
Bisulfite conversion was performed as previously described (Irizarry et al. 2009). Briefly, total genomic DNA was isolated from 5-azacytidine-pretreated MSCs using a DNeasy Tissue Kit (Qiagen). Two micrograms of genomic DNA were denatured in a volume of 50 μL by freshly prepared NaOH (final concentration 0.3 M) for 30 min at 42 °C. After denaturation, 30 μL freshly prepared Hydroquinone (10 mM) and 510 μL Sodium bisulfite (3.6 M, pH 5.0) were added, and incubated at 50 °C for 16 h. Modified DNA was purified using the DNeasy spin column (Qiagen) and eluted in 50 μL. This was followed by desulfonification by adding 5.5 μL NaOH (3 M) for 15 min at 37 °C. Samples were neutralized by adding 33 μL ammonium acetate (10 M, pH 7.0), followed by ethanol precipitation and resuspension in water. Nested primers (shown in Table 1) were used for PCR analysis, and the products were gel purified using the Qiaex II Gel Extraction kit (Qiagen). They were then cloned into pUCm-T vector (Sangon, China) and transformed into E. coli strain DH5a. DNA samples from six positive clones per original set of cells were sequenced. The dlx5 promoter sequence was analyzed at UCSC (http://genome.ucsc.edu/), and the patterns of methylation were evaluated using DNAMAN (Lynnon Corporation, Canada).
Statistical analysis
All data were presented as the mean value ± standard deviation (SD) of each group. Variation between groups was evaluated using the Student’s t-test, with a confidence level of 95% (P ≤ 0.05) being considered statistically significant.
Results
Characterization of the isolated MSCs
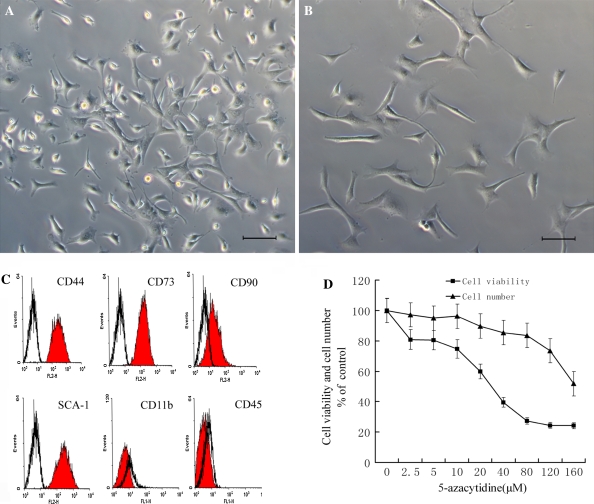
The freshly isolated MSCs from murine bone marrow consisted predominantly of round-shaped erythrocytes and nonadherent hematopoietic cells. After 24 h, nonadherent cells were removed by changing the culture medium. Cells with a spindle-shaped characteristic appeared to be predominant among the attached cells by day 5 (Fig. 1a). MSCs were subcultured when they had grown to 90% confluence. Most of the 3rd passage MSCs presented a fibroblast-like phenotype (Fig. 1b). In the immunophenotypic analyses, the majority of the cells at passage 3 were negative for CD11b and CD45, while positive for CD44, CD73 and SCA-1 (Fig. 1c), also known as the cell-surface antigens of MSCs (Dominici et al. 2006; Chen et al. 2008).
Fig. 1.
The identification and cell viability assay of MSCs. a Primary culture MSCs. b The 3rd passage MSCs with homogeneous fibroblast-like morphology. Scale bars are 20 μm. c Flow cytometry analysis of MSCs. Flow cytometry histograms demonstrate the typical expression pattern of surface antigens. The filled areas indicate the cells stained with PE-conjugated antibodies against CD 44, CD 73, CD 90, CD SCA-1 and FITC-conjugated antibodies against CD11b and CD45, whereas the empty areas indicate the isotype-matched monoclonal antibody control. d Cytotoxicity Assay of MSCs. Cells were treated with various concentrations of 5-azacytidine for 24 h, then cell viability was measured with MTT and cell numbers were counted. Relative levels of cell viability were expressed as percentage of the untreated control
It is shown that about 77% of MSCs in our research were CD90 positive cells, indicating that the MSCs were not a homogeneous population. MSC cultures have been reported to consist of two different cell types, i.e., slowly renewing MSCs (SR-MSCs) and rapidly renewing MSCs (RS-MSCs). The latter has little or no expression of CD90 (Delorme et al. 2006). Thus, the MSCs isolated in our experiment may contain both of these cell populations, although the majority seems to be SR-MSCs.
Cytotoxic effect of 5-azacytidine on MSCs
The cytotoxic effect of 5-azacytidine on MSCs was determined by cellular viability analysis and cell counting assays. Little decrease in cell viability or cell numbers was seen when MSCs were treated for 24 h with 5-azacytidine at concentrations below 10 μM (Fig. 1d). The 50% inhibitory concentration of 5-azacytidine for MSCs was about 40 μM, at which level the cellular viability was reduced by nearly 50% and the cell numbers were decreased by 15% when compared with the untreated control groups. Therefore, concentrations between 0 μM (control) and 40 μM were considered to be moderate and were chosen for use in subsequent experiments.
Alkaline phosphatase assay
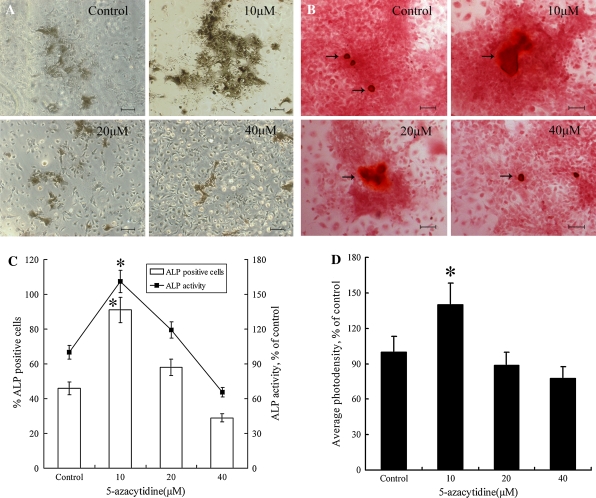
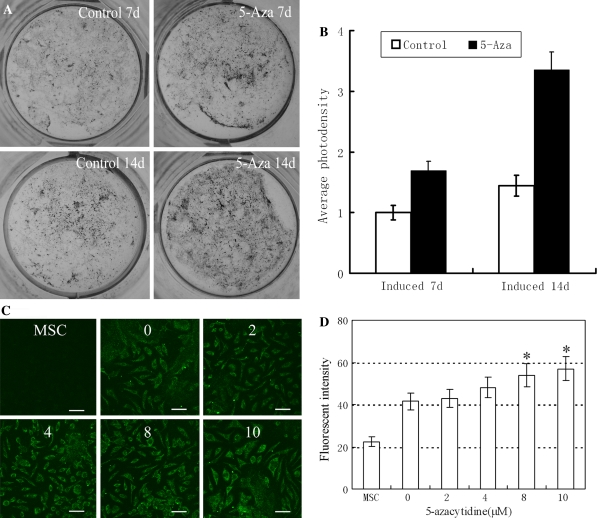
Alkaline phosphatase (ALP), a membrane-bound enzyme abundant in early bone formation, plays important roles in osteogenesis. As such, it is widely used as an differentiation marker associated with osteogenesis (Dimai et al. 1998; Avbersek-Luznik et al. 2007). In the present study, about 46% of the cells were capable of developing into ALP positive cells under normal inducing condition, without pre-treatment with 5-azacytidine. However, when cells were pretreated with 10 μM 5-azacytidine, the percentage of ALP positive cells was increased to 91%, which was significantly (P < 0.05) higher than the control groups (Fig. 2a, c). In parallel, the activity of ALP was significantly (P < 0.05) up-regulated in the 5-azacytidine pretreated group, which was in accordance with that of ALP positive cells (Fig. 2c). Treatment of cells with 5-azacytidine at concentrations higher than 10 μM decreased the percentages of ALP positive cells and ALP activity, indicating that 10 μM was an optimal dosage for the induction of osteogenic differentiation. In our experiment, two groups of MSCs were pretreated with 10 μM 5-azacytidine for 7 or 14 days. Expression of ALP was clearly increased by 5-azacytidine in both time treatments (Fig. 4a, b).
Fig. 2.
Analysis of ALP activity and mineralization of induced MSCs. MSCs were pretreated with different concentrations (0, 10, 20 or 40 μM) of 5-azacytidine for 24 h and analyzed at the 14th day after osteogenic induction. a Modified Gomori staining. b Analysis of mineralization by Alizarin red S staining. The black arrows indicate the mineralized nodules. c Quantitative analysis of ALP. The broken line represents the ALP activity in cell lysates and the columns represent the percentages of ALP staining positive cells. d Photodensity analysis of Alizarin red S staining. (*P ≤ 0.05). Scale bars are 50 μm
Fig. 4.
ALP Assay and Collagen type I immunofluorescence staining. a Modified Gomori staining, MSCs pretreated with 10 μM 5-azacytidine for 24 h were induced in an osteogenic system for 7 or 14 days. b Average photodensity analysis of (a). c MSCs pretreated with various concentrations (0, 2, 4, 8 or 10 μM) of 5-azacytidine for 24 h were induced in osteogenic system for 14 days before collagen type I immunofluorescence staining. The images were collected and analyzed with Zeiss LSM 510 laser scanning confocal microscope. Scale bars are 20 μm. d Fluorescence intensity assay of collagen type I (*P ≤ 0.05)
Calcium mineralization determination
Alizarin red S staining was used to determine the level of calcium mineralization, which was also an indicator of the degree of osteogenic differentiation. The presence of 10 μM 5-azacytidine resulted in a deeper intensity of alizarin red S staining, which indicated enhanced calcium mineralization compared with control groups (Fig. 2b, d). However, when the concentration reached 40 μM, the calcium mineralization intensity declined. This result was consistent with that obtained for ALP assays. Therefore, 10 μM 5-azacytidine was chosen to promote osteogenic differentiation in all subsequent experiments.
Changes in gene expression in 5-azacytidine pretreated MSCs
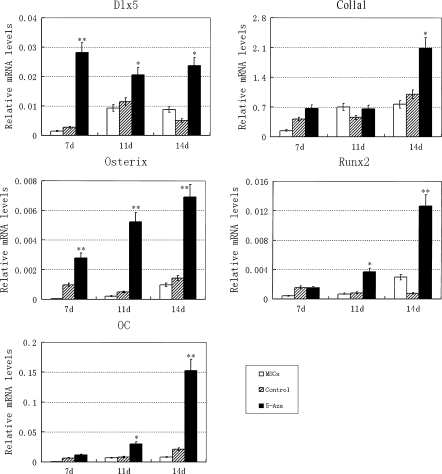
The expressions of runx2, col1a1, osterix, and osteocalcin are hallmarks of osteogenesis, while dlx5 is an important transcription factor that regulates runx2 and osterix expression. In our experiments, expressions of dlx5, runx2, col1a1, osterix and osteocalcin were increased following 5-azacytidine treatment at day 7, 11, or 14 (Fig. 3). Pretreatment of MSCs with a suitable concentration of 5-azacytidine therefore could promote osteogenic differentiation.
Fig. 3.
RT-QPCR analysis of osteogenic genes. MSCs pretreated with 10 μM 5-azacytidine for 24 h were induced for 7, 11 or 14 days in osteogenic medium before analysis. Each gene expression was normalized to GAPDH (*P ≤ 0.05, **P ≤ 0.01)
Dose-dependent effects
Osteogenic differentiation was evaluated by examining the levels of collagen type I, another hallmark of osteogenic differentiation. As shown in Fig. 4c, d, the fluorescence intensity of collagen type I increased in a dose-dependent manner in groups pretreated for 24 h with a range of 5-azacytidine concentrations between 2 and 10 μM.
Methylation analysis of genomic DNA
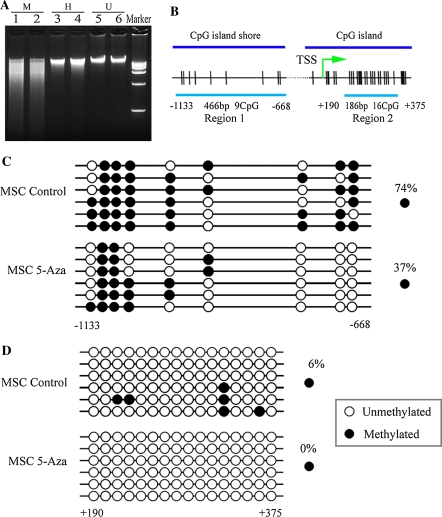
Genomic DNA was digested with a pair of isoschizomers, Msp I and Hpa II, which possess different sensitivities to DNA methylation. As shown in Fig. 5a, genomic DNA from both control and 5-azacytidine pretreated MSCs was completely digested by Msp I, but only partially digested by Hpa II. In the Hpa II digestion experiment, the DNA sample isolated from 5-azacytidine pretreated MSCs appeared to be more sensitive to Hpa II in comparison with that from the control cells. This suggested that methylation might be essential for the growth and maintenance of MSCs. Treatment of MSCs with 5-azacytidine substantially decreased the methylation level of genomic DNA, in agreement with previous observations that 5-azacytidine acts as a demethylating chemical in cancer cells (Christman 2002; Halaban et al. 2009). Hypomethylation of genomic DNA might therefore be involved in osteogenic differentiation.
Fig. 5.
Effect of 5-azacytidine on DNA methylation. a Methylation analysis of genomic DNA. MSCs were grown for 24 h in the absence (lanes 1, 3 and 5) or presence of 10 μM 5-azacytidine (lanes 2, 4 and 6). Lanes represent undigested (U), or digested with Msp I (M) or Hpa II (H). Tracks labeled kb contain kilobase DNA marker with standard bands of 15, 10, 7.5, 5.0, 2.5 and 1.0 kb. DNA was visualized on ethidium bromide-stained 0.8% agarose gel. b Map of CpG dinucleotides (tick marks) examined in two regions of dlx5 promoter. Region1 and Region2 represent the CpG island shore and the CpG island, respectively. Regions examined are indicated by the continuous line. Numbers are relative to the transcription start site (TSS). c, d Bisulfite sequencing of Region1 and Region2 sequences, respectively in the dlx5 promoter after 24 h of 5-azacytidine treatment. Control is the same region in untreated MSCs. Six bacterial clones of PCR products were sequenced. Each row represents one bacterial clone with one circle symbolizing one CpG. Closed circles, methylated CpG; open circles, unmethylated CpG. Positions relative to the transcription start are indicated
Bisulfite sequencing of the dlx5 promoter
To determine whether the increased expression of osteogenic genes in MSCs had an underlying epigenetic basis, the DNA methylation status in dlx5 promoter was examined by bisulfite sequencing. A schematic overview of the promoter structure is shown in Fig. 5b. Two regions in the promoter were selected, one (region1, −1,133 to −668) was in the CpG island shore and the other (region2, +190 to +375) was in the CpG island, which is located from −400 to +929 bp relative to the transcription start site. CpG hypermethylation was detected within the CpG island shore in control MSCs. After treatment with 10 μM 5-azacyticine for 24 h, the methylation level of this region was clearly reduced from 74 to 37% (Fig. 5c). Methylation was almost completely absent at region2 of the CpG island where CpG density is high, but DNA methylation in this region still decreased in response to 5-azacytidine (Fig. 5d).
Discussion
Differentiation involves a number of key cellular changes involving physiology, structural architecture, and function (Yeo et al. 2007). The derivation of specific somatic cells from pluripotent stem cells also occurs in a well organized and programmed manner. Every event in the course of differentiation should therefore be accompanied by coordinated expression and repression of different subsets of genes (Yeo et al. 2007). DNA methylation is one of epigenetic mechanisms known to regulate chromatin organization and gene expression, but is also a reversible process, which allows for differentiation-associated gene expression. Therefore, elucidating the mechanistic relationship between DNA methylation patterns and gene regulation during stem cell differentiation should improve our understanding of differentiation-associated cellular changes. This, in turn, may facilitate the development of manipulation procedures that will allow the control of stem cell differentiation into a desired cell type.
For studies of DNA methylation, 5-azacytidine is a very effective tool, as it can be incorporated into DNA to form covalent adducts with cellular DNA methyltransferase 1 (DNMT1). This results in a decrease in the activity of DNMT1 and causes demethylation of genomic DNA (Juttermann et al. 1994). At this point in time, 5-azacytidine is now widely used in cancer therapies as an anticancer agent. It is believed that hypomethylation evoked by 5-azacytidine can increase expression of a great number of genes, such as the tumor suppressor genes p16, p21 and p53, by changing the epigenetic modification patterns of the cellular DNA (Fang and Lu 2002; Egger et al. 2007; Wu et al. 2007). However, whether 5-azacytidine is directly involved in stem cell differentiation remains in question (Liu et al. 2003; Antonitsis et al. 2007; Burlacu et al. 2008). There is little documentation to show effects of 5-azacytidine on osteogenic differentiation or to explain its mode of action during this process. For example, 5-azacytidine has been reported to modulate the expression of alkaline phosphatase, but had no effect on the production of osteocalcin in human bone marrow fibroblasts or osteoblast-like cell (MG63) models (Locklin et al. 1998). In the present study, we investigated the effects of 5-azacytidine on the osteogenic differentiation of MSCs. Pretreatment of MSCs with an appropriate concentration of 5-azacytidine significantly promoted osteogenic differentiation, as determined by enhanced expression of dlx5, runx2, col1a1, osterix, and osteocalcin, by increased activity of ALP, and by facilitated mineralization of calcium. Furthermore, the 5-azacytidine-facilitated osteogenic differentiation was accompanied by the hypomethylation of genomic DNA, suggesting that epigenetic regulation mediated by DNA demethylation occur during osteogenic differentiation.
As further evidence, methylation patterns of an osteogenic differentiation-regulated gene (dlx5) were examined. The methylation changes of the dlx5 promoter occurred in both the “CpG island” region and the “CpG island shore” regions, the latter of which has been recently found to be important for the DNA methylation-mediated regulation of gene expression (Irizarry et al. 2009). In MSCs, CpG hypermethylation was present within the “CpG island shore” region, whereas little methylation was detected in the “CpG island” region (Fig. 5c, d). After treatment with 5-azacyticine, hypermethylation of the “CpG island shore” region was significantly reduced from 74 to 37%, while there was still little methylation in the “CpG island” region. This suggested that methylation in “CpG island shore”, rather than in “CpG island”, as previously presumed, was involved in the regulation of dlx5 expression. Methylation alteration in the “CpG island shore” caused by 5-azacytidine appeared to be a potential contributor to osteogenic differentiation.
Another possible mechanism for 5-azacytidine-induced cell differentiation may be that a cell subpopulation is selected for by the treatment of a cell population with this chemical (Halaban et al. 2009). However, the present study indicated that 5-azacytidine-elevated osteogenic gene expression and differentiation were accompanied by a decrease in DNA methylation, but not in cell numbers. Thus, DNA demethylation, rather than cell selection, may largely contribute to 5-azacytidine-promoted osteogenic differentiation.
In mammals, DNA methylation is restricted to the CpG dinucleotides, which are largely depleted from the genome except at short genomic CpG-rich regions called CpG islands that are commonly located at promoters. For a long period of time, it was generally believed that DNA methylation and its mediated epigenetic regulation occurred within the CpG islands (Issa 2004; Sato and Meltzer 2006; Teodoridis et al. 2008; Harder et al. 2009), and little attention was paid to the methylation in CpG islands ‘shores’. However, recent research has shown that most methylation alterations in colon cancer occur in CpG islands shores (Irizarry et al. 2009). Thus, the observation that methylation in CpG islands shores was involved in MSC gene regulation may provide new insights into the role of DNA methylation in stem cell differentiation.
Acknowledgments
This work was supported by grant J20020579-30116 from the Key Science and Technology Foundation of Zhejiang Province, People’s Republic of China.
Footnotes
Guo-Shun Zhou and Xiao-Lei Zhang contributed equally.
Contributor Information
Guo-Shun Zhou, Phone: +86-572-2023301, FAX: +86-572-2216961, Email: gfz@mail.huptt.zj.cn.
Jian-Zhong Shao, Phone: +86-571-88206582, FAX: +86-571-88206582, Email: shaojz@zju.edu.cn.
References
- Anjos-Afonso F, Siapati EK et al (2004) In vivo contribution of murine mesenchymal stem cells into multiple cell-types under minimal damage conditions. J Cell Sci 117:5655–5664. doi:10.1242/jcs.01488 [DOI] [PubMed]
- Antonitsis P, Ioannidou-Papagiannaki E et al (2007) In vitro cardiomyogenic differentiation of adult human bone marrow mesenchymal stem cells: the role of 5-azacytidine. Interact Cardiovasc Thorac Surg 6:593–597. doi:10.1510/icvts.2007.157875 [DOI] [PubMed]
- Avbersek-Luznik I, Gmeiner Stopar T et al (2007) Activity or mass concentration of bone-specific alkaline phosphatase as a marker of bone formation. Clin Chem Lab Med 45:1014–1018. doi:10.1515/CCLM.2007.186 [DOI] [PubMed]
- Burlacu A, Rosca AM et al (2008) Promoting effect of 5-azacytidine on the myogenic differentiation of bone marrow stromal cells. Eur J Cell Biol 87:173–184. doi:10.1016/j.ejcb.2007.09.003 [DOI] [PubMed]
- Chen J, Ghazawi FM et al (2006) Valproic acid and butyrate induce apoptosis in human cancer cells through inhibition of gene expression of Akt/protein kinase B. Mol Cancer 5:71. doi:10.1186/1476-4598-5-71 [DOI] [PMC free article] [PubMed]
- Chen TH, Chen WM et al (2007) Sodium butyrate activates ERK to regulate differentiation of mesenchymal stem cells. Biochem Biophys Res Commun 355:913–918. doi:10.1016/j.bbrc.2007.02.057 [DOI] [PubMed]
- Chen Y, Shao JZ et al (2008) Mesenchymal stem cells: a promising candidate in regenerative medicine. Int J Biochem Cell Biol 40:815–820. doi:10.1016/j.biocel.2008.01.007 [DOI] [PubMed]
- Christman JK (2002) 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21:5483–5495. doi:10.1038/sj.onc.1205699 [DOI] [PubMed]
- Delorme B, Chateauvieux S et al (2006) The concept of mesenchymal stem cells. Regen Med 1:497–509. doi:10.2217/17460751.1.4.497 [DOI] [PubMed]
- Dimai HP, Linkhart TA et al (1998) Alkaline phosphatase levels and osteoprogenitor cell numbers suggest bone formation may contribute to peak bone density differences between two inbred strains of mice. Bone 22:211–216. doi:10.1016/S8756-3282(97)00268-8 [DOI] [PubMed]
- Dominici M, Le Blanc K et al (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 8:315–317. doi:10.1080/14653240600855905 [DOI] [PubMed]
- Egger G, Aparicio AM et al (2007) Inhibition of histone deacetylation does not block resilencing of p16 after 5-aza-2′-deoxycytidine treatment. Cancer Res 67:346–353. doi:10.1158/0008-5472.CAN-06-2845 [DOI] [PubMed]
- Fang JY, Lu YY (2002) Effects of histone acetylation and DNA methylation on p21 (WAF1) regulation. World J Gastroenterol 8:400–405 [DOI] [PMC free article] [PubMed]
- Fouse SD, Shen Y et al (2008) Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2:160–169. doi:10.1016/j.stem.2007.12.011 [DOI] [PMC free article] [PubMed]
- Freimoser FM, Jakob CA et al (1999) The MTT [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide] assay is a fast and reliable method for colorimetric determination of fungal cell densities. Appl Environ Microbiol 65:3727–3729 [DOI] [PMC free article] [PubMed]
- Halaban R, Krauthammer M et al (2009) Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: clinical implications. PLoS ONE 4:e4563. doi:10.1371/journal.pone.0004563 [DOI] [PMC free article] [PubMed]
- Harder J, Engelstaedter V et al (2009) CpG-island methylation of the ER promoter in colorectal cancer: analysis of micrometastases in lymph nodes from UICC stage I and II patients. Br J Cancer 100:360–365. doi:10.1038/sj.bjc.6604859 [DOI] [PMC free article] [PubMed]
- Hosseinkhani M, Hasegawa K et al (2007) Trichostatin A induces myocardial differentiation of monkey ES cells. Biochem Biophys Res Commun 356:386–391. doi:10.1016/j.bbrc.2007.02.151 [DOI] [PubMed]
- Hsieh J, Nakashima K et al (2004) Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA 101:16659–16664. doi:10.1073/pnas.0407643101 [DOI] [PMC free article] [PubMed]
- Imre R, Fekete P (1983) A rapid technique for alkaline phosphatase enzyme activity in tissues. Acta Histochem 73:17–21 [DOI] [PubMed]
- Irizarry RA, Ladd-Acosta C et al (2009) The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41:178–186. doi:10.1038/ng.298 [DOI] [PMC free article] [PubMed]
- Issa JP (2004) CpG island methylator phenotype in cancer. Nat Rev 4:988–993 [DOI] [PubMed]
- Jaiswal RK, Jaiswal N et al (2000) Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen-activated protein kinase. J Biol Chem 275:9645–9652. doi:10.1074/jbc.275.13.9645 [DOI] [PubMed]
- Juttermann R, Li E et al (1994) Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA 91:11797–11801. doi:10.1073/pnas.91.25.11797 [DOI] [PMC free article] [PubMed]
- Koc ON, Lazarus HM (2001) Mesenchymal stem cells: heading into the clinic. Bone Marrow Transplant 27:235–239. doi:10.1038/sj.bmt.1702791 [DOI] [PubMed]
- Liu Y, Song J et al (2003) Growth and differentiation of rat bone marrow stromal cells: does 5-azacytidine trigger their cardiomyogenic differentiation? Cardiovasc Res 58:460–468. doi:10.1016/S0008-6363(03)00265-7 [DOI] [PubMed]
- Locklin RM, Oreffo RO et al (1998) Modulation of osteogenic differentiation in human skeletal cells in vitro by 5-azacytidine. Cell Biol Int 22:207–215. doi:10.1006/cbir.1998.0240 [DOI] [PubMed]
- Meijer GJ, de Bruijn JD et al (2007) Cell-based bone tissue engineering. PLoS Med 4:e9. doi:10.1371/journal.pmed.0040009 [DOI] [PMC free article] [PubMed]
- Sato F, Meltzer SJ (2006) CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer 106:483–493. doi:10.1002/cncr.21657 [DOI] [PubMed]
- Sulewska A, Niklinska W et al (2007) DNA methylation in states of cell physiology and pathology. Folia histochemica et cytobiologica / Polish Academy of Sciences. Pol Histochem Cytochem Soc 45:149–158 [PubMed]
- Teodoridis JM, Hardie C et al (2008) CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett 268:177–186. doi:10.1016/j.canlet.2008.03.022 [DOI] [PubMed]
- Wu YH, Tsai Chang JH et al (2007) Xeroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene 26:4761–4773. doi:10.1038/sj.onc.1210284 [DOI] [PubMed]
- Yeo S, Jeong S et al (2007) Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem Biophys Res Commun 359:536–542. doi:10.1016/j.bbrc.2007.05.120 [DOI] [PubMed]