Abstract
One of the central goals of human genetics is the identification of loci with alleles or genotypes that confer increased susceptibility. The availability of dense maps of single-nucleotide polymorphisms (SNPs) along with high-throughput genotyping technologies has set the stage for routine genome-wide association studies that are expected to significantly improve our ability to identify susceptibility loci. Before this promise can be realized, there are some significant challenges that need to be addressed. We address here the challenge of detecting epistasis or gene-gene interactions in genome-wide association studies. Discovering epistatic interactions in high dimensional datasets remains a challenge due to the computational complexity resulting from the analysis of all possible combinations of SNPs. One potential way to overcome the computational burden of a genome-wide epistasis analysis would be to devise a logical way to prioritize the many SNPs in a dataset so that the data may be analyzed more efficiently and yet still retain important biological information. One of the strongest demonstrations of the functional relationship between genes is protein-protein interaction. Thus, it is plausible that the expert knowledge extracted from protein interaction databases may allow for a more efficient analysis of genome-wide studies as well as facilitate the biological interpretation of the data. In this review we will discuss the challenges of detecting epistasis in genome-wide genetic studies and the means by which we propose to apply expert knowledge extracted from protein interaction databases to facilitate this process. We explore some of the fundamentals of protein interactions and the databases that are publicly available.
Keywords: protein-protein interaction, expert knowledge, epistasis, MDR, SNP
Introduction
Measuring a million or more single nucleotide polymorphisms (SNPs) across the entire human genome is now technically and economically possible. As a result, the domain of human genetics is experiencing an information explosion and, at the same time, an understanding implosion. That is, our ability to generate genetic data is far outpacing our ability to make sense of it. This is particularly true given the complexity of the genotype and phenotype mapping relationship that results from phenomena such as epistasis, or gene-gene interaction, and plastic reaction norms, or gene-environment interaction. There are several important challenges that must be overcome before we can take full advantage of genome wide data to decompose the genetic architecture of a complex trait into its component interacting parts (Moore and Ritchie 2004).
First, there is the need for powerful statistical methods to model the relationship between combinations of SNPs and disease susceptibility. Genotype combinations increase exponentially with the number of SNPs being analyzed, and there may not be enough subjects to represent each combination. As a result, traditional parametric statistical approaches, such as logistic regression, are not effective due to inaccurate parameter estimates. A second challenge is the selection of which SNPs should be included in the analysis. Common human diseases are likely to be the result of multiple interacting genetic factors that may not necessarily also exhibit independent effects. This means that methods that employ exhaustive searches are necessary to fully explore interactions in the absence of significant independent effects. Due to the computational intensity of analyzing all pair-wise and multiple-way SNP combinations, conducting genome wide genetic studies with thousands of SNPs is computationally infeasible. A third challenge is the interpretation of gene-gene interaction models. Although we are able to obtain a statistical model of interaction that confers risk for disease, we still need to be able to interpret these results in the context of human biology so that they may be used for the benefit of disease treatment and prevention.
The goal of this essay is to explore the challenges of detecting epistasis in genome-wide association studies and to explore the use of expert knowledge from public databases to ease the computational burden of detecting and characterizing interactions. It is already known that depending on where the SNPs occur in the genome they can have an effect on protein structure by changing amino acid sequences. These can be insertions, deletions, repetitions, non-sense, mis-sense, or silent mutations. Whether they are in a coding region of the genome or not, these SNPs may affect protein-protein interaction, protein expression, alternative splicing, stability, folding, ligand binding, or catalysis, thus inducing or influencing disease state in individuals (Wang and Moult 2001, Cavallo and Martin 2005). We focus here on the role of information extracted from protein-protein interaction databases for improving the computational efficiency of genome-wide studies of epistasis.
Challenges of Detecting Epistasis
Recognized for many years, epistasis has been described from two different perspectives, biological and statistical (Cordell 2002; Moore and Williams 2005). Biological epistasis, as defined by Bateson (1909) who coined the term, results from physical interactions among biomolecules in gene regulatory networks and biochemical pathways at the cellular level that are dependent on the genotypes of an individual. Statistical epistasis, as defined by Fisher (1918), is deviation from additivity in a linear mathematical model that describes the relationship between multilocus genotypes and phenotype variation at the population level. While determining the relationship between the two remains a challenge, it is an important endeavor if we wish to infer biological conclusions from statistical results.
Epistasis, along with other phenomena such as locus heterogeneity, phenocopy, and gene-environment interaction are major sources of complexity in the mapping relationship between genotype and phenotype in common human diseases. Detecting and characterizing epistasis to gain an understanding of the genetic susceptibility to these diseases is not a simple task. Traditional methods of analysis such as linear and logistic regression have had limited success due to the sparseness of data in high dimensions. For example, when interactions among multiple SNPs are considered, there are many multilocus genotype combinations that may have very few or no data points. This can lead to an increase in type I and type II errors due to parameter estimates with very large standard errors. It is evident that we need research strategies that embrace, rather than ignore, the complexity of the relationship between genotype and phenotype (Templeton 2000; Moore 2003; Sing et al. 2003; Thornton-Wells et al. 2004; Moore and Williams 2005; Rea et al. 2006).
Multifactor dimensionality reduction (MDR) was developed as a nonparametric and model-free data mining method for detecting, characterizing, and interpreting epistasis in the absence of significant independent effects in genetic and epidemiologic studies of complex traits such as disease susceptibility (Ritchie et al. 2001, 2003; Hahn et al. 2003; Hahn and Moore 2004; Moore 2004; Moore et al. 2006; Moore 2007). The goal of MDR is to change the representation of the data using a constructive induction algorithm to make non-additive interactions easier to detect using any classification method such as naïve Bayes or logistic regression (Moore et al. 2006; Moore 2007). This is accomplished by first labeling each genotype combination as high-risk or low-risk using some function of a discrete endpoint such as case-control status. A new MDR variable with two levels is constructed by pooling all high-risk genotype combinations into one group and all low-risk combinations into another group. Traditionally, MDR constructed variables have been evaluated with a probabilistic naïve Bayes classifier that is combined with 10-fold cross validation to obtain an estimate of predictive accuracy or generalizability of epistasis models. While the MDR method has proven to be an effective way of detecting epistasis in a number of diseases, to analyze all of the combinations of SNP interactions in large datasets or genome-wide studies would be impractical even with access to the largest and most powerful computers available.
To illustrate the scope of such an analysis, consider a recent report from the International HapMap Consortium (Altshuler et al.2005) that suggests that approximately 300,000 carefully selected SNPs may suffice to represent all of the relevant genetic variation across the human Caucasian genome. If this is to be regarded as the lower limit of a genome-wide association study, then approximately 4.5 ×1010 pairwise combinations (300,000 choose 2) and 4.5 × 1015 three-way combinations (300,000 choose 3) would need to be exhaustively analyzed to detect low-order epistasis using MDR. If 106 MDR evaluations can be computed each second, then evaluation of each individual SNP would require less than one second of computer time. However, computing all two-way and three-way MDR models would require more than 52,000 days of computer time. Access to a 1,000 processor supercomputer might reduce this to 52 days which is within the realm of possibility. However, extending this to all four-way combinations is not computationally feasible. This adds to the many challenges of detecting epistasis on a genome-wide scale (Moore and Ritchie 2004).
As previously mentioned, an important and understandably difficult goal in human genetics is to determine which of the many thousands of SNPs are useful for predicting who is at risk for common diseases. It was nearly a decade ago that Risch and Merikangas first seriously proposed the testing of all known SNPs in the human genome for disease association either directly or by linkage disequilibrium with other SNPs (Risch and Merikangas 1996), and today this ‘genome-wide’ approach is expected to revolutionize the genetic analysis of common human diseases (Hirschhorn and Daly 2005; Wang et al. 2005). Currently, it is possible to measure more than a million SNPs with a genome-wide human SNP array available from Affymetrix, and Illumina has released the Human1M DNA Analysis BeadChip that is capable of profiling 1,000,000 SNPs on a single array across the human genome.
Certainly, with these technologies now available to the scientific community at more affordable costs, it’s clear to see how such large amounts of data are being rapidly produced. Unfortunately, due to the lack of logical methods to summarize this quantity of information within a biological context, investigators are at a loss when they reach the analysis stage in their research. In fact, our ability to measure genetic information, and biological information in general, is far outpacing our ability to interpret it. It is probable that most of these large scale studies harbor a wealth of information about susceptibility genes that can be used to improve the prevention, diagnosis, and treatment of common diseases. However, to access this information to our full advantage, we need to address the specific technical challenges that confront researchers in the analysis process, such as the computational limitation of a large-scale genetic analysis with methods such as MDR and our ability to interpret it. We believe that we can overcome this limitation by utilizing expert knowledge about our data to guide our analysis as well as the biological interpretation of our results.
Expert Knowledge
Expert knowledge can be defined as existing biological or statistical information about the problem at hand that can be incorporated into the analysis process to guide an algorithm in a more directed fashion. For example, when considering SNPs or genes in a genetic analysis, biological expert knowledge may be derived from what is known about the function of biochemical pathways, Gene Ontology (GO), or expression information for that gene. Some software packages make explicit use of expert knowledge from multiple sources. For example, the bioPIXIE system makes use of existing biological expert knowledge to improve the accuracy of its process-specific network prediction in S. cerevisiae (Myers et al. 2005). This system takes user input from the biologist who enters proteins they want to evaluate for functional relationships or that they expect to play a role in the same biological process. The query is directed to a confidence-weighted network based on integrated data for additional related proteins thus outputting a predicted network that is probabilistically biased towards the biological processes represented in the initial of query proteins. bioPIXIE integrates gene expression data, physical and genetic interaction data, and sequence and literature data with a Bayesian network. Biological expert knowledge, such as that implemented in bioPIXIE, can also be extracted from a number of other sources such as from pathway and gene information from Kyoto Encyclopedia of Genes and Genomes (KEGG) or the Gene Ontology database (GO), from literature based information by mining PubMed abstracts, or from interaction information found in protein interaction databases, which we will focus on in the remainder of this review.
In addition to biological information, it is also useful to use prior statistical knowledge to help guide an epistasis analysis. For example, LOD scores from a prior linkage analysis could be used to weight SNPs in certain chromosomal regions higher during a combinatorial epistasis analysis. That is, SNPs from a certain pathway or chromosomal region would be evaluated for interactions with a higher probability than others in the dataset. Statistical knowledge could also come from filter algorithms that explicitly assess the quality of a SNP based on their relationship with the clinical endpoint. The Tuned ReliefF (TuRF) algorithm is an example of an algorithm that can assign high quality scores to SNPs involved in complex interactions (Moore and White 2007). The TuRF algorithm uses a nearest neighbor approach to assess SNP quality and thus doesn’t suffer from the computational limitations of an algorithm that explicitly considers combinations of SNPs. As such, it is very useful for preprocessing the data prior to analysis. Once computed, the TuRF scores can be used to select some reduced number of SNPs for combinatorial analysis or can be used to help guide a computational search algorithm.
Protein-Protein Interactions
Proteomics and the study of protein-protein interactions (PPI’s) are becoming increasingly important in our effort to understand human diseases on a system wide level. While mass spectrometry has been a useful technology applied to the discovery of components in protein complexes (Yates 2000), PPI’s have traditionally been measured using a variety of assays such as immunoprecipitation and yeast two-hybrid (Y2-H). To reconstruct the entire network of protein-protein interactions within cells remains a challenge, yet this is becoming a more approachable problem. The field of proteomics is advancing and the aforementioned techniques to detect PPI’s have been scaled up to measure interactions on a genome-wide level. High-throughput techniques have also been developed to identify protein complexes using affinity pull-down followed by mass spectrometry (Pelligrini et al. 2004), and systematically constructed double knockout strains in yeast have proven to be useful for constructing a large-scale view of genetic interaction networks (Tong et al. 2001). To complement these experimental techniques, a number of computational methods have been developed that are capable of identifying pairs of proteins that have co-evolved, suggesting that these proteins may interact within that cell (Pelligrini et al. 1999, Pelligrini et al. 2004, Tan et al. 2004). Thanks to the development and curation of protein interaction databases, which will be elaborated on later, up-to-date information on these interaction networks is accessible and publicly available to the scientific community.
Some of these techniques have been actively employed in the study of the pathogenesis of Huntington’s disease (HD), an autosomal neurodegenerative disorder that causes cognitive impairment, psychiatric problems, and motor dysfunction. This inherited disease is caused by the expansion of a polyglutamine (polyQ) tract in the huntingtin (htt) protein. Although this protein was discovered over a decade ago, constructing the protein interaction network that it belongs to is still an ongoing process that is providing clues about the function of htt and its role in HD pathogenesis (Li, S. and Li, X. 2004). Many interaction partners for mutant and wild-type htt have been elucidated over the past decade by Y2-H, affinity chromatography, and immunoprecipitation. To follow these studies, recently, it was hypothesized that genetic modifiers of HD nuerodegeneration should be enriched among htt protein interactors and to test this, both high-throughput Y2-H screening and affinity pull-down followed by mass spectrometry were utilized (Kaltenbach et al. 2007). This group was able to identify 104 htt interactions with Y2-H and 130 interactions with their pull-down method. To elucidate the biological relevance of these interactions, with a high-content validation assay, they also tested a set of 60 genes encoding interacting htt proteins for their ability to act as genetic modifiers of nuerodegeneration in the HD Drosophila model. Results showed that 45% of these genes were high confidence genetic modifiers, much higher than the 1%–4% observed in unbiased genetic screens, and that these genes were similarly represented among proteins discovered with their Y2-H and pull-down/mass spectrometry methods. These results demonstrate that these methods are equally useful for identifying biologically relevant interactions (Kaltenbach et al. 2007).
Making use of information available in PPI databases such as KEGG and the Human Protein Reference Database (HPRD) along with PubMed publications, another group evaluated the commonality of molecular pathogenic mechanisms of neurodegenerative disorders including HD along with Alzheimer’s disease (AD), Parkinson’s disease (PD), dentatorubral-pallidoluysian atrophy (DRPLA) and prion disease (PRION), and amyotrophic lateral sclerosis (ALS). They examined the PPI networks associated with causative proteins, such as htt, and found 19 proteins common to all diseases from literature as well as 81 new common proteins from their network constructed using database information. Many of these proteins identified were previously characterized as being associated with the respective diseases. A relatively high correlation between all diseases for all of their analysis was seen including commonality in characteristic protein domains. They concluded that the interactions found in this study in silico may serve to function in the common pathogeneic mechanisms among neurodegenerative disorders (Limviphuvadh et al. 2007).
As the effort continues to reconstruct the entire proteome, it would be to our advantage to exploit the breadth of knowledge contained in PPI databases. Not only will we gain a greater understanding of numerous biological processes, but also presumably be able to apply this knowledge to advance other fields of research such as drug discovery, disease prognosis, and the study of disease susceptibility. High-throughput molecular profiling approaches, such as microarray technology, have already been successful in the advancement of these fields, yet, likewise, a rate limiting step has been the ability to interpret the biological meaning of the data. Often this problem has been approached from a pathway perspective that involves investigating which pathways are perturbed in a case-control population, which pathways determine a good or bad prognosis, or which pathways are activated or repressed in response to a certain stimuli or compounds (Pelligrini et al. 2004). Such an approach can simplify the analysis and interpretation of genome-wide or large-scale datasets. We wish to seek similar solutions to the challenges we encounter when attempting to detect epistatic interactions in large datasets with methods such as MDR.
Human Protein Interaction Databases
Currently, there exist numerous publicly available protein interaction databases that contain information about human specific interactions (Table 1). The majority of PPI’s in these databases are from curation of the literature by biologists, however some are incorporated by direct deposit prior to publication by the investigator (Mathivanan et al. 2006). In a majority of the PPI databases, the user will enter a protein of choice either by protein name or accession number according to RefSeq, Genbank, OMIM, SwissProt, or Entrez Gene, and in return will receive a list of protein interactors, information pertaining to the experimental evidence for that interaction, as well as information about the protein itself. Another common feature of most databases is the ability to visualize the network of the queried protein and its interactors. We will touch on some of the key features of certain databases. For a more comprehensive review and additional information on these databases please see (Mathivanan et al. 2006).
Table 1.
Lists the protein interaction databases reviewed, citations for these databases, and the website from which they can be accessed.
| PPI Database | Citation | Website |
|---|---|---|
| HPRD | Mishra G et al. (2006) Nucleic Acids Res 34: D411–D414 | http://www.hprd.org |
| BioGRID | Breitkreutz BJ et al. (2008). Nucleic Acids Res 36: D637–D640 | http://www.thebiogrid.org |
| BIND | Alfarano C et al. (2005) Nucleic Acids Res 33: D418–D424 | http://bind.ca |
| MINT | Chatr-aryamontri A et al. (2007) Nucleic Acids Res 35: D572–D574 | http://mint.bio.uniroma2.it |
| DIP | Salwinski L et al. (2004) Nucleic Acids Res 32: D449–D451 | http://dip.doe-mbi.ucla.edu |
| Reactome | Vastrik I et al. (2007) Genome Biol 8: R39 | http://www.reactome.org |
| STRING | von Mering C et al. (2007) Nucleic Acids Res 35: D358–D362 | http://string.embl.de |
| GeneNetwork | Franke L et al. (2006) Am J Hum Genet 78: 1011–1025 | http://www.genenetwork.nl |
| UniHI | Chaurisa G et al.(2007) Nucleic Acids Res 35: D580–4 | http://www.mdc-berlin.de/unihi |
One of the largest publicly available databases is the Human Protein Reference Database (HPRD), which to date has over 38,000 PPIs, over 270,000 pub-med links, access to curated pathways, as well as information about post-translational modifications (PTMs), domain architecture, protein functions, enzyme–substrate relationships, subcellular localization, tissue expression, and disease association of genes. An interesting feature of this database is the Protein Distributed Annotation System that enables researchers to annotate proteomic information in the context of HPRD data so that it is easily shared with the rest of the scientific community. Another large and growing database that has similar components is BioGrid, which currently houses approximately 42,800 human PPI’s, but altogether contains > 200,000 interactions from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Mus musculus, and Drosophila melanogaster in addition to Homo Sapiens.
Other available databases that are smaller than the HPRD and BioGrid yet offer additional unique features are The Biomolecular Interaction Network Database (BIND), the Molecular Interaction database (MINT), the Database of Interacting Proteins (DIP), and Reactome. For example, BIND and MINT provide confidence scores for each interaction, specifically for Y2-H experiments in BIND. In MINT, this score is based off of the number of interactions, the number of citations, and the type of experiment conducted to detect that interaction, while in BIND the score is based off of shared or related GO annotations, phenotypic profiling, homologous interactions, domain structure, and the number publications (Mathivanan et al., 2006). MINT also contains information pertaining to protein interactions with promoter regions and mRNA. Unique to DIP, the user can select to have certain PPIs evaluated based on paralogous interactions, common expression profiles of interactors, or through domain interaction preferences.
Reactome is not specifically a PPI database, but a curated resource for human pathway data based on biologic reaction networks. Reactome reactions are described as taking place between ‘physical entities’ which include not only proteins, but also nucleic acids, single small molecules, macromolecular complexes, and even subatomic particles. All proteins, genes, and reactions are cross-referenced to a variety of widely used databases such as Entrez Gene, Online Mendelian Inheritance in Man (OMIM), and KEGG, and each reaction is supported by evidence from biomedical literature as well as documented with approved citations (Vastrik et al., 2007). The user has the ability to search the database using a reaction name, gene name, protein name, or any of several alternative identifiers. Reactions in the output are represented graphically, and the user has the option to click on ‘top level’ pathways to delve deeper into the hierarchy with increasing detail at each level. Additionally, one can select for non-human species and all accession numbers for all genes and proteins involved can be downloaded. For a more in-depth review of this database please see (Vastrik et al. 2007).
Resources such as the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), Unified Human Interactome (UniHI), and GeneNetwork access a number of the reviewed databases to integrate protein interaction information. STRING incorporates PPI information from HPRD, BioGrid, MINT, BIND, DIP, as well as imports known reactions from Reactome and KEGG pathways. Automated textmining of PubMed abstracts and information from other databases such as the Saccharomyces Genome Database (SGD) and WormBase supplement this information. For interactions in organisms that have not been confirmed experimentally, STRING is capable of running a set of prediction algorithms and transferring known interactions from model organisms to other species based on the prediction orthology for those proteins (von Merring et al. 2007). The user however, has the option to select which organism the queried protein and its interactors will pertain to. Each interaction is given a numerical confidence score based on the experimental evidence and orthological evidence behind that interaction, which allows the user to filter networks according to a desired confidence threshold.
UniHI integrates protein-protein interactions not only from large Y2-H screens and curated databases such as HPRD, DIP, BIND, Reactome, and others we haven’t discussed but also predicts interactions based on orthology and computational textmining approaches. This database also provides detailed information about each interaction including statistical interaction validation by gene co-expression data and validation by shared path length according to GO co-annotation hierarchy. The source of the interactions are also documented and provided along with links to access more about that particular source of information. A useful feature of UniHI is that it allows for a highly target search by which the user can exclude certain mapping approaches like Y2-H, display only proteins that are common interaction partners to multiple proteins in a query, display only interactions in that occur in multiple maps, or display only direct interactions (Chaurasia et al. 2007).
GeneNetwork is comprised of known interactions from BIND, HPRD, Reactome, and KEGG. Similar to STRING, GeneNetwork supplies predicted interactions based on biological process and molecular function annotation from the GO database. Additional experimental data is incorporated such as co-expression data from ~450 microarrays from the Stanford Microarray Database (SMD) and the NCBI Gene Expression Omnibus (GEO). Human yeast two-hybrid and interactions based on orthologous high-throughput protein-protein interactions from lower eukaryotes are also included. After submitting a query for a given gene, the user is returned a list of interactors each of which have an overall likelihood score along with likelihood scores for that interaction based on microarray co-expression, human PPI prediction, and orthologous PPI predictions. Positive evidence of known interactions from HPRD, BIND, KEGG, and Reactome is indicated in additional columns. A recent study used this database to rank the best positional candidates in susceptibility loci on the basis of their interactions using a method they developed known as the “Prioritizer” (Franke et al. 2006).
A recent study examining protein-protein interaction networks for human inherited neurodegenerative disorders characterized by ataxia (i.e loss of balance or coordination) illustrates how these databases have been used to help us better understand pathogenic mechanisms underlying human diseases. Lim et al., (2006) examined protein interaction networks involved in cerebellar Purkinje cell degeneration which is the primary cause of coordination and balance loss in inherited ataxias. They developed a network for 54 proteins involved in 23 ataxias first by Y2-H screens and then expanded this network based on information from literature-curated and evolutionarily conserved interactions. Relevant direct PPI’s were added from available interaction networks developed by Rual et al., 2005, and Stelzl et al., 2005, and binary interactions were indentified for the 54 ataxia associated baits and 561 interacting prey proteins using literature based information from, BIND, HPRD, DIP, MINT, and the mammalian protein-protein interaction database (MIPS). Also 1527 potential human interlogs (i.e. potentially evolutionarily conserved interactions) were indentified from more than one species using the InParanoid database. Since 68% and 63% of literature-curated and interlog interactions are annotated to similar GO compartments, this group suggests that these identified interactions are of similar quality to interactions they identified in their Y2-H screens. The network demonstrated that several ataxia proteins interact and that there are shared pathways and mechanisms in this class of diseases. This study by Lim et al. will hopefully be able to provide additional knowledge about individual protein function and candidate genes for other diseases with similar phenotypes.
These are just a few of the more widely used publicly available databases that provide information on protein-protein interactions. Certainly, each one has unique features that allow researchers to gain access to vast amounts of useful biological information that can be broadly applied. In order to utilize this abundance of information as expert knowledge, we need to identify the important information we want to extract, which database or databases will provide us with this, and the method by which we will extract and apply that information.
Expert Knowledge from PPI Databases
We wish to exploit protein interaction data to improve the genetic analysis of common human diseases. As we have illustrated, the information available to the scientific community in curated protein interaction databases is abundant, and there are specific issues that need to be addressed in order to use this information as expert knowledge and apply it to the genetic analysis of genome-wide studies.
One possible method would be to identify all of the genes associated with the SNPs in a dataset for whose protein products have evidence of direct interaction with each other and filter that dataset accordingly. Filtering based on the direct interactions may prove to be a simple solution, but doing so may ignore potentially important biological information. Interactions do not have to be direct, and it may be beneficial to include the SNPs and genes to a certain level according to their indirect interactions, in other words, by taking a more pathway based approach. Another or an additional option would be to utilize or develop a confidence score for present interactions based on information available from a PPI database or even multiple databases. As mentioned, MINT, BIND, STRING, and GeneNetwork all provide a confidence score for interactions based on information such as the type of experiment conducted to detect that interaction and the supporting literature for that interaction. Specific metrics could be developed that would allow all SNPs or genes to be prioritized or weighted based on biological information about their interactions or allow investigators to filter SNPs or genes based on a determined interaction confidence threshold.
If one were to take any of these approaches, it would appear that the vital information to extract from these databases would be the direct and indirect interaction partners found in the data set (to a certain level) as well as the evidence or confidence score to support those interactions. While this also seems to be a rather simple approach, one needs to consider that the number of databases available that could provide this information is abundant and that this information may not be consistent between databases. For example, Mathivanan et al. (2006), thoroughly reviews the features of a number of databases including MINT, BIND, HPRD, DIP, and Reactome and concludes that while there may be good overlap at the protein level between these databases, the level of overlap between PPI’s is not as great. They also find that for PPI’s that do overlap between databases, there exists a difference in annotation partly on account of differences that arise according to how biologists interpret the experimental results. This presents an obstacle when attempting to apply this expert knowledge from multiple databases and may lead to the exclusion of important interactions or the inclusion of non-influential interactions in a dataset. Considering this, it may be beneficial to use an integrated database such as STRING or GeneNetwork which have, in their respective ways, brought together the various information in a number of databases.
Other issues that arise concern the bias that may exist across all databases, or the fact that genes and SNPs in the dataset may be unannotated or anonymous. How does one deal with anonymous SNPs or ones that are not in coding regions? It may be that a researcher wishes to consider this SNP as part of the gene that it is closest to or perhaps they may consider what annotated SNPs are in linkage disequilibrium (LD) with it. Also, there are many proteins in the human proteome that have not been studied thoroughly or even studied at all and may be underrepresented or non-existent in these databases. To add to this, there exists a bias of experimental methods for capturing certain interactions, for example, Y2-H experiments are not entirely adequate to detect interactions with integral membrane proteins (Mathivanan et al. 2006). Keep in mind that just because an interaction is not detected on the biological level, it does not mean that this interaction does not exist or will not be seen at the statistical level, and alternatively, what is detected statistically, may not have any biological relevance (Moore and Williams 2005). Therefore we must be aware of and concerned about the amount of important information we may potentially be missing due to bias and lack of annotation and what types of studies this type of expert knowledge is appropriate for.
Likewise, some may argue that using expert knowledge is a bias in and of itself despite bias in the databases. The ability to conduct a genome wide association study has been said to ‘relax’ the need for a strong prior hypothesis because the whole genome can be analyzed at once (Chanock et al. 2007). Those in support of ‘genomic agnosticism’ believe that when conducting genome wide analysis, they will assume every SNP in the genome to be equally functional (Carlson 2006). This brings us back to the issue of what information we may be missing by applying expert knowledge. While the benefits of conducting an unbiased GWAS study are valid, such as having no prior hypothesis, elimination of bias, and inclusion of all information, we are still at a loss for computational power to conduct these studies and fully explore all interactions.
These issues demonstrate the need for a method to evaluate the metrics and information we extract from these databases. Making use of data that is available and that has already been evaluated for interactions would aid in this process. With access to data where the biological importance of the results is known and the interactions are known, we could determine if this same information is retained after applying expert knowledge. Therefore we may be able to gage the amount of information we gain or, on the other hand, that we may lose by applying expert knowledge. Since we are also concerned with the efficiency of evaluating genome-wide studies, we may want to explore simulated large scale datasets that are already imbedded with known epistatic interactions, both physical and statistical. We would then be able to compare metrics we develop based on information from these databases. This will hopefully allow us to achieve the most meaningful results in the most efficient manner.
Conclusion/Outlook
The expanding field of human genetics and the availability of high-dimensional datasets from genome-wide studies make it computationally expensive and impractical to carry out a genetic analysis study utilizing data mining methods such as MDR. However, to complement this expansion, there has also been much advancement in the field of proteomics with the availability of high throughput methods to detect and characterize protein interactions and with the development of curated protein interaction databases to provide the scientific community with access to this information. Since there is no indication that technological advancements in either field will come to a halt anytime soon, the amount of valuable genetic and proteomic data produced will continue to grow.
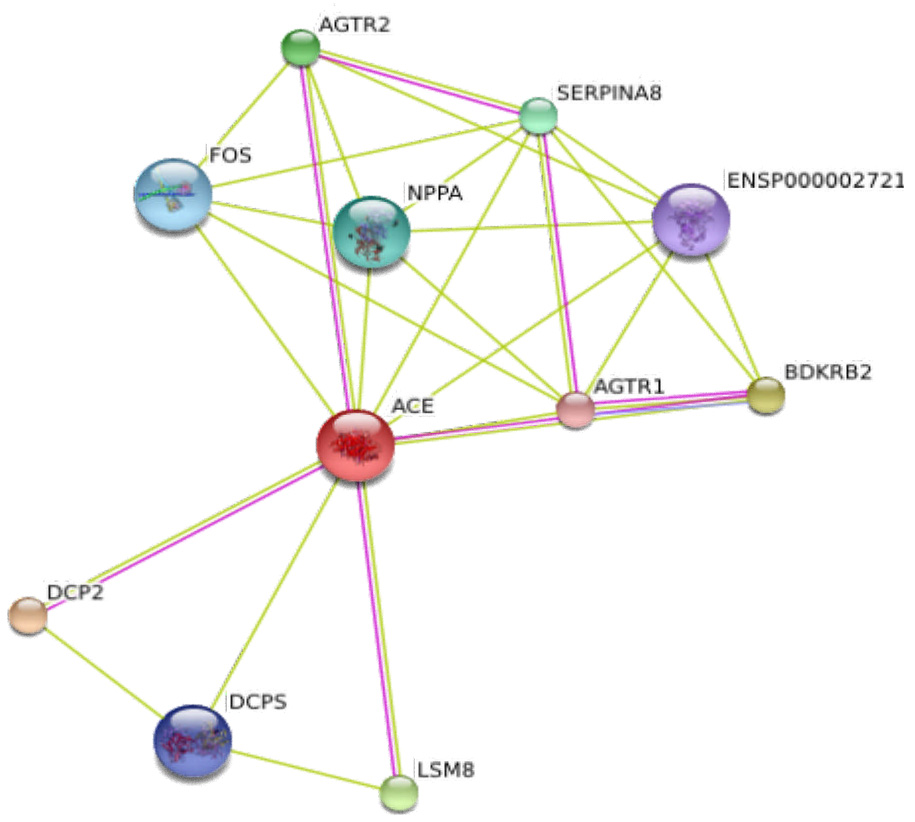
We have proposed that expert knowledge extracted from protein interaction databases may reduce the computational burden of large-scale and genome-wide studies as well as facilitate the biological interpretation of the data. It is important to keep in mind that SNPs are not the only form of genetic variation that can influence disease susceptibility. Copy number variation and sequence repeats have been shown to be influential as well. For example, a triplication of a gene known as α-synuclein, was shown to cause Parkinson’s disease (Singleton et al. 2003). This evidence would have been missed in a typical SNP analysis. Like the tools for conducting genome-wide studies of SNPs, technology to investigate these other types of variations are also advancing. Since we suspect that studies involving other forms of genetic variation will run into similar problems with large scale data analysis, we hope that our methods may in some way be applicable to facilitate such studies. We also hope that exploiting expert knowledge helps us in understanding the relationship between biological and statistical epistasis. We can begin to dissect this relationship by simply examining if epistatic interactions detected statistically, such as with MDR or other methods, are also found to exist at the protein level in the interaction databases mentioned. To illustrate this, we used a number or databases to query the protein interactions represented by significant SNP interactions in two genetic association studies. Coutinho et al. (2007) used MDR to analyze seven candidate genes in the serotonin metabolic and neurotransmission pathways mapping autism linkage regions and reported a significant interaction between polymorphisms in the 5-hydroxytryptamine (serotonin) receptor 5A (HTR5A), integrin beta 3 precursor (ITGB3), and sodium dependent serotonin transporter (SLC6A4) (P=0.001). Evidence for physical interactions between SLC6A4 and HTR5A is found for these genes in the STRING database when querying HTR5A and is based on evidence from textmining. When querying SLC6A4 or ITGB3 in STRING, evidence for interaction between these two genes is provided and is also based on information from textmining. Both interactions use this specific paper along with others as sources of evidence. Another genetic study analyzed interactions in polymorphisms influencing levels of tissue plasminogen activator (t-PA) and plasminogen activator inhibitor 1(PAI-1), which influence risk of arterial thrombosis (Asselbergs et al. 2007). Using a 2-way ANOVA statistical test, they found significant interactions between polymorphism in the bradykinin B2 gene and ACE (p=0.003) on t-PA in females and between polymorphisms in bradykinin B2 and angiotensin II type 1 receptor (AT1R/AGT1R) on t-PA in males (p=0.006). This latter interaction was also significant for PAI-1 levels in both males and females. Strong evidence for interaction for all three of these genes is seen when querying STRING (Figure 1.) and is supported by both experimental and textmining evidence. One or more of these interactions are found in the databases that STRING integrates such as HPRD, BIND, Reactome, MINT, BioGrid, DIP, KEGG annotated pathways, and DIP.
Figure 1.
Illustrates the protein interaction network as displayed in STRING when querying ACE. Interactions between ACE, BDKRB2, and AGTR1 are seen. Evidence for these interactions is derived from both experimental evidence (purple lines) and text mining evidence (green lines).
Certainly, biological interactions representative of statistical interactions are present in these databases, but we need to get to be able to exploit these resources to their full potential. To do so we need to develop a logical method to evaluate the information in these databases and also the metrics developed from this information in order to incorporate this type of expert knowledge into our analysis. While we don’t expect this to be a simple task, success in similar endeavors (Moore and White 2007; Myers et al. 2005) have assured us that it is an import and worthwhile one that needs to be explored.
Once we have been able to successfully develop these methods, not only will we improve the ease at which we will be able to identify important epistatic interactions in genome-wide studies, but we will gain an understanding of the physical biology that underlies these interactions and perhaps their role in a given disease. We expect these expert knowledge-based methods to enhance our comprehension of common human diseases and eventually lead to an improvement in the prevention, treatment, and diagnosis these diseases.
Acknowledgments
This publication was funded in part by National Institute of Health grants LM009012 and AI59694. We would like to thank Drs. Scott Gerber, David Jewell, Dean Madden and Mike Whitfield for helpful discussions that lead to some of the ideas in this paper.
References
- Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Ouellette BFF, Hogue CWV, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly MJ. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergs FW, Williams SM, Hebert PR, Coffey CS, Hillege HL, Navis G, Vaughan DE, van Gilst WH, Moore JH. Epistatic effects of polymorphisms in genes from the renin-angiotensin, bradykinin, and fibrinolytic systems on plasma t-PA and PAI-1 levels. Genomics. 2007;89(3):362–369. doi: 10.1016/j.ygeno.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W. Mendel’s Principles of Heredity. Cambridge: Cambridge University Press; 1909. [Google Scholar]
- Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J, Wood V, Dolinski K, Tyers M. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS. Agnosticism and equity in genome-wide association studies. Nat Genet. 2006;38(6):605–606. doi: 10.1038/ng0606-605. [DOI] [PubMed] [Google Scholar]
- Cavallo A, Martin AC. Mapping SNPs to protein sequence and structure data. Bioinformatics. 2005;21(8):1443–1450. doi: 10.1093/bioinformatics/bti220. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, et al. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia G, Yasir I, Hanig C, Herzel H, Wanker EE, Futschik ME. UniHI: an entry gate to the human protein interactome. Nucleic Acids Res. 2007;35:D590–D594. doi: 10.1093/nar/gkl817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Coutinho AM, Sousa I, Martins M, Correia C, Morgadinho T, Bento C, Marques C, Ataide A, Miguel TS, Moore JH, Oliveira G, Vicente AM. Evidence for epistasis between SLC6A4 and ITGB3 in autism etiology and in the determination of platelet serotonin levels. Hum Genet. 2007;121:243–256. doi: 10.1007/s00439-006-0301-3. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinburgh. 1918;52:399–433. [Google Scholar]
- Franke L, van-Bakel H, Fokkens L, de-Jong ED, Egmont-Petersen M, Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Hahn LW, Moore JH. Ideal discrimination of discrete clinical endpoints using multilocus genotypes. In Silico Biol. 2004;4:0016. [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Kaltenbach LS, Romero E, Becklin RR, Chettier R, Bell R, Phansalkar A, Strand A, Torcassi C, Savage J, Hurlburt A, Cha G-H, Ukani L, Chepanoske CL, Zhen Y, Sahasrabuhde S, Olson J, Kurschner C, Ellerby LM, Peltier JM, Botas J, Hughes RE. Huntingtin Interacting Proteins are Genetic Modifiers of Neurodegeneration. PloS Genetics. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington’s disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabási AL, Vidal M, Zoghbi HY. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125(4):801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Mathivanan S, Periaswamy B, Gandi T, Kandasamy K, Suresh S, Mohmood R. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;7 Suppl 5:S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra G, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivkumar K, Prasad TSK, Pandey A, et al. Human Protein Reference Database - 2006 Update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Moore JH. Computational analysis of gene-gene interactions in common human diseases using multifactor dimensionality reduction. Expert Rev Mol Diagn. 2004;4:795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- Moore JH. A global view of epistasis. Nat Genet. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- Moore JH. Genome-wide analysis of epistasis using multifactor dimensionality reduction: feature selection and construction in the domain of human genetics. In: Zhu X, Davidson I, editors. Knowledge Discovery and Data Mining: Challenges and Realities with Real World Data. Hershey: IGI Press; 2007. pp. 17–30. [Google Scholar]
- Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden W, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Moore JH, Ritchie MD. The challenges of whole-genome approaches to common diseases. JAMA. 2004;291:1642–1643. doi: 10.1001/jama.291.13.1642. [DOI] [PubMed] [Google Scholar]
- Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: Systems biology and a more modern synthesis. BioEssays. 2005;27:637–646. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- Moore JH, White BC. Tuning ReliefF for genome-wide genetic analysis. In: Marchiori E, Moore JH, Rajapakse J, editors. Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics: Lecture Notes in Computer Science. Vol. 4447. 2007. pp. 166–175. [Google Scholar]
- Myers CL, Robson D, Wible A, Hibbs MA, Chiriac C, Theesfeld CL, Dolinski K, Troyanskaya OG. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Haynor D, Johnson JM. Protein Interaction Networks. Expert Rev Proteomics. 2004;1:239–249. doi: 10.1586/14789450.1.2.239. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Rea TJ, Brown CM, Sing CF. Complex adaptive system models and the genetic analysis of plasma HDL-cholesterol concentration. Perspect Biol Med. (4) 2006;49:490–503. doi: 10.1353/pbm.2006.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor dimensionality reduction reveals high-order interactions among estrogen metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Risch NJ, Merikangas KR. The future of genetic studies of complex human disease. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing CF, Stengard JH, Kardia SL. Genes, environment, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2003;23:1190–1196. doi: 10.1161/01.ATV.0000075081.51227.86. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, TyersStark M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Tan SH, Zhang Z, Ng SK. ADVICE: Automated Detection and Validation of Interaction by Co-Evolution. Nucleic Acids Res. 2004;32:W69–W72. doi: 10.1093/nar/gkh471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Epistasis and complex traits. In: Wade M, Brodie B III, Wolf J, editors. Epistasis and Evolutionary Process. New York: Oxford University Press; 2000. [Google Scholar]
- Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics, and human disease: Analytical retooling for complexity. Trends Genet. 2004;20:640–647. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CWV, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. STRING 7: recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: Theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;4:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Willis RC, Hoque CW. Searching, viewing, and visualizing data in the Biomolecular Interaction Network Database (BIND). Chapt 8.8.9. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0809s12. [DOI] [PubMed] [Google Scholar]