Abstract
Fusariumproliferatum, F. subglutinans, and F. verticillioides are known causes of ear and kernel rot in maize worldwide. In Mexico, only F. verticillioides and F. subglutinans, have been reported previously as causal agents of this disease. However, Fusarium isolates with different morphological characteristics to the species that are known to cause this disease were obtained in the Highland-Valley region of this country from symptomatic and symptomless ears of native and commercial maize genotypes. Moreover, while the morphological studies were not sufficient to identify the correct taxonomic position at the species level, analyses based in the Internal Transcribed Spacer region and the Nuclear Large Subunit Ribosomal partial sequences allowed for the identification of F. subglutinans, F. solani, and F. verticillioides, as well as four species (F. chlamydosporum, F. napiforme, F. poae, and F. pseudonygamai) that had not previously been reported to be associated with ear rot. In addition, F. napiforme and F. solani were absent from symptomless kernels. Phylogenetic analysis showed genetic changes in F. napiforme, and F. pseudonygamai isolates because they were not true clones, and probably constitute separate sibling species. The results of this study suggest that the biodiversity of Fusarium species involved in ear rot in Mexico is greater than that reported previously in other places in the world. This new knowledge will permit a better understanding of the relationship between all the species involved in ear rot disease and their relationship with maize.
Keywords: fungus, ITS, nLSU, seed, Zea mays
Introduction
Maize (Zea mays L.) is one of the four basic food staples of the world population [1] and at the same time is the crop that has the first place in production volume in the world. In developed countries most of the maize produced is used for animal feed and industrial uses other than human food [2]. In Mexico maize is the main human food source and per capita consumption is estimated to be 328 g day−1 [3].
The majority of the reported maize diseases, affecting roots, stalks, ears, and kernels, are caused by fungi [4]. Among these diseases, ear rot is one of the most important in all the countries where this cereal is grown. In addition to reduced crop yield, ear rot adverasely affects the physical, physiological, and phytosanitary qualities of the seed [5, 6]. The causal agents reported worldwide as responsible for ear rot are Fusarium proliferatum, F. subglutinans, and F. verticillioides [4]. These pathogens survive in the soil, in infected plant debris, and inside apparently healthy seed and can affect the embryo and pericarp without visible symptoms. Infection can be seedborne and systemic in the crop from seedling to harvest, or starting during the pollination where the silks are infected by the airborne conidia. During harvest, ear rot appears as individual rotted kernels or as randomly scattered groups of rotted kernels [4, 7]. In addition, depending on the specific fungus responsible for the ear rot, the production of mycotoxins can be an important source of contamination.
The three Fusarium species involved in ear rot disease are included in the Gibberella fujikuroi complex [7, 8]. Morphological differences between the three species are an important step in the classification, however, currently there is no consensus definition among research groups that will allow definitive identification based on morphology alone. Revisions in recent years have asserted that the first criteria to define a Fusarium species [7], was the use of the biological species concept, but it has some limitations. Some researchers have adopted the concept of phylogenetic species which considers the use of one or more conserved genes or sequences to define a species of this genus [9], such as the Internal Transcribed Spacer (ITS) region of the ribosomal genes 18S-5.8S-28S [10].
In Mexico, the species that have been found to cause ear rot are F. verticillioides [11] and F. subglutinans [12, 13], and since this disease is the most important in the Highland Valley, based on reduced yield and grain quality [8], the aim of this research was to determine the biodiversity of the Fusarium species associated with ear rot in this geographical area of the country utilizing both a morphological and phylogenetic approach.
Materials and methods
During the fall-winter of 2002, at the Colegio de Postgraduados’ plots, located in Montecillo-Highland-Valley (elevation 2250 m), in the central part of Mexico, 10 symptomatic and two symptomless ears from each one of 28 native maize genotypes were collected (Table 1).
Table 1.
Relation of 28 native maize genotypes and their origin in the Highland of Mexicoa
| Genotypes | Origin |
|---|---|
| ASV11 | Sierra Purepecha, Michoacan |
| ASV34 | Sierra Purepecha, Michoacan |
| ASV36 | Sierra Purepecha, Michoacan |
| ASV45 | Sierra Purepecha, Michoacan |
| ASV49 | Sierra Purepecha, Michoacan |
| ASV64 | Sierra Purepecha, Michoacan |
| ASV71 | Sierra Purepecha, Michoacan |
| ASV76 | Sierra Purepecha, Michoacan |
| ASV84 | Sierra Purepecha, Michoacan |
| ASV86 | Sierra Purepecha, Michoacan |
| ASV87 | Sierra Purepecha, Michoacan |
| ASV102 | Sierra Purepecha, Michoacan |
| ASV111 | Sierra Purepecha, Michoacan |
| ASV112 | Sierra Purepecha, Michoacan |
| Qro-21 | Sierra Purepecha, Michoacan |
| (Chalqueño Qro) | |
| Zac-66 | Sierra Purepecha, Michoacan |
| (Chalqueño Dgo-Zac) | |
| Hgo-88 | Sierra Purepecha, Michoacan |
| (Chalqueño Hgo) | |
| Pedro Cruz | Sierra Purepecha, Michoacan |
| (7a CSM) | |
| (Chalco Crema) | |
| Santos Altamirano | Valle de Chalco, Edo. de |
| 6a CSM | Mexico |
| (Chalco palomo) | |
| Comp. Familias | Valle de Chalco, Edo. de |
| (Chalqueño Cajetes) | Mexico |
| Manuel Montes de Oca | Valle de Chalco, Edo. de |
| (Ciclo 8) | Mexico |
| David Rivera Enrrique | Valle de Chalco, Edo. de |
| (1a CSM) | Mexico |
| Pedro Cruz Linares | Valle de Chalco, Edo. de |
| (8a CSM) | Mexico |
| Ignácio Rosas | Valle de Chalco, Edo. de |
| (7a CSM) | Mexico |
| David Rivera Reyes | Valle de Chalco, Edo. de |
| (2a CSM) | Mexico |
| Pedro Hernandez | Valle de Chalco, Edo. de |
| (Ciclo 8) | Mexico |
| Santos Altamirano | Valle de Chalco, Edo. de |
| (7a CSM) | Mexico |
| Oaxaca 492 | Oaxaca |
aThe native maize genotypes are grown from 1900 to 2700 m elevation.
Symptomatic and symptomless ear isolates
The ears of all the genotypes with visible rot symptoms were grouped according to the mycelium colour. Of each group, a fungal sample grown on the top of the kernels was taken to obtain monoconidial cultures. To isolate the fungi that were within the symptomless kernels, hundred of seeds were randomly chosen from the apparently healthy ears and mixed. The seeds were disinfested using a 3:1 solution of sodium hypochlorite 1.5 %, and ethanol 25% for 2 min. After that, they were rinsed three times with sterile distilled water. Internal fungi were obtained through the Blotter method [14]. The colonies that developed over the seed were examined microscopically. Fusarium colonies were selected according to the mycelium colour.
Monoconidial cultures
The mycelium of each group selected from symptomatic and symptomless kernels was placed into 10 ml test tubes with 5 ml of sterile distilled water, and shaken in a Mini Vortex (VWR, USA). The content of individual tubes was poured into Petri dishes with water-agar (WA) (18 g of agar/l) and allowed to stay on the plates for 10 s and then the excess was removed. The Petri dishes were maintained at room temperature (22–24°C) for 24 h. The germinated conidia were transferred to new Petri dishes containing Potato Carrot Agar medium (PCA) (20 g of potato, 20 g of carrot, and 15 g of agar/l) and kept at room temperature with constant white light for seven days. The conidia were transferred into sterile glycerol 25%, and stored in Eppendorf tubes at −84°C.
Morphological characterization
To identify the fungi at the genus level, the Manual of Barnett and Hunter [15] was used and the determination of the species was according to Booth [16], Nelson et al. [17], and Burgess et al. [18]. Also the descriptions of Marasas et al. [19], Pascoe [20], and Nirenberg and O’Donnell [21] were referred to when necessary.
Colour of the colony
The Fusarium cultures were placed into slant tubes containing Potato Dextrose Agar medium (PDA) (250 g of potato, 20 g of dextrose, and 20 g of agar/l) amended with a half-cup of potato pulp. To promote sporulation and colouring, the tubes were maintained at 20°C with continuous white light for 10 days [17]. Finally, the colour of the mycelia was evaluated on both sides, the tubes were grouped according to the colour, and only one isolate was selected from each group for further morphological and phylogenetic analysis.
Structure formation
To set up microcultures in humidity chambers, Petri dishes were used [22]. Aluminium foil, a microscope slide, and a PDA square of 5 mm3 were placed in the plate, in this order one over the other, and a portion of mycelia was taken and placed on each PDA square. A cover slide was placed over the inoculated PDA squares. The Petri dishes were sealed and maintained at±24°C under white (12 h) and black light (12 h) (365 nm-General Electric 40 W F40SL, Mexico) for seven days. After the mycelia colonized the cover slide it was removed with sterile forceps and permanently mounted on a microscope slide with a drop of acidified glycerol. The size, shape and other key characteristics of each structure were recorded and photomicrographs were taken with a digital camera (Nikon COOLPIX 5000, Japan).
Obtaining of macroconidia and napiforme microconidia
The isolates were placed on Carnation-Leaf-Agar medium (CLA) (18 g of agar/l), and 10 carnation leaves were amended to the medium before the agar solidified [23], the leaves were irradiated previously with gamma rays at the Nuclear Centre of Mexico. The Petri dishes were kept at room temperature (22–24°C) with white light for 25 days, and then permanently mounted for microscopic evaluation.
Formation of microconidia into chains
To stimulate this structure isolates were grown in KCl medium (8 g of KCl, and 14 g of agar/l) amended with 10 carnation leaves. The Petri dishes were kept at room temperature (22—24°C) under black light for 10 days, and the aerial mycelium was microscopically examined.
Formation of chlamydospores
This conserved structure was induced in PDA medium, and the Petri dishes were kept at room temperature (22–24°C) with white light for 14 days.
Phylogenetic analysis
The DNA of 21 monosporic cultures was obtained with the protocol of Ahrens and Seemüller [24], and quality was verified by electrophoresis on a 1% agarose gel (Ultrapure, Gibco, USA) using TBE buffer (Tris-Borate-EDTA pH 8.0). The gel was stained with ethidium bromide and the amplicons were visualized in a transilluminator (Gel Doc 2000, BIO RAD®, USA). The DNA concentration was quantified in a Perkin-Elmer spectrophotometer (Lambda BIO 10, USA).
To amplify the ITS region ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [10], primers were used, and to amplify a partial sequence of 28S rRNA gene, ITS5 and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) primers were used [10, 11].PCR master mix was prepared in a final volume of 25 µl containing 1× of Taq DNA buffer, 0.13 mM of MgCl2, 16 µMof dNTPs, 20 qmole of each primer, 0.4 U of Taq-DNA polymerase (University Biotecnologies ®, Mexico), and 80 ng of DNA. PCR was carried out in a Perkin-Elmer thermocycler (CT 2400 ICA, USA) with an initial denaturing at 95°C for 2 min; 30 cycles of denaturing at 95°Cfor 1 min, annealing at 50°Cfor 30 s, and extension at 72°C for 2 min; afterwards at 72°C for 10 min as single final extension cycle. The PCR product was purified using a QIAquick kit (Qiagen, USA) according to the instructions given by the manufacturer. The fragment amplified was verified on a 0.8% agarose gel, and the gel was stained as above. The remaining PCR-products were sequenced in both directions in an ABI PRISM 3700 (Applied Biosystems, USA). The sequences were edited with Lasergene 2001, V.5 Software (DNASTAR Inc., USA), and the most related sequence was obtained using GenBank-Blast (NCBI-National Centre for Biotechnology Information). DNA sequences were aligned with the profile mode of ClustalW, and an evolutionary distances matrix was generated with Kimura 2 parameter substitution model. The evolutionary tree for the datasets was constructed with the neighbour-joining algorithm and the confidence of the tree was assessed by bootstrap analysis based on 5000 boot strap replications using MEGA 3.1 software [25].
Results and discussion
Isolates obtained
Of the symptomatic and symptomless kernels, eight and 13 isolates were obtained, respectively, on the basis of colour in PDA slant tubes (Table 2).
Table 2.
Morphological comparison of the structures of seven Fusarium species associated with ear rot of maize obtained from symptomatic and symptomless ears during fall to winter 2002 in Montecillo, Mexico
| Morphological characteristics | Gibberella fujikuroi complex | F. poae*,** | F. solani** | ||||
|---|---|---|---|---|---|---|---|
| F. napiforme** | F. pseudonygamai*,** | F. subglutinans*,** | F. verticillioides*,** | F. chlamydosporum*,** | |||
| Microconidia (length × width in μm) | 9.8 × 5.0a, 10.2 × 2.4b, 19.25 × 3.9c | 11.1 × 2.42, 5.0−13.8 × 1.9−4.0 | 11.32 × 3.1, 8−12 × 2.5–3 | 11.8 × 2.4, 5.0−12.0 × 1.5−2.5 | 9.7 × 3.3 | 11.46 × 7.0, 8−12 × 7−10d | 9.5 × 3, 8−16 × 2−4 |
| Microconidia in chains | + | + (short) | − | + | − | − | − |
| False heads microconidia | + | + | + | + | − | − | + |
| Mesoconidia | − | − | 1−3 septate | − | 0−3 septate.O | − | − |
| Shape and size (length × width in µm) | 17.0 × 4.5 | 11.0 × 4 | |||||
| Macroconidia (length × width in µm | 43.5 × 3.8 | 39.0 × 4.0 | 37.4 × 4.2 | 31.0 × 3.7 | 40.4 × 3.5 | 22.1 × 3.6 | 50.3 × 5.0 |
| Conidiophore | Branched | Single or branched | Branched | Single or branched | Branched | Branched | Branched |
| Phialides (length × width in µm | Monophialide | Monophialide | Monophialide | Monophialide | Monophialide Polyphialide | Monophialide | Monophialide |
| 27.0 × 3.0 | 26.0 × 2.43 | 23.5 × 2.7 | 22.0 × 3.5 | 26.7 × 2.7 | 14.0 × 6.0 | 45.0−73.0 × 2.4 | |
| Polyphialide | Polyphialide | Polyphialide | |||||
| Chlamydospores | + | − | − | − | + | − | + |
| Coloure | White/purple | White/orange, gray | White purple/pink-purple | Purple gray/pink-purple | brown/brown | White pink/orange | Light cream/light cream |
| Isolates | 2, 4, 6 | 10B, 13A, 13B, 18, 19 | 9, 11, 14 | 5, 10A | 16, 17 | 12, 15A, 15B | 3, 7, 8 |
*Isolates obtained from 100 symptomless kernels.
** isolates obtained from symptomatic kernels.
aNapiform conidia with 0–1 septate.
bOvoid conidia with 0–1 septate.
cOvoid conidia with 1–3 septate.
dAmpuliform conidia
eSlant tubes containing PDA with amended potato pulp.
Morphological characterization
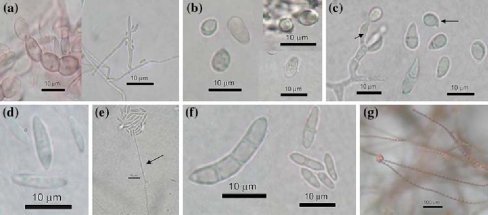
Typical Fusarium structures were observed in the 21 isolates selected. In general, we observed noncoloured septate mycelium, globular to fusiform microconidia with zero to three septate; falcate to cylindric macroconidia with seven septate according to the species; and the mesoconidia (the third type of Fusarium conidia) with zero to three septate [21], some species had brown coloured chlamydospores (Figure 1).
Figure 1.
Morphological differential features of Fusarium species. (a) F. chlamydosporum: brown chlamydospores in chains and polyphialide, (b) F. napiforme: napiform microconidia (globose with a small protuberance), (c) F. poae: ampuliform microconidia with papilla, 0–1 septate and monophialide, (d). F. pseudonygamai: large microconidia aseptate, (e) F. solani: false heads microconidia, and large monophialide, (f) F. subglutinans: large mesoconidia, fusiform, straight 1–3 septate, and G. F. verticillioides: microconidia in large chains observed on Petri dish under the low power of the microscope 10tx.
The species identified morphologically were F. chlamydosporum (9.52%), F. napiforme (14.29%), F. poae (14.29%), F. pseudonygamai (23.8%), F. solani (14.29%), F. subglutinans (14.29%), and F. verticillioides (9.52%) (Table 2).
Four species of Fusarium; F. napiforme (37.5%), F. solani (37.5%), F. subglutinans (12.5%), and F. verticillioides (12.5%), were obtained from symptomatic kernels, however from apparently healthy kernels five species were isolated out of which only two were present in both symptomatic and symptomless kernels; F. subglutinans (15.4%) and F. verticillioides (7.7%). The remaining Fusarium colonies belonged to three different species; F. chlamydosporum (15.4%), F. poae (23%), and F. pseudonygamai (38.5%). These results indicate that these species of Fusarium are latent inside the seed as reported by White and Munkvold and Desjardins [4, 5], and when the phytopathogenic species have appropriate environmental conditions they grow systemically and affect the crop, thus, becoming sources of dissemination of these pathogens to other areas.
The seven species found are associated with other maize diseases, and were found throughout the plants. F. proliferatum, F. subgutinans, and F verticillioides have been isolated in leaf axil and other maize tissues during ear fill, while F. solani was only reported associated with maize in India [26, 27]. F. poae, and F. solani, which cause head blight and root rot in maize, are transmitted by seed [16, 28]. F. chlamydosporum, F. poae, and F. pseudonygamai have been associated with other gramineous species such as Pennisetum typhoides and Sorghum caffrorum [19].
Five of the species identified produce mycotoxins such as fusaric acid and/or fumonisin (F. napiforme, F. subglutinans, and F. verticillioides); fusarin (F. poae, and F. verticillioides), moniliformin (F. chlamydosporum, F. napiforme, F. subglutinans, and F. verticillioides); trichothecenes ({ogF. poae}) and naphthazarin (F. verticillioides) inmaize kernels [29]. F. pseudonygamai, and F. solani have not been reported to produce mycotoxins [21, 29, 30].
Phylogenetic analysis
Analysis of ITS and nLSU regions
PCR was successfully performed and bands ((550 bp) were obtained. Each sequence was compared to sequences of the same species deposited at GenBank. Similarities ranged from 99.6% to 100%. Total nucleotides amplified were 470–533 bp and corresponded to the ITS1 and ITS2 complete regions; the 3′ portion of the 18S gene, 5.8S complete sequence, and the 5′ end of the 28S gene. PCR-products corresponding to nLSU gene were approximately 600 bp. With respect to ITS sequences, the isolate identified morphologically as F. chlamydosporum did not have any ITS sequence corresponding to the species deposited at GenBank database, so its closest relative was Fusarium spp. (AF158314) with a percentage of similarity of 98.6% (Table 3).
Table 3.
Molecular characterization of 21 Fusarium isolates from symptomatic and symptomless ear rot of maize using ITS region in Mexico
| Isolates | Morphological identification | Accession most related (Blast-GenBank) | Value of Aa | Similarity indexb | Difference in nucleotides | Proceeding |
|---|---|---|---|---|---|---|
| 16 | F. chlamydosporum DQ297572 | Fusarium sp. AY213706 | 1001 | 100 | 0 | USA |
| 17 | F. chlamydosporum DQ297573 | Fusarium sp. AY213706 | 1001 | 100 | 0 | USA |
| 2 | F. napiforme DQ297553 | F. napiforme X94175 | 1005 | 99.8 | 1 | Netherlands |
| 4 | F. napiforme DQ297554 | F. napiforme X94175 | 1021 | 99.6 | 2 | Netherlands |
| 6 | F. napiforme DQ297555 | F. napiforme X94175 | 1021 | 100 | 0 | Netherlands |
| 12 | F. poae DQ297556 | F. poae AY053440 | 963 | 100 | 0 | Norway |
| 15A | F. poae DQ297557 | F. poae AY053440 | 942 | 99.8 | 0 | Norway |
| 15B | F. poae DQ297558 | F. poae AY053440 | 942 | 100 | 0 | Norway |
| 10B | F. pseudonygamai DQ297559 | F. pseudonygamai U34563 | 985 | 99.8 | 1 | USA |
| 13A | F. pseudonygamai DQ297560 | F. pseudonygamai U34563 | 1005 | 100 | 0 | USA |
| 13B | F. pseudonygamai DQ297561 | F. pseudonygamai U34563 | 1005 | 100 | 0 | USA |
| 18 | F. pseudonygamai DQ297562 | F. pseudonygamai U34563 | 985 | 100 | 0 | USA |
| 19 | F. pseudonygamai DQ297563 | F. pseudonygamai U34563 | 985 | 100 | 0 | USA |
| 3 | F. solani DQ297564 | F. solani AY755617 | 977 | 99.6 | 2 | Mexico |
| 7 | F. solani DQ297565 | F. solani AY755617 | 989 | 99.6 | 2 | Mexico |
| 8 | F. solani DQ297566 | F. solani AY755617 | 1019 | 99.6 | 2 | Mexico |
| 9 | F. subglutinans DQ297567 | F. subglutinans X94167 | 993 | 99.8 | 1 | Netherlands |
| 11 | F. subglutinans DQ297568 | F. subglutinans X94167 | 999 | 99.8 | 1 | Netherlands |
| 14 | F. subglutinans DQ297569 | F. subglutinans X94167 | 1019 | 99.8 | 1 | Netherlands |
| 5 | F. verticillioides DQ297570 | F. verticillioides AY533376 | 1043 | 99.8 | 1 | Austria |
| 10A | F. verticillioides DQ297571 | F. verticillioides AY533376 | 999 | 99.8 | 1 | Austria |
aAlignment.
bAlignment done with Lasergene 2001 V.5 software (DNASTAR, Inc. Madison, USA).
The 28S rRNA gene partial sequences showed the same results as the ITS region. However, in the case of the isolate identified morphologically as F. chlamydosporum it was 98.6% equal to F. chlamydosporum (accession number AY213706). This result could be explained considering that for some species of Gibberella fujikuroi complex, the 28S gene gives a better resolution than ITS region. The ITS and nLSU sequences were deposited at Genbank NCBI (Table 3). Also, all isolates were placed in the Fungal Collection of Colegio de Postgraduados (registration on World Data Centre for Microorganisms in process).
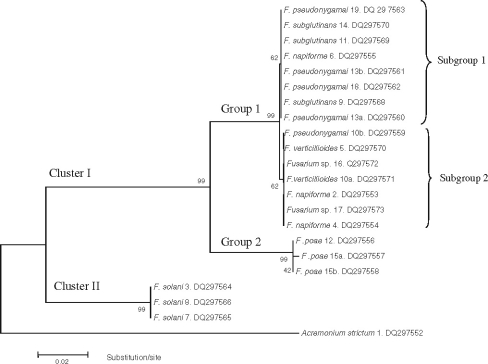
The species formed two clusters (Cluster I and II) (Figure 2). Cluster I had two groups (Group 1 and 2). Group 1 corresponded to two of the 16 sections as proposed by Wollenweber and Reinking [17] and both sections were in the G. fujikuroi complex [21]; Liseola section with F. pseudonygamai, F. subglutinans, and F. verticillioides, and Dlaminia section with F. napiforme. Group 2 also had two subgroups as evidenced by nucleotidic differences in the sequences of the isolates (Figure 3). F. pseudonygamai (DQ297559) had a cytosine at base 99 of ITS1, and not a thiamine as the other isolates of this species and F. napiforme (DQ297555) had a thiamine at base 99 of ITS1, and not a cytosine as in the isolate DQ297553. The isolate DQ 297554 of this species had a guanine at base 29, which corresponded to the beginning of the 28S gene. It also did not have a cytosine as in the other two isolates analysed in this same region (Figure 3). These results clearly show that these two species, as well as F. subglutinans, are separated into reproductively isolated populations that probably constitute separate sibling species [7], and in the future they may form different monophyletic species.
Figure 2.
Phylogenetic tree based on ITS region of 21 Mexican isolates of Fusarium species obtained from ear rot (symptomatic and symptomless kernels) using Kimura 2 parameter substitution method. The evolutionary scheme was constructed with the neighbor-joining algorithm using MEGA 3.1 software. The confidence of the tree was assessed by bootstrap analysis based on 5000 replications.
Figure 3.
Alignment showing differences in nucleotides: (A) ITS1 of Fusarium pseudonygamai, base 99 (asterisk). (B) ITS1 of F. napiforme, base 99 (asterisk). (C) 28S gene of F. napiforme, base 29 (asterisk).
Group 2 included three isolates of F. poae, of the Sporotrichiella section. Isolate DQ297557 had a thiamine at the 438 base of ITS2, and not an adenine as in the other two isolates.
In the cluster II there were the three F. solani isolates that have been grouped by Snyder and Hansen [18] in the Martiella y Ventricosum section of the F. solani complex [31]. Acremonium strictum was considered as and outgroup (Figure 3).
The position of species in the Liseola and Dlaminia sections in the same group can be explained by the fact that both had Gibberella as the teleomorph, are part of the same complex, and produce moniliformin. The isolates identified as F. chlamydosporum, according to its morphology and considered as Fusarium spp. by ITS Blast search, included in the same group was unexpected because its teleomorph is unknown. However, it produces moniliformin [29], and this fact might reflect its relation with the other species.
Through morphological and phylogenetic analysis, seven species of Fusarium identified as F. chlamydosporum, F. napiforme, F. poae, F. pseudonygamai, F. solani, F. subglutinans, and F. verticillioides, were found to be associated with ear rot disease, and F. chlamydosporum, F. poae, F. pseudonygamai, F. subglutinans, and F. verticillioides were found within asymptomatic kernels as well. Four of the identified species have not been reported previously to be associated with ear rot in Mexico. For this reason the biodiversity of Fusarium species involved in ear rot in Mexico appears to be greater than that reported previously in other places in the world. This new knowledge will permit a better understanding of the relationship between all the species involved in ear rot disease and their relationship with maize.
Acknowledgement
The first author wishes to express her thanks to Universidad Autonoma del Estado de Hidalgo, as well as to PROMEP-Program, for support of doctoral studies at the Colegio de Postgraduados in Mexico. All authors are grateful to CONACYT for financial support of this research through Grant 38409-V. We also thank Dr. Leobigildo Córdova-Téllez for his help in the field experiment, and MC Lily Zelaya-Molina for her comments during the writing of this paper.
References
- 1.Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004;19:535–544. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 2.Farnham DE, Benson GO, Pearce RB. Corn perspective, culture. In: White JP, Jonson AL, editors. Corn Chemistry and Technology. 2. St. Paul, MN: American Association of Cereal Chemists Inc.; 2003. pp. 1–34. [Google Scholar]
- 3.Figueroa de J D. La tortilla vitaminada. Avance y perspectiva 1999; 18: 149-58
- 4.White GD. Compendium of corn diseases. 3. St. Paul MN: APS Press; 2000. p. 78. [Google Scholar]
- 5.Munkvold GP, Desjardins AE. Fumonisins in maize, can we reduce their ocurrence? Plant. Dis. 1997;81:556–565. doi: 10.1094/PDIS.1997.81.6.556. [DOI] [PubMed] [Google Scholar]
- 6.Vigier B, Reid LM, Seifert KA, Stewart DW, Halminton RI. Distribution and prediction of Fusarium species associated with maize ear rot in Ontario. Can. J. Plant Pathol. 1997;19:60–65. [Google Scholar]
- 7.Steenkamp ET, Wingfield BD, Desjardins AE, Marasas WFO, Wingfield MJ. Cryptic speciation in Fusarium subglutinans. Mycologia. 2002;94:1032–1043. doi: 10.2307/3761868. [DOI] [PubMed] [Google Scholar]
- 8.Félix R, Romero S. Etiología de la germinación prematura del maíz en Huamantla, Tlaxcala. Agrociencia. 1981;43:81–87. [Google Scholar]
- 9.O’Donnell K. Ribosomal DNA internal transcribed spacer are highly divergent in the phytopathogenic ascomycete Fusariumsambucinum (Gibberella pulicaries) Curr. Genet. 1992;22:213–220. doi: 10.1007/BF00351728. [DOI] [PubMed] [Google Scholar]
- 10.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press, ; 1990. pp. 315–322. [Google Scholar]
- 11.Perez-Brito D, Jeffers D, Gonzales-de-Leon D, Khairallah M, Cortes-Cruz M, Velasquez-Cardelas G, Aspiroz-Rivero S, Srinivasam G. QTL mapping of Fusarium moniliforme ear rot resistance in highland maize, Mexico. Agrociencia. 2001;35:181–196. [Google Scholar]
- 12.Desjardins AE, Plattner RD, Gordon TR. Gibberella fujikuroi mating population A and Fusarium subglutinans from teosinte species and maize from Mexico and Central America. Mycol. Res. 2000;104:865–872. doi: 10.1017/S0953756299002002. [DOI] [Google Scholar]
- 13.Steenkamp ET, Coutinho TA, Desjardins AE, Wingfield BD, Marasas WFO, Wingfield MJ. Gibberella fujikuroi mating population E is associated with maize and teosinte. Mol. Plant Pathol. 2001;2:215–221. doi: 10.1046/j.1464-6722.2001.00072.x. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal VK, Sinclair JB. Principles of Seed Pathology. Boca Raton, Florida, USA: CRC Press, Inc.; 1987. [Google Scholar]
- 15.Barnett HL, Hunter BB. Illustrated Genera of Imperfect Fungi. 4. St. Paul MN: APS Press; 1998. p. 218. [Google Scholar]
- 16.Booth C. The Genus Fusarium. Commonwealth Mycological Institute. Kew, Surrey, England: CAB International; 1971. p. 237. [Google Scholar]
- 17.Nelson PE, Toussoun TA, Marasas WFO. Fusarium Species. An illustrated manual for identification. University Park and London. USA: The Pensylvania State University; 1983. p. 193. [Google Scholar]
- 18.Burgess LW, Summerell BA, Bullock S, Gott KP, Backhouse D. Laboratory Manual for Fusarium Research. 3. Sydney: University of Sydney; 1994. p. 133. [Google Scholar]
- 19.Marasas WFO, Rabie CJ, Lübben A, Nelson PE, Toussoun TA. Fusarium napiforme, a new species from millet and sorghum in southern Africa. Mycologia. 1987;76:910–914. doi: 10.2307/3807697. [DOI] [Google Scholar]
- 20.Pascoe IG. Fusarium morphology I: identification and characterization of a tirad conidia type, the mesoconidium. Mycotaxon. 1990;37:150–160. [Google Scholar]
- 21.Nirenberg HL, O’Donnell K. New Fusarium species and combination within the Gibberella fujikuroi species complex. Mycologia. 1998;90:434–458. doi: 10.2307/3761403. [DOI] [Google Scholar]
- 22.Onnions AHS. Preservation of fungi. In: Booth C, editor. Methods in Microbiology. New York: Academic Press; 1971. pp. 113–151. [Google Scholar]
- 23.Fisher NL, Marasas WFO, Toussoun TA. Taxonomic importance of microconidial chains in Fusarium section Liseola and effects of water potential in their formation. Mycologia. 1983;75:693–698. doi: 10.2307/3792999. [DOI] [Google Scholar]
- 24.Ahrens U, Seemüller E. Detection of DNA of plant pathogenic mycoplasma like organisms by polymerase chain reaction that amplifies a sequence of the 16S rRNA gene. Phytopathology. 1992;82:828–832. doi: 10.1094/Phyto-82-828. [DOI] [Google Scholar]
- 25.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 26.Dowd PF, Barnett CJ, Jonson ET, Beck JJ. Leaf axil sampling of midwest U.S. maize for mycotoxigenic Fusarium fungi using PCR analysis. Mycopathologia. 2004;158:431–440. doi: 10.1007/s11046-004-3862-5. [DOI] [PubMed] [Google Scholar]
- 27.Varshney JL. Pathogenic fungi recorded in maize crop raised from exotic germplasm. Indian Phytopathol. 1988;41:242–244. [Google Scholar]
- 28.Ocamb CM, Kommedahl T. Rhizosphere competence of Fusarium species colonizing corn roots. Phytopathology. 1994;84:166–172. doi: 10.1094/Phyto-84-166. [DOI] [Google Scholar]
- 29.Desjardins AE, Proctor RH. Biochemistry and genetics of Fusarium toxins. In: Summerell BA, Leslie JF, Backhouse D, Bryden WL, Burguess LW, editors. Fusarium Paul E. Nelson Memorial Symposium. St. Paul MN: APS Press; 2001. pp. 29–49. [Google Scholar]
- 30.Wayne LB, Logrieco A, Abbas HK, Porter JK, Vesonder RF, Richard JL, Cole RJ. Other significant Fusarium mycotoxins. In: Summerell BA, Leslie JF, Backhouse D, Bryden WL, Burguess LW, eds. Fusarium. Paul E. Nelson Memorial Symposium. St. Paul MN: APS Press, 2001: 360-92
- 31.Matuo T, Snyder WC. Use of morphology and mating populations in the identification of formae speciales in Fusarium solani. Phytopathology. 1973;63:562–565. [Google Scholar]