Abstract
The naked mole-rat is the longest living rodent with a maximum lifespan exceeding 28 years. In addition to its longevity, naked mole-rats have an extraordinary resistance to cancer as tumors have never been observed in these rodents. Furthermore, we show that a combination of activated Ras and SV40 LT fails to induce robust anchorage-independent growth in naked mole-rat cells, while it readily transforms mouse fibroblasts. The mechanisms responsible for the cancer resistance of naked mole-rats were unknown. Here we show that naked mole-rat fibroblasts display hypersensitivity to contact inhibition, a phenomenon we termed “early contact inhibition.” Contact inhibition is a key anticancer mechanism that arrests cell division when cells reach a high density. In cell culture, naked mole-rat fibroblasts arrest at a much lower density than those from a mouse. We demonstrate that early contact inhibition requires the activity of p53 and pRb tumor suppressor pathways. Inactivation of both p53 and pRb attenuates early contact inhibition. Contact inhibition in human and mouse is triggered by the induction of p27Kip1. In contrast, early contact inhibition in naked mole-rat is associated with the induction of p16Ink4a. Furthermore, we show that the roles of p16Ink4a and p27Kip1 in the control of contact inhibition became temporally separated in this species: the early contact inhibition is controlled by p16Ink4a, and regular contact inhibition is controlled by p27Kip1. We propose that the additional layer of protection conferred by two-tiered contact inhibition contributes to the remarkable tumor resistance of the naked mole-rat.
Keywords: longevity, p16Ink4a, p53, pRb, tumor suppressor
The naked mole-rat (Heterocephalus glaber) is one of the most extraordinary mammals. Naked mole-rats live in large subterranean colonies in Africa. The colonies have a eusocial structure with one breeding queen (1), similar to ants and bees. Naked mole-rats display exceptional longevity, with a maximum lifespan exceeding 28 years (2, 3). This is the longest lifespan for a rodent species, and is especially striking, considering the small, approximately 35 g, body mass of the naked mole-rat. In comparison, a similarly sized house mouse has maximum lifespan of 4 years (4, 5). It was suggested that naked mole-rats display negligible senescence (3), characterized by very slow or nonexistent changes in physiological parameters with age, and the lack of an age-related increase in mortality rate (6). Accordingly, naked mole-rats do not exhibit age-related changes in basal metabolism, body composition, or bone mineral density (3, 7). Furthermore, naked mole-rats reproduce prolifically until death (2, 3). These characteristics suggest that naked mole-rats have evolved efficient anti-aging defenses, making these rodents an invaluable model organism for aging research.
Cancer is a major age-related disease in humans, and accounts for approximately 23% of human mortality (8). In mice, cancer mortality is much higher, where it reaches 90% in some strains (9). Clearly, to achieve long life, species must possess efficient anticancer mechanisms. In addition to their other extraordinary characteristics, naked mole-rats show an unusual resistance to cancer. These animals have never been observed to develop any spontaneous neoplasms (3). So far anticancer strategies used by naked mole-rats have remained unknown.
Mammals have evolved complicated anti-cancer defenses consisting of cell cycle checkpoints, apoptosis, and replicative senescence controlled by a network of tumor-suppressor genes such as p53 and Rb. Anticancer mechanisms have been studied extensively in humans and mice. These studies revealed important differences between human and mouse anti-cancer defenses, which may explain the differences in cancer susceptibility between these species (10). Human cells use replicative senescence as an anticancer mechanism (11), while mouse somatic cells express telomerase (12) and are immortal in culture (13). Another difference is that in humans both the Rb and p53 tumor suppressors play important roles in controlling cell proliferation, creating backup barriers for cancer progression, while in the mouse Rb plays a relatively minor role (14, 15).
Contact inhibition is a process of arresting cell growth when cells come in contact with each other. As a result, normal cells stop proliferating when they form a monolayer in a culture dish. Contact inhibition is a powerful anticancer mechanism that is lost in cancer cells (16). Cancer cells do not arrest their growth when they fill a culture dish, but continue to proliferate, piling up on top of each other and forming multilayered foci. The growth arrest in contact inhibition is signaled by membrane proteins, and is mediated by elevated levels of p27Kip1 (p27) cyclin-dependent kinase inhibitor. p27 binds cyclin-CDK complexes and arrests the cells in the G1 phase of the cell cycle (17–20). The importance of contact inhibition for tumor suppression is underscored by the fact that the low levels of expression of p27 correlate with poor survival in cancer patients (21, 22). A cyclin-dependent kinase inhibitor, p16Ink4a (p16) has also been proposed to mediate contact inhibition, although its levels do not change appreciably in confluent human or mouse cultures (23, 24).
Our previous comparative analysis of cell telomerase activity and cell proliferation across 15 rodent species suggested that small long-lived rodent species evolve telomere-independent anticancer mechanisms (25, 26). Naked mole-rat fibroblasts do not display replicative senescence, but proliferate very slowly in culture, indicating the presence of an unknown mechanism that controls cell proliferation (26). This mechanism may contribute to the extraordinary tumor resistance of this species.
In the present study, we set out to identify and characterize the mechanisms that restrict cell proliferation in the naked mole-rat. We show that naked mole-rat fibroblasts become contact inhibited at a very low cell density, a phenomenon we termed “early contact inhibition.” We demonstrate that early contact inhibition requires the activity of both Rb and p53 pathways, and is associated with induction of p16Ink4a (p16). While p27Kip1 (p27) is not induced in cells undergoing early contact inhibition, but accumulates in transformed cells that lost early contact inhibition and arrest at high cell density. Thus, naked mole-rat cells possess two levels of contact inhibition, in contrast to the single level found in humans and mice. In summary, our study provides mechanistic clues to the unusual tumor resistance of the naked mole-rat.
Results
Oncogenic Ras and SV40 Large T do Not Induce Anchorage-Independent Growth of Naked Mole-Rat Fibroblasts.
Naked mole-rats have not been observed to develop cancer. Therefore, we tested the resistance of naked mole-rat cells to malignant transformation. A combination of oncogenic Ras and SV40 large T (LT) is sufficient to transform mouse fibroblasts (14). Therefore we tested whether the same combination of oncoproteins will confer anchorage-independent growth to naked mole-rat fibroblasts. We previously showed that naked mole-rat cells have detectable endogenous telomerase activity and are immortal in culture, therefore do not require telomerase for immortalization (26). Wild-type LT is a viral oncoprotein that binds and inactivates both p53 and pRb. The mutant derivative LTK1 inactivates only p53, while LTΔ434–444 inactivates only pRb and its family members (p107 and p130) (27). The functionality of SV40 LT and its derivatives in naked mole-rat cells was confirmed by co-transfecting them into naked mole-rat cells with the plasmids containing p53 or pRb response elements fused to firefly luciferase gene (Fig. S1).
Mouse skin fibroblasts transfected with V12 H-Ras and SV40 LT or its mutant derivatives formed large robust colonies (Fig. 1A), while naked mole-rat fibroblasts remained single-cells, or formed small abortive colonies (Fig. 1B). This result suggests that naked mole-rat cells require more “hits” than mouse cells for malignant transformation.
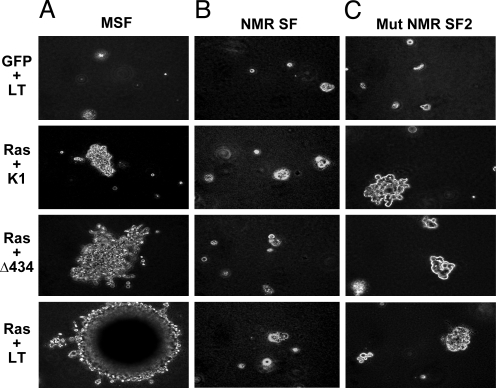
Fig. 1.
Assay of anchorage-independent growth in mouse and naked mole-rat skin fibroblasts. Cells were transfected with the vectors encoding different forms of LT antigen (LT, LTK1, and LTΔ434–44) and oncogenic Ras, and plated in soft agar. Figure shows representative microphotographs of colonies generated after 3 weeks at magnification ×20. (A) Mouse skin fibroblasts. (B) Naked mole-rat skin fibroblasts. (C) NMRSF2, a mutated line of naked mole-rat skin fibroblasts that lost early contact inhibition.
Naked Mole-Rat Cells Are Hypersensitive to Contact Inhibition.
In an effort to understand the basis of the extraordinary cancer resistance of the naked mole-rat, we examined the growth of naked mole-rat fibroblasts in vitro. Mouse as well as human fibroblasts are, typically, cultured by seeding 5 × 105 cells per 100-mm dish, and then splitting the cells when they reach confluence by forming a dense monolayer. When we attempted culturing naked mole-rat cells under the same regimen we could not obtain any cell proliferation. A solution to the problem was found when we plated naked mole-rat cells at a lower cell density (5 × 105 cells per 100-mm dish). When plated at a low cell density, naked mole-rat cells began to proliferate but reached confluence upon the formation of the first cell-cell contacts (Fig. 2A). This is in sharp contrast to mouse fibroblasts that form a dense monolayer of cells at confluence (Fig. 2B).
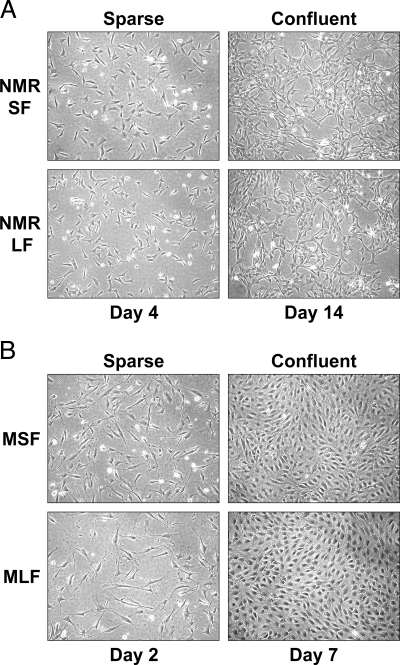
Fig. 2.
Early contact inhibition in naked mole-rat fibroblasts. Growing and confluent naked mole-rat (A) and mouse (B) fibroblasts. Naked mole-rat fibroblasts arrest cell proliferation when few cell-cell contacts are formed, a phenomenon we termed early contact inhibition. Naked mole-rat cells do not form a dense monolayer like the mouse cells. NMR SF, naked mole-rat skin fibroblasts; NMR LF, naked mole-rat lung fibroblasts; MSF, mouse skin fibroblasts, MLF, mouse lung fibroblasts.
To quantify this phenomena, cells were plated at low density on grided plates and counted daily (Fig. 3 A and B). The maximum cell density of naked mole-rat skin and lung fibroblasts was more than three times lower than that of mouse skin and lung fibroblasts, respectively (Fig. 3 A and B). Confluent naked mole-rat cell cultures had very low levels of thymidine incorporation, similar to confluent mouse cultures (Fig. 3C), indicating that the apparent growth arrest of naked mole-rat cells is associated with the arrest of DNA synthesis consistent with contact inhibition. We conclude that naked mole rat cells are hypersensitive to contact inhibition, and term this phenomenon “early contact inhibition.”
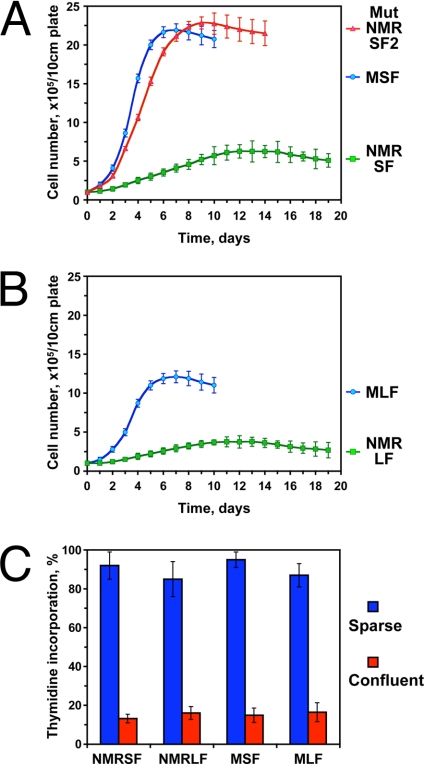
Fig. 3.
Naked mole-rat fibroblasts reach lower confluent cell density than mouse fibroblasts. (A) Comparison of cell densities attained by naked mole-rat and mouse skin fibroblasts. Cells were seeded on grided plates, and cell numbers were counted daily for each plate. MSF, mouse skin fibroblasts; NMRSF Mut, naked mole-rat skin fibroblast line that lost early contact inhibition; NMRSF, normal naked mole-rat skin fibroblasts. (B) Comparison of cell densities attained by naked mole-rat and mouse lung fibroblasts. MLF, mouse lung fibroblasts; NMRLF, naked mole-rat lung fibroblasts. Cells were seeded on grided plates and cell numbers were counted daily. The experiments were repeated at least four times, using fibroblast lines from different animals (except for NMRSF Mut). Error bars, standard deviations. (C) Analysis of DNA synthesis by thymidine incorporation in growing and confluent naked mole-rat and mouse cells. Early contact inhibited cells do not synthesize DNA. The experiments were repeated three times, and standard deviations are shown.
One naked mole-rat skin cell culture was kept on the same plate for four months, and gave rise to a fast growing clone (NMRSF Mut). These cells lost hypersensitivity to contact inhibition, and reached confluence at high cell density, similar to mouse cells (Fig. 3A). This observation indicates that a specific growth control mechanism restricts proliferation of naked mole-rat cells at higher cell densities, and when this mechanism is mutated or silenced, naked mole-rat cells are capable of reaching high cell densities under our standard culture conditions.
Early Contact Inhibition Is Mediated Predominantly by Cell-Cell Contact.
To determine whether early contact inhibition is caused by cell contact or by the accumulation of putative secreted factors we changed the growth media every 24 h and monitored cell density. The media was changed every 7 days for the control cultures. Frequent media change increased the maximum cell density of naked mole-rat skin fibroblasts (Fig. S2A); however, even with frequent media changes naked mole-rat cells did not reach cell densities of the mouse cells. The same treatment had no effect on the maximum cell density attained by naked mole-rat lung fibroblasts (Fig. S2B). Frequent media changes also slightly increased the cell density of mouse cells (Fig. S2 C and D). Furthermore, incubation of sparsely seeded naked mole-rat fibroblasts with the media taken from confluent naked mole-rat plates did not inhibit their growth (Fig. S2 E–H). As will be described below, plating of naked mole-rat cells at high density causes rapid onset of contact inhibition. Collectively, this data indicates that early contact inhibition of naked mole-rat cells is mediated, primarily, by cell-cell contact.
Early Contact Inhibition Requires the Activity of p53 and Rb Pathways.
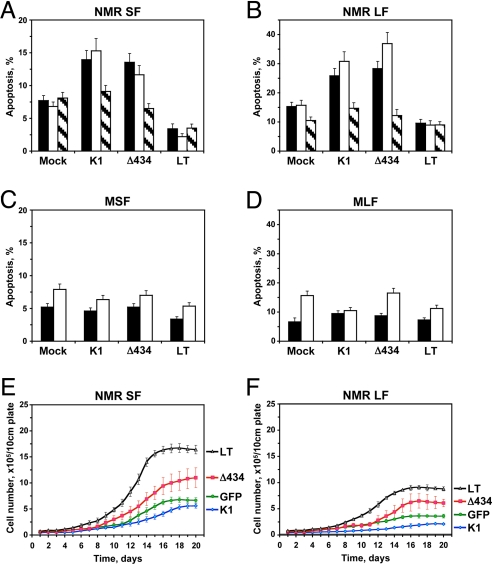
We then tested whether early contact inhibition is mediated by p53 or pRb signaling pathways. To do so we used plasmids encoding SV40 LT cDNA and its mutant derivatives LTK1 and LTΔ434–444. To test the effect of p53 or pRb depletion on the early contact inhibition, we transfected skin and lung fibroblasts from naked mole-rat and mouse with plasmids encoding LT, LTK1, LTΔ434–444, or GFP as a control. Cells were plated at low density on grided plates and the number of cells was counted daily (Fig. 4). Wild-type LT attenuated early contact inhibition in both naked mole-rat skin and naked mole-rat lung cells (Fig. 4 A and B) causing cells to reach higher confluent density. Interestingly, neither LTK1 nor LTΔ434–444 alone was able to attenuate early contact inhibition (Fig. 4 A and B). LT had no effect on the growth of mouse fibroblasts (Fig. 4 C and D). These results indicate that both p53 and pRb pathways must be perturbed to abolish early contact inhibition of naked mole-rat cells.
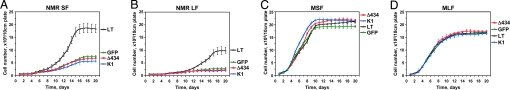
Fig. 4.
Inactivation of both p53 and pRb pathways is required to abolish early contact inhibition. Naked mole-rat skin (NMR SF), lung (NMR LF), mouse skin (MSF), and lung (MLF) fibroblasts were transfected with plasmids encoding SV40 Large T antigen (LT), or LT mutants LTK1 that targets only p53 family (K1), or LTΔ434 that targets only pRb-family (Δ434). Cells were seeded on grided plates, and cell numbers were counted daily. (A) SV40 Large T antigen but not K1 or Δ434 derivatives abrogates early contact inhibition in naked mole-rat skin fibroblasts (NMR SF) and causes the cells to grow to high density. (B) SV40 Large T antigen but not K1 or Δ434 derivatives abrogates early contact inhibition in naked mole-rat lung fibroblasts (NMR LF) and causes the cells to grow to high density. (C) SV40 Large T antigen and its mutants do not affect cell density of mouse lung fibroblasts (MLF). (D) SV40 Large T antigen and its mutants do not affect cell density of mouse skin fibroblasts (MSF). All of the experiments were repeated at least three times, and error bars show, standard deviations.
Early Contact Inhibition Is Safeguarded by Apoptotic Response.
To understand why inactivation of p53 or pRb alone did not abrogate early contact inhibition, we examined the levels of apoptosis in cells transfected with the oncoproteins. Naked mole-rat and mouse skin and lung fibroblasts were transfected with LT, LTK1, or LTΔ434–444 and the percent of apoptotic cells was measured by TUNEL on days 8 and 14 after plating (Fig. 5 A–D). Day 14 was chosen as naked mole-rat cells start to reach confluence on that day. Similar results were obtained using Annexin V staining.
Fig. 5.
Analysis of apoptosis in naked mole-rat cells transfected with SV40 large T antigen (LT) or its mutant derivatives K1, which binds p53 family of proteins, and Δ434, which binds pRb family members. (A) Apoptosis in naked mole-rat skin fibroblasts (NMR SF) transfected with the plasmids encoding large T antigen constructs. Apoptosis was assayed on days 8 (black bars) and 14 (white bars) after transfection, using TUNEL. Day 14 is the time when cells start entering early contact inhibition. Striped bars correspond to cells grown in the presence of 5 μM apoptosis inhibitor Z-VAD-FMK for 14 days. (B) Same experiment as in (A) done with naked mole-rat lung fibroblasts (NMR LF). (C) Apoptosis in mouse skin fibroblasts (MSF) transfected with the plasmids encoding large T antigen constructs. Apoptosis was assayed on days 8 (black bars) and 14 (white bars) after transfection, using TUNEL. (D) Same experiment as in (C) with mouse lung fibroblasts (MLF). (E) Analysis of cell proliferation in naked mole-rat skin fibroblasts in the presence of 5 μM apoptosis inhibitor Z-VAD-FMK. Corresponding growth curves without Z-VAD-FMK are shown in Fig. 3A. Cells were transfected with the plasmids encoding large T antigen constructs and seeded on grided plates. Cell numbers were counted daily. The experiments were repeated at least three times and error bars show standard deviations. (F) Analysis of cell proliferation in naked mole-rat lung fibroblasts in the presence of ZVAD-FMK as described in (E). Corresponding growth curves without ZVAD-FMK are shown in Fig. 3B. All experiments were repeated three times, and error bars, standard deviations.
In control naked mole-rat cells, the level of spontaneous apoptosis was low (7% in skin fibroblasts and 15% in lung fibroblasts). Remarkably, upon transfection with LTK1 or LTΔ434–444, the levels of apoptosis increased almost 2-fold (reaching 15% in skin fibroblasts and 35% in lung fibroblasts) (Fig. 5 A and B). In contrast, transfection with the wild-type LT reduced apoptosis below the control levels. The oncoproteins had little or no effect on spontaneous apoptosis in mouse cells (Fig. 5 C and D). Taken together, these data suggest that in the absence of either p53 or pRb activity, naked mole-rat cells proliferate beyond the confluence point, but then too close cell contact induces apoptosis in these cells. As a result, cell density remains low (Fig. 4 A and B). However, the wild-type LT inactivates both pRb and p53 pathways, therefore inhibiting the “backup” apoptotic response and leading to increased cell density. Loss of pRb is known to induce apoptosis in human cells and during mouse development (28, 29), however, induction of apoptosis due to the loss of p53 may be a unique property of the naked mole-rat.
To test this explanation, we cultured naked mole-rat cells transfected with oncoproteins in the presence of a broad-spectrum caspase inhibitor Z-VAD-FMK. The apoptosis inhibitor improved the growth of naked mole-rat fibroblasts transfected with LTΔ434–444, but had no effect on cells tranfected with the wild type LT or LTK1 (Fig. 5 E and F). LTΔ434–444 inactivates proteins of pRb family and is likely to impair cell cycle arrest, therefore a combination of the pRb inactivation and inhibition of apoptosis leads to higher cell density. LTK1 inactivates p53, leaving pRb-mediated cell cycle control intact. In the presence of the apoptosis inhibitor, LTK1-treated cells are still capable of inducing cell cycle arrest, and do not proliferate to higher density. Thus, early contact inhibition is mediated by cell cycle arrest via the pRb pathway, and is backed up by apoptotic response via the p53 pathway.
Early Contact Inhibition Is Associated with Induction of p16.
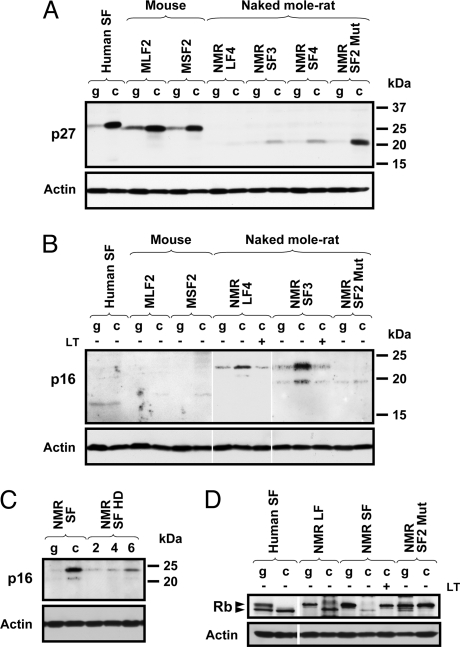
Cell cycle arrest in confluent mouse and human cells is mediated by the accumulation of p27 cyclin-dependent kinase inhibitor (18). To determine whether p27 accumulates in naked mole-rat cells undergoing early contact inhibition we examined the levels of p27 in growing and confluent human, mouse, and naked mole-rat fibroblasts by Western blot. p27 in naked mole-rat had a different molecular weight than mouse or human protein, but its identity was verified with two antibodies that recognize both human and mouse p27. As expected, confluent human and mouse cells accumulated high levels of p27 (Fig. 6A). Surprisingly, confluent naked mole-rat fibroblasts showed very slight induction of p27. However, the transformed naked mole-rat clone NMRSF2 Mut that lost early contact inhibition showed a strong increase in p27 levels. Thus, early contact inhibition is not mediated by p27, however, if early contact inhibition is perturbed, naked mole-rat cells undergo “regular” contact inhibition at high cell density associated with induction of p27.
Fig. 6.
Early contact inhibition in naked mole-rat cells is associated with accumulation of p16 while regular contact inhibition is associated with accumulation of p27. Human SF, human skin fibroblasts; MLF, mouse lung fibroblasts, MSF, mouse skin fibroblasts, NMR LF, naked mole-rat lung fibroblasts; NMR SF, naked mole-rat skin fibroblasts; NMR SF2 Mut, mutated naked mole-rat skin fibroblast line that lost early contact inhibition. The number at the end of the cell line name corresponds to the animal it was derived from. g, growing cells; c, confluent cells. (A) Western blot analysis of p27 in growing and confluent human, naked mole-rat, and mouse fibroblasts. p27 is strongly induced in cells undergoing regular contact inhibition such as human, mouse, or mutated naked mole-rat cells. The identity of the naked mole-rat protein was verified with two different antibodies. (B) Western blot analysis of p16 in growing and confluent human, naked mole-rat, and mouse fibroblasts. p16 accumulates in early contact inhibited naked mole-rat fibroblasts but not in human, mouse, or mutated naked mole-rat cells. The identity of the naked mole-rat protein was verified with two different antibodies. (C) p16 accumulates in naked mole-rat cells seeded at high density (HD). The first two lanes show growing cells (day 7) and confluent cells (day 20). The next three lanes correspond to naked mole-rat cells that were seeded at high density. The number above the lane indicates days after plating. (D) pRb becomes hypophosphorylated in early contact inhibited naked mole-rat cells.
We then examined whether another cyclin dependent kinase inhibitor, p16, is involved in early contact inhibition. Western blot was performed with antibodies to a conserved region of p16 that recognize both human and mouse protein, and the identity of the naked mole-rat p16 was verified with two different antibodies. Levels of p16 were not increased in confluent human or mouse fibroblasts (Fig. 6B). However, p16 was strongly induced in early contact inhibited naked mole-rat cells. The mRNA level of the naked mole-rat p16 was also increased in early contact inhibited cells (Fig. S3). Transfection with SV40 LT prevented accumulation of p16 (Fig. 6B). Remarkably, the levels of p16 were not increased in NMRSF2 Mut cell line that lost early contact inhibition (Fig. 6B). Furthermore, NMRSF2 cells have grown medium-sized colonies in soft agar (Fig. 1C), suggesting that the loss of early contact inhibition makes naked mole-rat cells more prone to anchorage-independent growth.
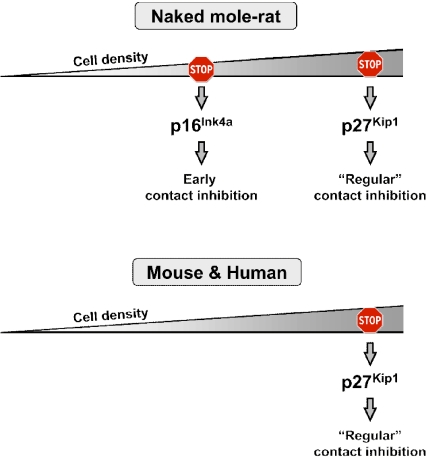
In the above experiments, naked mole-rat cells were seeded at low density and then incubated for 20 days. To test whether p16 will be induced by cell-cell contact without a prolonged incubation, we plated actively proliferating naked mole-rat cells at higher cell density. These cells stopped proliferation and rapidly induced p16 (Fig. 6C). Naked mole-rat fibroblasts displayed a shift in pRb phosphorylation and accumulated hypophosphorylated pRb as they entered confluence, which is consistent with the p16 activation (Fig. 6D). Collectively, these results indicate that naked mole-rat cells have two tiers of contact inhibition: early contact inhibition mediated by p16, and regular contact inhibition mediated by p27 (Fig. 7). These two tiers of contact inhibition may provide backup mechanisms that increase cancer resistance in vivo.
Fig. 7.
A model comparing contact inhibition in naked mole-rat to mouse and human. Naked mole-rat cells have two tiers of contact inhibition: early contact inhibition mediated by p16 and regular contact inhibition mediated by p27. In contrast, human and mouse only have regular contact inhibition. The presence of two-tiered contact inhibition may provide naked mole-rats an increased protection against tumor development.
Discussion
In this paper we describe a tumor-suppressor mechanism, early contact inhibition, that is present in the naked mole-rat but is lacking in all other mammalian species described to date. Our analysis also identifies the signaling pathways controlling early contact inhibition. We propose that naked mole-rats' extreme longevity has co-evolved with efficient anticancer adaptations.
How Does Early Contact Inhibition Compare to “Regular” Contact Inhibition?
Contact inhibition is a powerful tumor-suppressor mechanism that plays a fundamental role in regulating homeostasis in vivo and in vitro. Transformed cells lose contact inhibition and form multilayered foci (16). The antiproliferative signal is mediated by cell-cell contact, via surface receptors (17, 30–33). Our study demonstrates that naked mole-rat fibroblasts display an unusual, early contact inhibition. In culture, naked mole-rat fibroblasts never form a dense monolayer, like human or mouse cells. Instead, naked mole-rat fibroblasts arrest cell proliferation upon the formation of a loose network with a few cell-cell contacts. Similar to usual contact inhibition, early contact inhibition is mediated by cell-cell contacts. The difference between the two responses lies in the contact area between cells, which is required for induction of growth arrest. The difference, however, is not just quantitative. The two types of contact inhibition appear to be induced by somewhat different signaling pathways.
Regular contact inhibition is triggered by accumulation of p27, leading to inactivation of cyclin E-CDK2 complexes, which blocks phosphorylation of pRb (18, 34). In agreement with this we observed strong induction of p27 in confluent human and mouse fibroblasts (Fig. 6A). Some reports suggested the involvement of p16 in contact inhibition. However, in human cells the induction in p16 level was slight, and could only be detected by co-IP with cdk4 (19, 23). In mice, knockout of the entire INK4a locus caused mouse fibroblasts to grow to higher density (24); however, later studies where Arf and p16 were knocked-out separately seem to attribute most cell proliferation phenotypes of INK4a knockout MEFs to the loss of Arf (35–37). We did not observe induction of p16 in either human or mouse confluent fibroblasts (Fig. 6B). In contrast, p16 is strongly induced in naked mole-rat cells undergoing early contact inhibition (Fig. 6B). Thus, contact inhibition in human and mouse relies primarily on p27 and to a minor extent on p16, while early contact inhibition in naked mole-rat relies strongly on p16. Remarkably, naked mole-rat cells still maintain regular contact inhibition mediated by p27. Thus, the naked mole-rat may have the same genes, but the functions of p16 and p27 in contact inhibition became temporally separated. p16 is responsible for the early response, while p27 is induced as a backup mechanism when the p16-mediated protection had failed (Fig. 7). The presence of this two-tiered contact inhibition is likely to provide the naked mole-rat better protection against tumor development.
Evolutionarily, how do naked mole-rat cells maintain selection of regular-contact inhibition genes? It is possible that early contact inhibition is not fully active during embryogenesis, and this could be the time when p27-mediated contact inhibition plays a primary role. Another possibility is that mortality due to cancer provides selective pressure to maintain active regular-contact inhibition genes.
Species-Specific Differences in Anticancer Mechanisms.
The use of mouse models for human cancer research has yielded many important insights, however, significant differences exist in human and mouse carcinogenesis. In general, human cells are more resistant to malignant transformation, and require more genetic changes for malignant transformation than do mouse cells (14, 15). Perturbation of only two signaling pathways involving inactivation of p53 and constitutive activation of Raf (14) is required for the transformation of normal mouse fibroblasts. In contrast, six pathways (inactivation of p53, pRb, PP2A, and activation of telomerase, Raf, and Ral-GEFs) must be perturbed to malignantly transform human cells (14). Overall, it appears that human cells have more stringent senescence arrest, more parallel pathways for induction of senescence, and hence better anticancer mechanisms than mouse cells.
We found a combination of SV40 LT and activated Ras does not induce robust anchorage-independent growth of naked mole-rat cells. Naked mole-rat cells transfected with the oncogenes formed small abortive colonies in soft agar. We did not test tumorigenicity of these colonies in nude mice, however it is likely that that perturbation of a greater number of genetic pathways is required for transformation of naked mole-rat cells compared to mouse cells. Since naked mole-rats have not been observed to develop tumors, it is possible transformation of naked mole rat cells requires even more “hits” than transformation of human cells.
Relative contributions of p53 and pRb pathways differ between human and mouse (38). pRb function in regulating senescence and apoptosis appears to be more important in human cells than in mouse cells (39, 40). Interestingly, in naked mole-rat, the control of early contact inhibition depends heavily on p16. Thus, despite the fact that naked mole-rat is a rodent, it appears to have some human-like features in its anticancer apparatus. It is possible that the p16→pRb function becomes more important in long-lived, cancer-resistant species.
Our work shows that at least some of the pathways differ between human and naked mole-rat: human cells undergo replicative senescence, while naked mole-rat cells express telomerase and proliferate continuously. Interestingly, despite a close association between telomerase activity and cancer in humans, naked mole-rat cells remain resistant to malignant transformation in the presence of telomerase activity. How can this be achieved? Our present report identifies early contact inhibition as a property of naked mole-rat cells. Early contact inhibition may be a powerful alternative to replicative senescence, which protects telomerase positive naked mole-rat cells from malignant transformation.
Naked Mole-Rat, a Promising Model for Cancer Research.
Evolutionary increases in body mass or lifespan increase lifetime cancer risk. Large body mass elevates cancer risk, by increasing the total number of cells, hence the probability that each given cell could undergo malignant transformation. Long lifespan increases a risk of malignant transformation as a function of time. The increased mortality due to cancer is expected to drive adaptive evolution of anticancer mechanisms (41, 42). Our previous comparative analysis of multiple rodent species demonstrated that large-bodied species evolve repression of telomerase activity and replicative senescence, which is an important anticancer adaptation (25, 26). In contrast, long-lived small-bodied species do not have replicative senescence, but evolve other anticancer adaptations (26). The mechanisms of those adaptations remained unknown, as cancer research has traditionally focused on humans, which are large-bodied species displaying replicative senescence, or mice, which are small-bodied, short-lived, and cancer-prone, hence having fewer anticancer adaptations. Cancer-prone mouse models are valuable for development of cancer treatments. However, to find ways to prevent cancer before it occurs it would be extremely useful to study cancer-resistant models such as the naked mole-rat. Interestingly, unusual anticancer adaptations have been identified in another long-lived subterranean mole rat Spalax (43, 44). We anticipate that these unusual rodents have evolved multiple anticancer adaptations, which would pave the way for development of therapies for cancer treatment and prevention.
Experimental Procedures
Detailed experimental procedures including animals, cell isolation and culture, cell growth analysis, thymidine incorporation, transfections, Luciferase assays, analysis of apoptosis, RT-PCR, and Western blots are provided in the SI Text.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Institutes of Health and Ellison Medical Foundation (to V.G.), and Ellison Medical Foundation grant (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 19207.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905252106/DCSupplemental.
References
- 1.Jarvis JU. Eusociality in a mammal: Cooperative breeding in naked mole-rat colonies. Science. 1981;212:571–573. doi: 10.1126/science.7209555. [DOI] [PubMed] [Google Scholar]
- 2.Buffenstein R, Jarvis JU. The naked mole rat-a new record for the oldest living rodent. Sci Aging Knowledge Environ. 2002;2002:pe7. doi: 10.1126/sageke.2002.21.pe7. [DOI] [PubMed] [Google Scholar]
- 3.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: Insights from a successfully aging species. J Comp Physiol B. 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 4.Turturro A, et al. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- 5.de Magalhaes JP, Costa J, Toussaint O. HAGR: The human aging genomic resources. Nucleic Acids Res. 2005;33:D537–543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finch CE. Longevity, Senescence and the Genome. Chicago: Univ. of Chicago Press; 1990. [Google Scholar]
- 7.O'Connor TP, Lee A, Jarvis JU, Buffenstein R. Prolonged longevity in naked mole-rats: Age-related changes in metabolism, body composition and gastrointestinal function. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:835–842. doi: 10.1016/s1095-6433(02)00198-8. [DOI] [PubMed] [Google Scholar]
- 8.US Mortality Data 2005. National Center for Health Statistics, Centers for Disease Control and Prevention. 2008.
- 9.Lipman R, Galecki A, Burke DT, Miller RA. Genetic loci that influence cause of death in a heterogeneous mouse stock. J Gerontol A Biol Sci Med Sci. 2004;59:977–983. doi: 10.1093/gerona/59.10.B977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright W, Shay J. Telomere dynamics in cancer progression and prevention: Fundamental differences in human and mouse telomere biology. Nature Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- 11.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:S27–31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 12.Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parrinello S, et al. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: Modeling human cancer in mice. Nat Rev Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 16.Abercrombie M. Contact inhibition and malignancy. Nature. 1979;281:259–262. doi: 10.1038/281259a0. [DOI] [PubMed] [Google Scholar]
- 17.Levenberg S, Yarden A, Kam Z, Geiger B. p27 is involved in N-cadherin-mediated contact inhibition of cell growth and S-phase entry. Oncogene. 1999;18:869–876. doi: 10.1038/sj.onc.1202396. [DOI] [PubMed] [Google Scholar]
- 18.Polyak K, et al. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 19.Dietrich C, Wallenfang K, Oesch F, Wieser R. Differences in the mechanisms of growth control in contact-inhibited and serum-deprived human fibroblasts. Oncogene. 1997;15:2743–2747. doi: 10.1038/sj.onc.1201439. [DOI] [PubMed] [Google Scholar]
- 20.Kato A, Takahashi H, Takahashi Y, Matsushime H. Inactivation of the cyclin D-dependent kinase in the rat fibroblast cell line, 3Y1, induced by contact inhibition. J Biol Chem. 1997;272:8065–8070. doi: 10.1074/jbc.272.12.8065. [DOI] [PubMed] [Google Scholar]
- 21.Fuse T, et al. p27Kip1 expression by contact inhibition as a prognostic index of human glioma. J Neurochem. 2000;74:1393–1399. doi: 10.1046/j.1471-4159.2000.0741393.x. [DOI] [PubMed] [Google Scholar]
- 22.Sgambato A, Cittadini A, Faraglia B, Weinstein IB. Multiple functions of p27(Kip1) and its alterations in tumor cells: A review. J Cell Physiol. 2000;183:18–27. doi: 10.1002/(SICI)1097-4652(200004)183:1<18::AID-JCP3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 23.Wieser RJ, Faust D, Dietrich C, Oesch F. p16INK4 mediates contact-inhibition of growth. Oncogene. 1999;18:277–281. doi: 10.1038/sj.onc.1202270. [DOI] [PubMed] [Google Scholar]
- 24.Serrano M, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 25.Seluanov A, et al. Telomerase activity coevolves with body mass not lifespan. Aging Cell. 2007;6:45–52. doi: 10.1111/j.1474-9726.2006.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seluanov A, et al. Distinct tumor suppressor mechanisms evolve in rodent species that differ in size and lifespan. Aging Cell. 2008;7:813–823. doi: 10.1111/j.1474-9726.2008.00431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn WC, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martel C, Batsche E, Harper F, Cremisi C. Inactivation of retinoblastoma gene product RB or an RB-related protein by SV40 T antigen in MDCK epithelial cells results in massive apoptosis. Cell Death Differ. 1996;3:285–298. [PubMed] [Google Scholar]
- 29.Morgenbesser SD, Williams BO, Jacks T, DePinho RA. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- 30.Caveda L, et al. Inhibition of cultured cell growth by vascular endothelial cadherin (cadherin-5/VE-cadherin) J Clin Invest. 1996;98:886–893. doi: 10.1172/JCI118870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson PJ, Daniel TO. Emerging targets: Molecular mechanisms of cell contact-mediated growth control. Kidney Int. 2002;61:S99–105. doi: 10.1046/j.1523-1755.2002.0610s1099.x. [DOI] [PubMed] [Google Scholar]
- 32.Gradl G, Faust D, Oesch F, Wieser RJ. Density-dependent regulation of cell growth by contactinhibin and the contactinhibin receptor. Curr Biol. 1995;5:526–535. doi: 10.1016/s0960-9822(95)00105-9. [DOI] [PubMed] [Google Scholar]
- 33.Stockinger A, et al. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J Cell Biol. 2001;154:1185–1196. doi: 10.1083/jcb.200104036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.St Croix B, et al. E-Cadherin-dependent growth suppression is mediated by the cyclin-dependent kinase inhibitor p27(KIP1) J Cell Biol. 1998;142:557–571. doi: 10.1083/jcb.142.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krimpenfort P, et al. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 36.Sharpless NE, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 37.Sherr CJ. Parsing Ink4a/Arf: “Pure” p16-null mice. Cell. 2001;106:531–534. doi: 10.1016/s0092-8674(01)00486-x. [DOI] [PubMed] [Google Scholar]
- 38.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 39.Chautan M, et al. Interdigital cell death can occur through a necrotic and caspase- independent pathway. Curr Biol. 1999;9:967–970. doi: 10.1016/s0960-9822(99)80425-4. [DOI] [PubMed] [Google Scholar]
- 40.Chau BN, Wang JY. Coordinated regulation of life and death by RB. Nat Rev Cancer. 2003;3:130–138. doi: 10.1038/nrc993. [DOI] [PubMed] [Google Scholar]
- 41.Nunney L. Lineage selection and the evolution of multistage carcinogenesis. Proc Biol Sci. 1999;266:493–498. doi: 10.1098/rspb.1999.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leroi AM, Koufopanou V, Burt A. Cancer selection. Nat Rev Cancer. 2003;3:226–231. doi: 10.1038/nrc1016. [DOI] [PubMed] [Google Scholar]
- 43.Nasser NJ, et al. Alternatively spliced Spalax heparanase inhibits extracellular matrix degradation, tumor growth, and metastasis. Proc Natl Acad Sci USA. 2009;106:2253–2258. doi: 10.1073/pnas.0812846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avivi A, et al. P53 in blind subterranean mole rats–loss-of-function versus gain-of-function activities on newly cloned Spalax target genes. Oncogene. 2007;26:2507–2512. doi: 10.1038/sj.onc.1210045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.