Abstract
The coexistence of multiple gene translation machineries is a feature of eukaryotic cells and a result of the endosymbiotic events that gave rise to mitochondria, plastids, and other organelles. The conditions required for the integration of these apparatuses within a single cell are not understood, but current evidence indicates that complete ablation of the mitochondrial protein synthesis apparatus and its substitution by its cytosolic equivalent is not possible. Why certain mitochondrial components and not others can be substituted by cytosolic equivalents is not known. In trypanosomatids this situation reaches a limit, because certain aminoacyl-tRNA synthetases are mitochondrial specific despite the fact that all tRNAs in these organisms are shared between cytosol and mitochondria. Here we report that a mitochondria-specific lysyl-tRNA synthetase in Trypanosoma has evolved a mechanism to block the activity of the enzyme during its synthesis and translocation. Only when the enzyme reaches the mitochondria is it activated through the cleavage of a C-terminal structural extension, preventing the possibility of the enzyme being active in the cytosol.
Keywords: aminoacyl-tRNA synthetases, mitochondria, tRNA, trypanosoma
Aminoacyl-tRNA synthetases (ARS) and transfer RNAs (tRNAs) are central components of the genetic code (1). The aminoacylation reaction of tRNAs catalyzed by ARS ensures the right association between codons and amino acids in a mechanism at least as ancient as the genetic code itself (2, 3). The universal distribution and conservation of both molecules shows that, to a large extent, ARS and tRNAs had fully evolved by the time of the first major phylogenetic split between archaea and bacteria (4, 5).
Aminoacyl-tRNA synthetases evolved from single domain proteins that possibly formed heterodimeric complexes around tRNA molecules or their precursor minihelices (6, 7). Extant ARS are structurally divided into two symmetrical classes (class I and class II) that recognize two different, but equivalent, sets of amino acids. From the analysis of available crystal structures of tRNA-ARS complexes it is apparent that three ancestral modes of interaction between a class I and a class II active site domains may have existed around a single tRNA molecule (7, 8).
The complex set of recognition interactions required for faithful aminoacylation of tRNAs by ARS was mostly evolved before the endosymbiotic events that gave rise to the different forms of eukaryotic cells. Consequently, the genesis of mitochondria, chloroplasts, apicoplasts, and other cellular compartments brought together fully diverged sets of ARS and tRNAs (9). This process continued with the transfer of ARS genes from the mitochondria to the nucleus, and the selection of enzymes capable of working both in the cytosol and in organelles. However, this process has not completely eliminated the existence of duplicated ARS (10).
In most eukaryotic species mitochondria and chloroplasts contain a full or almost full complement of tRNAs that are exclusively used by the translation machinery of the organelle (11). It is unclear why all eukaryotic cells maintain separate translational apparatuses for the cytosol and the mitochondria. This situation is particularly intriguing among kinetoplastids, where only nuclear genes code for tRNAs which function indistinctively in the cytosol and in the mitochondria, but mitochondria-specific ARS still exist (12). It has been determined that at least some of the Trypanosoma and Leishmania tRNAs that are transported to the mitochondria become modified (13, 14). Differences between mitochondrial and cytosolic ribosomes may be responsible for the existence of these differentially modified tRNAs (15).
However, the reasons for the existence of segregated protein synthesis machineries inside Trypanosoma remain unclear. To investigate the factors responsible for the maintenance of these separate translational apparatuses, and the mechanisms that functionally separate them, we have focused on the two lysyl-tRNA synthetases (KRS) coded by the genome of Trypanosoma brucei. As mentioned above, all tRNALys in Trypanosoma are nuclear encoded making unclear the need for two independent KRS.
Here we report that, in Trypanosoma brucei, a strict functional segregation of the cytosolic and mitochondrial KRSs takes place through a regulatory mechanism in ARS. After translation the nuclear-encoded mitochondrial KRS (TbKRS2) is inactive due to the presence of a C-terminal extension that is cleaved when the enzyme reaches the lumen of the mitochondria. This C-terminal sequence is not necessary for mitochondrial transport, which requires a canonical N-terminal signal sequence. After mitochondrial import and cleavage of the N-terminal signal peptide the C-terminal sequence is also cleaved producing the mature and active form of the enzyme. Thus, in Trypanosoma, the functional segregation of the mitochondrial and cytosolic genetic codes is reinforced via a mechanism that prevents tRNA aminoacylation by mitochondrial ARS in the cytosol.
Results
Sequence Analysis of TbKRS and TbKRS2.
Two different coding sequences for KRS are annotated in the GeneDB T. brucei database. Tb927.8.1600 and Tb927.6.1510 code, respectively, for two proteins hereinafter named TbKRS1 and TbKRS2. Both predicted polypeptides contain an N-terminal oligonucleotide binding fold (OB fold) domain, and a C-terminal class II ARS-like catalytic core domain that includes the three signature sequences of class IIb ARS (16) (Fig. 1A).
Fig. 1.
KRS sequence alignments. Alignment of the carboxy terminus region of KRS sequences. Mitochondrial KRSs from Kinetoplastida order are boxed.
TbKRS1 is a 584-amino-acids-long protein that does not contain discernible signal peptides. It was thus predicted to act as a cytosolic KRS. The TbKRS2 sequence is 634 amino acids long and contains an N-terminal signal peptide predicted to act as a mitochondrial targeting sequence with high probability by different computational algorithms. Thus, TbKRS2 was predicted to act as a mitochondrial KRS. TbKRS2 also contains an unusual C-terminal extension of approximately 90 amino acids (Fig. 1B). This extension is specific to the order Kinetoplastida, has no significant similarity to known proteins, and is not predicted to be a localization signal sequence by any of the programs used in this work.
TbKRS2 Is the Mitochondrial lysyl-tRNA Synthetase.
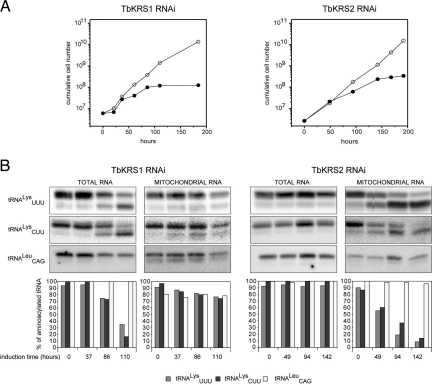
We first tested whether TbKRS1 and TbKRS2 were essential enzymes for T. brucei. RNA interference (RNAi) was used to knock-down TbKRS1 and TbKRS2 protein expression in vivo. Repression of expression of both enzymes led to growth arrest 3–4 days after induction of the RNAi treatment, followed by cell death (Fig. 2A). Thus, TbKRS1 and TbKRS2 are essential for T. brucei and are not interchangeable: the lack of TbKRS1 cannot be replaced by the activity of TbKRS2 and vice versa.
Fig. 2.
Effect of depletion of TbKRS1 and TbKRS2 by RNAi. (A) Effect of RNAi-mediated knockdown of TbKRS1 and TbKRS2 expression on T. brucei growth. Left panel shows the growth curve obtained for a representative clonal transgenic cell line in which TbKRS1 expression was repressed (●). As a control of the growth rate, uninduced cells from the same cell line (○) were grown in parallel. The panel on the right shows the growth curve of a representative clonal cell line in which TbKRS2 expression was repressed (●) in comparison to the uninduced cells. Induction of the interference started at time 0 h for both cell lines. (B) Effect of RNAi-mediated knockdown of TbKRS1 and TbKRS2 expression on tRNALys aminoacylation. Left panel shows TbKRS1 (Left) and TbKRS2 (Right) RNAi cell line Northern blots of total and mitochondrial RNA samples isolated and electrophoresed under acidic conditions. Time of induction is indicated at the bottom. The blots were probed for both tRNALys isoacceptors (CUU and UUU), as well as for a control tRNA (tRNACAGLeu). The bar graph shows the quantification of the results. Relative amounts of aminoacylated tRNAs are indicated for tRNAUUULys (light gray), tRNACUULys (dark gray), and tRNACAGLeu (white). For each lane, the sum of aminoacylated and deacylated RNA was set to 100%. Right panel shows the same information for the TbKRS2 RNAi cell line.
To confirm the effects of RNAi treatment we tested the levels of aminoacylated tRNALys in treated cells by acid gel electrophoresis (Fig. 2B). Knockdown of TbKRS1 resulted in the accumulation of uncharged tRNALys (both the CUU and the UUU isoacceptors) in the cytosolic fractions of treated organisms (Fig. 2B). Similarly, repression of TbKRS2 expression led to an accumulation of both tRNALys isoacceptors, but only in the isolated mitochondrial fractions. These results demonstrate that TbKRS1 is the enzyme responsible for the lysylation of cytosolic tRNALys and that TbKRS2 is responsible for mitochondrial tRNALys lysylation in T. brucei. We then determined the physical localization of the enzymes by immunoblot and immunofluorescence analysis.
Cellular Localization of TbKRS1 and TbKRS2.
The predicted cytosolic and mitochondrial localizations for TbKRS1 and TbKRS2, respectively, were studied by inmunofluorescence and inmunoblot analysis of T. brucei cellular fractions. To this end transgenic T. brucei cell lines expressing TbKRS1 and TbKRS2 fused to a Ty1 epitope were obtained.
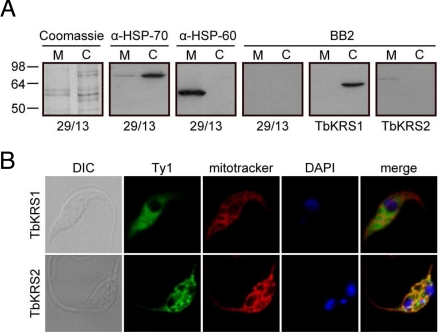
First we analyzed the localization of Ty1-tagged proteins by cellular fractionation studies. As expected, these experiments revealed a clear cytosolic distribution for TbKRS1 and a mitochondrial localization for TbKRS2 (Fig. 3A), confirming previous results. The distributions of the tagged proteins were also analyzed by means of confocal immunofluorescence using an anti-Ty1 monoclonal antibody. Confocal immunofluorescence analysis clearly confirmed the cytosolic localization of TbKRS1 and the mitochondrial distribution of TbKRS2 (Fig. 3B). Thus, signal peptide prediction, tRNALys in vivo aminoacylation analysis, and immunoblot and immunofluorescence data conclusively demonstrate a cytosolic localization for TbKRS1 and a mitochondrial localization for TbKRS2.
Fig. 3.
Subcellular localization of Trypanosomal KRSs. (A) Immunoblot analysis of TbKRS1 and TbKRS2. Crude cytosolic and crude mitochondrial extracts were analyzed for the presence of the Ty1 tagged TbKRS1 and TbKRS2. Quality of the fractionation was verified by direct visualization of the Coomassie staining and by immunoblot analysis of the cytosol specific HSP-70 and mitochondria specific HSP-60 markers. The positions of molecular size standards (in kDa) are shown in the left margin. (B) Confocal immunofluorescence analysis of TbKRS1 (Top) and TbKRS2 (Bottom) overexpressing cell line. Both proteins were expressed under the control of the tetracycline inducible PARP promoter and were carrying the Ty1 tag at their carboxy terminus. From left to right, cells were visualized by differential interference contrast (DIC), overexpressed protein was labeled with the BB2 monoclonal antibody against the Ty1 tag and the Alexa Fluor 488 antibody, mitochondria were stained with Mitotracker Red CMX Ros, DNA was stained with DAPI and finally green-red-blue channels were merged. The fluorescence fields shown were visualized independently by laser confocal microscopy at the appropriate wavelength.
TbKRS2 Localization and Function: Role of the Peptide Extensions.
We reasoned that the two idiosyncratic extensions displayed by the mitochondrial TbKRS2 may play a role in both the spatial and the functional separation of this protein from its cytosolic counterpart. To study their putative role as signal peptides two different approaches were taken.
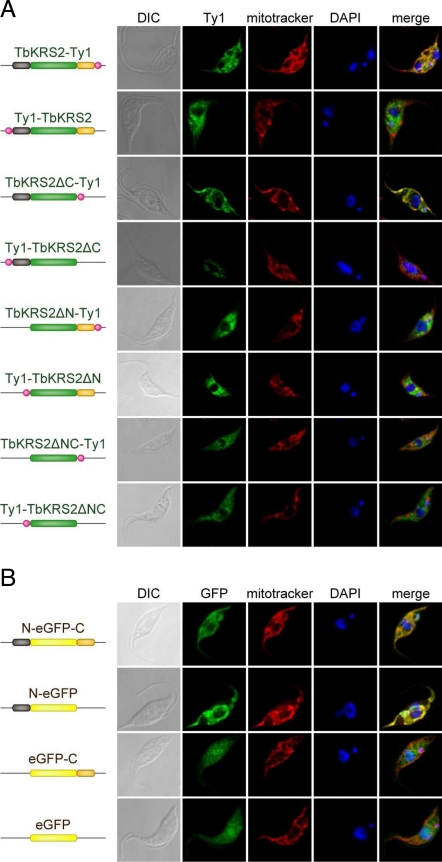
First, T. brucei cell lines expressing different versions of TbKRS2 lacking either or both extension sequences [TbKRS2ΔN (residues 27–634), TbKRS2ΔC (residues 1–546), and TbKRS2ΔNC (residues 27–546)] were created (Fig. 4). All proteins were tagged with a Ty1 epitope that was placed at either the amino or carboxy ends of each variant to prevent artifacts due to its presence. Second, a reporter system based on the green fluorescence protein (eGFP) was used. Stable transgenic T. brucei cell lines were constructed expressing eGFP fused to the carboxy end of the TbKRS2 N-terminal peptide (N-eGFP), the amino end of TbKRS2 C-terminal extension (eGFP-C), or between both extensions (N-eGFP-C) (Fig. 4).
Fig. 4.
Analysis of TbKRS2 import mechanism into the mitochondria. (A) Confocal immunofluorescence analysis of different TbKRS2 forms. From left to right, cells were visualized as described for Fig. 3B. (B) Confocal immunofluorescence analysis of full-length different eGFP fusion proteins. Cells were visualized as described for Fig. 3B, but overexpressed protein (green channel) was detected directly by GFP fluorescence.
Immunostaining results obtained with cells expressing different variants of TbKRS2 are shown in Fig. 4A. The pattern of TbKRS2 showed a staining pattern that matched precisely with the mitochondrial marker. Interestingly, an N-terminal Ty1 epitope blocked mitochondrial localization indicating that correct localization of the protein requires an intact N-terminal wild-type sequence but not a wild-type C-terminal sequence. TbKRS2ΔC showed a clear mitochondrial pattern, again blocked only by an N-terminal Ty1 epitope. Thus, the C-terminal peptide is dispensable for the mitochondrial localization of TbKRS2. Confirming this conclusion, TbKRS2ΔN and TbKRS2ΔNC generated a clear cytosolic pattern, independently of the Ty1 epitope position (Fig. 4A).
All of the results obtained with modified forms of TbKRS2 were confirmed with the eGFP constructions that contained all of the combinations of the N- and C-terminal peptides. Once again the N-terminal peptide was shown to be necessary and sufficient for the mitochondrial localization of eGFP, demonstrating that the idiosyncratic C-terminal extension of TbKRS2 is not required for its cellular distribution (Fig. 4B).
TbKRS2 Processing during Transport: Both Extensions Are Cleaved.
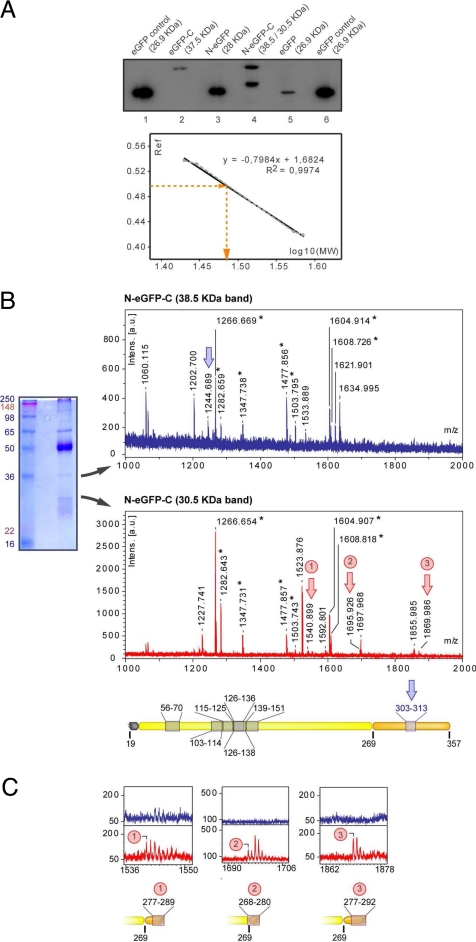
We used the cell lines expressing eGFP fused to the N- and/or C-terminal peptides to characterize the processing of these extensions during mitochondrial import. All forms of eGFP were purified from T. brucei total cell extracts by immunoprecipitation using a monoclonal antibody against eGFP. The immunoprecipitated protein was electrophoresed in high resolution SDS/PAGE gels, and the corresponding bands were visualized by Western blot with the same anti-eGFP antibody (Fig. 5).
Fig. 5.
TbKRS2 processing. (A) Immunoblot analysis of eGFP fusion proteins expressing cell lines. Total protein fractions of 8.5 × 106 cell equivalents of the different cell lines were electrophoresed on high resolution SDS/PAGE, transferred to a PVDF membrane and analyzed by immunoblot with α-GFP antibody. On the bottom, logarithm of known eGFP fusion proteins MW was plotted versus their relative mobility (Rf) to build a standard curve. Linear regression equation was used to determine the molecular weight of the lighter N-eGFP-C expressed product. (B) On the left, immunoprecipitation of N-eGFP-C expressing cell line with α-GFP. On the right, mass spectra obtained for the 38.5-kDa (top) and 30.5-kDa (bottom) immunoprecipitated proteins. The asterisk indicates the mass peaks that matched with the protein sequence. Blue and red arrows indicate mass peaks specific for 38.5- and 30.5-kDa proteins respectively. A reproduction to scale of N-eGFP-C is shown on the bottom, indicating the peptide matches. (C) Magnified view of the mass spectra obtained for the two proteins is compared on m/z regions corresponding to the 30.5-kDa protein specific mass peaks.
As can be seen in Fig. 5A, N-eGFP is detected as a single band of apparent molecular weight slightly larger than eGFP alone. Subsequent analysis of this band by Edman degradation revealed that TbKRS-2 is rapidly processed during import through the removal of the N-terminal sequence up to amino acid Q18.
Lane 2 of Fig. 5 shows that eGFP-C (previously shown to have a cytosolic distribution) is not processed and displays an apparent molecular weight corresponding to the complete fusion protein (37.46 kDa). By contrast, N-eGFP-C (localized to the mitochondria) is detected as two bands of equal intensity (Fig. 5, lane 4). One corresponds to the apparent molecular weight of eGFP-C, and the second band displays a lower approximate molecular weight of 30 kDa. No band with the expected molecular weight of N-eGFP-C can be detected. Analysis of both bands by mass spectrometry indicates that N-GFP-C undergoes two different steps of cleavage. On the one hand, the N-terminal peptide is cleaved, confirming the results obtained with N-eGFP and producing the high molecular weight band in the gel. On the other hand, the C-terminal peptide is cleaved during mitochondrial translocation, most likely between positions 280 and 292. The double cleavage of the N- and C-terminal extensions produces the lower band observed in the gel.
The fact that we could not detect full-length N-eGFP-C nor eGFP-C (a result of cleavage of the N-terminal peptide in N-eGFP-C) indicates that the N-terminal peptide is processed first, and the C-terminal peptide is cleaved later. Moreover, the fact that a significant amount of N-eGFP-C is found partially processed (eGFP-C) suggests that the processing of the C-terminal peptide is slower, supposedly when the protein has already entered the lumen of the mitochondria. It should be noticed that a recent analysis of the entire proteome of the insect stage of T. brucei (17) is consistent with our processing model for TbKRS2.
Our results show that both extensions in TbKRS2 are processed during mitochondrial transport. The N-terminal peptide is essential for the localization of the protein, and is cleaved first and rapidly. The C-terminal peptide is dispensable for mitochondrial localization and is cleaved after the protein has been translocated, possibly in the lumen of the organelle.
TbKRS2 Enzymatic Activity: Role of the Peptide Extensions.
The unusual C-terminal extension of the Trypanosoma mitochondrial KRS appears to have no function in the localization of the protein. We reasoned that the role of this extension may be linked to the enzymatic activity of TbKRS2. Additional domains that confer tRNA affinity are common in tRNA synthetases. These structures typically play no role in the aminoacylation reaction but provide the enzyme with additional binding energy or improve tRNA discrimination (18).
Expression plasmids were constructed for the expression of TbKRS2, TbKRS2ΔN, TbKRS2ΔC, and TbKRS2ΔNC in E. coli. A 6-histidine tag was added to all constructs to allow the purification of the proteins by affinity chromatography in Ni-NTA columns. TbKRS2, TbKRS2ΔN, and TbKRS2ΔNC could be purified and were used in subsequent aminoacylation and amino acid activation assays. None of the purified proteins had significant levels of activity with either transcripts of T. brucei tRNAUUULys, or the same tRNA purified from E. coli. This suggests that the natural modifications present in the mitochondrial substrate are essential for the aminoacylation of these tRNAs. Thus, we resorted to the analysis of the amino acid adenylation activities of the proteins to characterize the functional roles of the extensions in TbKRS2.
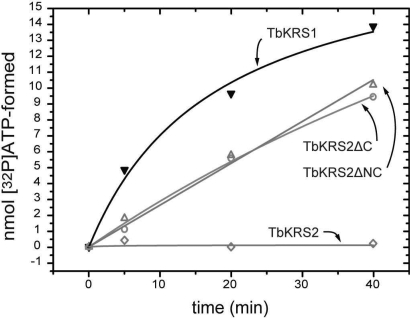
As can be seen in Fig. 6, full-length TbKRS2 has no detectable amino acid adenylation activity. Deletion of the C-terminal peptide (variant TbKRS2ΔC) generates an enzyme with levels of activity comparable to those of its cytosolic counterpart. TbKRS2ΔNC, has the same level of activity as TbKRS2ΔC, indicating that the presence or absence of the N-terminal extension is not important for the activation of the enzyme.
Fig. 6.
Role of N-terminal and C-terminal extensions in TbKRS2 on amino acid activation activity. ATP formation of the different TbKRS2 protein forms during the PPi exchange assay is compared to TbKRS1 in the same assay conditions. Full-length TbKRS2 was highly insoluble, and its soluble fraction showed no detectable activity in PPi exchange assays. Removal of the C-terminal extension restored activity of the enzyme to levels comparable to those of TbKRS1 at the same concentration. This recovery was independent of the presence or absence of the N-terminal extension of TbKRS2.
Thus, the C-terminal extension of TbKRS2 inhibits the activity of the protein, which is activated when the peptide is cleaved, while the N-terminal extension, crucial for mitochondrial targeting, has no effect on activity. This indicates that, during synthesis and translocation, the enzyme is prevented from catalyzing tRNA aminoacylation by the presence of the C-terminal extension. Once TbKRS2 reaches the interior of the mitochondria the C-terminal peptide is processed and the enzyme is activated.
Interestingly, despite several attempts, variant TbKRS2ΔN was impossible to purify because two different proteins eluted together out of the Ni-NTA resin. MS analysis demonstrated that both bands were variants of TbKRS2. One of them corresponded to the expected molecular weight of TbKRS2ΔN, while the second appeared to correspond to TbKRS2ΔNC. Thus, in the absence of the N-terminal peptide, the C-terminal extension of TbKRS2 is cleaved during the production or purification of the protein.
Discussion
Among the most remarkable features of the endosymbiotic events that drive the evolution of eukaryotes is the fact that these processes necessarily bring together completely different metabolisms and regulatory pathways. Indeed, symbiotic associations are characterized by the close interaction and coregulation of metabolic pathways (9), which undoubtedly favor the eventual integration of two different species.
However, the protein synthesis machinery most likely is inaccessible to this type of evolutionary coordination, as is evident from the very different nature of the extant translational machineries used in the mitochondria and cytosol of eukaryotic cells. An obvious reason for maintaining the merged translational apparatuses separate would be the prevention of tRNA misacylation or unproductive sequestering by noncognate ARS. In addition, segregation may also be of importance for the metabolic control of the different cellular compartments, which may depend on local amino acid concentrations for their regulation. Charged tRNAs and free amino acids are used by cells to gauge their nutritional balance, and these monitoring mechanisms could be altered by the combination of different tRNA populations.
An evolutionary trend toward unification of the mitochondrial and cytosolic translational apparatuses is apparent in metazoans where up to 17 aminoacyl-tRNA synthetases have evolved to act in both compartments (19). This seems to be accompanied by a process of size reduction in mitochondrial ARS (20, 21) and a dramatic divergence in the structure of tRNAs (22). The elimination of tRNA genes in Kinetoplastida mitochondria may be seen as another example of integration of the cell's genetic codes. In this context it is intriguing that Kinetoplastida still maintain several mitochondria-specific ARS, including KRS. In the process of characterizing the functional role of the sequence extensions found in TbKRS2 we have discovered that this enzyme is kept inactive by a C-terminal extension that is cleaved once the enzyme reaches the interior of the mitochondrion. This feature of TbKRS2 has a 2-fold interest. First, it is the first description of an ARS domain whose function appears to be the repression of the enzyme's activity until the right cellular localization is attained. Second, this mechanism for regulation may offer a glimpse into possible strategies that cells may use to functionally isolate their organelles.
Determining the mechanisms of cleavage of the C-terminal extension is beyond the scope of the present paper. However, our observations during the purification of TbKRS2ΔN indicate that this is an unstable form of TbKRS2 that is efficiently processed to TbKRS2ΔNC. Interestingly, TbKRS2 is not spontaneously processed. Thus, either the presence of the N-terminal peptide of TbKRS2 protects the C-terminal peptide against the action of proteases, or the cleavage of the N-terminal signal sequence triggers an autoproteolytic activity. The fact that the reporter eGFP-C is not processed in the cytosol of T. brucei (Fig. 5) suggests that the C-terminal peptide is processed by mitochondrial proteases. However, the purification protocols used for 6-his tagged proteins require abundant use of protease inhibitors, which would support the possibility that the enzyme undergoes self cleavage.
We cannot exclude additional functions for the C-terminal extension of TbKRS2. The peptide itself may have a function within the mitochondrion of Trypanosoma. It has been well reported that ARS domains can perform functions unrelated to gene translation after being excised from the original structure (23). In addition, the C-terminal extension may play a role in the transport of tRNALys from the cytosol to the lumen of the mitochondria, as has been reported in yeast (24). These additional activities, however, would be compatible with the regulatory function that we propose here.
Proper compartmentalization is essential for mitochondrial function, including signaling roles that regulate calcium homeostasis (25), control of apoptosis (26), or viral infection response mechanisms (27). In terms of protein synthesis, however, it is unclear to what extent, if any, does mitochondrial translation need to be separated from cytosolic protein synthesis. The trend among metazoa seems to be the reduction of ARS genes and the selection of dual-acting enzymes to the detriment of mitochondria-specific ARS. The strategy that we report here for the strict segregation of genetic code components may be an alternative to the fusion of both activities. Both evolutionary tactics, however, would have the same selective advantage: the prevention of deleterious interactions between coexisting protein synthesis machineries.
Materials and Methods
Bioinformatics.
TbKRS1 and TbKRS2 gene sequences (GeneDB accession no. Tb927.8.1600 and Tb927.6.1510 respectively) were obtained from the GeneDB database (http://www.genedb.org/) (28). BLASTP and BLASTN search tools (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to search for homologous protein and gene sequences (29). Primary sequence analysis was performed using the Expasy server (http://ca.expasy.org/tools/blast) (30). prediction of mitochondrial targeting sequences was performed with, Predotar (http://genoplante-info.infobiogen.fr/predotar/predotar.html), Mitoprot II (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html), TargetP (http://www.cbs.dtu.dk/services/TargetP/), SignalP (http://www.cbs.dtu.dk/services/SignalP/), and PSORT, PSORTII, iPSORT (www.psort.org). Structure-based alignments (31) were performed with CLUSTALX (32).
Cloning.
TbKRS1 and TbKRS2 were amplified from T. brucei genomic DNA and cloned into the pET30 vector for E. coli recombinant expression and protein purification. Deletion of TbKRS2 amino and carboxy terminus was achieved by site directed mutagenesis using QuikChange (Qiagen). Protein-expressing constructs designed for T. brucei expression were cloned into a derivative of pLEW100 plasmid (33). TbKRS1 and all TbKRS2 forms were fused to the Ty1 epitope, which is specifically recognized by BB2 antibody (34). RNAi constructs were built using stem loop constructs. TbRKS1 nucleotides 1280–1750 and TbKRS2 nucleotides 18–496 were amplified and cloned twice in antisense orientation in a derivative of pLEW100 plasmid (33).
T. brucei Culture and Transfection.
Procyclic T. brucei, strain 29/13 (33) was maintained at 2 × 105 to 3 × 107 cells/mL in MEM-Pros medium, supplemented with 15% FBS, 25 μg/mL hygromycin and 15 μg/mL G-418 at 27 °C. Cells were harvested at mid log phase for all of the applications. Cells were transfected on the Nucleofector system using Human T Cell Kit and program U-033 settings. Cells (2 × 107) were harvested, washed with 0.5% BSA in PBS, and resuspended in 100 μL of the transfection solution provided. Serial dilutions of the transfected culture were grown in conditioned medium to get clonal populations. Transfectants were selected and maintained with the addition of 2 μg/mL puromycin, and protein expression or RNAi induction of the transfected construct was induced with 10 μg/mL doxycycline.
Analysis of In Vivo Aminoacylation.
Mitochondrial and cytosolic crude fractions from RNAi cell lines were separated by digitonin extraction, as described elsewhere (35). RNA was extracted from the crude fractions using TRIzol. RNA samples from induced and uninduced cell lines was electrophoresed on high resolution acid gels, as described in (36) and transferred to a Hybond XL membrane by vacuum gel drying transfer (37). Percentage of aminoacylated tRNAUUULys, tRNACUULys and tRNACAGLeu was analyzed by Northern blot as already described (35) using the following probes: GATCCATGGCGCCTTCCGTGGGGATC for tRNAUUULys, GCACCCCCCGTGGGGCTCGAAC for tRNACUULys, and GCCTCCGGAGAGATGACGACC for tRNACAGLeu. The signals were digitized in the PhosphorImager™ through Typhoon Scanner Control software and quantified using ImageQuantTM TL software.
Immunoblot Analysis of Transfected Cell Lines.
TbKRS1 and TbKRS2 expressing cell lines were fractionated using digitonin extraction protocol described elsewhere (13). Thirty thousand cell equivalents per fraction were electrophoresed and analyzed by immunoblot using BB2 antibody. eGFP immunoblot analysis was performed on whole cell extracts. Protein samples from 0.85 × 107 cells were resolved by high resolution SDS/PAGE, and analyzed by immunoblot with α-GFP antibody (ImmunoKontact).
Immunofluorescence Analysis of Transgenic Cell Lines.
Fifty nanomolar MitoTracker was added to the cell cultures for 15 min before the immunostaining for the mitochondrion labeling. Cells were then harvested, washed with MEM-Pros medium, settled onto glass slides, fixed with 4% (wt/vol) paraformladehyde in PBS for 10 min, and permeabilized with 2% (wt/vol) Triton X-100 for 2 min. After washing in PBS, cells were incubated for 2 h with a 1/50 dilution of BB2 antibody, raised against the Ty1 tag, and for 1 h with Alexa Fluor 488 α-mouse antibody. BB2 and α-mouse antibody are only used for TbKRS1 and TbKRS2 expressing cell lines analysis. eGFP expressing cell lines were analyzed by direct fluorescence of the eGFP. Finally, DNA was labeled with 1 μg/mL DAPI in PBS for 2 min. Slides were mounted in Mowiol (prepared according to the product supplier's protocol) and examined on a Leica TCS SP5 confocal microscope. For the immunofluorescence analysis of eGFP expressing cell lines, neither the BB2 nor the α-mouse antibody were used.
Enzyme Overexpression and Purification.
BL21 DE3 cells (Novagen) transformed with pET30 Ek/LIC derived plasmids were grown to 37 °C to an optical density A600 = 1.5.AU. After a sudden decrease of temperature to 20 °C, protein expression was induced with 1 mM IPTG for 12 h at 20 °C. Protein purification on HisTrap nickel affinity columns (GE Healthcare) was performed according to the manufacture's protocol on an AKTA FPLC system. Eluted protein was dialyzed and kept in 50 mM Tris·HCl pH 8, 100 mM NaCl, 50% vol/vol glycerol at -20 °C. Enzyme concentrations were determined by active site titration (38).
Pyrophosphate (PPi) Exchange Assay.
In vitro reactions were performed at 37 °C in 50 mM HEPES·NaOH pH 7.2, 20 mM MgCl2, 1 mM DTT, 2 mM ATP, 4 mM lysine, 2 mM NaPPi, 2MBq/μmol [32P] NaPPi (PerkinElmer). Reactions were initiated with 100 nM enzyme, and quenched with 3.5% wt/vol perchloric acid, 3% wt/vol activated charcoal, 100 mM NaPPi. [32P]ATP formed during the exchange reaction and bound onto the charcoal is washed, resuspended in scintillation liquid, and detected on the scintillation counter with QuantaSmart Operating Software.
Proteomics.
MALDI-TOF mass spectrometry and Edman degradation analyses were performed in the Proteomics and Bioinformatics facility of the Universitat Autònoma de Barcelona, a member of the ProteoRed network.
Acknowledgments.
We thank Keith Matthews (University of Edinburgh, Edinburgh) for providing the BB2 antibody. This work was supported by Spanish Ministry of Science and Education Grant BIO2006–01558 (to L.R.d.P.), and Swiss National Foundation Grant 3100A0_121937 (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Di Giulio M. The evolution of aminoacyl-tRNA synthetases, the biosynthetic pathways of amino acids and the genetic code. Orig Life Evol Biosph. 1992;22:309–319. doi: 10.1007/BF01810859. [DOI] [PubMed] [Google Scholar]
- 3.Schimmel P, Giege R, Moras D, Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagel GM, Doolittle RF. Phylogenetic analysis of the aminoacyl-tRNA synthetases. J Mol Evol. 1995;40:487–498. doi: 10.1007/BF00166617. [DOI] [PubMed] [Google Scholar]
- 5.Ribas de Pouplana L, Turner RJ, Steer BA, Schimmel P. Genetic code origins: tRNAs older than their synthetases? Proc Natl Acad Sci USA. 1998;95:11295–11300. doi: 10.1073/pnas.95.19.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delarue M. Partition of aminoacyl-tRNA synthetases in two different structural classes dating back to early metabolism: Implications for the origin of the genetic code and the nature of protein sequences. J Mol Evol. 1995;41:703–711. doi: 10.1007/BF00173148. [DOI] [PubMed] [Google Scholar]
- 7.Ribas de Pouplana L, Schimmel P. Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell. 2001;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- 8.Schimmel P, Ribas de Pouplana L. In: The ribosome. Stillman B, editor. Cold Spring Harbor, NY: Cold Spring Harbor Symposium Quantum Biology; 2001. pp. 161–166. [Google Scholar]
- 9.Lopez-Garcia P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- 10.Schneider A. Does the evolutionary history of aminoacyl-tRNA synthetases explain the loss of mitochondrial tRNA genes? Trends Genet. 2001;17:557–559. doi: 10.1016/s0168-9525(01)02439-8. [DOI] [PubMed] [Google Scholar]
- 11.Duchene AM, Pujol C, Marechal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 12.Hauser R, Schneider A. tRNAs are imported into mitochondria of Trypanosoma brucei independently of their genomic context and genetic origin. EMBO J. 1995;14:4212–4220. doi: 10.1002/j.1460-2075.1995.tb00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charriere F, Helgadottir S, Horn EK, Söll D, Schneider A. Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc Natl Acad Sci USA. 2006;103:6847–6852. doi: 10.1073/pnas.0602362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charriere F, et al. Dual targeting of a tRNAAsp requires two different aspartyl-tRNA synthetases in trypanosoma brucei. J Biol Chem. 2009;284:16210–16217. doi: 10.1074/jbc.M109.005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan TH, Bochud-Allemann N, Horn EK, Schneider A. Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc Natl Acad Sci USA. 2002;99:1152–1157. doi: 10.1073/pnas.022522299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 17.Panigrahi AK, et al. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frugier M, Moulinier L, Giegé R. A domain in the N-terminal extension of class IIb eukaryotic aminoacyl- tRNA synthetases is important for tRNA binding. EMBO J. 2000;19:2371–2380. doi: 10.1093/emboj/19.10.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnefond L, et al. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: Characterization of AspRS and TyrRS. Biochemistry. 2005;44:4805–4816. doi: 10.1021/bi047527z. [DOI] [PubMed] [Google Scholar]
- 20.Aphasizhev R, et al. Conservation in evolution for a small monomeric phenylalanyl-tRNA synthetase of the tRNA(Phe) recognition nucleotides and initial aminoacylation site. Biochemistry. 1996;35:117–123. doi: 10.1021/bi9517998. [DOI] [PubMed] [Google Scholar]
- 21.Bullard JM, Cai YC, Demeler B, Spremulli LL. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J Mol Biol. 1999;288:567–577. doi: 10.1006/jmbi.1999.2708. [DOI] [PubMed] [Google Scholar]
- 22.Putz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. Rna. 2007;13:1184–1190. doi: 10.1261/rna.588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 24.Tarassov I, Entelis N, Martin R. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carafoli E. Historical review: Mitochondria and calcium: Ups and downs of an unusual relationship. Trends Biochem Sci. 2003;28:175–181. doi: 10.1016/S0968-0004(03)00053-7. [DOI] [PubMed] [Google Scholar]
- 26.Kroemer G. Mitochondrial control of apoptosis: An overview. Biochem Soc Symp. 1999;66:1–15. doi: 10.1042/bss0660001. [DOI] [PubMed] [Google Scholar]
- 27.Ohman T, Rintahaka J, Kalkkinen N, Matikainen S, Nyman TA. Actin and RIG-I/MAVS signaling components translocate to mitochondria upon influenza A virus infection of human primary macrophages. J Immunol. 2009;182:5682–5692. doi: 10.4049/jimmunol.0803093. [DOI] [PubMed] [Google Scholar]
- 28.Hertz-Fowler C., et al. GeneDB: A resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas de Pouplana L, Brown JR, Schimmel P. Structure-based phylogeny of class IIa tRNA synthetases in relation to an unusual biochemistry. J Mol Evol. 2001;53:261–268. doi: 10.1007/s002390010216. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 34.Bastin P, Bagherzadeh Z, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 35.Tan TH, Pach R, Crausaz A, Ivens A, Schneider A. tRNAs in Trypanosoma brucei: Genomic organization, expression, and mitochondrial import. Mol Cell Biol. 2002;22:3707–3717. doi: 10.1128/MCB.22.11.3707-3716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm ML, Baranowski W, Keith G, Wilhelm FX. Rapid transfer of small RNAs from a polyacrylamide gel onto a nylon membrane using a gel dryer. Nucleic Acids Res. 1992;20:4106. doi: 10.1093/nar/20.15.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fersht AR, et al. Active site titration and aminoacyl adenylate binding stoichiometry of aminoacyl-tRNA synthetases. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]