Abstract
Nitrogen acquisition and assimilation is a primary concern of insects feeding on diets largely composed of plant material. Reclaiming nitrogen from waste products provides a rich reserve for this limited resource, provided that recycling mechanisms are in place. Cockroaches, unlike most terrestrial insects, excrete waste nitrogen within their fat bodies as uric acids, postulated to be a supplement when dietary nitrogen is limited. The fat bodies of most cockroaches are inhabited by Blattabacterium, which are vertically transmitted, Gram-negative bacteria that have been hypothesized to participate in uric acid degradation, nitrogen assimilation, and nutrient provisioning. We have sequenced completely the Blattabacterium genome from Periplaneta americana. Genomic analysis confirms that Blattabacterium is a member of the Flavobacteriales (Bacteroidetes), with its closest known relative being the endosymbiont Sulcia muelleri, which is found in many sap-feeding insects. Metabolic reconstruction indicates that it lacks recognizable uricolytic enzymes, but it can recycle nitrogen from urea and ammonia, which are uric acid degradation products, into glutamate, using urease and glutamate dehydrogenase. Subsequently, Blattabacterium can produce all of the essential amino acids, various vitamins, and other required compounds from a limited palette of metabolic substrates. The ancient association with Blattabacterium has allowed cockroaches to subsist successfully on nitrogen-poor diets and to exploit nitrogenous wastes, capabilities that are critical to the ecological range and global distribution of cockroach species.
Keywords: Flavobacteria, insect endosymbiont, nitrogen conservation, uricolysis
Nitrogen acquisition for cellular metabolism is a primary concern of herbivorous animals. Cockroaches (Dictyoptera: Blattaria) typically consume decaying (nitrogen-poor) plant material but can be omnivorous, feeding opportunistically on decaying animal material and even (nitrogen-rich) conspecifics (1). Cockroaches respond to surplus nitrogen by concentrating and storing it as uric acid, which is excreted typically by terrestrial insects, within fat body cells (2). Increasing dietary nitrogen concentrations correlates with uric acid deposition in cockroach fat bodies (2, 3). These internal stores are thought to serve as metabolic reserves, providing an essential supplement when sufficient dietary nitrogen is lacking and a means for exploiting nitrogen-rich windfalls (4).
Although the ecological rationale for nitrogen conservation in phytophagous insects is clear, utilization of uric acid as a nitrogen reserve is not well understood. Many insects and other animals do not have the enzymatic capabilities required for producing ammonia from urate. Nitrogen recycling from stored uric acid in shield bugs and termites has been attributed to symbiotic microbes. The presence of Erwinia-like bacteria in Parastrachia japonensis, which are located exclusively in the gastric ceca, correlates with enzymatic uricolysis in the gastric ceca (5). Similarly, Reticulitermes flavipes hindgut bacteria ferment uric acid transported from the fat bodies, via the Malpighian tubules to the hindgut, into ammonia, CO2, and acetate (6–8). Cockroaches, close relatives of termites, harbor intracellular, maternally transmitted bacteria, called Blattabacterium sp., within their fat bodies, near the uric acid deposits (9), and they have been hypothesized to participate in nitrogen recycling from stored uric acid (10, 11). Periplaneta americana fat bodies are comprised of two distinct cell types: adipocytes, containing fat globules and uric acid crystals, and mycetocytes, which are filled densely with Blattabacterium (9, 11–13). After antibiotic treatment to remove Blattabacterium, mycetocytes appear to be highly degraded, uric acid accumulates significantly (13–15), and production of tyrosine, phenylalanine, isoleucine, valine, arginine, and threonine is reduced (16). Consequently, production of these amino acids has been attributed to Blattabacterium.
Phylogenetic analysis indicates that Blattabacterium is a member of the Flavobacteriales, where it clusters with other insect bacteriome-associated symbionts, Candidatus Sulcia muelleri (hereafter referred to as Sulcia, hosts in numerous sap-feeding insects within Hemiptera: Auchenorrhyncha) and Candidatus Uzinura diaspidicola (hosts in sap-feeding scale insects, family Diaspididae), and an insect host-manipulator, the male-killing ladybird beetle endosymbiont (17–20). Blattabacterium appears to have infected the common ancestor of cockroaches and termites >140 Mya (21, 22). It has been maintained in all cockroaches except the cave-dwelling Nocticola and lost in all termites except the basal species, Mastotermes darwiniensis (17, 22). Although the existence of Blattabacterium has been known for over 100 years, efforts to fully characterize it have been thwarted by its unsurprising recalcitrance to cultivation (23). Many beneficial bacterial symbionts with exclusively intracellular lifestyles (i.e., Buchnera aphidicola) have lost their ability to live outside their hosts but have free-living, cultivable relatives (e.g., Escherichia coli) (24). To elucidate further the role of Blattabacterium in nutrient provisioning, nitrogen conservation, and uric acid metabolism, we sequenced the genome of the P. americana endosymbiont Blattabacterium sp. BPLAN.
Results and Discussion
The genome of Blattabacterium sp. BPLAN is comprised of a 636,994-bp circular chromosome and a 3,448-bp plasmid with G + C contents of 28.2% and 28.5%, respectively. Thirty-three tRNAs corresponding to all of the amino acids are encoded on the chromosome. Additionally, a single 16S-23S-5S operon and a tmRNA gene are present. Complete transcriptional machinery, including the major sigma factor RpoD, is encoded in the genome. The origin of replication was predicted based on GC skew, and the first nucleotide in the nearest protein-coding region was designated as position 1 (Fig. S1). Blattabacterium encodes 581 ORFs, comprising 94% of the sequence, and 520 ORFs have an assigned function (Table 1). Four genes, encoding a putative oxidoreductase, two ABC transporters, and a bifunctional sulfate adenyltransferase, harbor internal stop codons and were annotated as pseudogenes.
Table 1.
Comparison of Blattabacterium sp. BPLAN genome with those of other Flavobacteriales
| Blattabacterium sp. BPLAN | Sulcia muelleri GWSS | Flavobacterium johnsonii UW101 | |
|---|---|---|---|
| Lifestyle | Intracellular symbiont | Intracellular symbiont | Free-living |
| Habitat | Cockroaches | Auchenorrhyncha | Soil and freshwater |
| Chromosome, bp | 636,994 | 245,530 | 6,096,872 |
| Plasmid [bp] | 1 [3,448] | 0 | 0 |
| G + C content [plasmid], % | 28.2 [28.5] | 22 | 34 |
| Total CDS | 581 | 227 | 5,017 |
| Average CDS length, bp | 1,031 | 1,006 | 1,057 |
| CDS density, % | 94 | 87 | 87 |
| CDS with assigned function | 520 | 210 | 3,022 |
| Pseudogenes | 4 | 0 | 39 |
| rRNAs | 3 | 3 | 18 |
| tRNAs | 33 | 31 | 62 |
General features of the Blattabacterium sp. BPLAN genome compared with those of other completely sequenced Flavobacteriales. CDS, protein coding sequences.
Overall, 54% of protein-encoding genes with assigned functions are involved in amino acid metabolism, translation and biogenesis, cell wall and outer membrane biogenesis, or coenzyme metabolism and energy production (Table S1). Complete de novo purine and pyrimidine biosynthetic pathways are encoded in the Blattabacterium genome. Interestingly, the ribonucleotide reductase (RNR), which performs the necessary reduction of ribonucleotides to deoxyribonucleotides, is comprised of an α subunit that is encoded by a chromosome-based gene and a β subunit that is encoded by a plasmid-based gene. The Blattabacterium plasmid does not carry genes involved in amino acid biosynthesis, as observed for plasmids of the aphid symbiont B. aphidicola. Thus, the sole function of the plasmid appears to be encoding the RNR β subunit, which is required for RNR function, but why this subunit has a plasmid location is not known. On the basis of its genome, Blattabacterium generates essential carbon precursor metabolites by gluconeogenesis, the citric acid cycle, and the pentose phosphate pathway. Adenosine 5′-triphosphate is generated aerobically using a cbb3-type cytochrome c oxidase-terminated electron transport chain.
Retention of genes encoding enzymes involved in vitamin and amino acid biosynthesis and loss of many genes encoding transcriptional regulatory functions and substrate-specific transporters are common features of obligate symbionts of insects (25). However, the exchange of molecules, including required metabolites, between symbiont and host cellular compartments is the foundation of the symbiotic interaction, implying that some transport mechanisms persist. In Blattabacterium, the NMN transporter PnuC is present and can facilitate exogenous pyridine uptake for NAD biosynthesis and recycling (26). Movement of proteins across the cell wall could be achieved by the Sec and twin-arginine targeting translocases encoded in the genome (27, 28). Blattabacterium also encodes multiple ATP-binding ABC transporter complexes. Although cation transport proteins are present in the Blattabacterium genome, few substrate-specific transporters, especially those involved in amino acid uptake or secretion, are identifiable, and this is consistent with other insect nutritional endosymbionts (18, 29).
Amino Acid Biosynthesis and Nitrogen Recycling.
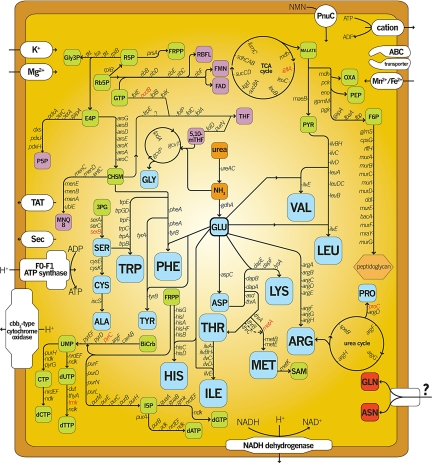
The central role hypothesized for Blattabacterium is amino acid biosynthesis and waste nitrogen recycling, and this role is supported by past experimental studies (13, 16, 30, 31). Metabolic pathway reconstruction from the genome sequence reveals complete biosynthetic pathways for all amino acids except asparagine, glutamine, methionine, and serine, implying that Blattabacterium can make most amino acids from a few substrates, such as glutamate, urea, and ammonia (Fig. 1). Genes involved in asparagine and glutamine biosynthesis are entirely absent. Previous experimental evidence suggests that Blattabacterium generates methionine by cysteine transsulfuration (30, 32). Genomic analysis reveals that all of the enzymes for methionine biosynthesis by this route are encoded by the genome except for the acetylhomoserine-yielding homoserine O-acetyltransferase. Alternatively, cystathionine γ-synthase produces homocysteine using cysteine and O-phosphohomoserine, a threonine biosynthesis metabolite, potentially circumventing the need for acetylhomoserine.
Fig. 1.
Predicted biosynthetic pathways. Blue, amino acids; green, metabolites; purple, vitamins. Red blocks are amino acids not produced by Blattabacterium. Genes in red italic type were not detected in the genome. 3PG, 3-phosphoglycerate; P5P, pyridoxal 5-phosphate; BiCrb, bicarbonate; R5P, ribose-5-phosphate; E4P, erythrose-4-phosphate; Rb5P, ribulose-5-phosphate; F6P, fructose-6-phosphate; FRPP, 5-phosphoribosyl 1-pyrophosphate; Gly3P, glyceraldehyde-3-phosphate.
Although genomic evidence for serine biosynthetic capability exists, Blattabacterium lacks the enzymes that catalyze the final serine-yielding reactions. Alternative mechanisms for endogenous methionine and serine biosynthesis may be used, or the metabolic intermediates or the actual amino acids are imported from the host cytoplasm.
Proline biosynthesis in some insects can proceed by arginine conversion in fat bodies, but enzymes for generating arginine from citrulline are thought to be absent in insects, implying an incomplete ornithine–urea cycle (33, 34). Blattabacterium can make arginine from glutamate, and it encodes the ornithine–urea cycle and enzymes to produce pyrroline-5-carboxylate, a proline biosynthetic intermediate that can be reduced further by a host-provided pyrroline-5-carboxylate reductase to yield proline. Blattabacterium must acquire proline from the cockroach, because it lacks the enzyme for the final biosynthetic step. Proline biosynthesis by arginine degradation also produces urea and ammonia, both of which can be used by the Blattabacterium-encoded urease and glutamate dehydrogenase to generate glutamate, effectively conserving nitrogen. Thus, our genomic data support the hypothesis that Blattabacterium recycles nitrogenous wastes and provides amino acids.
Opportunities for nitrogen scavenging and recycling are abundant due to various metabolic processes in Blattabacterium. Cystathionine γ-synthase and threonine dehydratase both produce ammonia while generating amino acid biosynthesis intermediates. Vitamin biosynthesis and modification processes also yield ammonia during biosynthesis. Enzymes involved in degradation and salvage pathways of ribonucleosides, ribonucleotides, pyridoxal 5′-phosphate, and nicotinamide produce ammonia as a by-product. Ammonia is toxic, but glutamate dehydrogenase (GdhA) can detoxify the intracellular environment and recycle ammonia into glutamate. Bacteria typically employ GdhA for nitrogen assimilation when ammonia is abundant and ATP is limited, because nitrogen assimilation by this route does not consume ATP and thus is physiologically advantageous (35–37).
Alternative Transcriptional Regulator, RpoN.
In view of evidence that Blattabacterium can recycle waste nitrogen, in the form of urea and ammonia, into amino acids and other metabolites, that this genome encodes the alternative sigma factor RpoN is intriguing. Bacteriome-associated endosymbionts typically lack regulatory genes, and Blattabacterium is the first to retain rpoN. In other bacteria, expression of genes involved in, but not limited to, nitrogen assimilation in bacteria can require RpoN, an enhancer-activated transcriptional regulator (38). We predicted two RpoN promoters within intergenic regions upstream of grpE and groES. GrpE is a co-chaperone of the stress-responsive DnaKJ chaperone complex (39). GroES and GroEL form a multimeric chaperone that assists protein folding (40). Similar RpoN promoter-like sequences were predicted in intergenic regions upstream of grpE in Bacteroides thetaiotaomicron and Flavobacterium psychrophilum and upstream of groES in B. thetaiotaomicron and Rhizobium etli. RpoN promoters are not observed upstream of these genes in E. coli or Bacillus subtilis.
RpoN-regulated gene expression is controlled tightly by enhancer-activator binding proteins that often bind >50 nt away from the RpoN promoter and must be brought in contact with the RpoN transcriptional holoenzyme by a DNA looping mechanism to initiate transcription (41). Blattabacterium does encode one RpoN enhancer-activator, FhlA, which, in E. coli, senses environmental formate and stimulates the transcription of genes involved in anaerobic ATP synthesis, but it lacks known FhlA-regulated genes (37, 42). Genomic evidence also indicates that Blattabacterium generates energy via aerobic respiration. Additionally, FhlA is not known to regulate either GroES or GrpE.
Uric Acid Metabolism.
“Storage excretion” of uric acid compounds in fat bodies is well documented in cockroaches and termites (2, 3, 43). In cockroaches, Blattabacterium-filled mycetocytes are adjacent to adipocytes containing uric acids (13) within the fat bodies, but little evidence indicates the role of Blattabacterium in this close physical association. Because a cultivation-dependent approach is nearly impossible, we used bioinformatics to explore the Blattabacterium genome for uricolytic capabilities. Uricase catalyzes the first step in aerobic uric acid catabolism to allantoin (44, 45), but no genes encoding a uricase were identified. A search of sequence databases for Bacteroidetes genes encoding uricases resulted in nine hits to genes encoding proteins annotated as uricase or as either hydroxyisourate hydrolase or transthyretin-like periplasmic protein. The latter proteins are homologous to B. subtilis PucM, which has been shown to catalyze a necessary intermediate uricolytic hydrolysis reaction (46). Alternatively, xanthine dehydrogenase (Xdh) degrades uric acid to xanthine under anaerobic conditions (8), but no Xdh homologue was detectable. Such homologues are widely present in other Bacteroidetes: A search of Bacteroidetes genomes for homologues of Xdh yielded >23 proteins annotated as xanthine dehydrogenase and an additional 53 proteins annotated as having conserved Xdh functional domains. Thus, although genes encoding uricolytic enzymes are harbored commonly by members of the Bacteroidetes, they appear to have been lost in Blattabacterium sp. BPLAN.
The hypothesis that uricolysis is carried out by Blattabacterium using proteins annotated as having an alternative or no known function cannot be ruled out entirely. A second possibility is that the insect host supplies uricolytic enzymes. A BLAST analysis of expressed sequences from two cockroach species (P. americana and Blattella germanica) reveals partial transcripts encoding homologues of proteins participating in uricolysis (GenBank accession nos. FG125694, FG128011, and FG132115). Xanthine dehydrogenase is variably present among animals but is found in most insects, based on genomes sequenced to date. An Xdh homologue is encoded in B. germanica (GenBank accession no. FG127839), based on blastx analysis of expressed sequences from B. germanica. Weighing against the hypothesis that the cockroach supplies the uricolytic enzymes are results showing that uricase activity is absent in termite tissues (43) and that antibiotics dramatically reduce uricolytic activity (8).
A third hypothesis is that uric acid is transported to the gut, where it is catabolized by uricolytic microorganisms. In R. flavipes, uric acid is transported actively from hemolymph, via the Malpighian tubules, to the hindgut, where anaerobic Bacteroides and Streptococcus degrade uric acid to ammonia (8). Cockroaches and termites are close relatives, with similar dietary preferences and excretory physiology (43). Uricolytic Clostridium have been isolated consistently from P. americana (47). Thus, members of the cockroach gut microflora can degrade uric acid (2). Similarly, antibiotic treatment of shield bugs (Insecta: Hemiptera) eliminates uricolytic bacteria from the gastric cecum and reduces uricolytic activity (5). If microbial degradation of uric acid occurs in the gut lumen of P. americana, then the role of Blattabacterium may be to use the products of uricolysis (urea and ammonia) for amino acid production.
Blattabacterium Phylogenetics and Comparative Genomics.
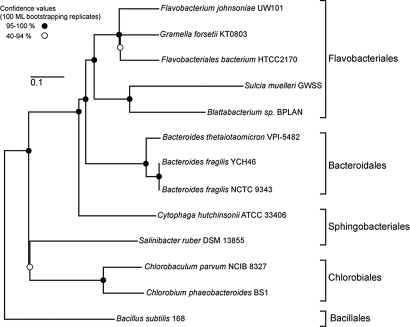
According to maximum likelihood trees of concatenated protein alignments from completely sequenced members of Bacteroidetes/Chlorobi, Blattabacterium sp. BPLAN belongs to the Flavobacteriales and is related most closely to Sulcia, another obligate insect symbiont of (Fig. 2). These results are consistent with 16S rRNA gene analyses that included more taxa (19, 21, 22). Many genes, including some expected to enable metabolic plasticity, are eliminated from obligate endosymbiont genomes (Table S1), leaving genes essential to maintaining the host–symbiont lifestyle (i.e., amino acid biosynthesis) (48). Blattabacterium is typical of insect nutritional symbionts in possessing a highly reduced genome at 637 kb; the even tinier Sulcia genome is one of the smallest yet discovered at 245 kb (18). Thus, Blattabacterium's genome provides clues to stages of genome reduction, potentially resembling an ancestral state for the tiny Sulcia genome. Consistent with this view, the Sulcia gene set is largely a subset of that of Blattabacterium. Only eight Sulcia ORFs lack homologues in Blattabacterium. Five are annotated as conserved hypothetical proteins of unknown function. The other three Sulcia-specific ORFs encode the α and β subunits of the ferredoxin oxidoreductase complex, which is involved in carbon dioxide assimilation, and the glutamate metabolic enzyme pyrroline-5-carboxylate dehydrogenase.
Fig. 2.
Phylogenetic relatedness of Blattabacterium sp. BPLAN to Cytophaga–Flavobacterium–Bacteroidetes bacteria with completely sequenced genomes. (Scale bar, 0.1 substitutions per site.)
Sulcia and Blattabacterium have similar roles within insect hosts. Both are capable of synthesizing essential amino acids, with Blattabacterium able to make all 10. Sulcia from the glassy-winged sharpshooter can make eight, excluding histidine and methionine, for which pathways are encoded by the co-resident symbiont, Baumannia cicadellinicola (18).
Comparisons of numbers of genes assigned to categories based on functional roles are also consistent with the view that Blattabacterium represents an intermediate stage between larger genomes of free-living bacteria, represented within the Bacteroidetes by Flavobacterium johnsonii, and the extremely tiny genomes of some obligate symbionts, represented by Sulcia. Blattabacterium and F. johnsonii show highly significant differences in distributions of genes among functional categories (χ2 = 330, df = 13, P < 0.00001). In Blattabacterium, genes involved in transcriptional regulation, signal transduction, secondary metabolites, and defensive activities are disproportionately lost, whereas those involved in translation, amino acid metabolism, and coenzyme metabolism are retained more often, a pattern resembling that noted previously for symbiotic Gammaproteobacteria (25, 48). Blattabacterium and Sulcia also differ in representation of different functional categories (χ2 = 49, df = 13, P < 0.00001), and this comparison illuminates the patterns of gene loss during the transition from small (Blattabacterium) to tiny (Sulcia) genomes. Sulcia has retained differentially genes in the categories corresponding to translation and amino acid biosynthesis and has differentially eliminated genes in the categories of unknown function, biosynthesis of lipids, and cell wall components.
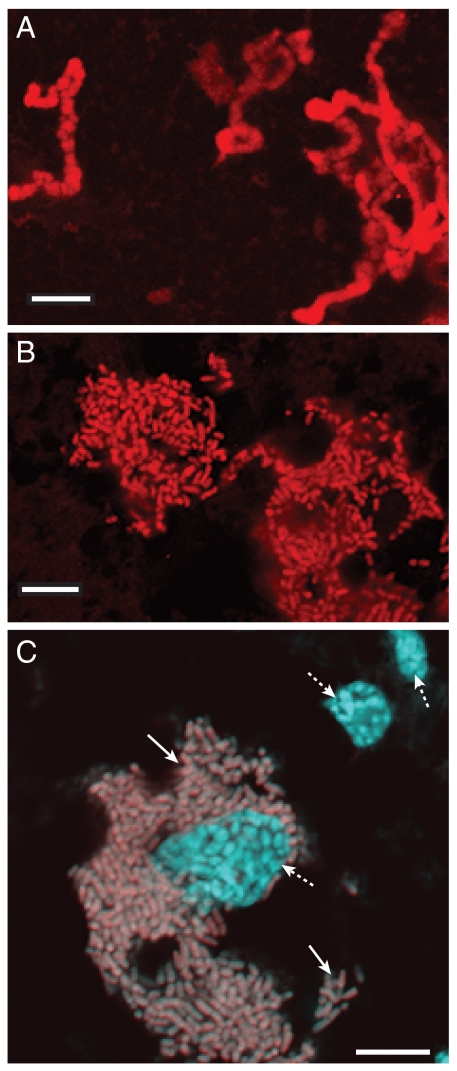
Loss of genes determining cell size and shape are likely to contribute to morphological changes associated with the most extreme stages of genome reduction. Sulcia's small genome does not encode the rod-shape-determining proteins MreB and MreC or the pathways for producing a cell wall, and these losses correlate with irregular cell morphology (Fig. 3A). This cell morphology and lack of genes for synthesis of cell wall components also are observed in Carsonella ruddii (Gammaproteobacteria) and Hodgkinia cicadicola (Alphaproteobacteria), two unrelated symbionts with even tinier genomes (145 and 160 kb) (49, 50). Microscopy confirms that Blattabacterium alone inhabits cockroach fat bodies, where it populates the cytoplasm (Fig. 3C). Additionally, it has a well-defined, rod-shaped cellular morphology and an intact cell wall (Fig. 3B) (13, 51), consistent with the presence of MreB and MreC and a complete peptidoglycan biosynthetic pathway.
Fig. 3.
Comparison of the cell morphology of Blattabacterium sp. BPLAN and Sulcia muelleri. Confocal micrographs of Cy3-labeled S. muelleri (A) and Blattabacterium sp. BPLAN (B). (C) A FISH micrograph of Blattabacterium sp. BPLAN within Periplaneta americana. A composite image of P. americana abdominal tissue hybridized with a Cy3-labeled Blattabacterium-specific probe and counterstained with DAPI. Dashed arrows, insect nuclei; solid arrows, Blattabacterium sp. BPLAN cells. (Scale bar, 10 μm.)
An analysis of the gene order of homologous proteins indicates little conservation of gene order between Sulcia and Blattabacterium (Fig. S2). Thus, one or both of these lineages continued to undergo extensive chromosomal rearrangements after their divergence.
Conclusions
The Blattabacterium genome confirms and elucidates its central role in nitrogen metabolism within the cockroach system, including nitrogen recycling from ammonia and urea and extensive capabilities for amino acid provisioning. These metabolic capabilities are likely to have been important in allowing cockroaches to expand their ecological niche by exploiting food sources in which nitrogen sources are variable and often limited. The first step in the utilization of uric acid as a nitrogen storage compound appears to be provided not by Blattabacterium, which lacks a uricase homologue, but by either the insect host or, as known to occur in termites, bacteria in the gut lumen. Previous hypotheses suggesting that Blattabacterium is capable of uricolysis are not supported by the genomic evidence, raising the question of why Blattabacterium is located in such close proximity to uric acid deposits in fat bodies. Possibly, a Blattabacterium ancestor was uricolytic but, in the course of genome reduction, lost its uricase, transferring this function to uric acid degraders among the gut microflora. Blattabacterium resembles other insect nutritional symbionts in having lost almost all specific transporters and regulatory genes but is unique for insect symbionts in retaining the transcriptional regulator RpoN.
Materials and Methods
DNA Preparation.
Whole fat bodies were dissected from five adult P. americana (Carolina Biological Supply) in 1× PBS and lightly homogenized by hand in 200 μL of 1× PBS. Homogenate was passed through 20- and 11-μm filters and centrifuged at 8,000 × g at 4 °C for 10 min. The pellet was resuspended in 200 μL of Buffer AL from the Qiagen DNEasy kit, and DNA was prepared according to the manufacturer's protocols. Extracted DNA was suspended in 1× PBS to a concentration of 104 ng/μL. Quantitative PCR using the bacterial 16S rRNA and host EF1-α genes was performed to determine the ratio of Blattabacterium-to-cockroach total DNA. The final ratio was ≈610:1. We confirmed that Blattabacterium was the only bacterium represented in the sample by sequencing amplicons obtained from a PCR using universal 16S rDNA bacterial primers. Five micrograms of total DNA was submitted to the University of South Carolina and sequenced using a Roche/454 GS-FLX 100.
Genome Assembly.
Two sequencing runs yielded 225,047 reads with an average read length of 255 nt. Sequence reads were assembled into 11,737 contigs by the Newbler assembler, version 1.1.03, and visualized with Consed (52) and SeqMan (DNASTAR). Fourteen contigs were distinct in having greater length and greater read average depth of coverage. Thirteen contigs, ranging from 1,019 to 146,043 nt and together comprising 110,232 reads (49% of all reads), were ordered, using “-to” and “-from” information appended to the trace filenames by the Newbler assembler, into a putative circular genome of 636,578 nt with an average 28.4% G + C content and average read depth of 44.2×. Sanger sequencing across gaps between the 13 contigs enabled assembly of a single circular contig of 636,994 nt with a 28.2% G + C content. The remaining 3,249 nt contig displayed no significant match to any other contig over its entire length and had an average read depth of coverage of 78.1×, which was almost 2× that of the genome. ORF Finder was used to search for ORFs, followed by blastp search of the putative ORFs against the GenBank nr database, yielding regions encoding four proteins with high similarity to bacterial proteins. Given the read depth, its lack of assembly with the rest of the genome, and the presence of bacterial genes, we hypothesized that this fragment corresponded to a plasmid. By designing outward-oriented PCR primers based on the sequences at the ends of this contig, we obtained a single product that linked the ends of this fragment and confirmed a plasmid.
Genome Annotation.
Prodigal was used to predict protein-coding regions. Functional annotation of the coding regions proceeded by combining results of blastp searches against the GenBank nr (downloaded November 20, 2008) and COG myva (53) databases and hmmpfam (HMMER, version 2.3.2) searches against TIGRFAM, version 8.0 (54), and Pfam, version 23.0 (55), databases. The 16S rRNA coding operon was identified by analyzing intergenic regions by blastn searches against GenBank. The blastx searches of the intergenic regions against GenBank nr did not yield additional protein-coding regions. The tRNAs were identified by tRNAscan-SE, version 1.23 (56), using the bacterial model. The tmRNAs were identified using BRUCE (57) and confirmed by blastn search against GenBank. The origin of replication was predicted using originx (58). Metabolic pathway prediction was performed using Pathologic within Pathway Tools, version 13.0 (59). Circular chromosome representation was constructed using DNAPlotter, version. 1.1 (60). Promoter prediction was performed using the promscan Perl script (61) to score 16-mers using a weight matrix generated from 186 RpoN-dependent promoters (41).
Comparative Genomic Analyses.
The Blattabacterium genome was compared with that of F. johnsonii, a related free-living bacterium, and to S. muelleri, a related obligately symbiotic bacterium with a genome of 225 kb. Genes were assigned to COG functional categories (53) for each genome (Table S1), and significance of differences was evaluated using χ2 goodness-of-fit tests for the pairwise comparisons of Flavobacterium–Blattabacterium and Blattabacterium–Sulcia. To reduce the number of cells with low expected values, we combined categories corresponding to poorly characterized genes (general predicted function and unknown function) and defense and secondary metabolites, and we eliminated remaining categories in which the expected number of genes was <100 for the largest (Flavobacterium johnsonii) genome. This resulted in 14 functional categories, corresponding to COGs C, E, G–M, O, P, T, R + S, and Q + V. Two-tailed tests were performed. Gene synteny between the Sulcia and Blattabacterium genomes was evaluated by plotting the genomic locations of Sulcia genes against the locations of their Blattabacterium homologues.
Phylogenetic Analysis.
We used MAFFT, version 6.624b, with the L-INS-i algorithm (62), to align individual protein datasets. Customized Perl scripts were used to remove columns within the alignments containing gap characters and concatenate all of the aligned protein sequence regions before phylogenetic analysis. A maximum likelihood tree, based on 100 bootstrap replications, was constructed using PhyML, version 2.4.4, with the JTT model of amino acid substitution (63). The proteins used for the alignments and the genomes from which they were obtained can be found in SI Text.
Fluorescence in Situ Hybridization.
A third-instar P. americana specimen was euthanized and then beheaded to facilitate reagent infiltration. The specimen was fixed in 4% paraformaldehyde and embedded in paraffin before sectioning (64). In situ hybridization was performed on 4-μm sagittal tissue sections affixed to microscope slides with a cyanine 3 fluorophore (Cy3)-labeled Blattabacterium-specific oligonucleotide probe, BLATTA303 (CCAGTGTGGGGGATCAC). 4′,6-Diamidino-2-phenylindole was used as a counterstain. Sulcia muelleri cells were obtained from Clastoptera arizonana bacteriomes and hybridized with a Cy3-labeled S. muelleri-specific oligonucleotide probe, SULCIA303 (CCAATGTGGGGGWACGC). Protocols for tissue preparation and FISH hybridization were adapted from ref. 65. Images were obtained using a Zeiss LSM 501 META laser confocal microscope.
Supplementary Material
Acknowledgments.
We thank Joe Jones at the University of South Carolina for 454 sequencing, Sheena Brown for cockroach specimens, and John McCutcheon for technical assistance. This work was funded by the National Science Foundation Grant 0626716 (to N.A.M.). and the University of Arizona Center for Insect Science through National Institutes of Health Training Grant 2 K12 GM000708 (to Z.L.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CP001429 and CP001430).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907504106/DCSupplemental.
References
- 1.Bell WJ, Roth LM, Nalepa CA. Cockroaches: Ecology, Behavior, and Natural History. Baltimore: Johns Hopkins Univ Press; 2007. [Google Scholar]
- 2.Mullins DE, Cochran DG. Nitrogen metabolism in American cockroach. I. An examination of positive nitrogen-balance with respect to uric-acid stores. Comp Biochem Physiol. 1975;50:489–500. [Google Scholar]
- 3.Mullins DE, Cochran DG. Nitrogen metabolism in the American cockroach: An examination of whole body and fat body regulation of cations in response to nitrogen balance. J Exp Biol. 1974;61:557–570. doi: 10.1242/jeb.61.3.557. [DOI] [PubMed] [Google Scholar]
- 4.Cochran DG. Nitrogen excretion in cockroaches. Annu Rev Entomol. 1985;30:29–49. [Google Scholar]
- 5.Kashima T, Nakamura T, Tojo S. Uric acid recycling in the shield bug, Parastrachia japonensis (Hemiptera: Parastrachiidae) during diapause. J Insect Physiol. 2006;52:816–825. doi: 10.1016/j.jinsphys.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Potrikus CJ, Breznak JA. Anaerobic degradation of uric-acid by gut bacteria of termites. Appl Environ Microbiol. 1980;40:125–132. doi: 10.1128/aem.40.1.125-132.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potrikus CJ, Breznak JA. Uric acid-degrading bacteria in guts of termites [Reticulitermes flavipes (Kollar)] Appl Environ Microbiol. 1980;40:117–124. doi: 10.1128/aem.40.1.117-124.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potrikus CJ, Breznak JA. Gut bacteria recycle uric-acid nitrogen in termites-a strategy for nutrient conservation. Proc Natl Acad Sci USA. 1981;78:4601–4605. doi: 10.1073/pnas.78.7.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gier HT. The morphology and behavior of the intracellular bacteroids of roaches. Biol Bull. 1936;71:433–452. [Google Scholar]
- 10.Donnella JF, Kilby BA. Uric acid metabolism by symbiotic bacteria from the fat body of Periplaneta americana. Comp Biochem Physiol. 1967;22:235–252. doi: 10.1016/0010-406x(67)90184-3. [DOI] [PubMed] [Google Scholar]
- 11.Lanham UN. The Blochmann bodies: Hereditary intracellular symbionts of insects. Biol Rev Camb Philos Soc. 1968;43:269–289. doi: 10.1111/j.1469-185x.1968.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 12.Blochmann F. Occurrence of bacteria-like particles in tissues and eggs of different insects. Bioi Zentralbl. 1887;7:606–608. German. [Google Scholar]
- 13.Brooks MA. Comments on classification of intracellular symbiotes of cockroaches and description of the species. J Invertebr Pathol. 1970;16:249–258. [Google Scholar]
- 14.Brooks MA, Richards AG. Intracellular symbiosis in cockroaches. I. Production of aposymbiotic cockroaches. Biol Bull. 1955;109:22–39. [Google Scholar]
- 15.Malke H. Production of aposymbiotic cockroaches by means of lysozyme. Nature. 1964;204:1223–1224. doi: 10.1038/2041223a0. [DOI] [PubMed] [Google Scholar]
- 16.Henry SM. The significance of microorganisms in nutrition of insects. Trans NY Acad Sci. 1962;24:676–683. [Google Scholar]
- 17.Bandi C, et al. The establishment of intracellular symbiosis in an ancestor of cockroaches and termites. Proc R Soc London Ser B. 1995;259:293–299. doi: 10.1098/rspb.1995.0043. [DOI] [PubMed] [Google Scholar]
- 18.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruwell ME, Morse GE, Normark BB. Phylogenetic congruence of armored scale insects (Hemiptera: Diaspididae) and their primary endosymbionts from the phylum Bacteroidetes. Mol Phylogenet Evol. 2007;44:267–280. doi: 10.1016/j.ympev.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Majerus TMO, et al. Extreme variation in the prevalence of inherited male-killing microorganisms between three populations of Harmonia axyridis (Coleoptera: Coccinellidae) Heredity. 1998;81:683–691. [Google Scholar]
- 21.Clark JW, Hossain S, Burnside CA, Kambhampati S. Coevolution between a cockroach and its bacterial endosymbiont: A biogeographical perspective. Proc R Soc London Ser B. 2001;268:393–398. doi: 10.1098/rspb.2000.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol Biol Evol. 2003;20:907–913. doi: 10.1093/molbev/msg097. [DOI] [PubMed] [Google Scholar]
- 23.Brooks MA, Richards K. On the in vitro culture of intracellular symbiotes of cockroaches. J Invertebr Pathol. 1966;8:150–157. doi: 10.1016/0022-2011(66)90123-6. [DOI] [PubMed] [Google Scholar]
- 24.Ochman H, Moran NA. Genes lost and genes found: Evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 25.Wernegreen JJ. For better or worse: Genomic consequences of intracellular mutualism and parasitism. Curr Opin Genet Dev. 2005;15:572–583. doi: 10.1016/j.gde.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Grose JH, et al. Assimilation of nicotinamide mononucleotide requires periplasmic AphA phosphatase in Salmonella enterica. J Bacteriol. 2005;187:4521–4530. doi: 10.1128/JB.187.13.4521-4530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee PA, Tullman-Ercek D, Georgiou G. The bacterial twin-arginine translocation pathway. Annu Rev Microbiol. 2006;60:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn A. From the Sec complex to the membrane insertase YidC. Biol Chem. 2009;390:701–706. doi: 10.1515/BC.2009.059. [DOI] [PubMed] [Google Scholar]
- 29.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 30.Block RJ, Henry SM. Metabolism of sulphur amino-acids and of sulphate in Blattella germanica. Nature. 1961;191:392–393. [Google Scholar]
- 31.Douglas AE. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 1989;64:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 32.Henry SM, Block RJ. Sulfur metabolism of insects. 6. Metabolism of sulfur amino acids and related compounds in German cockroach, Blattella germanica (L.) Contrib Boyce Thompson Inst. 1961;21:129–145. [Google Scholar]
- 33.Inokuchi T, Horie Y, Ito T. Urea cycle in silkworm Bombyx mori. Biochem Biophys Res Commun. 1969;35:783–787. doi: 10.1016/0006-291x(69)90691-3. [DOI] [PubMed] [Google Scholar]
- 34.Reddy SR, Campbell JW. Enzymic basis for the nutritional requirement of arginine in insects. Experientia. 1977;33:160–161. doi: 10.1007/BF02124040. [DOI] [PubMed] [Google Scholar]
- 35.Helling RB. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J Bacteriol. 1994;176:4664–4668. doi: 10.1128/jb.176.15.4664-4668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helling RB. Pathway choice in glutamate synthesis in Escherichia coli. J Bacteriol. 1998;180:4571–4575. doi: 10.1128/jb.180.17.4571-4575.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 38.Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent σ54 (σN) transcription factor. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabo A, et al. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horwich AL, Farr GW, Fenton WA. GroEL–GroES-mediated protein folding. Chem Rev. 2006;106:1917–1930. doi: 10.1021/cr040435v. [DOI] [PubMed] [Google Scholar]
- 41.Dombrecht B, Marchal K, Vanderleyden J, Michiels J. Prediction and overview of the RpoN-regulon in closely related species of the Rhizobiales. Genome Biol. 2002;3:RESEARCH0076. doi: 10.1186/gb-2002-3-12-research0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skibinski DAG, et al. Regulation of the hydrogenase-4 operon of Escherichia coli by the σ54-dependent transcriptional activators FhlA and HyfR. J Bacteriol. 2002;184:6642–6653. doi: 10.1128/JB.184.23.6642-6653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potrikus CJ, Breznak JA. Uric acid in wood-eating termites. Insect Biochem. 1980;10:19–27. [Google Scholar]
- 44.Wu XW, Lee CC, Muzny DM, Caskey CT. Urate oxidase: Primary structure and evolutionary implications. Proc Natl Acad Sci USA. 1989;86:9412–9416. doi: 10.1073/pnas.86.23.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol. 2006;2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y, et al. Transthyretin-related proteins function to facilitate the hydrolysis of 5-hydroxyisourate, the end product of the uricase reaction. FEBS Lett. 579:4769–4774. doi: 10.1016/j.febslet.2005.07.056. [DOI] [PubMed] [Google Scholar]
- 47.Cruden DL, Markovetz AJ. Microbial ecology of the cockroach gut. Annu Rev Microbiol. 1987;41:617–643. doi: 10.1146/annurev.mi.41.100187.003153. [DOI] [PubMed] [Google Scholar]
- 48.Moran NA. Microbial minimalism: Genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 49.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 50.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radek R. Light and electron microscopic study of a Rickettsiella species from the cockroach Blatta orientalis. J Invertebr Pathol. 2000;76:249–256. doi: 10.1006/jipa.2000.4984. [DOI] [PubMed] [Google Scholar]
- 52.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 53.Tatusov RL, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haft DH, et al. TIGRFAMs: A protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laslett D, Canback B, Andersson S. BRUCE: A program for the detection of transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res. 2002;30:3449–3453. doi: 10.1093/nar/gkf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Worning P, Jensen LJ, Hallin PF, Staerfeldt HH, Ussery DW. Origin of replication in circular prokaryotic chromosomes. Environ Microbiol. 2006;8:353–361. doi: 10.1111/j.1462-2920.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 59.Keseler IM, et al. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Studholme DJ, Dixon R. Domain architectures of σ54-dependent transcriptional activators. J Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 63.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 64.Barbosa P. Manual of Basic Techniques in Insect Histology. Amherst, MA: Autumn Publishers; 1974. [Google Scholar]
- 65.Daims H, Stoecker K, Wagner M. In: Molecular Microbial Ecology. Osborn AM, Smith C, editors. London: BIOS Scientific Publications; 2005. pp. 213–240. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.