Abstract
A hallmark of the NF-κB transcription response to inflammatory cytokines is the remarkably rapid rate of robust activation and subsequent signal repression. Although the rapidity of postinduction repression is explained partly by the fact that the gene for IκBα is strongly induced by NF-κB, the newly synthesized IκBα still must enter the nucleus and compete for binding to NF-κB with the very large number of κB sites in the DNA. We present results from real-time binding kinetic experiments, demonstrating that IκBα increases the dissociation rate of NF-κB from the DNA in a highly efficient kinetic process. Analysis of various IκB mutant proteins shows that this process requires the C-terminal PEST sequence and the weakly folded fifth and sixth ankyrin repeats of IκBα. Mutational stabilization of these repeats reduces the efficiency with which IκBα enhances the dissociation rate.
Keywords: binding kinetics, disordered proteins, protein–DNA interaction, transcription, surface plasmon resonance
The NF-κB transcription factors play key roles in normal growth and development, in inflammatory and immune responses, and in numerous human diseases (1, 2). The most abundant NF-κB is the p50/p65 heterodimer, but other homo- and heterodimers of p65 (RelA), RelB, c-Rel, p50, and p52 subunits are present also (1). Specific inhibitors of NF-κB transcription, including IκBα, IκBβ, and IκBε, block the transcriptional activity of p65- and c-Rel-containing NF-κB dimers (3). In resting cells, NF-κB transcriptional activity is strongly inhibited by IκBs that keep the NF-κB in the cytoplasm, preventing its nuclear localization and association with DNA (4, 5). Stress signals induce activation of IκB kinase, which phosphorylates the N-terminal signal response domain of NF-κB–bound IκBα, leading to subsequent ubiquitination and degradation by the proteasome (6). NF-κB then enters the nucleus, binds DNA, and regulates transcription of its numerous target genes (7).
DNA-bound NF-κB has been detected at hundreds of genes (8) with a loosely defined consensus sequence, based on NF-κB–responsive genes, called a “κB site” (1). Thousands of such sites are present in the DNA (9, 10). NF-κBs use the Rel homology domain to recognize κB sites in the DNA, but only p65, c-Rel, and RelB have transcription activation domains (11). The large number of genes that are activated by NF-κBs show widely varying levels and kinetics of transcription activation and postinduction repression, but the mechanism of this diversity still is not understood (12). DNA binding specificity may not be able to explain the wide variations in transcription responses. Although NF-κB family members can form homo- and heterodimers, and purified dimers are able selectively to bind various oligonucleotides corresponding to diverse κB sites (13), crystal structures of several NF-κB homo- and heterodimers with various κB sites show few specific base contacts (14–17). Early experiments measuring DNA binding were done under nonphysiological conditions in which the binding affinity of the NF-κB for DNA was shown to be extremely high, 10−10 M (18). A wide range of binding affinities, from 10−10M (18) to 5 × 10−7M, for the same κB site binding to the same NF-κB (p50/p65) heterodimer have been reported in the literature (19). Recent intracellular photobleaching experiments suggest that NF-κB dissociates from the DNA at a surprisingly rapid rate (20).
We previously showed that IκBα binds tightly to NF-κB with a KD of 40 pM at 37 °C as a consequence of a slow dissociation rate constant on the order of 10−4 s−1, which translates into a half-life of 2 h (5). The measured intracellular half-life also is on the order of several hours (21). Previously reported binding affinities in the nanomolar range for IκBα binding to NF-κB(p50/p65), obtained from gel shift and competition assays, are inconsistent with the extremely long intracellular half-life of the complex (19) as well as with the direct binding experiments (5).
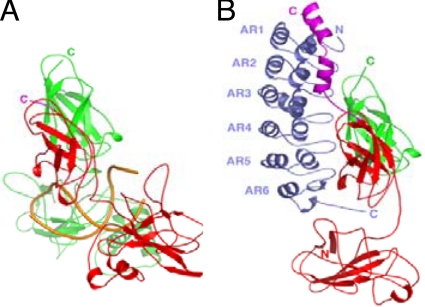
Crystal structures of DNA-bound NF-κB(p50/p65) and IκBα-bound NF-κB(p50/p65) show overlapping but nonidentical binding surfaces (15, 22–24). DNA contacts the loops protruding from the dimerization and N-terminal domains of the Rel homology domain and the linker between them, whereas IκBα mainly contacts the dimerization domain and helix3-NLS-helix4 at the C terminus of the Rel homology domain of p65 (Fig. 1). The ankyrin repeat domain (ARD) of IκBα forms the main interaction surface with the dimerization domains of NF-κB (23, 24).
Fig. 1.
(A) The crystal structure of NF-κB (p50 shown in green; p65 shown in red) bound to κB-site DNA (shown in gold) (22). (B) The crystal structure of IκBα (shown in blue) bound to NF-κB (p50 shown in green; p65 shown in red; p65 NLS shown in purple) (24). The figure was prepared using PyMOL (DeLano Scientific).
The IκBα gene is strongly activated by NF-κB, resulting in new synthesis of IκBα (25–27). The newly synthesized IκBα enters the nucleus and is responsible for rapid postinduction repression of NF-κB transcriptional activity (28, 29). Early experiments with IκBα showed that it was very efficient in competing with DNA for binding NF-κB (30). These results led to the suggestion that IκBα can enter the nucleus and remove NF-κB from the DNA by an “active dissociation” mechanism. However, the term “active” implies that IκBα increases the rate of dissociation of NF-κB from the DNA, but the equilibrium competition experiments cannot prove this effect (4). Rapid replacement of DNA with IκBα would be expected, based on the already rapid binding kinetics of NF-κB with DNA and the tighter binding affinity of IκBα with NF-κB, without any need to invoke an active dissociation mechanism (5).
Here, we have undertaken biophysical experiments designed to measure the association and dissociation kinetics of NF-κB binding to single κB sites in the DNA under physiological conditions. The dissociation rates recapitulate the in vivo photobleaching kinetic results (20). Surprisingly, 2 different kinetic experiments also show that IκBα increases the dissociation rate of NF-κB(p50/p65) from the DNA in a highly efficient and concentration-dependent manner. Thus, the previously proposed active dissociation mechanism, which was based on insufficient experimental evidence, does, in fact, occur. Experiments using mutants of NF-κB and IκBα suggest a mechanism in which the weakly folded ankyrin repeats (ARs) 5 and 6 of the IκBα ARD are required for this phenomenon.
Results
DNA Binding to NF-κB Is Rapid and Reversible.
Previously, binding thermodynamics measurements of single κB-site DNA molecules binding to various homo- and heterodimers of NF-κB family members have shown a broad range of binding affinities from picomolar (18) to nanomolar (31) to almost micromolar (19) levels. Recently, in vivo fluorescence recovery after photobleaching experiments showed rapid kinetics of exchange of NF-κB from the DNA with a half-life of the bound NF-κB on the order of 30 s (20). We carried out real-time binding kinetics measurements using surface plasmon resonance (SPR) with purified NF-κB homo- and heterodimers of p50 and p65 and oligonucleotides containing a single κB site at physiological salt concentrations and 37 °C. We chose 6 different DNA sequences based on previous reports that different κB sites show very different kinetics of transcription activation and postinduction repression (12). Except for the urokinase promoter sequence, the NF-κB(p50/p65) heterodimers bound with observed KDs in the nanomolar range. Lower affinities were observed for the NF-κB(p65/p65) homodimers, and these homodimers also showed more variable binding affinities (supporting information (SI) Table S1). The nanomolar binding affinities to specific κB DNA sites are the result of rapid association (ka = 1 × 106 M−1s−1) and dissociation (kd = 1.7 × 10−2s−1) rates at 37 °C under physiological conditions. To validate the SPR measurements, equilibrium and stopped-flow fluorescence was used to determine the binding association and dissociation kinetics for NF-κB(p50/p65) heterodimers binding to the IgκB site contained in a hairpin with a pyrene label at the 5′ end. Equilibrium binding experiments in which the change in fluorescence intensity was measured, carried out at 25 °C, gave a Kd of 10 nM (Fig. S1). The stopped-flow kinetics experiments gave a ka of 1.2 × 108 M−1s−1 and a kd of 0.4 s−1, resulting in an observed Kd of 3 nM. It is relatively common for both ka and kd values to be higher when determined by fluorescence kinetics than when determined by SPR, but for overall Kd values to be similar (32). A random sequence of DNA showed no binding by SPR and a binding affinity of >50 nM by stopped-flow fluorescence. The dissociation rates, determined at physiological ionic strength and temperature, translate to a half-life of 1.5 s (fluorescence) to 40 s (SPR), results that are similar to the half-life of 10–30 s measured in vivo by fluorescence bleaching experiments (20).
IκBα Increases the Rate of Dissociation of NF-κB from Promoter Sites.
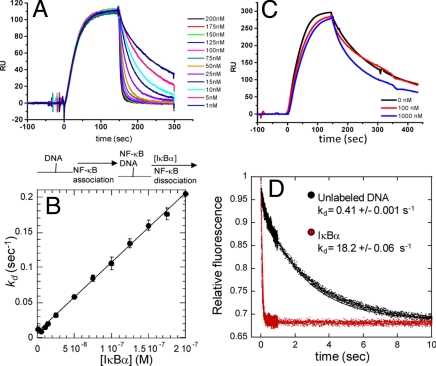
A flowing, real-time kinetics experiment using SPR was designed to probe whether IκBα increases the rate of dissociation of NF-κB from the DNA. In this experiment, κB site-containing oligonucleotides were captured via a biotin–streptavidin linkage onto the sensor chip. NF-κB(p50(39–376)/p65(1–325)) was allowed to bind and reach pseudoflowing equilibrium. The flow then was switched rapidly to the second sample loop (using the co-inject feature of the Biacore 3000) either to buffer or to buffer containing a low concentration of IκBα to monitor dissociation at different IκBα concentrations. Low surface density of immobilized oligonucleotide, rapid flow rates (40 μL/min), and physiological salt concentrations excluded the potential complication of re-binding of the NF-κB. Under these conditions, the exponential dissociation rate constant (kd) is measured directly (33). Fig. 2A shows the results of such an experiment and demonstrates that IκBα dramatically increases the rate of dissociation of NF-κB from the DNA. In these kinetic experiments, the dissociation phase can be described by Eq. 1, where kdDNA is the first-order rate at which NF-κB dissociates from the DNA in the absence of IκBα, kdi is the second-order rate constant at which IκBα increases the dissociation, and the observed dissociation rate is (kdDNA + kdi[I]) where [I] is the concentration of IκBα in the dissociation phase.
The IκBα-independent dissociation rate, kdDNA, already was measured at 0.007 s−1 at 25 °C (Table S1). The rate of IκBα-mediated dissociation NF-κB from the DNA (kdi) was determined from the slope of the plot of the observed dissociation rate vs. the concentration of IκBα. For wild-type IκBα, this value is nearly 106 M−1s−1, indicating that IκBα-mediated dissociation is a highly efficient process (Fig. 2B).
Fig. 2.
(A) Real-time binding and dissociation experiment monitored by SPR. Biotinylated κB-site DNA was bound to the streptavidin chip (t = 0). NF-κB(p50(19–363)/p65(1–325)) was allowed to associate with the DNA until a pseudoflowing equilibrium was reached (t = 100 s). Varying concentrations of IκBα then were injected through the second sample loop (co-inject experiment), and the dissociation rate constant (kd) was measured. A schematic of the binding events is shown below the graph. (B) Plot of the dissociation rate constant determined from 4 independent experiments, performed as described in (A), as a function of IκBα concentration. The slope of the line (106 M−1s−1), which is the pseudosecond-order rate constant for active dissociation, indicates that IκBα-enhanced dissociation is highly efficient. (C) A control experiment, performed as described in (A), in which varying concentrations of DNA (red, 100 nM; blue, 1000 nM) were used instead of IκBα in the dissociation step. Even at 1000 nM DNA, only a slight difference in the dissociation rate was observed. (D) Stopped-flow fluorescence experiment in which pyrene-labeled hairpin DNA (0.25 μM) complexed to NF-κB(p50(19–363)/p65(1–325)) (0.5 μM) in syringe 1 was mixed rapidly with a 50-fold excess (relative to NF-κB) of either unlabeled hairpin DNA (black, kd = 0.41 s−1) or IκBα (red, kd = 18.2 s−1).
When κB-site DNA was co-injected at up to micromolar concentrations, the dissociation rate did not change (Fig. 2C). In addition, when NF-κB was immobilized and IκBα was bound, no effect on the IκBα dissociation rate was observed even at high concentrations of κB-site DNA. Stopped-flow fluorescence experiments also were performed in which the NF-κB·pyrene hairpin DNA complex was dissociated with a 50-fold molar excess of either unlabeled DNA or IκBα. Again, IκBα caused a dramatic 45-fold increase in the dissociation rate from 0.4 s−1 to 18 s−1, whereas the unlabeled DNA had no effect (Fig. 2D).
Residues 305–325 of NF-κB (Helix 4 of the NLS Extension) Are Required for IκBα-Mediated Dissociation.
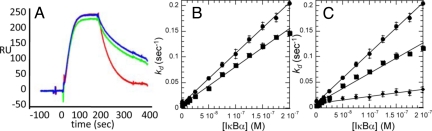
Truncation mutants were constructed to ascertain which parts of the NF-κB and IκBα molecules were important for IκBα-mediated dissociation. Initially, the NF-κB (p65) was truncated at residue 304, thus deleting helix 4 just after the NLS sequence. Although this protein formed heterodimers and bound to κB-site DNA with the same affinity as wild type, IκBα did not enhance dissociation if residues 305–325 were missing from p65 of the NF-κB heterodimer (Fig. 3A). This result was not satisfying, however, because deletion of residues 305–325 causes a dramatic loss of binding affinity of NF-κB for IκBα (5). To probe more subtly the role of the NF-κB(p65) NLS interaction with IκBα in facilitating dissociation, Arg 304 in the p65 NLS was mutated to Ala. This mutation reduced IκBα·NF-κB binding affinity by 2.2-fold and reduced the IκBα-mediated dissociation by 1.5-fold (Fig. 3B).
Fig. 3.
(A) Truncated NF-κB(p50(19–363)/p65(1–304)) was bound to the DNA, and in this case co-injection of IκBα during the dissociation step is ineffective. (B) Plot of kd vs. concentration of IκBα in the active dissociation of NF-κB(p50(19–363)/p65(1–325)) (•) or NF-κB(p50(19–363)/p65(1–325)R304A) (■) from κB-site DNA measured as described in Fig. 2. (C) Truncation of IκBα(67–287) (•) at the PEST sequence results in decreased ability to enhance dissociation. (■), IκBα (67–281); (♦), IκBα (67–275) (♦).
We previously had shown that truncation of residues 288–317 of IκBα had no effect on NF-κB binding nor did phosphorylation of the PEST sequence (5, 34). As expected, IκBα (67–317) and PEST-phosphorylated IκBα (67–317) also efficiently mediated dissociation. Deletion of residues 282–287 to truncate partially the IκBα PEST sequence significantly reduced the efficiency of IκBα-mediated dissociation of NF-κB from the DNA. Deletion of residues 275–287 to remove the PEST sequence completely had an even larger effect (Table S2A and Fig. 3C). These results strongly implicate the negatively charged PEST sequence in removal of NF-κB from the DNA.
The Weakly-Folded Fifth and Sixth Repeats Are Critical for IκBα-Mediated Dissociation.
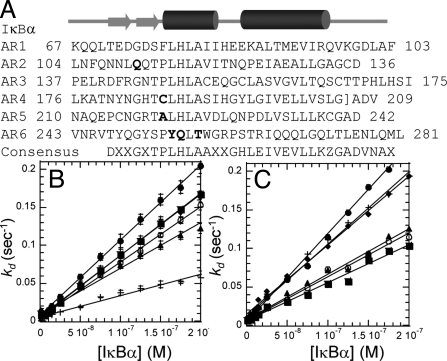
IκBα does not conform to the consensus for stable ARDs, and we have shown previously that mutation back to the consensus sequence for stable ARDs stabilizes IκBα (35, 36). All the stabilized mutants were less able to increase the dissociation of NF-κB(p50/p65) from the DNA despite widely varying NF-κB binding affinities (Table S2B and Fig. 4). Mutation of Y254L/Q255H weakened the binding by 100-fold, but this protein still was effective in increasing the dissociation of NF-κB from the DNA (Table S2B). In contrast, mutation of Y254L/T257A, which prefolds the sixth repeat, weakened binding only 30-fold but was much less efficient in increasing dissociation (36). The C186P/A220P mutant bound to NF-κB with the same binding kinetics and affinity (Table S2B) but was less efficient at increasing dissociation of NF-κB. The C186P/A220P mutant was less able to enhance dissociation on 3 different κB promoter sequences, indicating that enhanced dissociation is DNA sequence-independent (Fig. 4C).
Fig. 4.
(A) Sequence of IκBα; the sites of stabilizing mutations are shown in bold. (B) Stabilized IκBα variants wild-type IκBα (•), IκBα (C186P, A220P) (■), IκBα (Q111G) (▴), IκBα (Y254L/Q255H) (○), and IκBα (Y254L/T257A) (+) were less able to enhance dissociation. (C) For 3 different promoter sequences (HIV-κB, MIP2, RANTES), decreases in the rate of active dissociation for IκBα (C186P, A220P) (○, HIV-κB; ▴, MIP2; ■, RANTES) were similar to those observed with wild-type IκBα (•, HIV-κB; +, MIP2; ♦, RANTES).
Discussion
NF-κB·DNA Binding Constants.
As cited in the Introduction, several widely varying values have been reported for the affinity of the interaction between NF-κB and DNA and between NF-κB and IκBα. Although it is not possible to replicate physiological conditions exactly, it now has become clear that the binding affinity of NF-κB for a single canonical κB site is 3–10 nM. Protein–DNA interactions are very sensitive to ionic strength (37), and the anomalously low Kd of 0.1 nM probably reflects the low salt concentration used in these experiments (18). We have no explanation for the much weaker binding affinities measured by fluorescence anisotropy (19). The nanomolar binding affinity for κB-site DNA is derived from fast-on, fast-off kinetics, and these results are fully consistent with intracellular measurements (20). Given that the dissociation of NF-κB from the DNA already is relatively rapid, it was surprising that IκBα significantly enhances the dissociation rate even further. Why and how IκBα markedly increases the NF-κB·DNA dissociation rate is discussed further in the following sections.
NF-κB Has Overlapping but Nonidentical Binding Sites for DNA and IκBα.
Although several different mechanisms of transcriptional inhibition have been proposed, including the formation of an inhibited ternary complex, IκBα does not work by such a mechanism. Crystal structures of NF-κB(p50/p65) bound with either DNA or IκBα show that the binding sites on NF-κB for DNA and for IκBα are overlapping but are not identical, and binding of DNA or IκBα to NF-κB is known to be mutually exclusive. IκBα mainly contacts the NLS and the dimerization domains, whereas DNA mainly contacts the interface between the dimerization domains and the N-terminal domains (Fig. 1). The very high binding affinity appears to be almost entirely the result of small regions of the binding interface at either end of the elongated contact surface, resulting in a large folding enthalpy contribution to the binding affinity (5, 34). Truncation of the NLS domain of NF-κB(p65) (residues 291–325) results in a 5,000-fold decrease in binding, and this polypeptide alone binds to IκBα with an affinity of 1 μM (34). At the other end of the interface, ARs 5 and 6 are not fully folded in free IκBα, but they fold upon binding to NF-κB (38). In addition, the PEST region, which also is critical for binding, is negatively charged, like the DNA, and might enhance DNA dissociation from NF-κB partly via electrostatic repulsion.
Structural Model of IκBα-Mediated Active Dissociation.
A possible model of the enhanced dissociation process was developed based on the mutational studies (Fig. 5). In this model, when IκBα approaches the DNA-bound NF-κB, it interacts first with the p65 NLS, which does not participate in binding to DNA and is thus available for ternary complex formation (14–16, 22). Deletion of residues 305–321 of NF-κB(p65) did not affect NF-κB·DNA binding but abolished the ability of IκBα to enhance NF-κB dissociation from the DNA. Theoretical studies predicted that mutation of Arg-304 to Ala in the NLS would reduce IκBα·NF-κB binding affinity (39), and indeed, this mutation reduced affinity by approximately the same amount that it reduced the active dissociation. These observations support the hypothesis that interaction with the NLS is the first step (Fig. 5). With its 1-μM binding affinity, the interaction of the first and second ARs of IκBα with this “NLS polypeptide” region of NF-κB(p65) would be sufficient to form a small amount of short-lived ternary complex. The formation of ternary complexes between other IκB proteins and their respective NF-κBs and DNA has been predicted and observed (40, 41). Subsequently, IκBα may dissociate from the ternary complex, or it may interact further with NF-κB causing its dissociation from the DNA.
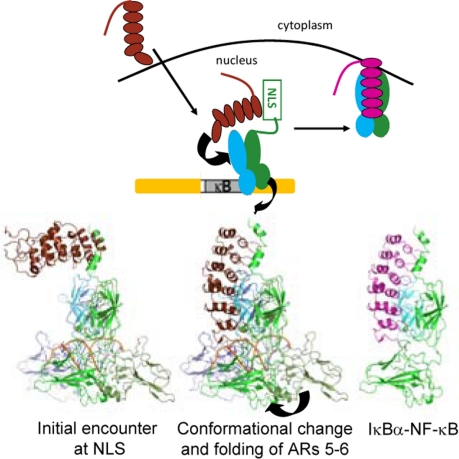
Fig. 5.
Schematic diagram depicting IκBα entering the nucleus and facilitating dissociation of NF-κB from the DNA. A possible structural model showing putative intermediates in the process is shown below. The helix3-NLS-helix4 segment of NF-κB is disordered in the DNA-bound NF-κB, and it is likely that IκBα first encounters this segment and binds transiently. Formation of the final IκBα-bound NF-κB requires folding of ARs 5 and 6 and dissociation of the DNA and probably requires a conformational change of the N-terminal domains of NF-κB. In the last step, the NF-κB/IκBα complex is formed and exits the nucleus. Because the complex has an extremely slow dissociation rate, enhanced dissociation rapidly results in complete removal of NF-κB from the DNA even when only small concentrations of IκBα are present.
The weakly folded C-terminal part of the IκBα ARD is critical to its ability to promote dissociation of NF-κB from the DNA. This region folds onto the dimerization domains of the NF-κB (p50/p65) and shares a binding interface with the DNA. We previously showed that ARs 5 and 6 of IκBα are weakly folded (38, 42, 43) and that mutations that restore the consensus for stable ARs can promote folding of this region (35, 36). The stabilizing mutations had varied effects on NF-κB·IκBα binding, but all showed reduced ability to promote dissociation of NF-κB from the DNA. The C186P/A220P mutation bound to NF-κB with the same affinity but was significantly less able to enhance NF-κB dissociation from the DNA.
Kinetic Model of IκBα-Mediated Active Dissociation.
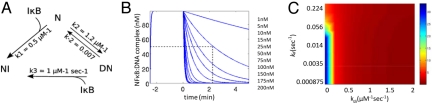
To explore whether kinetic enhancement would be important under physiological conditions, we built an ordinary differential equation model using some parameters derived from a validated computational model of NF-κB signaling (29). Quantitative models of NF-κB signaling have been built, but these models do not explicitly consider the κB-site-NFκB interaction because the kinetic rate constants remain uncertain (29). We assumed that there is an excess of κB DNA binding sites in the genome such that all nuclear NF-κB can bind to DNA, as is observed experimentally. This situation is, in fact, mimicked by the experimental conditions of the SPR experiments described in earlier sections. The model contains the 3 species: NF-κB, DNA, and IκBα. NF-κB can bind to the DNA, unbind, and form a complex with IκBα. Additionally, IκBα can bind to the NF-κB·DNA complex forming a transient ternary complex (estimated Kd = 1 μM), but this model gave results equivalent to the simpler model shown in Fig. 6A. The active removal of NF-κB from the DNA complex by IκBα is determined by the parameter, k3.
Fig. 6.
(A) Schematic diagram of the reactions governing the NF-κB·DNA interaction. NFκB (N) binds DNA (D) to form a DNA·NF-κB complex (DN), which may dissociate spontaneously or via an IκBα that the forms an NF-κB·IκBα complex (NI). (B) Simulations of IκBα-mediated dissociation of the DNA·NF-κB complex. Following formation of the DNA·NF-κB complex, a flow of IκBα at indicated concentrations was added to the system at time 0. The DNA·NF-κB complex dissociates at an effective rate that is dependent on the concentration of IκBα. (C) The half-life of the DNA·NF-κB complex (at the indicated color scale) as a function of the spontaneous DNA·NF-κB kd and the IκBα-dependent DNA·NF-κB kdi. IκBα had a profound effect on the effective DNA·NF-κB half-life over a wide range of spontaneous DNA·NF-κB kd values; at the experimentally measured NF-κB kd of 0.007 s−1, IκBα-enhanced dissociation is very important for ensuring a short complex half-life. For all simulations, NF-κB = 100 nM; κB sites = 400 nM; flux of IκB = 10 nM/s.
The rate constants governing the NF-κB·DNA complex (ka, kd) were determined by SPR (5) and were validated by intracellular experiments (21). Simulation of the SPR experiment itself, following the addition of 100 nM of NFκB to the DNA, resulted in rapid formation of an NFκB·DNA complex. At time 0, a specific concentration flux of IκBα was added to the system akin to the flowing IκBα into the flowcell of the Biacore instrument. The model simulations recapitulate the experimental observation that IκBα accelerates the dissociation of the NF-κB·IκBα complex in a concentration-dependent manner (Fig. 6B). The half-life of the NF-κB·DNA complex was derived from simulations carried out using physiological concentrations of DNA and proteins, and a color scale plot was generated to reveal the dissociation characteristics of the NFκB·DNA complex as a function of the IκBα-dependent and IκBα-independent dissociation rate constants (Fig. 6C). The model simulations show that active dissociation by IκBα has significant impact on the effective NFκB·DNA complex half-life for a wide range of NFκB·DNA dissociation rates, even when the IκB-independent off rate is as much as 16 times faster than the experimentally measured kd. The apparent robustness underscores the likely physiological importance of IκBα-enhanced dissociation of NFκB from κB sites.
Kinetic enhancement of dissociation is expected to increase dramatically the effectiveness of newly synthesized IκBα in causing transcriptional repression before an equilibrium concentration is reached that is sufficiently high to compete effectively for κB sites in the DNA. Thus, new synthesis of even a small amount of IκBα would be expected to reduce NF-κB transcriptional activity significantly. Enhanced dissociation by IκBα may play an even more critical role in turning off transcription from genes that have multiple κB sites (45) or when transcription co-activators increase the affinity of transcription factors for some sites in the DNA (46). To understand the extent to which active dissociation operates under physiological conditions, measurements of IκBα-mediated active dissociation of NF-κB from specific promoters in cells will be required.
Experimental Procedures
Protein Expression and Purification.
Human wild-type IκBα67–287 and mutant and truncated forms introduced by QuikChange (Stratagene) mutagenesis were expressed at 20 °C in Escherichia coli BL21 DE3 cells and purified using a Hi-load Q Sepharose (GE Healthcare) followed by a Superdex 75 column (GE Healthcare), as described previously (5, 43). The protein concentrations were determined by spectrophotometry, using a molar absorptivity of 12,090 M−1cm−1.
NF-κB proteins were expressed in E. coli BL21 DE3 cells at room temperature and were purified by a tandem Q then S Sepharose column (GE Healthcare) and finally by size exclusion on an S-200 column (GE Healthcare) as previously described (5). Protein concentrations were determined as described previously (5). For the fluorescence experiments, N-terminal hexahistidine-NF-κB(His-p50(39–350)/p65(10–321)) heterodimer was prepared using a coexpression method described previously (47) and was purified by nickel affinity chromatography, cation exchange on MonoS (GE Healthcare), and finally by size exclusion on an S-200 column (GE Healthcare). Proteins purified by the 2 different methods were confirmed by mass spectrometry.
SPR Experiments.
Sensorgrams were recorded on a Biacore 3000 (GE Healthcare) using streptavidin chips. For IκBα binding experiments, biotinylated NF-κB (p65) was prepared as already described (5). Homo- and heterodimers were formed by incubating the biotinylated NF-κB(p65) with a large excess of the other, un-biotinylated subunit in vitro for 1 h at 25 °C and overnight at 4 °C and were captured on the surface of the streptavidin sensor chip as previously described (5). IκB-binding data were collected in150 mM NaCl, 10 mM Tris (pH 7.5), 10% (wt/vol) glycerol, 3 mM dithiothreitol, 0.5 mM sodium azide, 0.2 mM EDTA, and 0.005% P20 running buffer at temperatures between 25 °C and 37 °C.
For the NF-κB DNA binding studies, oligonucleotides were synthesized and purified by RP HPLC (IDT Technologies). The coding strand contained a 5′ biotin tag followed by a triethlylene glycol linker and was hybridized with its complementary strand in vitro by heating to 95 °C followed by slow cooling to room temperature in 150 mM NaCl, 10 mM MOPS (pH 7.5). The coding strand sequences were as follows:
HIV-κB/Igκ: 5′ biotin-TEG-TCTGAGGGACTTTCCTGATC3′ (48)
MIP2: 5′ biotin-TEG-GCTCAGGGAATTTCCCTGGT3′
RANTES :5′ biotin-TEG-GCTTGGGGAGTTTCCACAAA3′
Urokinase promoter: 5′ biotin-TEG-TGCTGGGGAAAGTACAAGTG3′
IFN: 5′ biotin-TEG-CGCTGGGGAAATTCCAGGGA3′
Random: 5′ biotin-TEG-TCTGAGTAGACGTGCTGATC3′.
Low surface densities (5–20 RU) of the double-stranded oligonucleotides in 500 mM NaCl, 10 mM MOPS (pH 7.5), 0.5 mM EDTA, 0.5 mM sodium azide, 0.005% P20 (GE Healthcare) were captured on an streptavidin chip using manual inject. Sensorgrams were run at 50 μL/min and referenced to an ummodified surface. NF-κB binding data were collected in 150 mM NaCl, 10 mM MOPS (pH 7.5), 0.005% P20 at 25 °C, with regeneration by a 1-min pulse of 2M NaCl followed by an extra clean step after each injection. Association and dissociation rate constants were obtained by global fitting of the real-time kinetic data using the BiaEvaluation 4.1 software and a simple 1:1 binding model.
Dissociation rates in the presence of varying concentrations of DNA or IκBα were measured at 25 °C using the co-inject method. An injection of 100 μL of 10 μM NF-κB was followed by a 100-μL injection of IκBα at 40 μL/min. IκBα concentrations were 200, 175, 150, 125, 100, 75, 50, 25, 15, 10, 5, and 1 nM, with regeneration at the end of each co-inject by a 0.5-min pulse of 2M NaCl followed by an extra clean step. The dissociation phases were fit using the BiaEvaluation software to obtain an apparent dissociation rate for a given concentration of IκBα.
Fluorescence Studies.
All fluorescence experiments used a pyrene-labeled hairpin DNA: 5′-AmMC6/GGGAAATTCCTCCCCCAGGAATTTCCC-3′ (IDT Technologies) corresponding to the IFN-κB site (GGGAAATTCC). The hairpin DNA [20 nmol in 75 μL of 0.1 M sodium tetraborate (borax), pH 8.5] was labeled at the 5′ end with 1-pyrenebutyric acid N-hydroxyl succinimide ester (14 μL of a 9-mg/mL solution in DMSO; Sigma-Aldrich) at the AmMC6 group at 25 °C for 6 h. The reaction was quenched by addition of 1 mL ethanol, and the labeled DNA was purified by C18 RP-HPLC in 20 mM ammonium acetate, pH 6.5, with a 60-min gradient from 0 to 60% acetonitrile. The oligonucleotide and the protein were dissolved in 25 mM Tris, 150 mM NaCl, 0.5 mM EDTA, and 1 mM DTT at pH 7.5 and 25 °C.
DNA·NF-κB binding constant titration measurements were performed using a photon-counting fluorometer (FluoroMax-P). Samples were incubated at 25 °C for 3 min before the start of each experiment. KD determination was performed with a constant final concentration of the pyrene-DNA in a 1-mL cuvette (5 nM in 1-mL final volume) to which various concentrations of NF-κB were added. The sample was exited at 346 nm (2-nm slit), and the emission was monitored at 377 nM (5-nm slit) with an integration time of 2 s and a 3-min equilibration time. For each NF-κB concentration, the fluorescence intensity of a blank sample in Tris buffer was subtracted from the labeled DNA-containing sample. The data were fitted (Kaleidagraph software 4.0, Synergy, Inc.) to Y = m1*([m2+m3+X)-sqrt[(m2+m3+X)2̂–4*m3*X])/(2*m3), where Y corresponds to the maximum fluorescence, m1 is the amplitude, m2 is the KD, m3 is the pyrene-DNA concentration, and X is the NF-κB concentration.
Rapid kinetics experiments were performed at 25 °C on a Biologic SFM-20 stopped-flow fluorimeter. The mixing volume was 120 μL with a sampling period of 200 μs to 5 ms. The association kinetics for the pyrene-DNA were measured in triplicate at 10 concentrations of NF-κB heterodimer (0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, 1, 1.5, and 3 μM) maintaining a constant concentration of pyrene-DNA (0.1 μM before mixing). The association rate constant was obtained from a single exponential fit (Kaleidagraph, Synergy, Inc.).
The dissociation rate constant of the NF-κB·pyrene DNA complex was measured at a fixed concentration of the NF-κB·pyrene DNA complex (in a 2:1 ratio; final NF-κB concentration of 50 nM) by adding an excess of unlabeled pyrene DNA (1:10, 1:50, and 1:100, NF-κB·pyrene DNA). The experiment was repeated using IκBα67–287 instead of unlabeled DNA hairpin in the same concentrations and ratios. For both experiments, the data were fit (Kaleidagraph) to the equation m1 + m2*exp(-m3*x) where m1 = end point, m2 = amplitude, and m3 = Kd.
Supplementary Material
Acknowledgments.
We thank Diego U. Ferreiro for many helpful discussions and for making the YQLH mutant. Research funding was provided by National Institutes of Health Grant P01 GM071862.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908797106/DCSupplemental.
References
- 1.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Takada Y, Boriek AM, Aggarwal BB. Nuclear factor-kappaB: Its role in health and disease. Journal of Molecular Medicine. 2004;82:434–448. doi: 10.1007/s00109-004-0555-y. [DOI] [PubMed] [Google Scholar]
- 3.Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: Intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle PA. IkB-NF-kB structures: At the interface of inflammation control. Cell. 1998;95:729–731. doi: 10.1016/s0092-8674(00)81694-3. [DOI] [PubMed] [Google Scholar]
- 5.Bergqvist S, Croy CH, Kjaergaard M, Huxford T, Ghosh G, Komives EA. Thermodynamics reveal that helix four in the NLS of NF-kappaB p65 anchors IkappaBalpha, forming a very stable complex. J Mol Biol. 2006;360:421–434. doi: 10.1016/j.jmb.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traenckner EB, Baeuerle PA. Appearance of apparently ubiquitin-conjugated I kappa B-alpha during its phosphorylation-induced degradation in intact cells. J Cell Sci Suppl. 1995;19:79–84. doi: 10.1242/jcs.1995.supplement_19.11. [DOI] [PubMed] [Google Scholar]
- 7.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 8.Schreiber J, et al. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci USA. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martone R, et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc Natl Acad Sci USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-kappaB with chromatin: The art of being at the right place at the right time. Nat Immunol. 2005;6:439–445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 11.Huxford T, Malek S, Ghosh G. Structure and mechanism in NF-kappa B/I kappa B signaling. Cold Spring Harb Symp Quant Biol. 1999;64:533–540. doi: 10.1101/sqb.1999.64.533. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunsch C, Ruben SM, Rosen CA. Selection of optimal kappa B/Rel DNA-binding motifs: Interaction of both subunits of NF-kappa B with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh G, van Duyne G, Ghosh S, Sigler PB. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 15.Müller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature. 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 16.Berkowitz B, Huang DB, Chen-Park FE, Sigler PB, Ghosh G. The x-ray crystal structure of the NF-kappa B p50. p65 heterodimer bound to the interferon beta-kappa B site. J Biol Chem. 2002;277:24694–24700. doi: 10.1074/jbc.M200006200. [DOI] [PubMed] [Google Scholar]
- 17.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 18.Baeuerle PA, Baltimore D. I kappa B: A specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 19.Phelps CB, Sengchanthalangsy LL, Huxford T, Ghosh G. Mechanism of I kappa B alpha binding to NF-kappa B dimers. J Biol Chem. 2000;275:29840–29846. doi: 10.1074/jbc.M004899200. [DOI] [PubMed] [Google Scholar]
- 20.Bosisio D, et al. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Dea EL, et al. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Molecular Systems Biology. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 23.Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 25.Brown K, Park S, Kanno T, Franzoso G, Siebenlist U. Mutual regulation of the transcriptional activator NF-kappa B and its inhibitor, I kappa B-alpha. Proc Natl Acad Sci USA. 1993;90:2532–2536. doi: 10.1073/pnas.90.6.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 27.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: Evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 28.Arenzana-Seisdedos F, et al. Nuclear localization of IkBa promotes active transport of NF-kB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 30.Zabel U, Baeuerle PA. Purified human I kappa B can rapidly dissociate the complex of the NF-kappa B transcription factor with its cognate DNA. Cell. 1990;61:255–265. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]
- 31.Chen-Park FE, Huang DB, Noro B, Thanos D, Ghosh G. The kappa B DNA sequence from the HIV long terminal repeat functions as an allosteric regulator of HIV transcription. J Biol Chem. 2002;277:24701–24708. doi: 10.1074/jbc.M200007200. [DOI] [PubMed] [Google Scholar]
- 32.Iakoucheva LM, Walker RK, van Houten B, Ackerman EJ. Equilibrium and stop-flow kinetic studies of fluorescently labeled DNA substrates with DNA repair proteins XPA and replication protein A. Biochemistry. 2002;41:131–143. doi: 10.1021/bi011041q. [DOI] [PubMed] [Google Scholar]
- 33.Karlsson R, Michaelsson A, Mattsson L. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 34.Bergqvist S, Ghosh G, Komives EA. The IkBa/NF-kB complex has two hot-spots, one at either end of the interface. Protein Sci. 2008;17:2051–2058. doi: 10.1110/ps.037481.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreiro DU, et al. Stabilizing IkappaBalpha by “consensus” design. J Mol Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truhlar SME, Mathes E, Cervantes CF, Ghosh G, Komives EA. Pre-folding IkappaBalpha alters control of NF-kappaB signaling. J Mol Biol. 2008;380:67–82. doi: 10.1016/j.jmb.2008.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Record MT, Lohman TM, de Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 38.Truhlar SM, Torpey JW, Komives EA. Regions of IkappaBalpha that are critical for its inhibition of NF-kappaB·DNA interaction fold upon binding to NF-kappaB. Proc Natl Acad Sci USA. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latzer J, Papoian GA, Prentiss MC, Komives EA, Wolynes PG. Induced fit, folding, and recognition of the NF-kappaB-nuclear localization signals by IkappaBalpha and IkappaBbeta. J Mol Biol. 2007;367:262–274. doi: 10.1016/j.jmb.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Malek S, Huang DB, Huxford T, Ghosh S, Ghosh G. X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex. J Biol Chem. 2003;278:23094–23100. doi: 10.1074/jbc.M301022200. [DOI] [PubMed] [Google Scholar]
- 41.Trinh DV, Zhu N, Farhang G, Kim BJ, Huxford T. The nuclear I kappaB protein I kappaB zeta specifically binds NF-kappaB p50 homodimers and forms a ternary complex on kappaB DNA. J Mol Biol. 2008;379:122–135. doi: 10.1016/j.jmb.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 42.Ferreiro DU, Cho SS, Komives EA, Wolynes PG. The energy landscape of modular repeat proteins: Topology determines folding mechanism in the ankyrin family. J Mol Biol. 2005;354:679–692. doi: 10.1016/j.jmb.2005.09.078. [DOI] [PubMed] [Google Scholar]
- 43.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 45.Ito CY, Kazantsev AG, Baldwin AS. Three NF-kappa B sites in the I kappa B-alpha promoter are required for induction of gene expression by TNF alpha. Nucleic Acids Res. 1994;22:3787–3792. doi: 10.1093/nar/22.18.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nalley K, Johnston SA, Kodadek T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature. 2006;442:1054–1057. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- 47.Sue SC, Cervantes C, Komives EA, Dyson HJ. Transfer of flexibility between ankyrin repeats in IkappaBalpha upon formation of the NF-kappaB complex. J Mol Biol. 2008;380:917–931. doi: 10.1016/j.jmb.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moorthy AK, Huang DB, Wang VY, Vu D, Ghosh G. X-ray structure of a NF-kappaB p50/RelB/DNA complex reveals assembly of multiple dimers on tandem kappaB sites. J Mol Biol. 2007;373:723–734. doi: 10.1016/j.jmb.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.