Abstract
Retinal cone photoreceptors mediate fine visual acuity, daylight vision, and color vision. Congenital hereditary conditions in which there is a lack of cone function in humans cause achromatopsia, an autosomal recessive trait, characterized by low vision, photophobia, and lack of color discrimination. Herein we report the identification of mutations in the PDE6C gene encoding the catalytic subunit of the cone photoreceptor phosphodiesterase as a cause of autosomal recessive achromatopsia. Moreover, we show that the spontaneous mouse mutant cpfl1 that features a lack of cone function and rapid degeneration of the cone photoreceptors represents a homologous mouse model for PDE6C associated achromatopsia.
Keywords: cone photoreceptor, hereditary retinal disorder, phosphodiesterase
The vertebrate retina contains two types of photoreceptors, rods and cones, that are capable of transducing physical light stimuli into a neuronal signal that forms the basis of vision and visual perception. Whereas rods are highly sensitive to light and active only under dim light conditions, cones are responsible for daylight vision and high acuity vision. Moreover, cones provide color discrimination based on the presence of distinct cone subtypes that express photopigments with different absorption spectra that eventually transmit their signals into separate neural circuits (color channels).
While the basic molecular mechanism of phototransduction follows a common principle in rods and cones, there is considerable difference in the response kinetics and adaptation capacities. Cones, for example, can adapt to light stimuli over several magnitudes of light intensities; they recover with much faster kinetics and also differ in their signal versus response relationship. This is at least in part due to differences in the biochemical and biophysical properties of the homologous proteins in rods and cones (1).
Achromatopsia can be considered as the prototype of cone malfunction disorders in humans. Achromatopsia is a congenital and mostly stationary retinal dystrophy characterized by low visual acuity, photophobia, nystagmus, and the complete disability to discriminate between colors (2). Electroretinographic recordings (ERG) in achromatopsia patients show complete absence or severely reduced cone photoreceptor function, while rod function is mostly normal (3, 4) or slightly reduced (5).
Achromatopsia is inherited as an autosomal recessive trait and up to now mutations in three genes, CNGA3, CNGB3, and GNAT2, have been identified to cause clinically indistinguishable forms of the disease (6–10). Moreover, further genetic heterogeneity of achromatopsia has been suggested (11).
The fate of cone photoreceptors in subjects with achromatopsia is of considerable interest for the development of therapeutic strategies, yet few mammalian models for achromatopsia have been characterized. These include a Cnga3 knock-out mouse (12) and a recently identified flock of Awassi sheep segregating a CNGA3 mutation (13), two dog breeds with mutations in CNGB3 (14), and the cpfl3 mutant mouse with a mutation in Gnat2 (15). While cone photoreceptors do not immediately degenerate in the cpfl3 retina, early progressive loss of cones has been observed in the Cnga3 knock-out mouse and the CNGB3 mutant dogs indicating alternative cellular responses resulting from the primary defects in cone phototransduction effectors.
In this study, we report mutations in the PDE6C gene encoding the catalytic alpha' subunit of the cone photoreceptor phosphodiesterase as a cause of autosomal recessive achromatopsia. We also show that the spontaneous mouse mutant cpfl1, featuring a lack of cone function and rapid cone photoreceptor degeneration, represents a homologous mouse model for PDE6C associated achromatopsia.
Results
Origin and Phenotype of the Mouse cpfl1 Mutant.
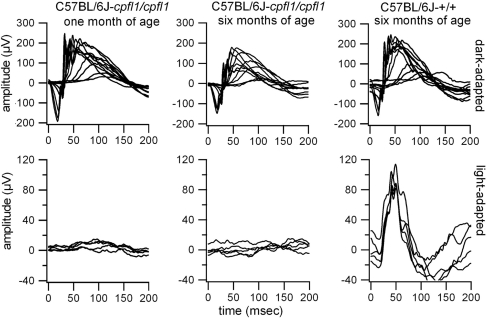
The cone photoreceptor function loss 1 (cpfl1) mouse mutant was discovered in the recombinant inbred strain CXB-1 (established from a BALB/cBy and C57BL/6By intercross), as part of an eye mutant resource screening program at The Jackson Laboratory (16). Subsequently, cpfl1 stock has been maintained by repeated backcrossing to C57BL/6J to make a congenic inbred strain, hereafter referred to as B6.Cg-cpfl1, or simply the cpfl1 strain. The phenotype can be easily typed by electroretinography (ERG) as early as 3 weeks of age. While dark-adapted ERG responses are normal, light-adapted responses representing cone function are virtually absent (Fig. 1). Histology of cpfl1 retinae revealed grossly normal morphology and layering (Fig. 2). However, as early as 3 weeks of age, there was vacuolization of a small subset of cells in the photoreceptor layer with subsequent rapid, progressive depletion of cone photoreceptors (Fig. 2). In addition, we also observed some swollen and pyknotic nuclei in the inner nuclear layer. Loss of cones further proceeds so that only very few could be detected in retinal sections of 5-month-old animals (Fig. S1).
Fig. 1.
Absence of cone function in the cpfl1 mutant. The dark-adapted (Top) and light-adapted (Bottom) ERGs of 1-month-old (Left) and 6-month-old (Center) cpfl1 mutants in comparison with C57BL/6J control mice (Right) demonstrate an early onset cone photoreceptor function loss.
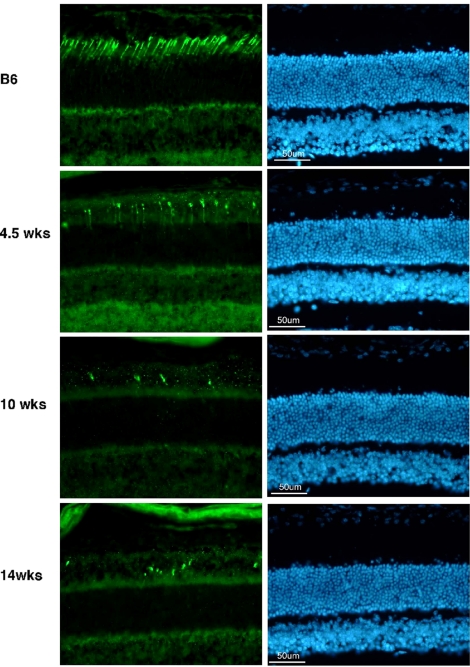
Fig. 2.
Selective loss of cone photoreceptors in the cpfl1 retina. Retinal sections of 4.5-, 10-, and 14-week-old cpfl1 mutants and C57BL/6J control animals immunostained with an antibody against cone transducin (Left) or stained with bisbenzimide for cell nuclei labeling (Right) showed a rapid loss of cones while the gross structure and layering of the retina is normal.
Linkage Mapping and Mutation Analysis in cpfl1.
In outcrosses, cpfl1 segregated as an autosomal recessively inherited trait. Applying linkage analyses with F2 animals generated from an outcross to CAST/Ei and subsequent backcross with cpfl1 mice, we initially mapped the cpfl1 locus to an interval of 3.2 ± 1.6 cM on mouse chromosome 19 between D19Mit99 and D19Mit46 (Fig. S2). To narrow down the interval, we genotyped a total of 378 F2 mice from a cpfl1× CAST/Ei intercross. This led to a refined genetic interval of 0.7 cM between D19Mit118 and D19Mit101. Marker D19Mit20 which localizes between these two latter markers, did not recombine with cpfl1. Nine genes mapped to this interval including Rbp4 (plasma retinol binding protein) and Pde6c (cone photoreceptor cGMP phosphodiesterase catalytic alpha' subunit) as primary candidates. To evaluate the congruency between the genetic and physical map, we performed STS content mapping on the YAC clone 398-A-5 and confirmed the presence of STSs reported in the MIT STS content map as well as the candidate gene Pde6c on this YAC clone.
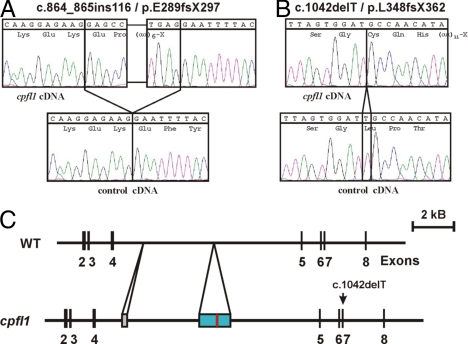
We performed comparative cDNA analysis of the Pde6c gene with retinal RNA obtained from C57BL/6J wild-type and cpfl1 mutant mice. This revealed the presence of a 116-bp insertion between exon 4 and 5 (c.864_865ins116), and an additional 1-bp deletion in exon 7 (c.1042delT) in the cpfl1 mutant. Both mutations result in frame-shifts introducing premature termination codons (p.E289fsX297 and p.L348fsX362, respectively) (Fig. 3 A and B).
Fig. 3.
Mutations in Pde6c in the cpfl1 mutant. Comparative sequencing of the Pde6c cDNA revealed a 116-bp insertion between exons 4 and 5 (A) and a 1-bp deletion in exon 7 (B) in the cpfl1 mutant in comparison with wild-type C57BL/6J sequences. A scheme showing the genomic organization of parts of Pde6C gene in C57BL/6J (WT) and cpfl1 is depicted in section C: the 116-bp cDNA insertion (red bar) represents a part of a 1,522-bp insertion (blue bar) into intron 4 of the Pde6c gene. The sequence of the genomic insertion is given in Fig. S3. The cpfl1 allele also contains an expanded hexanucleotide repeat sequence (gray bar). Note the second mutation, a 1-bp deletion, in exon 7 in the cpfl1 allele.
Comparative Southern blot and sequence analyses of intron 4 of the Pde6c gene between cpfl1 mutants and C57BL/6J mice showed that the cpfl1 allele contains an approximately 250 bp long expansion of a −GTGTCT-hexanucleotide repeat at the 5′ portion of the intron and a 1,522-bp insertion in the middle of intron 4 (Fig. 3C and Figs. S3 and S4). The 1,522-bp insertion is mainly composed of a truncated LINE 1 repeat element and a unique segment that is homologous to intron sequences of the murine diaphanous gene locus. The 116-bp cDNA insertion represents an internal part of the transposed diaphanous locus sequence that is flanked by canonical 5′ GT and 3′ AG intron junction dinucleotides. However the sequence of the splice acceptor is unusual in lacking the typical pyrimidine nucleotide stretch. In fact, RT-PCR experiments showed that splicing of this newly acquired exon is not fully efficient since also a small amount of correctly spliced Pde6c cDNA was observed. Yet this correctly spliced cDNA still carried the c.1042delT mutation. Based upon the genetic data, we conclude that the cpfl1 phenotype is caused by a lack of cGMP phosphodiesterase activity in cone photoreceptors.
Identification of Achromatopsia Subjects with Mutations in PDE6C.
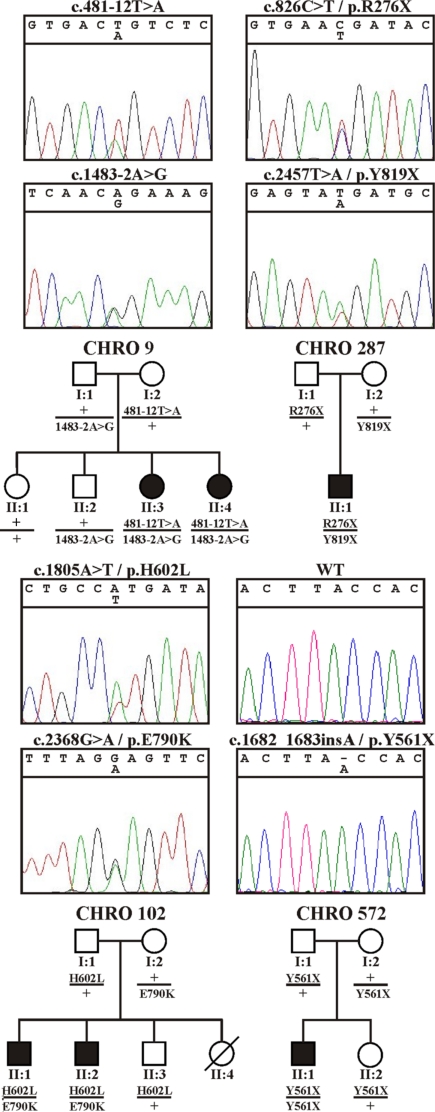
The lack of photopic ERG responses in the cpfl1 mutant follows the same clinical pattern as achromatopsia in humans. We therefore selected four families with at least two affected siblings, that did not carry mutations in the known achromatopsia genes (CNGA3, CNGB3, and GNAT2), for segregation analysis with four microsatellite markers flanking the human PDE6C locus on Chromosome 10q24. In two families we observed a segregation pattern compatible with linkage assuming recessive inheritance. Screening of the PDE6C gene by sequencing all coding exons in the index patient of these two families revealed the presence of compound heterozygous mutations in both families: c.481–12T>A and c.1483–2A>G in family CHRO 9, and c.1805A>T/p.His602Leu and c.2368G>A/p.Glu790Lys in family CHRO 102, respectively (Fig. 4). We then extended our mutation screening study to include 24 additional simplex achromatopsia patients that were also prescreened and did not carry mutations in the known achromatopsia genes. Among those 24 patients, we identified one further subject (CHRO 287) with two heterozygous nonsense mutations, c.826C>T/p.Arg276X and c.2457T>A/p.Tyr819X, and another subject (CHRO 572) with a homozygous 1-bp insertion, c.1682_83insA/p.Tyr561X in PDE6C. All mutations showed consistent independent segregation in the respective families (Fig. 4). The two missense mutations and the c.481–12T>A mutation were excluded in 100 controls (=200 chromosomes).
Fig. 4.
PDE6C mutations in achromatopsia patients and segregation analysis. Sequence traces of all identified mutations (Top) and their segregation in the respective families (Bottom). The segregation analyses confirmed compound heterozygosity in the affected patients in families CHRO 9, CHRO 287, and CHRO 102, and true homozygosity for the mutation in the affected patient in family CHRO 572.
Functional Analyses of Splicing Mutations and Missense Mutations.
We reasoned that c.481–12T>A and more apparently c.1483–2A>G might affect correct processing of PDE6C transcripts. Splicing might also be impaired in the case of c.2368G>A, which represents a substitution of the first nucleotide of exon 21. For the experimental evaluation of these potential splicing mutations we generated minigene constructs of PDE6C gene fragments that covered the mutated splice acceptor sequences, the complete intron and the flanking exon sequences. Upon transient expression of wild-type and mutant constructs in COS7 cells, we could show that the c.1483–2A>G mutation causes a complete loss of exon 12 at the cDNA level, which predicts an in-frame deletion of 49 amino acid residues (p.495_543del49) in the polypeptide (Fig. S5B). The c.481–12T>A mutation, on the other hand, activated a cryptic splice site 10 nucleotides upstream of the genuine 3′ splice acceptor site, which results in a frame-shift and the introduction of a premature termination codon (p.Asn161fsX169) (Fig. S5A). While those two mutations displayed fully penetrant splice defects in the minigene assay, we observed that the c.2368G>A mutation induced a partial skipping of exon 21 that predicts a frame-shift mutation (p.Glu790fsX17). Yet approximately 60% of the mutant transcripts were correctly spliced, harboring the c.2368G>A missense substitution (Fig. S5C).
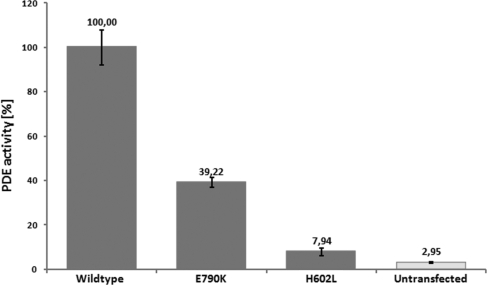
We therefore analyzed whether the p.Glu790Lys (as a result of the c.2368G>A mutation in correctly spliced transcripts) and the p.His602Leu substitutions (both encountered in CHRO 102) affect the catalytic activity of the enzyme. For this purpose, we applied the strategy of expressing a PDE5/PDE6 chimeric protein in Sf9 insect cells (17), which circumvents the failure to express type 6 phosphodiesterases in various cell types. Both mutations were introduced into a “humanized” PDE5/PDE6 construct, expressed in Sf9 cells and purified proteins assayed for cGMP hydrolysis activity. While the p.His602Leu mutant showed only baseline activity not significantly different from the untransfected control, we found that the p.Glu790Lys mutant still had considerable residual catalytic activity that reached approximately 40% of the wild-type enzyme activity (Fig. 5).
Fig. 5.
Reduction or loss of catalytic activity of p.His602Leu and p.Glu790Lys mutant phosphodiesterases, respectively. Wild-type and mutant PDE5/PDE6C chimeras were expressed in Sf9 cells and equal amounts of purified protein assayed for cGMP hydrolysis activity. Activities were normalized to the wild-type enzyme. Processed lysates from non-transfected Sf9 cells were used as controls. The catalytic activity of the p.Glu790Lys mutant was reduced to approximately 40% of the wild-type level, whereas the activity of the p.His602Leu mutant was essentially abolished.
Patient Phenotypes.
Patients with PDE6C mutations presented with a clinical picture typical for complete achromatopsia as exemplified for the two affected sisters in family CHRO 9, which have been followed up for 7 years (Fig. 6). Both patients, 24 and 26 years of age at the last clinical examination, reported photophobia and reduced visual acuity (VA) since earliest infancy with recent VA values of 0.08/0.1 (OD/OS) and 0.08/0.08, respectively. A congenital nystagmus was reported that still persists. Both patients were myopic with a refraction error (SE) of −8.00/−7.38 (OD/OS) and −7.38/−7.38, respectively. Psychophysical testing revealed a complete lack of color discrimination on various test platforms and a central scotoma, which is consistent with a loss of multifocal ERG responses for central test fields. ERG recordings showed extinguished responses under photopic and 30-Hz flicker stimulation while scotopic ERG responses were essentially normal. Funduscopy revealed an atrophy of the retinal pigment epithelium in the macula, which is an otherwise atypical feature in patients with achromatopsia (Fig. S6), but rather a clinical feature in patients with progressive cone dystrophy (e.g. 18).
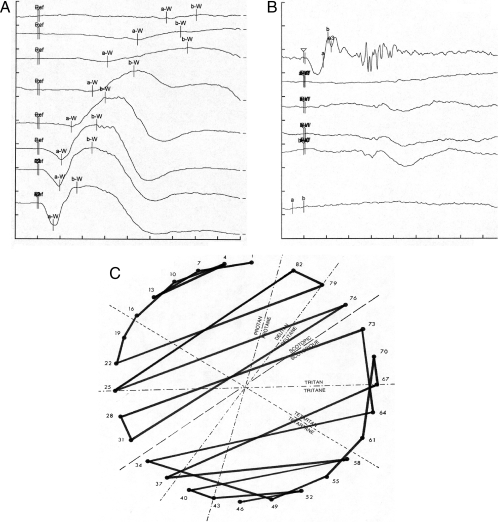
Fig. 6.
Clinical features of achromatopsia patients with PDE6C mutations. Ganzfeld ERG recordings under dark-adapted (A) and light-adapted (B) conditions. (A) Under scotopic conditions, the evoked responses were within the normal range. Only high intensity stimuli evoked responses within the lower normal range (series of stimuli with an intensity of 1, 2.5, 10, 100, 1,000, 2,500, and 10,000 mcds/m2 from top to bottom, respectively; x axis 20 ms/division; y axis 40 microvolt/division). (B) Oscillatory potentials (top row; x axis 20 ms/div.; y axis 100 microvolt/div.) and the recordings under photopic conditions revealed non-detectable responses to single (middle rows; x axis 20 ms/div.; y axis 100 microvolt/div.) and 30-Hz-Flicker stimuli (bottom: x axis 20 ms/div.; y axis 80 microvolt/div.). The responses were evoked by stimuli with an intensity of 2,500 mcd/m2, 200, 2,000, 2,500, 20,000, and 1,000 mcd/m2 from the top to the bottom, respectively. (C) Results of the FM 28 HUE color vision test with multiple permutations principally along the scotopic axis.
Discussion
In this study, we provide evidence from both the genetic data and functional investigations that mutations in PDE6C cause autosomal recessively inherited achromatopsia. In total, we identified seven different mutations in PDE6C including two nonsense mutations, one single base pair insertion, two splicing mutations, and two missense mutations. We confirmed the functional defect of the splicing mutations by minigene assays and the missense mutations by measuring the catalytic activity of recombinantly expressed proteins. His602Leu, which abolished cGMP hydrolysis activity, affects the first histidine residue of the second metal binding motif H-D-X-X-H that is strictly conserved across all PDE subfamilies. PDEs require divalent cations for enzymatic activity, and structural analyses of the PDE catalytic domains showed that the histidine residue corresponding to His602 in human PDE6C coordinates Zn2+. A tightly bound Zn2+ ion is an integral part of the PDE catalytic site (19, 20). Glu790 localizes to the C terminus of the catalytic domain in the vicinity of the inhibitory PDEγ subunit-binding site (21). A glutamic acid residue at this position is conserved among cone and rod PDEs but not in other PDE subfamilies. Structural predication with the Karplus-Schulz algorithm suggests that the Glu790Lys substitution might destabilize the α-helical structure of this part of the protein.
Interestingly we found a dual effect of the c.2368G>A/p.Glu790Lys mutation. On one hand, it induces a defect in transcript splicing that is, however, not fully penetrant, at least in our heterologous assay system. On the other hand, we found that the Glu790Lys substitution induces a reduction in the catalytic activity of the enzyme to about 40% of the wild-type level. Since both defects are quantitative, we reason that the pathogenicity of this mutation results from an additive effect at the transcript and the protein levels.
All genes currently known to cause achromatopsia encode for principle components of the phototransduction cascade: the core subunit of the G protein transducin, the two subunits of the cGMP-gated channel, and the catalytic subunit of the phosphodiesterase, as shown in this paper. The absence of transducin and phosphodiesterase activity can be considered functionally analogous in that the transduction cascade is interrupted and the photoreceptor remains in a permanent dark state. In contrast, a defect or lack of the cGMP-gated channel already impairs the resting state dark current and the establishment of a proper membrane potential. Despite this mechanistic distinction, the phenotypic outcome is rather indifferent. The clinical presentation of patients with PDE6C mutations was mostly typical for complete achromatopsia. Aspects like macular atrophy or slightly reduced scotopic ERG responses observed in some of the patients have also been reported in studies on patients with the more common CNGA3 or CNGB3 mutations (5, 22, 23). The lack of cone responses as well as the results from the color vision testing also demonstrates that the PDE6C mutations impair the function of all three types of cones in the human retina. Conversely, the defect specifically affects cone function while rod function is essentially normal.
We have demonstrated that the cpfl1 mouse mutant is a homologous natural model for PDE6C associated achromatopsia, which allows the investigation of the disorder at the histological and cellular level. cpfl1 mutants show pronounced loss of cone photoreceptors at early stages although few cones still persist for up to several months. The degeneration of cone photoreceptors is very selective since the morphology and layering of the retina appeared mostly normal. These findings are in sharp contrast to a pde6c mutant zebrafish in which loss of cones is followed by structural alterations of rod outer segments, a gradual degeneration of rods most prominently in the center of the retina, and dystrophic changes in the outer plexiform layer and the inner nuclear layer of the retina (24). In fact, rod ERG responses were strongly reduced in 20-day-old larvae. However the loss of central rods was transient and the number of rods and their morphology appeared normal at the age of 3 months. This recovery was attributed to the ongoing generation of new photoreceptors thoughout lifetime and regenerative capacity of amphibians and some teleosts including zebrafish. While newly generated cones continue to degenerate soon after formation, newly formed rods are capable to integrate into the retina and to replace lost photoreceptors in this mutant. More recently another zebrafish mutant with a missense substitution in pde6c has been reported that shows a slower degeneration of cones (25). In this mutant, rods were irregularly shaped and showed mislocalization of rhodopsin but they appeared to survive and to regain mostly normal morphology. The findings of these two studies suggest that rods do not per se require cones for their survival, even in the cone-rich retina of zebrafish, and that the morphological alteration or degeneration of rods as well as the structural changes in the outer plexiform layer at early stages result from the massive initial loss of cones.
The alterations of the Pde6c gene in the cpfl1 mouse mutant are predicted to result in a truncated polypeptide that lacks the catalytic domain and thus most likely represents a null allele. This allele carries two distinguishable mutations, an insertion in intron 4 that induces the inclusion of a foreign exon in the mRNA and furthermore a single nucleotide deletion in exon 7. The nature of the co-existence of the two mutations is currently unknown. They may have been introduced either sequentially or simultaneously through the course of the proposed retrotransposition of sequences into the murine Pde6c locus.
Mutations in PDE6C are functionally analogous to defects in the rod phosphodiesterase that cause retinitis pigmentosa or rarely congenital stationary nightblindness in humans (26, 27) and the extensively studied murine rd1 and rd10 phenotypes (28, 29).
Photoreceptor phosphodiesterases catalyze the hydrolysis of cGMP upon light stimulation. Reduced levels of cGMP subsequently lead to a closure of cGMP-gated channels and membrane hyperpolarization. It has been proposed that the lack of phosphodiesterase activity may lead to increased levels of cGMP and a toxic overload with Ca2+ due to the steady influx of Ca2+ through the cGMP-gated channel. In fact, highly elevated cGMP and Ca2+ levels have been reported in rod photoreceptors of rd1 mutant mice that harbor a mutation in the homologous Pde6b gene (30, 31). While DNA fragmentation and TUNEL staining clearly indicate loss of photoreceptors in rd1 mutants through apoptosis (32, 33), the exact mechanism underlying the execution of apoptosis is still an issue of ongoing research (34, 35). It will be interesting to study whether cone and rod degeneration caused by a primary analogous defect do also share signaling cascades trigging cell death, and even more importantly may both benefit from neuroprotective treatment such as HDAC4 overexpression as shown very recently for the rd1 mutant (36).
Experimental Procedures
Patients and Clinical Examination.
Achromatopsia patients underwent standardized ophthalmological examination including visual acuity testing, determination of refractive errors, slit lamp biomicroscopy, funduscopy, scotopic and photopic electroretinographic recordings, perimetry, and color vision testing. Venous blood was taken from patients and family members after informed consent. Total genomic DNA was extracted according to standard procedures. The study followed the tenets of the Declaration of Helsinki and the genetic investigations have been approved by the local ethical committee.
Mouse Breeding and Phenotyping.
The mice in this study were bred and maintained under standardized conditions in the research animal facility at The Jackson Laboratory. All experiments were approved by the Institutional Animal Care and Use Committee and conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
All mice in the characterization studies had pupils dilated with 1% atropine ophthalmic drops (Bausch and Lomb Pharmaceuticals Inc.) and were evaluated by indirect ophthalmoscopy with a 78 diopter lens for signs of retinal degeneration, such as vessel attenuation and alterations in the retinal pigment epithelium. Fundus photography was performed with a Kowa Genesis small animal fundus camera (37) Dark-adapted and light-adapted electroretinograms (ERG) were recorded as previously described (15). Briefly, after 2 h of dark adaptation, mice were anesthetized with an i.p. injection of ketamine (15 mg/g) and xylazine (7 mg/g body wt), and ERGs were recorded from the corneal surface of one eye after pupil dilation (1% atropine sulfate). Rod-dominated responses were recorded to short-wavelength (λmax = 470 nm; Wratten 47A filter) flashes of light over a 4.0-log unit range of intensities (0.3 log unit steps) up to the maximum allowable by the photopic stimulator. Cone-dominated responses were obtained with white flashes (0.3 steps) on the rod-saturating background after 10 min of exposure to the background light to allow complete light adaptation.
Retinal Histology and Immunohistochemistry.
Eyes for light microscopy were immersed in cold fixative (1% paraformaldehyde, 2% glutaraldehyde, and 0.1 M cacodylate buffer), left in fixative for 24 h, and then transferred to cold 0.1 M cacodylate buffer solution for an additional 24 h. Samples were embedded in methacrylate historesin or cryo-embedded in Tissue Tek (Leica), and sections were cut and stained with hematoxylin and eosin (HE) or bisbenzimide. Immunostaining was performed with antibodies against cone transducin, M opsin or HRP-conjugated peanut agglutinin.
Genetic Mapping and Molecular Genetic Analysis.
Linkage mapping of cpfl1.
To determine the chromosomal localization of the cpfl1 gene we mated B6.Cg-cpfl1 mice to CAST/Ei mice. F1 mice were backcrossed to B6.Cg-cpfl1 mice or were intercrossed to produce F2 mice. DNA was isolated from tail tips and used for PCR amplifications with 25 ng DNA in a 10-μL volume containing 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 2.5 mM MgCl2, 0.2 mM oligonucleotides, 200 μM dNTP, and 0.02 U AmpliTaq DNA polymerase. PCR products were separated on 3% MetaPhor (FMC) agarose gels and visualized under UV light after ethidium bromide staining. A genome scan of microsatellite (Mit) DNA markers was conducted on pooled DNA samples. After detection of linkage to chromosome 19, the microsatellite markers D19Mit19, D19Mit20, and D19Mit82 were scored on individual DNA samples.
cDNA Analysis of the Pde6c gene.
Total RNA was prepared from whole murine retinae by homogenization and extraction with TRIZOL reagent (Invitrogen Life Technologies). Total RNA was reverse-transcribed with oligo(dT) primers applying the SuperScript II First Strand cDNA Synthesis Kit (Invitrogen). Overlapping fragments of the Pde6c cDNA were amplified by PCR, purified on spin columns and used for sequencing applying BigDye Terminator Chemistry on an ABI 3100 DNA sequencer (Applied Biosystems).
Genomic DNA analysis.
Mouse genomic DNA was purified from homogenized mouse liver according to standard procedures. For Southern blot analysis, 15 μg genomic DNA was digested with 2 × 20 U BamHI, NcoI, or XbaI, respectively, overnight at 37 °C, extracted with phenol/chloroform, precipitated and separated on a 0.8% agarose gel. The DNA was transferred onto a nylon membrane by pressure blotting and fixed by UV light. A murine Pde6c cDNA fragment covering exons 3–7 was labeled by random priming with α-32P-dCTP and α-32P-dATP and used for Southern Blot hybridization in Hybrisol II solution (Oncor) for 18 h at 65 °C. Blots were washed in 2× SSC, 0.1% SDS at room temperature and finally in 0.1× SSC, 0.1% SDS at 55–65 °C. Blots were exposed to X-ray film for 12–36 h. For comparative genomic sequencing, overlapping fragments of intron 4 of Pde6c were amplified by PCR and sequenced by primer walking.
Genetic analysis of achromatopsia patients.
For human chromosome 10q24 segregation analysis, microsatellite markers were genotyped by PCR amplification and sized on an ABI 377 DNA sequencer (Applied Biosystems). Mutation screening was performed by PCR amplification of coding exons and flanking intron sequences, and subsequent DNA sequencing of PCR products as described above. Sequences were analyzed either manually or with the aid of the SeqMan software (Lasergene). Sequences of the primers and PCR conditions are available from B.W. upon request.
Heterologous Splicing Assay.
Segments covering exons 1–3, 11–14, and 19–21, including the entire internal introns as well as parts of the flanking intron sequences, respectively, of PDE6C were amplified from genomic DNA applying Pfu Turbo polymerase (Stratagene) and blunt-end cloned using the pCR-Script PCR Cloning Kit (Stratagene). Inserts were excised by restriction enzyme digestion and reinserted into pSPL3_2096 [a derivate of pSPL3 (Invitrogen) with a stuffer fragment cloned into the original NotI site]. Constructs were partially sequenced, including the vector/insert boundaries and all exon/intron boundaries. COS7 cells were cultured in DMEM supplemented with 5% (vol/vol) non-essential amino acids and 10% FCS. Cells were transfected with pSPL3-PDE6C constructs using Lipofectamine (Invitrogen). Total RNA was prepared 36 h post-transfection using TRIZOL reagent (Invitrogen), reverse transcribed with either oligo(dT) or SA2 primer (a reverse primer located in the 3′ tat exon of the pSPL3 vector) and AMV reverse transcriptase [Takara RT-PCR Kit (Takara)] and the cDNA amplified with pSPL3 exon primers. RT-PCR products were either directly sequenced or sequenced after an intermitting cloning step.
Recombinant Expression and Functional Analysis of PDE6C Mutants.
Constructs to express human chimeric PDE5/PDE6C with the baculovirus/Sf9 insect cell system were generated as described (17). The humanized PDE5/PDE6 construct contains residues 1–445 and 741–790 of human PDE6C, and residues 516–781 and 831–845 of human PDE5.
The mutations p.His602Leu and p.Glu790Lys were introduced by in vitro mutagenesis (Quik Change in vitro Mutagenesis Kit, Stratagene). For protein expression, Sf9 cells (3 × 106 cells/mL, in a total volume of 400 mL) were infected with recombinant baculoviruses at MOI of 3–5. Recombinant proteins were purified by affinity chromatography on a His-bind resin (Novagen). cGMP hydrolysis was measured using [3H]-labeled cGMP and 5 μg purified protein as described (38). The reaction was stopped after 10 min by addition of AG1-X2 anion exchange resin (Bio-Rad). The radioactivity in the supernatant was measured with a scintillation counter. Experiments were done in triplicates and cGMP hydrolysis activity normalized to the wild-type PDE5/PDE6C chimera activity.
Supplementary Material
Acknowledgments.
We thank Norm Hawes of The Jackson Laboratory (who recently passed away) for many helpful discussions; his selfless attitude toward science and kindness were a joy and inspiration. We also thank Norman Rieger for animal care and genotyping; Mathias Seeliger, Naoyuki Tanimoto, Susanne Beck, Michael Bonin, Karin Schäferhoff, Francois Paquet-Durand, and Dragana Trifunovic for fruitful discussions; Melissa Berry for proofreading of the manuscript; and Jeremy Nathans for providing antibodies against M and L cone opsins. This work was supported by National Institutes of Health Grants EY07758, EY11996, and RR01183 and Deutsche Forschungsgemeinschaft Grant Wi1189/6-1 and KFO134-Ko2176/1-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907720106/DCSupplemental.
References
- 1.Kawamura S, Tachibanaki S. Rod and cone photoreceptors: Molecular basis of the difference in their physiology. Comp Biochem Physiol A Mol Integr Physiol. 2008;150:369–377. doi: 10.1016/j.cbpa.2008.04.600. [DOI] [PubMed] [Google Scholar]
- 2.Hess RF, Sharpe LT, Nordby K. Cambridge: Cambridge Univ Press; 1990. Night vision: Basic, clinical and applied aspects. [Google Scholar]
- 3.Eksandh L, Kohl S, Wissinger B. Clinical features of achromatopsia in Swedish patients with defined genotypes. Ophthalmic Genet. 2002;23:109–120. doi: 10.1076/opge.23.2.109.2210. [DOI] [PubMed] [Google Scholar]
- 4.Kellner U, Wissinger B, Kohl S, Kraus H, Foerster MH. Molecular genetic findings in patients with congenital cone dysfunction: Mutations in the CNGA3, CNGB3, or GNAT2 genes. Ophthalmologe. 2004;101:830–835. doi: 10.1007/s00347-003-0976-y. [DOI] [PubMed] [Google Scholar]
- 5.Khan NW, Wissinger B, Kohl S, Sieving PA. CNGB3 achromatopsia with progressive loss of residual cone function and impaired rod-mediated function. Invest Ophthalmol Vis Sci. 2007;48:3864–3871. doi: 10.1167/iovs.06-1521. [DOI] [PubMed] [Google Scholar]
- 6.Kohl S, et al. Total colorblindness is caused by mutations in the gene encoding the α-subunit of the cone photoreceptor cGMP-gated cation channel. Nat Genet. 1998;19:257–259. doi: 10.1038/935. [DOI] [PubMed] [Google Scholar]
- 7.Kohl S, et al. Mutations in the CNGB3 gene encoding the β-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum Mol Genet. 2000;9:2107–2116. doi: 10.1093/hmg/9.14.2107. [DOI] [PubMed] [Google Scholar]
- 8.Sundin OH, et al. Genetic basis of total colorblindness among the Pingelapese islanders. Nat Genet. 2000;25:289–293. doi: 10.1038/77162. [DOI] [PubMed] [Google Scholar]
- 9.Kohl S, et al. Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am J Hum Genet. 2002;71:422–425. doi: 10.1086/341835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aligianis IA, et al. Mapping of a novel locus for achromatopsia (ACHM4) to 1p and identification of a germline mutation in the alpha subunit of cone transducin (GNAT2) J Med Genet. 2002;39:656–660. doi: 10.1136/jmg.39.9.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohl S, et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur J Hum Genet. 2005;13:302–308. doi: 10.1038/sj.ejhg.5201269. [DOI] [PubMed] [Google Scholar]
- 12.Biel M, et al. Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc Natl Acad Sci USA. 1999;96:7553–7557. doi: 10.1073/pnas.96.13.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banin E, et al. Cone dysfunction and congenital day blindness in Awassi sheep is caused by a mutation in the CNGA3 gene. Invest Ophthalmol Vis Sci. 2009;50:E4145. [Google Scholar]
- 14.Sidjanin DJ, et al. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum Mol Genet. 2002;11:1823–1833. doi: 10.1093/hmg/11.16.1823. [DOI] [PubMed] [Google Scholar]
- 15.Chang B, et al. Cone photoreceptor function loss-3, a novel mouse model of achromatopsia due to a mutation in Gnat2. Invest Ophthalmol Vis Sci. 2006;47:5017–5021. doi: 10.1167/iovs.05-1468. [DOI] [PubMed] [Google Scholar]
- 16.Chang B, et al. Retinal degeneration mutants in the mouse. Vision Res. 2002;42:517–525. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 17.Granovsky AE, et al. Probing domain functions of chimeric PDE6alpha'/PDE5 cGMP-phosphodiesterase. J Biol Chem. 1998;273:24485–24490. doi: 10.1074/jbc.273.38.24485. [DOI] [PubMed] [Google Scholar]
- 18.Small KW, Silva-Garcia R, Udar N, Nguyen EV, Heckenlively JR. New mutation, P575L, in the GUCY2D gene in a family with autosomal dominant progressive cone degeneration. Arch Ophthalmol. 2008;126:397–403. doi: 10.1001/archopht.126.3.397. [DOI] [PubMed] [Google Scholar]
- 19.Xu RX, et al. Atomic structure of PDE4: Insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- 20.Sung BJ, et al. Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature. 2003;425:98–102. doi: 10.1038/nature01914. [DOI] [PubMed] [Google Scholar]
- 21.Granovsky AE, Artemyev NO. Identification of the gamma subunit-interacting residues on photoreceptor cGMP phosphodiesterase, PDE6alpha. J Biol Chem. 2000;275:41258–41262. doi: 10.1074/jbc.M008094200. [DOI] [PubMed] [Google Scholar]
- 22.Wissinger B, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69:722–737. doi: 10.1086/323613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaelides M, et al. Progressive cone dystrophy associated with mutation in CNGB3. Invest Ophthalmol Vis Sci. 2004;45:1975–1982. doi: 10.1167/iovs.03-0898. [DOI] [PubMed] [Google Scholar]
- 24.Stearns G, Evangelista M, Fadool JM, Brockerhoff SE. A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J Neurosci. 2007;27:13866–13874. doi: 10.1523/JNEUROSCI.3136-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishiwaki Y, et al. Mutation of cGMP phosphodiesterase 6alpha'-subunit gene causes progressive degeneration of cone photoreceptors in zebrafish. Mech Dev. 2008;125:932–946. doi: 10.1016/j.mod.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 26.McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci USA. 1995;92:3249–3253. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gal A, Orth U, Baehr W, Schwinger E, Rosenberg T. Heterozygous missense mutation in the rod cGMP phosphodiesterase beta-subunit gene in autosomal dominant stationary night blindness. Nat Genet. 1994;7:64–68. doi: 10.1038/ng0594-64. [DOI] [PubMed] [Google Scholar]
- 28.Bowes C, et al. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990;347:677–680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- 29.Gargini C, Terzibasi E, Mazzoni F, Strettoi E. Retinal organization in the retinal degeneration 10 (rd10) mutant mouse: A morphological and ERG study. J Comp Neurol. 2007;500:222–238. doi: 10.1002/cne.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farber DB, Lolley RN. Cyclic guanosine monophosphate: Elevation in degenerating photoreceptor cells of the C3H mouse retina. Science. 1974;186:449–451. doi: 10.1126/science.186.4162.449. [DOI] [PubMed] [Google Scholar]
- 31.Fox DA, Poblenz AT, He L. Calcium overload triggers rod photoreceptor apoptotic cell death in chemical-induced and inherited retinal degenerations. Ann N Y Acad Sci. 1999;893:282–285. doi: 10.1111/j.1749-6632.1999.tb07837.x. [DOI] [PubMed] [Google Scholar]
- 32.Chang GQ, Hao Y, Wong F. Apoptosis: Final common pathway of photoreceptor death in rd, rds, and rhodopsin mutant mice. Neuron. 1993;11:595–605. doi: 10.1016/0896-6273(93)90072-y. [DOI] [PubMed] [Google Scholar]
- 33.Portera-Cailliau C, Sung CH, Nathans J, Adler R. Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. Proc Natl Acad Sci USA. 1994;91:974–978. doi: 10.1073/pnas.91.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doonan F, Donovan M, Cotter TG. Caspase-independent photoreceptor apoptosis in mouse models of retinal degeneration. J Neurosci. 2003;23:5723–5731. doi: 10.1523/JNEUROSCI.23-13-05723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sancho-Pelluz J, et al. Photoreceptor cell death mechanisms in inherited retinal degeneration. Mol Neurobiol. 2008;38:253–269. doi: 10.1007/s12035-008-8045-9. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawes NL, et al. Mouse fundus photography and angiography: A catalogue of normal and mutant phenotypes. Mol Vis. 1999;5:22. [PubMed] [Google Scholar]
- 38.Natochin M, Artemyev NO. Mutational analysis of functional interfaces of transducin. Methods Enzymol. 2000;315:539–554. doi: 10.1016/s0076-6879(00)15866-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.