Abstract
Telomeres must be capped to preserve chromosomal stability. The conserved Stn1 and Ten1 proteins are required for proper capping of the telomere, although the mechanistic details of how they contribute to telomere maintenance are unclear. Here, we report the crystal structures of the C-terminal domain of the Saccharomyces cerevisiae Stn1 and the Schizosaccharomyces pombe Ten1 proteins. These structures reveal striking similarities to corresponding subunits in the replication protein A complex, further supporting an evolutionary link between telomere maintenance proteins and DNA repair complexes. Our structural and in vivo data of Stn1 identify a new domain that has evolved to support a telomere-specific role in chromosome maintenance. These findings endorse a model of an evolutionarily conserved mechanism of DNA maintenance that has developed as a result of increased chromosomal structural complexity.
Keywords: end capping, Stn1, Ten1, Cdc13, t-RPA
The protection of chromosome ends is a central problem in telomere biology. Originally observed as DNA ends that are refractory to X-ray-induced damage (1), telomere capping is now defined as a mechanism that safeguards the natural ends of chromosomes from recognition by the DNA damage machinery (2–4). A number of telomere proteins, bound to either the duplex region or the single-strand extension of telomeres contribute, either directly or indirectly, to chromosome end protection (5, 6). End protection is essential to chromosomal stability: loss of this activity leads to end-to-end fusions, nucleolytic degradation, and/or recognition and aberrant processing by the DNA damage machinery.
End protection activity is conferred by a widely conserved telomere end-binding complex, referred to as POT1-TPP1 in most species and TEBPα/TEBPβ in the ciliates (6). This complex specifically recognizes the telomeric G-rich ssDNA overhang universally present at telomeres (7–9). In the case of TEBPα/TEBPβ, the high-resolution structure of the complex bound to ssDNA revealed that the 3′ end of the DNA is completely buried from solvent exposure through interactions with multiple copies of the oligonucleotide/oligosaccharide binding motif (OB fold), providing a clear mechanism for end protection (10). Although the structures of POT1 and TPP1 in isolation suggest structural homology to the TEBPα/TEBPβ complex, their organization on the DNA is distinct, suggesting an alternate mode of action (8, 11). In both mammalian cells and fission yeast, the POT1-TPP1 heterodimer is part of a larger complex, called shelterin, which performs both end protection and telomere length regulation (6).
Increasingly, evidence indicates that a second end-binding complex, minimally composed of two proteins called Stn1 and Ten1, also protects chromosome termini. First discovered in budding yeast (12, 13), the Stn1-Ten1 complex is broadly distributed across eukaryotic phyla (14–16). In budding yeast, fission yeast, and plants, null alleles result in severe and catastrophic consequences for chromosome ends (12, 13, 15, 16). These proteins also participate in telomere length regulation, but their roles in capping and length maintenance are poorly understood (17).
The Stn1 and Ten1 proteins have been most extensively studied in budding yeast, performing their telomere-related functions in association with the telomere-binding protein Cdc13 (17). All three are essential for viability, as depletion of any one of these proteins results in rapid and extensive nucleolytic degradation of the C-rich strand of the telomere (12, 13, 18). The resulting elongated single-stranded region (which can extend for >10 kb) (19) signals the DNA damage machinery, resulting in a RAD9-dependent cell cycle arrest (13, 20). In vitro studies show Ten1 copurifies with the predicted N-terminal OB fold domain of Stn1 (14). Similar results are observed in fission yeast, where Stn1 and Ten1 interact by yeast two-hybrid analysis and stn1-Δ and ten1-Δ mutants suffer extensive loss of terminal DNA, with rare survivors arising only as a result of chromosome circularization (15).
How Cdc13, Stn1, and Ten1 protect chromosome termini from resection is a poorly understood process. However, the parallels between the unregulated 5′ → 3′ resection that occurs at unprotected yeast termini and the transient appearance of ssDNA at newly generated double-strand breaks (DSBs) may be instructive (21–23). This comparison, as well as numerous other observations, has supported an emerging model that telomere maintenance shares an evolutionary relationship with the DNA repair machinery. A recent study suggesting that Cdc13, Stn1, and Ten1 form a heterotrimeric complex that bears a number of similarities to the replication protein A complex (RPA; which is a heterotrimer consisting of RPA70, RPA32, and RPA14 subunits in humans; these are Rpa1, Rpa2, and Rpa3, respectively, in yeast) provides additional support for this proposed evolutionary link (14).
RPA, which is the major ssDNA-binding protein complex in eukaryotic cells, has a central role in multiple aspects of DNA metabolism, including repair of DSBs (24, 25). The premise that the ends of chromosomes employ a telomere-specific t-RPA is based on several points of comparison between these two complexes. The large subunits of both complexes—Cdc13 and RPA70/Rpa1—employ OB folds for high affinity, ssDNA binding (25, 26). Although little is known about the domain structure of Cdc13 outside of the DNA-binding domain, it is predicted to contain at least one additional OB fold (27). Secondary and tertiary structure predictions suggest that the N-terminal domain of Stn1 adopts an OB fold similar to that of the OB-fold domain of RPA32. This prediction is strengthened by the observation that the Rpa2 OB fold can be substituted with the Saccharomyces cerevisiae Stn1 putative OB fold in vivo (14). Although an OB fold was also weakly predicted for Ten1, there is no experimental evidence yet supporting this prediction (15). No reliable predictions can be made about the C-terminal domain of Stn1, leaving the potential contributions of this domain to telomere function unresolved.
To better define the parallels between Cdc13, Stn1, and Ten1 and the RPA complex, we have solved the crystal structures of the C-terminal domain of the S. cerevisiae Stn1 protein and the full-length Schizosaccharomyces pombe Ten1 protein. These structures both exhibit striking similarity to their corresponding domains in the RPA32 and RPA14 proteins, respectively. This study also reveals one key difference: the Stn1-C domain is composed of tandem winged helix-turn-helix (wHTH) motifs, whereas the C-terminal domain of RPA32 possesses only one such motif. In vivo analysis, using structure-based site-directed mutagenesis, reveals that this second Stn1 wHTH motif performs a telomere-specific function. These observations establish that Stn1 and Ten1 are in fact distantly related to subunits of the RPA complex. Also, they suggest that the proposed t-RPA complex has acquired at least one domain that confers a specialized function specific for chromosome termini.
Results
Structure of the C-Terminal Domain of Stn1 Reveals Tandem wHTH Motifs.
Previous work suggests that the N-terminal domain of Stn1 adopts an OB fold; however, sequence analysis and structure prediction provide no insights into the structure or function of the C-terminal domain. As a first step in addressing the function of the C-terminal Stn1 domain, we have solved the crystal structure of residues 313–491 of the S. cerevisiae Stn1 protein to 2.5 Å (R = 24.6, Rfree = 30.1). A preliminary electron density map was achieved via single isomorphous replacement with anomalous scattering (SIRAS) with native and gold-derivatized crystals (Fig. 1A; Fig. S1A and Table S1). A higher resolution native dataset was used for refinement to improve the resolution (Fig. S1B and Table S1). This structure reveals an overall topology consisting of two lobes composed of nonsuperimposable, wHTH motifs (Fig. 1A). The canonical wHTH motif is characterized by three α-helices (H1-H3), three β-strands (β1-β3), and two loops or “wings” (W1 and W2), with the β-strands forming a typical twisted antiparallel β-sheet (28, 29). Relative to the canonical wHTH topology, an additional α-helix (H3) is present in the N-terminal wHTH domain, with three consecutive α helices between β1 and β2. Also, the W2 element is not present in this domain, which connects directly into the second wHTH after β3. The C-terminal wHTH domain adopts a wHTH structure with no additional structural elements. The β-strands, however, are significantly longer than usual, and the wings in this lobe are flexible and not defined by the electron density.
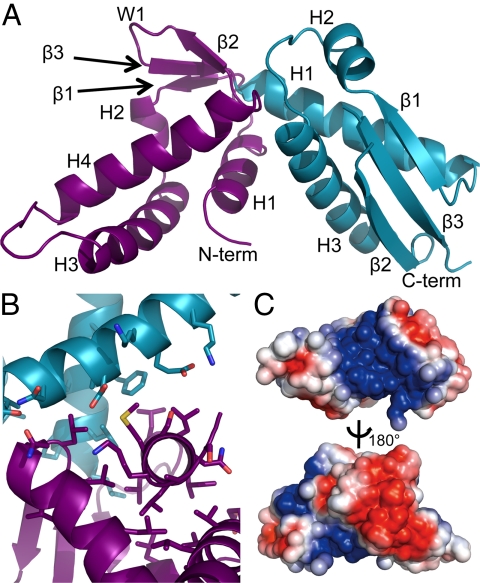
Fig. 1.
Crystal structure and electrostatic surface representations of the C-terminal domain of S. cerevisiae Stn1. (A) The 2.5-Å crystal structure of Stn1-C contains tandem wHTH motifs (N-terminal wHTH, purple; C-terminal wHTH motif, teal). The secondary structure elements defining each wHTH motif are labeled. (B) The tight packing of hydrophobic residues at the interface of the N- and C-terminal wHTH motifs restricts the flexibility of the protein. View is looking down the H1 helix at the N terminus of the structure, with helices H4 and H3 of the N- and C-terminal wHTH motifs, respectively, in the forefront. (C) Electrostatic surface representations of the Stn1-C structure. (Upper) Same orientation as in A showing the patch of basic charge at the lobe-lobe interface; (Lower) 180° along the indicated axis from Upper, showing a clustering of acidic residues on and around the β-sheet of the N-terminal wHTH motif. Blue, positive charge; red, negative charge. All figures prepared using PyMOL (36).
The interface between the wHTH lobes is composed of H1 and the loop between H4 and β2 of the N-terminal wHTH and H1 and H3 of the C-terminal wHTH. This interface buries 253 Å2 of surface area and is composed primarily of hydrophobic residues (Fig. 1B). Also, the individual helices of each wHTH lobe form a compact structure with residues from the antiparallel β-sheets contributing to the hydrophobic core. An angle of ≈90° is created between the wHTH lobes at the interface where H4 on the N-terminal wHTH and H3 on the C-terminal wHTH meet (Fig. 1B). The helices at the cleft created between the N- and C-terminal wHTH lobes are flanked by three loops of 6–8 residues each. The loop between H3 and H4 of the N-terminal wHTH lobe contains solvent exposed hydrophobic and charged residues, and appears to be flexible. However, the loops flanking both ends of H3 of the C-terminal lobe contain hydrophobic residues that contribute to the core packing, suggesting limited flexibility in this region. Also, the tight packing of these helices suggests that there is little flexibility of the structure at the lobe-lobe interface (Fig. 1B). An electrostatic surface representation of this interface shows that it is highly basic, due primarily to five positively charged solvent exposed residues on H3 on the C-terminal wHTH as well as one residue each on H1 and H4 of the N-terminal wHTH (Fig. 1C Upper). Conversely, the N-terminal wHTH domain on the opposite face is highly acidic due to a clustering of negatively charged residues on the β-sheet and on H2, H3, and H4 (Fig. 1C Lower).
The individual wHTH domains of Stn1-C were submitted separately to the DALI server (30), because no structures were found that matched the full C-terminal domain of Stn1. Of the top ten hits to the N-terminal lobe of Stn1-C (Z scores > 5.3), seven structures belong to the wHTH superfamily. Among these top structural matches was the C-terminal domain of RPA32 (RPA32-C; Z score = 5.9) (31). A superposition of the N-terminal wHTH lobe of Stn1-C with RPA32-C illustrates the high degree of similarity between the structures, which superimpose with an rmsd of 1.9 Å over 58 atoms (Fig. 2A). One notable difference is the additional helix, H3, present in the Stn1-C structure that is not found in RPA32-C. The supplementary helix in Stn1-C does not affect the packing angles of the other three helices relative to each other, because the orientation of the secondary structure elements in the superposition of the two wHTH domains is maintained between structures.
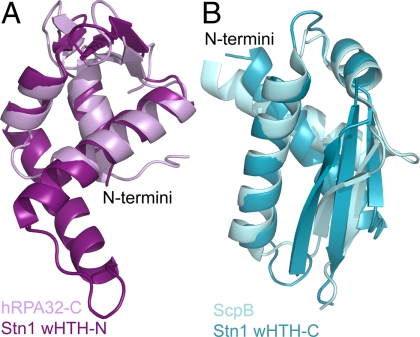
Fig. 2.
Superposition of the N- and C-teminal lobes of Stn1-C with RPA32 and ScpB, respectively. (A) The N-terminal lobe of Stn1-C superpositions with RPA32-C (31) with an rmsd of 1.9 Å over 58 atoms. The C-terminal domain of RPA32 contains three α helices which superposition with helices H1, H2, and H4 of Stn1-C. Helix H3 of Stn1-C is not included in the superposition. (B) The C-terminal lobe of Stn1-C superpositions with residues 1–75 of the ScpB protein with an rmsd of 2.8 Å over 72 atoms. Figures were prepared using PyMOL (36).
In contrast, the top ten DALI hits to the C-terminal wHTH domain of Stn1-C were more diverse, including two hits to the winged helix superfamily, two hits to the R3H superfamily, and several hits not belonging to any superfamily. However, the top hit (Z score = 6.4), the bacterial ScpB protein involved in chromosome partitioning, is a wHTH protein (32). Residues 1–75 of the ScpB structure superpositions with the C-terminal wHTH of Stn1-C with an rmsd of 2.8 Å over 72 atoms (Fig. 2B). Thus, we conclude that the C-terminal domain is also a wHTH domain.
C-Terminal wHTH Motif Confers a Telomere-Specific Function on Stn1.
To assess the potential role of the two wHTH motifs of Stn1-C in telomere function, missense mutations were introduced in solvent-exposed residues of each wHTH lobe and examined for in vivo phenotypes, with residues mutated to alanine, as well as to a charged residue in most cases. We focused our attention on the β-sheet of each lobe, because prior work had suggested that the comparable surface on RPA32 provides a site for protein interaction(s) (31). Plasmids bearing individual stn1 mutations were introduced into a stn1-Δ strain, using standard yeast genetic techniques, to generate strains expressing each stn1 mutation under control of the native STN1 promoter, in the absence of the wild-type STN1 gene. The resulting set of strains were examined for viability and telomere length, as well as sensitivity to DNA damage.
Mutations were introduced in six residues that targeted the β-sheet region of the N-terminal wHTH motif (E330, K389, D390, K391, E394, and D397). Notably, disruption of these residues had essentially no effect on telomere function. Mutations in D397 exhibited a modest telomere elongation phenotype, whereas the rest of the mutations had little or no effect (Fig. S2 A and B). These stn1− strains also displayed a growth rate that was indistinguishable from that of the parental STN1 strain, as might be predicted from the prior demonstration that the C terminus of Stn1 is dispensable for viability (14), and did not exhibit enhanced sensitivity to DNA damage. These observations argue that the β-sheet region of the N-terminal wHTH motif of Stn1 does not interact with a telomere length regulator.
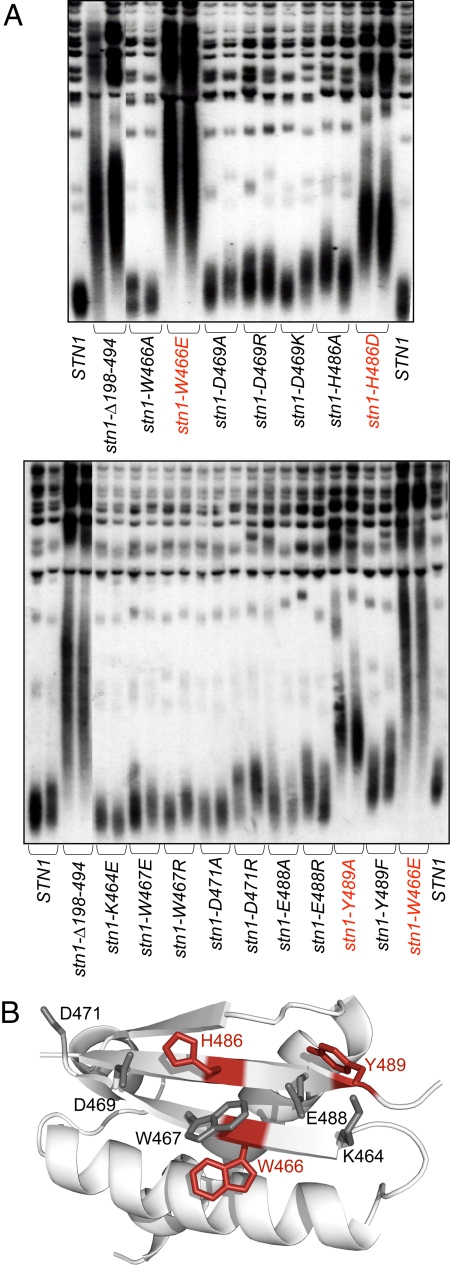
In contrast, in vivo analysis of the C-terminal wHTH motif revealed that this second lobe performs a telomere-specific function. Once again, mutations were introduced into a total of eight residues on the surface of the β-sheet. Mutations in a cluster of three aromatic residues (W466, H486, and Y489) had substantial effects on telomere length regulation (Fig. 3A). This phenotype did not appear to be due to a general destabilization of this second wHTH lobe, because mutations (including charge swap mutations) introduced into other residues on the surface of this sheet (R464, W467, D469, and E488) had little, or no, impact on telomere length. Also, this difference was not simply the consequence of destabilization, because Western blot analysis demonstrated that proteins with mutations W466 and H486 were expressed at levels that were indistinguishable from that of wild-type Stn1 protein (Fig. S3A). Telomeres in the stn1-W466E and stn1-W466R strains were elongated to a greater extent than that of the stn1-Δ198–494 strain (Fig. 3A; Fig. S3B), and this extreme telomere length dysregulation correlated with substantial defects in cell cycle progression. Although the stn1-W466E strain had a growth rate that was only slightly impaired, relative to wild type, FACS analysis revealed a delay in progression through the G2/M phase of the cell cycle (Fig. S4). The stn1-W466R strain was even more severely affected: The strain exhibited a notable growth defect, with single colony formation taking ≈2 days longer than the isogenic wild-type strain. This severe reduction in growth rate was accompanied by a pronounced accumulation in the G2 phase of the cell cycle. The cell cycle profile exhibited by these two mutant strains was suggestive of DNA damage. Consistent with this prediction, introduction of a rad9-Δ or rad24-Δ mutation partially, or fully, relieved the block to progression through the cell cycle in the stn1-W466E and stn1-W466R strains, respectively (Fig. S4). These results demonstrate that a determinant for telomere maintenance in the C-terminal domain of Stn1 stems from residues clustered on the β-sheet of the second wHTH motif (Fig. 3B).
Fig. 3.
Negative regulation of telomere length by the C-terminal wHTH lobe of Stn1. (A) Telomere length of the indicated stn1 mutant strains, derived from structure-based mutagenesis. (B) Residues tested for effects on telomere length are mapped on the C-terminal wHTH lobe of Stn1-C; the residues exhibiting the most severe defect (indicated in red) are located on the β-sheet, whereas residues that when mutated result in little or no phenotype are in gray. The N-terminal lobe is removed for ease of viewing. Figure prepared using PyMOL (36).
Ten1 Is a Paralog of RPA14.
The small Ten1 protein is a rapidly diverging protein whose role at the telomere is poorly understood. Although it was possible to predict with reasonable confidence that the Stn1 N-terminal domain adopts an OB fold that is similar to that of RPA32, no comparable structural predictions were possible for the budding yeast Ten1 protein (14). A subsequent study predicted that the fission yeast Ten1 adopts an OB fold based on sequence analysis, but with notable low confidence (15). Thus, an unequivocal determination of the potential evolutionary relationship between RPA14 and Ten1 requires high-resolution structural analysis. To address this relationship, we have solved the crystal structure of the full length, 11.5-kDa S. pombe Ten1 protein (R = 18.8, Rfree = 23.8). Phases for an initial electron density map were obtained by means of single-wavelength anomalous dispersion (SAD), with an iodide derivative crystal diffracting to 1.7 Å (Fig. S5 A and B and Table S2). The structure of Ten1 is comprised of the antiparallel five-stranded Greek key motif characteristic of the OB fold superfamily (Fig. 4A) (33). At the N terminus of the protein, a short α-helix (H1) caps the top of the β-barrel, lying nearly perpendicular to the long axis of the barrel. H1 is followed by five consecutive strands comprising the β-barrel (β1-β5) that is formed by two, three-stranded antiparallel β-sheets. As is typical of the β-barrels of OB folds, β1 wraps around the structure such that it participates in both three-stranded antiparallel β-sheets. This feature of β1 is accommodated by a conserved glycine (G25 in the Ten1 structure) in the middle of the β1 strand that results in the distinctive kink, thereby allowing the strand to wrap around the β-barrel. Also, a long irregularly structured loop of 13 residues is found between strands β3 and β4. After strand β5, the C terminus of the protein contains a second α-helix (H2) that runs parallel to the entire length of the long axis of the β-barrel. Although the telomere-binding OB fold proteins frequently have an α-helix at the C terminus (7), the C-terminal α-helix in Ten1 is unusual in its orientation, parallel rather than perpendicular, to the β-barrel (Fig. 4A). In the OB folds typical of telomere end-binding proteins, the C-terminal α-helix appears to serve as a structural support; however, we speculate the α-helix in the Ten1 protein has an alternative function, further detailed in the discussion section of this text.
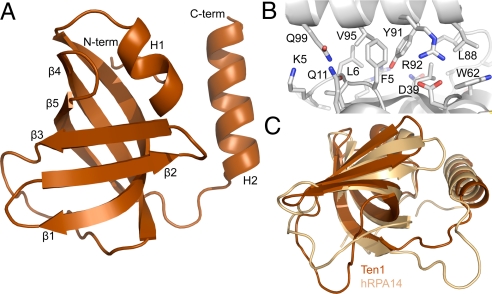
Fig. 4.
The crystal structure of the full length S. pombe Ten1 protein reveals structural similarities to RPA14. (A) The 1.7-Å refined crystal structure of Ten1 is an OB fold motif shown with the secondary structure elements indicated. (B) The C-terminal α-helix of Ten1 packs against elements of the β-barrel as well as the N-terminal irregularly structured loop creating a tightly associated interface. (C) The N- and C-terminal helices as well as the β-barrel of the Ten1 and RPA14 (34) OB folds superposition with an rmsd of 1.96 Å over 94 atoms. Figures prepared using PyMOL (36).
A compact core of hydrophobic residues is observed throughout the interior of the β-barrel, which is capped at on end by hydrophobic residues on one face of the H1 helix. At the opposite end of the β-barrel is the long loop connecting β3 and β4, which seals the hydrophobic core with residues at the N- and C-terminal ends of the loop. The B-factors for this loop are significantly higher than the rest of the protein, perhaps because the solvent exposed charged and hydrophobic residues have some flexibility. The C-terminal α-helix (H2) is tightly associated with residues on the outer surface of β1, β4, and loop β2-β3 of the β-barrel, forming a hydrophobic interface that buries 1,077 Å2 of surface area. There is one salt bridge at this interface between Arg-92 on H2 and Asp-39 in the loop between β2 and β3 (Fig. 4B). The six irregularly structured but experimentally well-defined N-terminal residues also participate in the packing of the C-terminal helix and the hydrophobic core. Residues on the loop connecting β5 to the C-terminal helix further stabilize the hydrophobic interface.
Structural relatives of the Ten1 OB fold were identified through the DALI server (30). Although Ten1 adopts the common OB fold topology, the DALI matches were overwhelmingly the OB folds of RPA32 and RPA14, Z scores of 12.8 and 12.6, respectively (34). It was not surprising that both RPA32 and RPA14 were the top hits in the DALI search, because these two structures are quite similar to one another. The Ten1 structure superpositions with RPA14 with an rmsd of 1.96 Å over 94 atoms (Fig. 4C). All of the secondary structural elements of Ten1 (H1, β1-β5, and H2) are present in RPA14, which contains no supplementary structural components relative to Ten1. Also, the unstructured regions (N terminus) and loops (β3 to β4 and β5 to H2) of Ten1 are similarly arranged in the RPA14 structure. Notably, the orientation of the helix (H2) of RPA14, which forms the interaction surface with RPA32, is preserved in Ten1.
Discussion
Stn1-C Is a Telomere-Specific Interaction Domain.
Identifying the structural similarities between RPA32 and Stn1 allows us to consider the extensive biochemical and structural data available for this RPA protein in the context of the Stn1 protein. The RPA32/Rpa2 protein participates in numerous DNA repair pathways, including nucleotide excision repair, DSB homologous recombination, as well as postreplicative base excision repair. The critical role of RPA32/Rpa2 in these pathways has been attributed to protein–protein interactions between the wHTH domain of RPA32/Rpa2 and DNA repair enzymes (25). The solution structure of the wHTH of RPA32-C was solved in complex with a peptide of the UNG protein, an enzyme involved in base excision repair (31). The RPA32-C/UNG peptide interaction is mediated primarily through an acidic patch of residues proximal to the β-sheet of RPA32-C that align with basic residues on the induced helix UNG peptide. Stn1-C possesses a similar large acidic patch on the N-terminal wHTH, stemming from residues clustered on the β-sheet and the N terminus of H4 (Fig. 1C Lower). Strikingly, however, mutation of this acidic patch indicates that this surface is not used by Stn1 for telomere length regulation (Fig. S2).
In contrast, the β-sheet of the second wHTH domain is clearly required for telomere function, because mutations in three of the eight amino acids that comprise this surface conferred substantial telomere dysfunction (Fig. 3 A and B). In particular, mutations in W466 resulted in both a telomere elongation phenotype, as well as cell cycle progression defect, phenotypes that were in fact more severe than that exhibited by a strain expressing a version of Stn1 that was completely deleted for the C terminus. Thus, this in vivo analysis indicates that Stn1 is distinguished from RPA32/Rpa2 through the acquisition of a second wHTH domain that confers a telomere-specific function on Stn1.
The wHTH motifs frequently also exhibit sequence specific dsDNA binding activity. Recognition of dsDNA by wHTH domains is primarily mediated through the “recognition helix,” H3, which in the C-terminal wHTH of Stn1-C is highly basic (Fig. 1C Upper). Despite this similarity, gel shift and NMR studies revealed no detectable binding of the Stn1-C domain to double- or single-stranded, telomeric and nontelomeric DNA oligonucleotides. This result is not particularly surprising, because both RPA32-C (31), a close structural relative of the N-terminal wHTH, and ScpB (32), the closest structural relative to the C-terminal wHTH, also do not bind DNA.
S. pombe Ten1 Contains a Conserved DNA Maintenance Motif.
The crystal structure of the S. pombe Ten1 protein and its structural relationship to RPA14 suggest Ten1 may function in a similar fashion as RPA14. The C-terminal helix of RPA14 serves as an interaction surface for RPA subunit assembly, and notably, this helix is also present in our Ten1 structure. Although the C-terminal helix of Ten1 is involved in crystal packing, the unusual orientation noted for this helix is also observed in the corresponding C-terminal α-helix of RPA14 when bound to RPA32 (35). Also, the C-terminal α-helix of Ten1 is tightly associated with the β-barrel and the irregularly structured N-terminal loop, suggesting the orientation of H2 in Ten1 parallel to the β-barrel is not merely an artifact of crystal packing. Yeast two-hybrid data, colocalization experiments, and ChIP assays with S. pombe Stn1 and Ten1 indicate that these proteins interact with the telomeres of fission yeast (15). Because the S. pombe Stn1 protein is predicted to contain an N-terminal OB fold analogous to that of RPA32, it is likely these proteins interact in a similar manner as RPA14 and RPA32. This idea is supported by the observation of potentially analogous interaction sites in the C-terminal α-helix of Ten1 (I86, Y91, and R98). The presence of an RPA70-like subunit within the S. pombe complex has not been identified.
The structural relationship between Stn1/Ten1 and RPA32/RPA14 implies that telomere-specific proteins have evolved from the DNA DSB repair machinery, suggesting a common mechanism exists for DNA maintenance. This mechanism appears to be conserved throughout eukaryotes, although the specific proteins that perform these functions have diverged, as is evident by the very weak sequence similarity between the S. pombe and S. cerevisiae Ten1 proteins. Regardless of the sequence divergence, these two proteins are predicted to be structural orthologs, and the current data indicate they perform their essential function similarly by interacting with Stn1 proteins through conserved OB fold motifs to protect telomere ends.
Cdc13/Stn1/Ten1 Proteins Are Distantly Related to the RPA Complex.
The relationship between Cdc13, Stn1, and Ten1 and the RPA proteins was previously inferred from structure predictions, genetic experiments, and the demonstration of a direct Stn1/Ten1 interaction (14). The crystallographic studies presented in this work of the C-terminal domain of S. cerevisiae Stn1 and of the S. pombe Ten1 proteins provide direct evidence for an evolutionary relationship between the telomere specific end protection proteins and the heterotrimeric RPA complex. Whereas these proteins have diverged at the sequence level, the conserved structures imply functional preservation, with the specific roles of each complex having evolved to maintain different regions of the chromosome (Fig. 5). This parallel is perhaps most evident in the presence of a second wHTH motif in Stn1-C relative to RPA32. As biochemical data of the capping proteins emerges, it is becoming more evident that the relationship between Cdc13, Stn1, and Ten1 and the RPA proteins extends beyond structure.
Fig. 5.
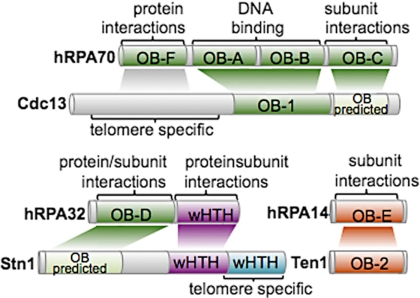
Similarities between the RPA proteins and Cdc13, Stn1, and Ten1 are evident at both the structural and functional levels. The OB fold of Cdc13, like OB-A/B of RPA70, is required for high affinity DNA binding. In vivo data supports the role of the N terminus of Cdc13 being involved in protein interactions, although the structure predictions for this region are uncertain. A second OB fold is predicted at the C-terminal end of Cdc13, much like that in RPA70 (27). An OB fold is also predicted for the N terminus of Stn1. The N-terminal lobe of the C-terminal domain of Stn1 confirms the evolutionary relationship between RPA32 and Stn1-C, whereas the C-terminal lobe has a telomere specific role. The strong structural relationship between Ten1 and RPA14 that this work identified implies this protein, like RPA14, functions as structural support, potentially interacting with Stn1 in a similar manner as RPA14 interacts with RPA32.
Materials and Methods
Protein Expression, Purification, and Crystallography.
Standard cloning and expression techniques were used to obtain purified Stn1-C and Ten1 proteins. Briefly, proteins were individually expressed in Escherichia coli from the pETDUET vector (MCS1) and purified using affinity chromatography and size exclusion chromatography. Optimal crystallization conditions were established using the hanging drop technique. Crystallography datasets were collected on a homesource CuKα X-ray beam as well as on beamline X29A of the National Synchrotron Light Source (Brookhaven National Laboratories). Experimental phasing was achieved via SIRAS using a gold derivative crystal for Stn1-C, and SAD using an iodine derivative crystal for Ten1. All data processing and structure refinement was performed using publicly available software. For detailed protein expression, purification, and crystallization, see SI Materials and Methods.
In Vivo Analysis of stn1− Missense Mutations.
All stn1− plasmids were derived from pVL1492, which contains 3.16 kb of genomic DNA encompassing the STN1 gene in YCplac111. Mutations were introduced using site-directed mutagenesis; constructs were either completely sequenced across the STN1 gene and/or subcloned back into an unmutagenized backbone. Plasmids bearing missense mutations in STN1 were transformed into YVL2394 (MATa stn1-Δ::KANMX6 ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1/pVL1046), and transformants were streaked for single colonies on 5-FOA-containing media at 23 °C, to identify isolates that had lost the covering wild type STN1 plasmid (pVL1046). Strains that had lost pVL1046 but retained the pVL1492 derivative were subsequently propagated on rich media for telomere length, DNA damage sensitivity, and FACS analysis.
Supplementary Material
Acknowledgments.
We thank Dr. David McKay for assistance with crystallography, Dr. Peter Baumann for providing a S. pombe cDNA library, and Sarah Altschuler for valuable comments on the manuscript. This work was supported by the National Science Foundation Grant 0617956 (to D.S.W.), University of Colorado Cancer Center Pilot Award (to D.S.W.), National Institutes of Health (NIH) Grant GM083953 (to R.T.B.), the G. Harold and Leila Y. Mathers Charitable Foundation (to V.L.), and NIH Training Appointment T32 GM-008732 (to A.D.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID code 3K10 (for Stn1-C) and 3K0X (for Ten1)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0909203106/DCSupplemental.
References
- 1.McClintock B. The behavior in successive nuclear divisons of a chromosome broken at meiosis. Proc Natl Acad Sci USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn E. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 4.Grandin N, Charbonneau M. Protection against chromosome degradation at the telomeres. Biochimie. 2008;90:41–59. doi: 10.1016/j.biochi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 6.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 7.Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 9.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 10.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 11.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 12.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 13.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao H, Cervantes R, Mandell E, Otero J, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 15.Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song X, et al. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lustig A. Cdc13 subcomplexes regulate multiple telomere functions. Nat Struct Biol. 2001;8:297–299. doi: 10.1038/86157. [DOI] [PubMed] [Google Scholar]
- 18.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13–1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 23.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 24.Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 25.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 27.Theobald DL, Wuttke DS. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mer G, et al. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 32.Kim JS, et al. Crystal structure of ScpB from Chlorobium tepidum, a protein involved in chromosome partitioning. Proteins. 2006;62:322–328. doi: 10.1002/prot.20751. [DOI] [PubMed] [Google Scholar]
- 33.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng X, et al. Structure of the full-length RPA 14/32 complex gives insights into the mechanism of DNA binding and complex formation. J Mol Biol. 2007;374:865–876. doi: 10.1016/j.jmb.2007.09.074. [DOI] [PubMed] [Google Scholar]
- 36.DeLano W. The PyMOL User's Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.