Abstract
Proteolysis has major roles in diverse biologic processes and regulates the activity, localization, and intracellular levels of proteins. Linking signaling pathways and physiologic processes to specific proteolytic processing events is a major challenge in signal transduction research. Here, we describe N-CLAP (N-terminalomics by chemical labeling of the α-amine of proteins), a general approach for profiling protein N-termini and identifying protein cleavage sites during cellular signaling. In N-CLAP, simple and readily available reagents are used to selectively affinity label the α-amine that characterizes the protein N terminus over the more highly abundant ε-amine on lysine residues. Protein cleavage sites are deduced by identifying the corresponding N-CLAP peptides, which are derived from the N-termini of proteins, including the N-termini of the newly formed polypeptide products of proteolytic cleavage. Through selective affinity purification and tandem mass spectrometry analysis of 278 N-CLAP peptides, we characterized proteolytic cleavage events associated with methionine aminopeptidases and signal peptide peptidases, as well as proteins that are proteolytically cleaved after cisplatin-induced apoptosis. Many of the protein cleavage sites that are elicited during apoptotic signaling are consistent with caspase-dependent cleavage. These data demonstrate the utility of N-CLAP for proteomic profiling of protein cleavage sites that are generated during cellular signaling.
Keywords: caspase, N-CLAP, N-terminalomics, tandem mass spectrometry
Proteolytic processing of proteins is a major regulatory mechanism in signal transduction (1). Proteases regulate signaling pathways by altering protein function. For example, proteases can either convert an inactive proenzyme to an active enzyme or inactivate specific proteins (1). The human genome is predicted to contain 561 proteases or protease-like enzymes, suggesting a potentially broad role for proteases (2). However, in the vast majority of cases, the specific proteins that are targeted by a protease are unknown. Similarly, the proteins and the sites that are cleaved during a signaling event are difficult to identify.
To characterize protein processing on a proteomic scale, one strategy is to systematically profile protein N-termini in tissue or cellular samples. “N-terminalomics” is motivated by the idea that the population of N-termini changes after protease activation. This is because any single protein cleavage event results in the formation of an “internal” N terminus. Proteomic approaches typically use protein samples that have been digested with proteases, such as trypsin, to generate peptides suitable for tandem mass spectrometry (MS/MS) analysis. Selective enrichment of N-terminal peptides (i.e., the peptides derived from the N-termini of proteins) would be required to identify protein N-termini and proteolytic cleavage sites. Thus, there is considerable interest in the development of strategies that selectively enrich for N-terminal peptides before MS analysis.
Several techniques to enrich for N-terminal peptides rely on “negative selection,” in which non–N-terminal peptides are removed from a sample, leaving N-terminal peptides (3–6). However, because most peptides are not N-terminal peptides, the efficiency needs to be very high to obtain a preparation enriched in N-terminal peptides. In contrast, “positive selection” involves the purification of N-terminal peptides from a mixture of peptides. Selective labeling of the N-termini of proteins would allow N-terminal peptides to be readily purified after protease digestion (7, 8). The challenge of positive selection is to selectively recognize the α-amino group, which is present exclusively on the N-termini of proteins, but not the ε-amino group, which is present at high levels in proteins on lysine side chains.
Here we describe a proteomic strategy, N-terminalomics by chemical labeling of the α-amine of proteins (N-CLAP). N-CLAP utilizes features of Edman chemistry to enable the selective labeling and capture of N-terminal peptides from proteins in complex biological samples. We demonstrate that N-CLAP can be used to characterize the diversity of N-terminal processing as well as signal peptidase specificity. Using this method, we also identify protein processing that occurs during apoptosis and identify several unique potential targets of caspase processing. Together, these data demonstrate that N-CLAP can be used to identify cleavage sites in proteins and identify proteolytic cleavage events that occur during cell signaling.
Results
A Chemoselective Strategy for Labeling and Enriching N-Terminal Peptides.
The N terminus of proteins is characterized by an α-amine, as opposed to the ε-amines that are on lysine side chains. The α-amine (the pKa of its ammonium ion ≈8.95) is notable for its slightly reduced pKa relative to that of ε-amines (the pKa of its ammonium ion ≈10.53) (9). However, given this narrow pKa window, there is no pH at which the α-amine can be selectively labeled with an amine-labeling reagent, because ε-amines, albeit less reactive, are typically much more abundant (10). The circumstances are further complicated by the fact that the pKa of the ammonium ion for the α-amine and the ε-amine can be affected by the side chain of the adjacent amino acids, hydrogen bonding, and other intramolecular interactions (11, 12).
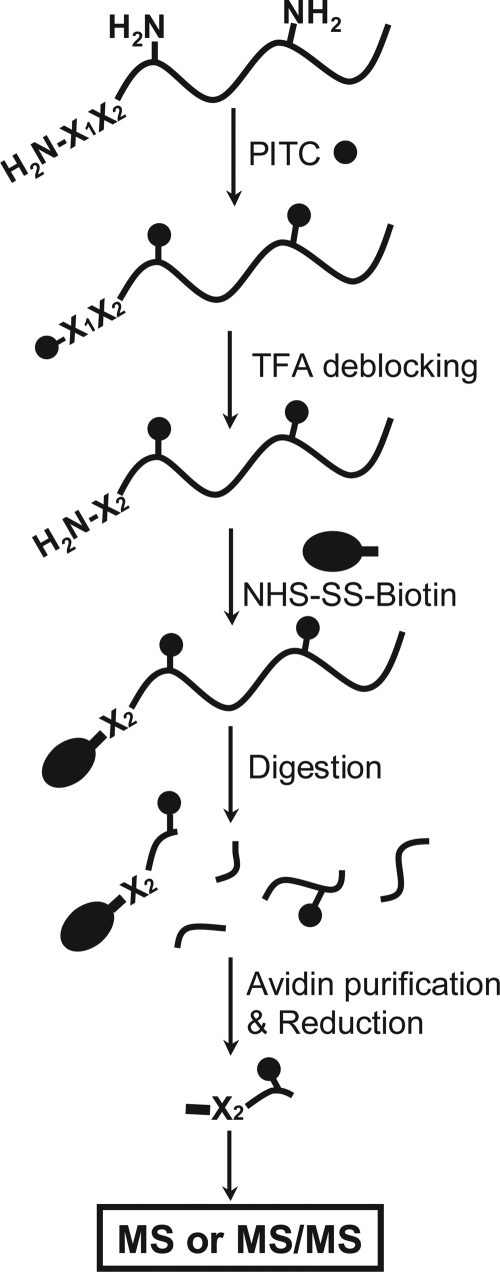
To address these issues, we have developed a chemical labeling strategy, N-CLAP, that chemoselectively labels the N terminus. This labeling strategy is based on the chemistry of Edman degradation, which is used for N-terminal amino acid sequencing (13) [SI Appendix, Fig. S1A]. In N-CLAP, the amine-reactive Edman reagent, phenyl isothiocyanate (PITC), is used to block all of the amines in proteins (Fig. 1). TFA is then used to trigger an intramolecular cyclization of the PITC-modified α-amine, which results in cleavage of the peptide bond between the first and second amino acid. This intramolecular cyclization does not occur for the PITC adduct on the ε-amine because an energetically favorable five-membered ring cannot form at this position (14) and leaves the PITC-modified ε-amine on the lysines intact (15). Therefore, the PITC adducts on the N-termini are selectively “deblocked.” After TFA treatment, the protein is shortened by one amino acid, and importantly, contains only a single α-amine, whereas all lysines remain blocked. The α-amine is then labeled using an amine-reactive tagging reagent, such as a biotinylating agent, resulting in selective tagging of the N terminus of each protein. Proteins are digested, and tagged peptides are purified, for example using avidin agarose. N-terminal peptides are eluted and identified by MS (Fig. 1).
Fig. 1.
Strategy for N-CLAP. After PITC treatment, all amines in a protein are blocked (filled circle). Reaction with TFA selectively deblocks the N terminus but not PITC-modified lysines. The newly generated amino groups at the N-termini are then reacted with an amine-specific labeling reagent, such as EZ-Link Sulfo-NHS-SS-biotin (oval). After protein digestion, the N-terminal peptides are recovered using avidin-based resins, eluted with a reducing agent, and then identified by MS.
We first addressed whether N-terminal peptides generated by N-CLAP would be sufficiently unique to identify parent proteins. The majority (74%) of N-CLAP peptides prepared by virtual trypsin digestion of proteins listed in the Swiss-Prot human protein database (v55.6, July 1, 2008) (16) are at least 5 aa long, a length that may provide enough sequence information for protein identification. Of these peptides, 68% identify a single protein in the database (SI Appendix, Fig. S1 B–D). Most of the peptides that match more than one protein match a family of highly similar isoforms. Thus, for N-CLAP peptides prepared with trypsin, 54% of the human proteome could be unambiguously detected. To increase the coverage, the results from parallel experiments, each using a separate protease, could be merged. For example, using trypsin and Glu-C would increase coverage of the proteome to 96.6% (SI Appendix, Fig. S1E). Thus, a significant fraction of potential N-CLAP peptides would provide unambiguous identification of parent proteins. In some cases, the masses of N-CLAP peptides alone could be used to identify many proteins. This would be most useful when analyzing small proteomes with instrumentation capable of providing highly accurate masses (SI Appendix, Fig. S1F).
Selective Labeling of Peptide N-Termini.
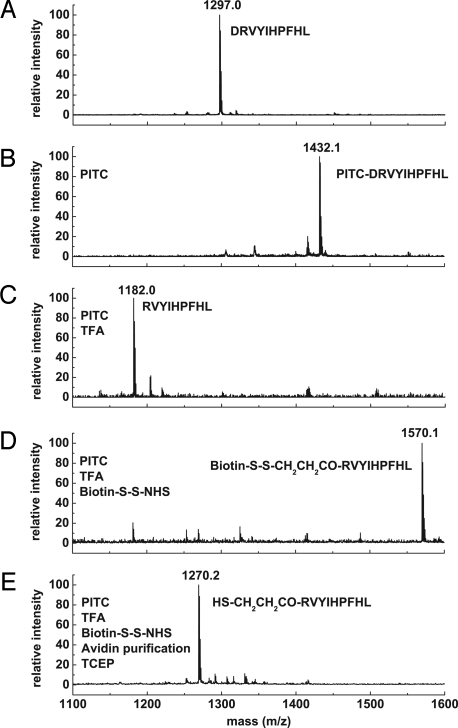
To evaluate N-CLAP, we first characterized the efficiency of each chemical reaction using a model peptide, angiotensin I, which contains a single amine at its N terminus. After reaction of angiotensin I with PITC, the apparent mass increased by the expected 135 Da (Fig. 2 A and B). After TFA treatment, the peptide mass is reduced by 250 Da, which corresponds to the mass of the N-terminal residue, aspartic acid, modified with PITC (Fig. 2C). A mass increase of 388 Da was detected after labeling the peptide with the amine-reactive cleavable labeling reagent EZ-Link Sulfo-NHS-SS-biotin (Fig. 2D). The final step, removal of the biotin by Tris(2-carboxyethyl)phosphine (TCEP)-mediated reduction of the disulfide linker, resulted in the expected loss of 300 Da, corresponding to the removal of the thiol-containing biotin moiety (Fig. 2E). Similar results were obtained with another peptide, derived from β-catenin (SI Appendix, Fig. S2 A–F).
Fig. 2.
Selective and efficient labeling of angiotensin I using N-CLAP. (A) MALDI-TOF-MS of angiotensin I results in a major peptide ion at 1297.0. (B) PITC-treated angiotensin I results in a major peptide ion reflecting the expected thiocarbamoyl PITC modification. (C) TFA treatment of PITC-modified angiotensin I results in a single prominent peptide ion with a mass corresponding to angiotensin I missing the first N-terminal residue. (D) The deblocked angiotensin I was reacted with EZ-Link Sulfo-NHS-SS-biotin. MALDI-TOF-MS revealed a single major peak corresponding to the modified peptide. (E) Avidin-purified and TCEP-eluted sample results in a 1,270.2-Da peptide ion, corresponding to angiotensin I peptide without N-terminal aspartic acid and with a remnant from the cleavable biotin tag. The treatments, peptide sequence, and the chemical modification for each sample are indicated.
We next performed a similar analysis using the c-Myc peptide, which contains both an α-amine and an ε-amine from an internal lysine (SI Appendix, Fig. S2 G–K). As with angiotensin I, the c-Myc peptide was readily modified by PITC, and the first amino acid was readily removed with TFA. Biotinylation resulted in selective labeling of the α-amine, whereas the PITC-blocked ε-amine was not labeled (SI Appendix, Fig. S2J). Notably, the PITC modification did not impair peptide fragmentation, indicating that MS/MS spectra obtained from PITC-modified peptides could be used for peptide identification. Together, these data suggested that each step of N-CLAP can occur with high efficiency and with negligible side reactions.
Selective Enrichment of N-Terminal Peptides from Proteins.
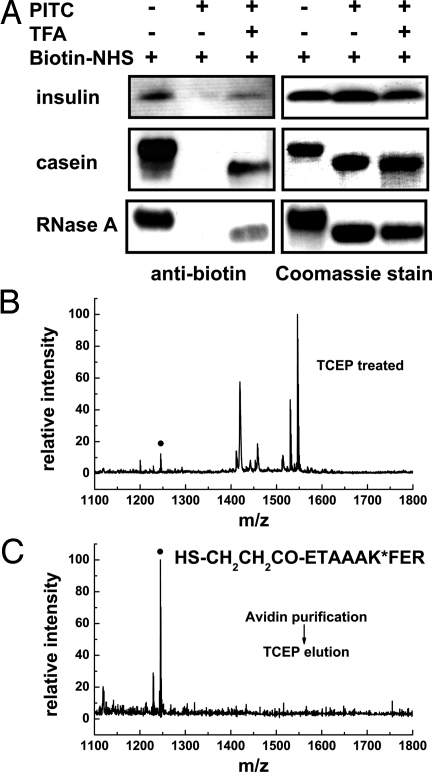
We next applied N-CLAP to individual proteins. To monitor the completeness of each step, an assay for amines in proteins was developed. Amines were labeled with biotin N-hydroxysuccinimide ester (biotin-NHS). Signals obtained after Western blotting with a biotin antibody indicate the presence of unmodified amines. Incubation of insulin, casein, or RNase A with PITC resulted in complete loss of amine reactivity (Fig. 3A). After TFA treatment, amine reactivity was recovered, consistent with selective deblocking of the N terminus. In the case of insulin, the recovered signal is approximately half as intense as the signal for the unmodified protein (Fig. 3A, Top), consistent with the presence of two amines in the heavy chain of insulin, one α-amine and one ε-amine at its sole lysine residue. However, the recovered signal is much weaker for casein and RNase A (Fig. 3A, Middle and Bottom) than that of the original samples, which is expected because these two proteins have many lysines (12 and 7 for casein and RNase A, respectively).
Fig. 3.
Assessment of the N-CLAP for enrichment of the N-terminal peptide from individual proteins. (A) PITC completely modifies amines in proteins, and TFA generates new N-terminal amines. The presence of amines in the unreacted, PITC-treated, and both PITC- and TFA-treated samples (insulin, α-casein, and RNase A) was measured by reaction with biotin-NHS and visualized by Western blotting with a biotin antibody. The band intensity reflects the relative amount of free amines. Coomassie staining confirms comparable loading in each lane. A slight increase in the mobility of PITC-modified proteins is typically detected and likely reflects the reduced positive charge on the protein due to lysine modification. (B) MALDI-TOF-MS spectrum of the tryptic peptides generated after RNase A was subjected to N-CLAP, digested, and reduced (without avidin purification). MALDI-TOF reveals multiple peptides, including a peptide consistent with the N-terminal RNase A N-CLAP peptide (filled circle). (C) N-CLAP enriches for N-terminal peptides. After RNase A is subjected to N-CLAP, the tryptic peptides are purified on avidin agarose and eluted with TCEP. MALDI-TOF-MS spectrum of the TCEP eluate reveals a predominant peak. This peptide corresponds to the N-CLAP peptide from the RNase A N terminus, containing a PITC adduct on an internal lysine (asterisk) and an N-terminal mercaptoethanoate amine adduct (shown in sequence) derived from the cleaved biotinylation reagent.
We next addressed whether the N-terminal tryptic peptide could be enriched using N-CLAP. After PITC and TFA treatment, RNase A was labeled with EZ-Link Sulfo-NHS-SS-biotin. MALDI-TOF-MS of the tryptic digest reduced by TCEP revealed numerous peptides, including a relatively low abundance peak with a mass corresponding to the expected mass of the N-terminal peptide containing a PITC-modified lysine and a mercaptoethanoate (HSCH2CH2CO–) amine adduct that remains after the cleavage of the biotin tag (Fig. 3B). However, after avidin affinity purification, this peak was the predominant peak (Fig. 3C and SI Appendix, Fig. S3). Additionally, a peak representing a peptide in which the PITC modification on lysine is hydrolyzed, which would appear as a 1,196-Da peak, was not detected, confirming the stability of the PITC adduct. Together, these data demonstrate the usefulness of N-CLAP for positive purification of N-terminal peptides.
Characterization of the Diversity of N-Terminal Methionine Processing.
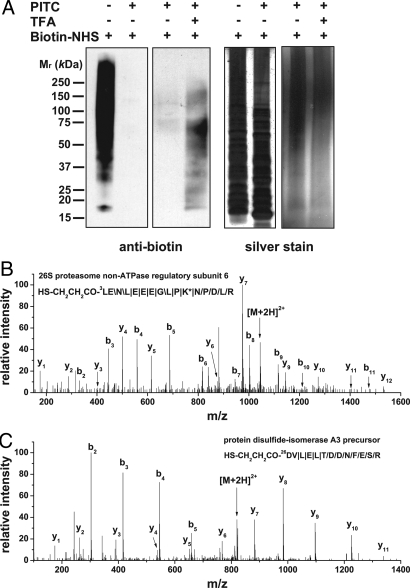
We next examined the ability of N-CLAP to generate N-terminal peptides from cellular lysates. Cell lysate was prepared from human Jurkat T cells and subjected to N-CLAP. As with the individual proteins, PITC effectively blocked the amines in the lysate (Fig. 4A), and TFA resulted in the reappearance of a fraction of the amine reactivity. Amines were biotinylated, proteins were digested, and N-terminal peptides were affinity isolated. A total of 80 N-terminal peptides were identified using liquid chromatography (LC) quadrupole (Q)-TOF MS/MS (SI Appendix, Fig. S4 and Table S1). As an example, the MS/MS spectrum of the N-terminal peptide from 26S proteasome non-ATPase regulatory subunit 6 is shown (Fig. 4B). As with many N-CLAP peptides, it starts at the third amino acid predicted from the coding sequence. This reflects the removal of the original N-terminal methionine by endogenous methionine aminopeptidases (MetAPs) and the loss of the second amino acid during the TFA deblocking step in the N-CLAP procedure. The identity of the residue lost during N-CLAP can be extrapolated from protein sequence databases.
Fig. 4.
Identification of protein N-termini from a complex mixture using N-CLAP. (A) Evaluation of the completeness of PITC and TFA treatment on Jurkat T cell lysate. The amine content of a Jurkat T cell lysate is monitored by Western blotting with a biotin antibody at different stages in the N-CLAP protocol. Note that the exposure time for the right panel is much longer than for the left panel to detect the recovered signal after TFA treatment. Silver staining showed similar loading. (B and C) Representative MS/MS spectra of 2 N-CLAP peptides from Jurkat T cell lysate. The sequence map of the peptides, the N-terminal modification, the position of the first amino acid in the coding sequences, and the protein names are indicated in the spectra. PITC modification on lysine is indicated by an asterisk. In B, the N-CLAP peptide starts from the third amino acid from the coding sequence, which indicates that the initial methionine is removed from the nascent protein. However, in C the N-CLAP peptide starts from the 26th amino acid from the coding sequence, which indicates that the first 24 aa are removed in the nascent protein because the N-CLAP removes one additional N-terminal amino acid. The symbols \, /, and | represent b-ions, y-ions, and both b-ions and y-ions, respectively.
Many of the N-CLAP peptides detected in Jurkat T cell lysate were derived from the N-termini of proteins lacking the N-terminal methionine. However, 27.2% of peptides were derived from proteins that retained the N-terminal methionine. This likely reflects inefficient removal of methionine by MetAPs, as has been described for methionine residues followed by charged or bulky amino acids (17). MS/MS analysis of N-CLAP peptides also indicated that numerous proteins exist in the cytosol as a heterogeneous population, with a fraction containing the N-terminal methionine and another fraction missing the N-terminal methionine. Some proteins (16.2%) were the source of three N-CLAP peptides, reflecting variable trimming of N-terminal residues. These data indicate that the identity of the N-terminal residue can be considerably different from what is predicted by sequence databases.
Determination of Organelle-Targeting Peptide Cleavage Sites Using N-CLAP.
Many N-CLAP peptides were derived from proteins that were processed to remove the first 15 or more N-terminal amino acids (Table S2). Some N-CLAP peptides were derived from transmembrane or secreted proteins, in which signal peptides are removed by signal peptide peptidases (SPPs) (18). Other N-CLAP peptides were derived from mitochondrial proteins, which are processed by mitochondrial processing peptidases (MPPs) to remove the transit peptides (19). As an example, the N-CLAP peptide derived from protein disulfide-isomerase A3 indicates that the first 24 aa of the nascent protein is removed (Fig. 4C), consistent with previous MS results (3).
Signal peptides lack simple consensus sequences can possibly be predicted using computational approaches (20). Because different algorithms predict different cleavage sites, the precise cleavage sites have to be experimentally verified. Signal peptides typically show 3 distinct regions: a positively charged N-terminal region, a hydrophobic region, and a C-terminal uncharged polar region. Examination of N-CLAP peptides (SI Appendix, Fig. S4 and Table S2) indicates that only 57%, 93%, and 28% of signal peptides contain a positively charged N terminus, a hydrophobic region, and a C-terminal uncharged polar region, respectively, and only 14% of signal peptides have all three regions. These results indicate considerable diversity in signal peptides and confirm previous studies reporting that signal peptides do not conform to a simple consensus sequence (20, 21).
All of the N-CLAP–identified putative mitochondrial transit peptides, except one, were enriched in Arg, Ser, and Ala and deficient in negatively charged residues, consistent with proposed characteristics for these sequences (22, 23). Most (68%) have a conserved Arg close to the cleavage site. However, there was also heterogeneity in cleavage sites, even in the same protein. For example, several distinct N-CLAP peptides were identified from ATPase inhibitor, reflecting distinct pools of protein cleaved after the 24th, 25th, and 26th amino acid (Table S2). These noncanonical cleavage events may reflect broad cleavage specificity of MPPs or may reflect additional N-terminal processing by distinct peptidases. More than half (52.6%) of the signal or transit sequences and the cleavage sites identified in our analysis had not been previously experimentally validated, according to the annotation in the Swiss-Prot database (16).
Identification of Caspase Cleavage Sites Using N-CLAP.
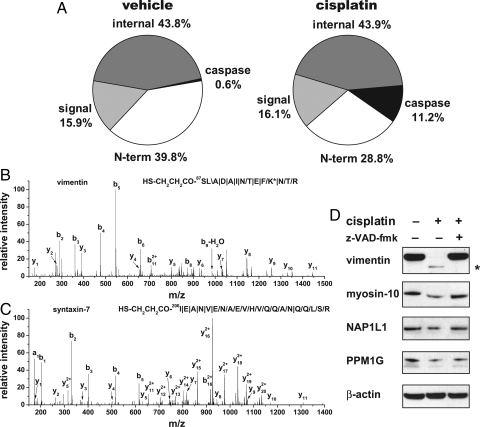
We next examined proteolytic processing events elicited by cisplatin, a chemotherapeutic agent. On the basis of a time course analysis of cisplatin-induced apoptosis in Jurkat T cells (SI Appendix, Fig. S5), cells were harvested after treatment with vehicle or cisplatin for 8 h and then processed using the N-CLAP procedure. The majority of N-CLAP peptides from both vehicle and cisplatin-treated Jurkat T cells were derived from the N terminus of proteins, from internal cleavages, or from the N terminus of proteins that were generated after the removal of signal sequences (Fig. 5A). However, after cisplatin treatment, many additional internal N-terminal peptides were also identified (Fig. 5 B and C), some of which were derived from proteins cleaved after well-established consensus caspase cleavage sites, such as DEXD and (I/L/V)EXD (24). Other proteins were cleaved at sites that did not resemble canonical caspase cleavage sites but occurred after aspartate, the minimal feature for caspase cleavage (SI Appendix, Fig. S4 and Table S3) (7, 24). Among the putative caspase substrates, only 17% had canonical caspase cleavage sites, whereas nearly half (48%) had the consensus sequence of DXXD. These data suggest physiologic utilization of heterogeneous cleavage sites by caspases.
Fig. 5.
Identification of caspase cleavage sites induced by apoptosis by N-CLAP. (A) Distribution of N-CLAP peptides prepared from vehicle- and cisplatin-treated Jurkat T cells. The N-CLAP peptides are classified into 4 categories: (i) N-terminal peptides (N-term), which comprise N-CLAP peptides starting from the 2nd, 3rd, and 4th amino acid from the coding sequence; (ii) N-terminal peptides from proteins with signal or transit peptides removed (signal); (iii) N-terminal peptides derived from internal N-termini of proteins cleaved in a manner consistent with caspase cleavage (caspase); and (iv) N-terminal peptides derived from internal N-termini of proteins cleaved at sites that do not resemble caspase cleavage sites (internal). (B and C) Representative MS/MS spectra of the N-CLAP peptides derived from the internal N-termini of vimentin and syntaxin-7. Jurkat T cells were treated with cisplatin (200 μM for 8 h). (D) Biochemical validation of caspase substrates. Western blot analysis of vimentin, myosin-10, NAP1L1 (nucleosome assembly protein 1-like 1), PPM1G (protein phosphatase 1G), and β-actin, a negative control, was used to assess the caspase dependence of their cleavage after cisplatin treatment. Jurkat T cells were treated with either DMSO, 200 μM cisplatin, or 200 μM cisplatin after 2 h pretreatment with z-VAD-fmk (20 μM). The levels of each of these proteins were reduced after cisplatin treatment, and the effect was blocked by inclusion of the caspase inhibitor. The size of the vimentin cleavage product (43 kDa, asterisk) is consistent with cleavage after Asp-85, which is indicated by the vimentin N-CLAP peptide.
Most of the proteins cleaved after cisplatin treatment were previously identified as caspase targets using various approaches (7, 25, 26). However, some of them, such as protein phosphatase 1G, myosin-10, HERV-H_3q26, and pyridoxal-dependent decarboxylase domain-containing protein 1, have not been previously reported as being cleaved during apoptosis. Overall, 56% of the cleavage sites have not been previously reported. To confirm that these cleavage events were caspase dependent, Jurkat T cells were treated either with vehicle, cisplatin, or cisplatin and z-VAD-fmk, a pan-caspase inhibitor. We selected proteins that included previously reported and newly discovered caspase substrates for validation, in part on the basis of commercial availability of antibodies. In each case, the selected proteins were either completely or partially degraded after the addition of cisplatin, and the cleavage was blocked by the caspase inhibitor, confirming that the cleavage events were indeed caspase dependent (Fig. 5D).
In addition to caspase cleavage sites, additional classes of cleavage sites were identified (SI Appendix, Fig. S4 and Table S4), consistent with findings that cisplatin induces the activation of proteases other than caspases (27). In some cases, N-CLAP identified proteins that had previously been shown to be cleaved during apoptosis, but the specific cleavage sites was not identified (28, 29). Together, these results demonstrate the ability of N-CLAP to identify numerous caspase-dependent and caspase-independent cleavage events after cisplatin treatment.
Discussion
Profiling the N-termini of proteins in complex mixtures is a powerful approach for elucidating proteolysis signaling pathways. Obtaining N-terminal peptides for N-terminalomics is challenging because the α-amine at the protein N terminus is nearly chemically indistinguishable from the ε-amines on lysines. We have developed a strategy for chemoselective labeling of the N terminus, N-CLAP, which has resulted in the identification of 278 N-CLAP peptides in normal and apoptotic Jurkat T cells. N-CLAP provides a straightforward chemical strategy for selective N-terminal labeling of proteins and for enrichment of N-terminal peptides.
Using N-CLAP to characterize protein N-termini in cellular lysates reveals a surprising degree of trimming of the protein N terminus, considering that MetAPs are the major enzymes that have been linked to N-terminal processing. It is not clear whether amino acid cleavage after the removal of the initial methionine is also mediated by MetAPs or whether other aminopeptidases are involved. Conceivably, the differentially processed N-terminal forms of these proteins have different stabilities, due to effects of N-terminal residues on protein stability (30), or may endow the protein with different functions. Similarly, the N-termini of membrane proteins revealed considerable variability, suggesting heterogeneity in the specificity of SPPs and MPPs or secondary protein processing events after initial cleavage.
Characterization of protease processing induced by cisplatin reveals cleavage at numerous different types of sites, including consensus caspase cleavage sites. Many of these sites have been previously described, validating the accuracy of N-CLAP. Additionally, we biochemically validated the caspase dependence of the cleavage of several proteins. In numerous cases, N-CLAP identified proteins that were already known to be cleaved during apoptosis, but the specific sites were not known. Thus, N-CLAP provides a strategy to readily identify cleavage sites in proteins, thereby permitting mutagenesis and the generation of cleavage-resistant proteins for mechanistic studies. Of note, we identified fewer cleavage sites than other studies using N-terminal labeling approaches (7). This likely reflects the limited number of N-CLAP samples subjected to MS and differences in MS instrumentation, peptide identification algorithms, and settings and parameters used for positive identification, as well as different apoptotic stimuli. These differences are unlikely to reflect an intrinsic inability to detect certain peptides that have been processed in the N-CLAP procedure by MS/MS. Indeed, a β-catenin peptide that was not detected in Jurkat T cell lysates in our initial N-CLAP experiments, but was detected in earlier studies (7), is detectable when a synthetic peptides is used (SI Appendix, Fig. S2 A–F).
Another potential application of N-CLAP is to identify the substrates of a specific protease. Comparison of N-CLAP peptides derived from cells expressing different levels of specific proteases or treated with an inhibitor of a protease of interest would reveal protease substrates. These analyses would be facilitated by tagging N-CLAP peptides using standard quantitative proteomic approaches (31).
In addition to monitoring protein processing, N-terminalomics can be used to enhance the sensitivity of quantitative proteomics. The large number of peptides obtained after tryptic digestion can prevent thorough identification of a significant fraction of the proteins in the sample owing to instrument duty cycle, ion suppression, and dynamic range. Although this is partially alleviated using techniques such as isotope-coded affinity tags, in which only cysteine-containing peptides are analyzed (32), N-CLAP can provide further simplification of a peptide mixture by providing a single peptide per protein. Additionally, using this approach, peptide identification can be enhanced by searching databases solely comprising N-terminal peptides (6) against MS/MS spectra, or highly accurate MS spectra when examining small proteomes (Fig. S1).
A major limitation of protocols that add tags to N-terminal amines is that, in some cases, the N terminus of proteins are “blocked” (e.g., acetylated). Although estimates of the frequency of blocked N-termini vary (33), blocked proteins would not be detectable in these approaches because they cannot be labeled on the N terminus. This may be addressed by using enzymes or chemical methods to remove N-terminal blocking groups (34). On the other hand, the pool of proteins that have not yet been blocked may be detectable using N-CLAP, depending on the sensitivity of the MS instrumentation. Indeed, we detected N-terminal peptides from several proteins, such as CDKN2AIP N-terminal-like protein and peptidyl-prolyl isomerase A, which have been reported to be acetylated on the N terminus (16). Similarly, in experiments meant to identify processing events during cellular signaling, the nascent internal N-termini may not be blocked during the time course of an experiment. N-terminal blocking may also not be a significant problem when examining secreted or extracellular proteins, because this process is generally thought to use acetylating agents, such as acetyl-CoA, which are typically cytosolic (35). However, certain N-terminal modifications, such as methylation, are expected to be detectable, on the basis of their known compatibility with Edman degradation (36).
In the N-CLAP procedure, all peptides contain a C-terminal arginine because trypsin does not cleave after the PITC-modified lysines that are generated during the N-CLAP procedure. Thus the y1-ion is always 175 m/z, which can be used to confirm the specificity of peptides identified using the N-CLAP approach. The only exceptions are peptides that derive from the C terminus of proteins, which end in the native C-terminal residue. Interestingly, we also find that many peptides obtained using the N-CLAP methodology have intense b-ions (see Figs. 4 and 5). This feature could be used in database searching to improve the identification of the N-terminal peptides.
Recently, an approach combining gel fractionation, MS, and an innovative bioinformatic analysis has been developed to identify protease substrates during apoptosis (37). This method determines the identity of proteins that are cleaved and is not designed to identify the specific cleavage sites. Strategies for the positive selection of N-terminal peptides and subsequent identification of cleavage sites have previously been described. In one approach, O-methylisourea is used to block ε-amines before α-amine labeling with conventional amine-reactive labeling reagents (8). Although O-methylisourea preferentially modifies the ε-amine of lysines over the α-amine, the reaction may be problematic owing to the absence of absolute specificity for the ε-amine of lysines and the incompletion of the reaction for proteins (8, 38). In a second approach, an engineered enzyme, subtiligase, is used to covalently link a cleavable glycolate ester-derivatized peptide tag to the N-termini of proteins, thereby facilitating the purification and subsequent protease-dependent release of N-terminal peptides (7). However, this approach requires specialized reagents that are not routinely available. An attractive feature of N-CLAP is that it can be performed using widely available and inexpensive laboratory reagents.
Materials and Methods
Sample Preparation and MS Analysis.
Samples were prepared by in-gel trypsin digestion, and N-CLAP peptides were purified using neutravidin agarose. Peptide samples were analyzed either by MALDI-TOF-MS (Applied Biosystems) or by LC/electrospray ionization (ESI) MS/MS on an Agilent 6250 Q TOF MS. Spectrum Mill was used for N-CLAP peptide identification by searching against a Swiss-Prot human database.
The materials used and detailed methods are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Yuliang Ma and Mr. Albert Morrishow at Weill Cornell Medical College (WCMC) Mass Spectrometry Core Facility for their assistance in LC/ESI-MS/MS and MALDI-TOF-MS analysis. Mass spectrometry was performed at the WCMC MS Core Facility using instrumentation supported by National Institutes of Health Grants RR19355 and RR22615. This work was supported by National Institute of Allergy and Infectious Diseases Grant AI068639 (to S.R.J.), the Dorothy Rodbell Sarcoma Foundation (S.R.J.), and Training Grant T32CA062948 from the National Cancer Institute (to G.X. and S.Y.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908958106/DCSupplemental.
References
- 1.Ehrmann M, Clausen T. Proteolysis as a regulatory mechanism. Annu Rev Genet. 2004;38:709–724. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- 2.Puente XS, Lopez-Otin C. A genomic analysis of rat proteases and protease inhibitors. Genome Res. 2004;14:609–622. doi: 10.1101/gr.1946304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gevaert K, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 4.Staes A, et al. Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC) Proteomics. 2008;8:1362–1370. doi: 10.1002/pmic.200700950. [DOI] [PubMed] [Google Scholar]
- 5.McDonald L, Robertson DH, Hurst JL, Beynon RJ. Positional proteomics: Selective recovery and analysis of N-terminal proteolytic peptides. Nat Methods. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- 6.McDonald L, Beynon RJ. Positional proteomics: Preparation of amino-terminal peptides as a strategy for proteome simplification and characterization. Nat Protoc. 2006;1:1790–1798. doi: 10.1038/nprot.2006.317. [DOI] [PubMed] [Google Scholar]
- 7.Mahrus S, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timmer JC, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SA. Quantitation of amino acids as 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate derivatives. In: Molnár-Perl I, editor. Quantitation of Amino Acids and Amines by Chromatography: Methods and Protocols. Vol 70. Amsterdam: Elsevier; 2005. p. 246. [Google Scholar]
- 10.Strickberger MW. Evolution. 3rd Ed. Sudbury, MA: Jones and Bartlett; 2000. [Google Scholar]
- 11.Stites WE, Gittis AG, Lattman EE, Shortle D. In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core. J Mol Biol. 1991;221:7–14. doi: 10.1016/0022-2836(91)80195-z. [DOI] [PubMed] [Google Scholar]
- 12.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 13.Edman P. Mechanism of the phenyl isothiocyanate degradation of peptides. Nature. 1956;177:667–668. [Google Scholar]
- 14.Bailey JM. Chemical methods of protein sequence analysis. J Chromatogr A. 1995;705:47–65. doi: 10.1016/0021-9673(94)01250-i. [DOI] [PubMed] [Google Scholar]
- 15.Jay DG. A general procedure for the end labeling of proteins and positioning of amino acids in the sequence. J Biol Chem. 1984;259:15572–15578. [PubMed] [Google Scholar]
- 16.Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JY, et al. Mutations at the S1 sites of methionine aminopeptidases from Escherichia coli and Homo sapiens reveal the residues critical for substrate specificity. J Biol Chem. 2004;279:21128–21134. doi: 10.1074/jbc.M401679200. [DOI] [PubMed] [Google Scholar]
- 18.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 19.Schneider H, Arretz M, Wachter E, Neupert W. Matrix processing peptidase of mitochondria. Structure-function relationships. J Biol Chem. 1990;265:9881–9887. [PubMed] [Google Scholar]
- 20.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 21.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 22.Schneider G, et al. Feature-extraction from endopeptidase cleavage sites in mitochondrial targeting peptides. Proteins. 1998;30:49–60. [PubMed] [Google Scholar]
- 23.Emanuelsson O, von Heijne G, Schneider G. Analysis and prediction of mitochondrial targeting peptides. Methods Cell Biol. 2001;65:175–187. doi: 10.1016/s0091-679x(01)65011-8. [DOI] [PubMed] [Google Scholar]
- 24.Thornberry NA, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 25.Van Damme P, et al. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- 26.Wu YH, Shih SF, Lin JY. Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. J Biol Chem. 2004;279:19264–19275. doi: 10.1074/jbc.M307049200. [DOI] [PubMed] [Google Scholar]
- 27.Pabla N, Dong Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008;73:994–1007. doi: 10.1038/sj.ki.5002786. [DOI] [PubMed] [Google Scholar]
- 28.Yuan X, et al. Nuclear protein profiling of Jurkat cells during heat stress-induced apoptosis by 2-DE and MS/MS. Electrophoresis. 2007;28:2018–2026. doi: 10.1002/elps.200600821. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt F, et al. Quantitative proteome analysis of cisplatin-induced apoptotic Jurkat T cells by stable isotope labeling with amino acids in cell culture, SDS-PAGE, and LC-MALDI-TOF/TOF MS. Electrophoresis. 2007;28:4359–4368. doi: 10.1002/elps.200700119. [DOI] [PubMed] [Google Scholar]
- 30.Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- 31.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 32.Gygi SP, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 33.Meinnel T, Peynot P, Giglione C. Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie. 2005;87:701–712. doi: 10.1016/j.biochi.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 34.Hirano H, Kamp RM. Deblocking of N-terminally modified proteins. Methods Mol Biol. 2003;211:355–363. doi: 10.1385/1-59259-342-9:355. [DOI] [PubMed] [Google Scholar]
- 35.Polevoda B, Sherman F. Nα-terminal acetylation of eukaryotic proteins. J Biol Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 36.Chen R, Brosius J, Wittmann-Liebold B. Occurrence of methylated amino acids as N-termini of proteins from Escherichia coli ribosomes. J Mol Biol. 1977;111:173–181. doi: 10.1016/s0022-2836(77)80121-6. [DOI] [PubMed] [Google Scholar]
- 37.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen LA. Group-specific reagents in protein chemistry. Annu Rev Biochem. 1968;37:695–726. doi: 10.1146/annurev.bi.37.070168.003403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.