Abstract
Natural killer T cells expressing an invariant T-cell receptor (iNKT) regulate activation of both innate and adaptive immunity in many contexts. iNKT cells accumulate in the liver and rapidly produce prodigious amounts of numerous cytokines upon activation, impacting the immune response to viral infection, immunosurveillance for malignant cells, and liver regeneration. However, little is known about the factors controlling iNKT homeostasis, survival and hepatic localization. Here, we report that the absence of the transcriptional regulator Id2 resulted in a severe, intrinsic defect in the accumulation of hepatic iNKT cells. Id2-deficient iNKT cells showed increased cell death in the liver, although migration and functional activity were not impaired in comparison to Id2-expressing iNKT cells. Id2-deficient iNKT cells exhibited diminished expression of CXCR6, a critical determinant of iNKT cell accumulation in the liver, and of the anti-apoptotic molecules bcl-2 and bcl-XL, compared to Id2-sufficient iNKT cells. Furthermore, survival and accumulation of iNKT cells lacking Id2 expression was rescued by deficiency in bim, a key pro-apoptotic molecule. Thus, Id2 was necessary to establish a hepatic iNKT cell population, defining a role for Id2 and implicating the Id targets, E protein transcription factors, in the regulation of iNKT cell homeostasis.
Keywords: E protein, homeostasis, Inhibitor of DNA binding, transcription factor
Natural killer T (NKT) cells are a distinct T lymphocyte lineage that are rapidly activated to produce cytokines that influence many cell types either upon recognition of antigen or in inflammatory settings, allowing them to functionally link the innate and adaptive immune responses (1–3). NKT cells develop in the thymus from CD4+CD8+ progenitors and pass through three developmental stages defined by expression of CD44 and NK1.1 (1, 2). The last maturation step for many NKT cells involves up-regulation of NK1.1 and usually occurs in the periphery after CD44hi NK1.1− NKT cells have exited the thymus (1, 2). During maturation, NKT cells up-regulate expression of many markers of T cell activation (including CD44, CD69, and CD122) and receptors normally expressed by NK cells (such as KLRG1 and NK1.1) (3). The NKT cell population can be divided into three main subsets: Type I are the well-studied Vα14 invariant population (iNKT); Type II include CD1d-reactive NKT cells with diverse non-Vα14 T cell receptors; and Type III are CD1d-independent NKT cells (1, 2, 4). The majority of iNKT cells are activated through interaction of their TCR with glycolipid antigens, such as alpha-galactosylceramide (αGalCer), presented by the MHC class I-like molecule CD1d on antigen presenting cells and can be detected with αGalCer-loaded CD1d tetramers (1, 3, 5). Several of these glycolipids are derived from microorganisms and mediate iNKT cell activation during infection (6). Upon activation, iNKT cells rapidly secrete a diverse set of cytokines representative of multiple CD4+ helper T cell subsets, notably including IFN-γ and IL-4, and therefore influence a wide range of immune responses and disease states (3).
Many signaling molecules, transcription factors, cytokines, and chemokines are involved in iNKT cell development, survival, and trafficking (1). Notably, mutations in the transcription factor T-bet and IL-15 both disrupt iNKT cells late in their maturation coinciding with their acquisition of effector functions (7, 8). Mature iNKT cells show a pattern of localization distinct from other T cells and are found preferentially in the liver as well as spleen, bone marrow, and thymus, but less so in the lymph nodes (5). CXCR3 and its ligand, CXCL9, are particularly important for iNKT cell trafficking to the periphery from the thymus (9). The chemokine receptor, CXCR6, and its transmembrane ligand, CXCL16, have been shown to be important for iNKT cell accumulation in the liver, either by affecting maturation or provision of an essential survival signal (10–13). LFA-1, a member of the β2 integrin family of adhesion molecules, has also been shown to influence accumulation of iNKT cells in liver (14, 15). However, which specific signals preferentially recruit iNKT cells to the liver or support their survival and maturation are not well-elucidated.
We noted that: 1) iNKT cells share phenotypic and functional properties with CD8+ memory T cells and NK cells, 2) T-bet and IL-15 are required for NK, CD8+ T cell and iNKT cell maturation as each cell type acquires effector function, and 3) deficiency in the transcriptional regulator Inhibitor of DNA-binding-2 (Id2) results in a development block in the transition from NK precursor to a mature NK cells and in CD8+ T cell effector/memory formation at a similar point in maturation (16–18). Thus, we hypothesized that Id2 would also regulate iNKT cell homeostasis/maturation.
The E/Id protein family of transcriptional regulators has been implicated in many aspects of lymphocyte development (19–21). E proteins are basic helix-loop-helix transcriptional activators/repressors that regulate lymphocyte development by binding to DNA at E-box sites, regulating expression of genes crucial to developmental progression and enforcing key developmental checkpoints (19–21). E protein DNA-binding activity can be negatively regulated by Id proteins, which heterodimerize with E proteins and prevent binding to target sequences (19). One of the Id proteins, Id2, has been shown to be crucial for development of multiple immune cell types (16–18). Id2-deficient mice lack Peyer's patches, peripheral lymph nodes, mature NK cells, and show diminished numbers of CD8α+ dendritic cells, TCR αβ IELs, and Langerhans cells (16–18). Recently, we reported that Id2 plays key role in regulating the CD8+ T cell response to infection, where Id2-deficiency resulted in increased apoptosis of effector cells and reduced formation of CD8+ memory T cells (17). Id2 mRNA was found to be expressed at an approximately 5-fold higher level in mature iNKT cells compared to CD4+ T cells, suggesting that it may function in this cell type (22). Despite the many known effects of Id2 on lymphoid cells, its function in iNKT cells was not known.
Results and Discussion
Id2-Deficient NKT Cells Fail To Accumulate in Liver and Bone Marrow.
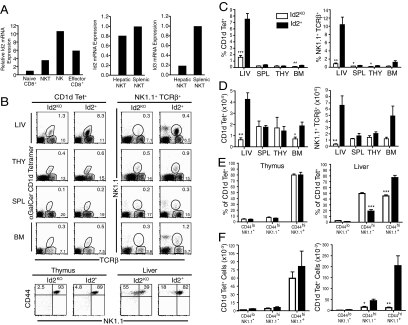
We compared the levels of Id2 mRNA expression by quantitiative PCR (qPCR) among purified populations of naive and effector CD8+ T cells, NK cells, and total NK1.1+TCRβ+ NKT cells sorted from spleen. Id2 mRNA expression was elevated in NK, NKT, and effector cells compared to naïve CD8+ T cells (Fig. 1A). As Id2 is a key factor in terminal maturation of NK cells and survival of effector CD8+ T cells (16–18, 23), the high mRNA expression in NKT cells suggested that it could also influence NKT cell development or survival. Interestingly when we compared Id2 mRNA levels between hepatic and splenic NKT cells, expression was similar between the two populations, while Id3 mRNA was dramatically lower in hepatic NKT populations (Fig. 1A). Thus, mRNA expression suggested that Id2 would be the relevant Id family member for hepatic NKT cells.
Fig. 1.
Id2KO chimeras have fewer NKT cells in the liver and bone marrow. (A) qPCR was performed on sorted populations of naïve CD8+ T cells, NK cells, NK1.1+ TCRβ+ NKT cells, and effector CD8+ T cells (Kb-OVAp+) on day 7 of infection with Lm-OVA. Data are representative of two independent experiments. (B) Representative flow cytometry plots identifying NKT cells as CD45.2+ NK1.1+TCRβ+ or CD1d tetramer+TCRβ+ lymphocytes from indicated tissues of Id2KO or Id2+ chimeras (upper). Representative flow cytometry plots showing CD44 and NK1.1 expression by CD1d tetramer+ Id2KO or Id2+ NKT cells from thymus and liver (lower). Data are representative of all Id2KO and Id2+ chimeras analyzed in at least four experiments. (C) Average (± SEM) percentage of NK1.1+ TCRβ+ or CD1d tetramer+ TCRβ+ NKT cells among donor-gated lymphocytes recovered from indicated tissues harvested from 6–14 Id2KO and Id2+ chimeras. (D) Absolute numbers of NK1.1+ TCRβ+ or CD1d tetramer+ NKT cells recovered from indicated tissues of Id2KO and Id2+ chimeras. Data are the average (± SEM) of at least four Id2KO and Id2+ pairs of chimeras. The P value for the NK1.1+ TCRβ+ percentages and absolute cell numbers was P = 0.06 for bone marrow. (E) Percent and (F) total number of cells for indicated developmental stages of NKT cells determined by assessing expression of NK1.1 and CD44 on TCRβ+CD1d tetramer+ lymphocytes recovered from chimeras. The average (± SEM) of at least five Id2KO and Id2+ chimeras analyzed in two experiments are graphed. Statistical significance was determined using unpaired two-tailed t-test where *, P < 0.05, **, P < 0.005, ***, P < 0.0005.
To determine the impact of Id2 deficiency on the NKT cell compartment, we analyzed fetal liver or bone marrow chimeras reconstituted with either Id2-deficient (Id2KO) or Id2-sufficent (Id2+) hematopoietic cells. We generated chimeras to study the hematopoietic system as Id2-deficiency leads to severe runting and neonatal death, perhaps due to defects in development of adipose tissue (17, 24). Examination of Id2+/− and Id2+/+ reconstituted chimeras showed no differences in their NKT populations (Fig. S1), thus experiments included both compared to Id2KO chimeras. NKT cell populations were identified by expression of the donor congenic marker (CD45.2), NK1.1, and TCRβ (referred to as NKT), or by staining with CD1d tetramers (3), which identify the NKT cell subset using the canonical Vα14 TCR (referred to as iNKT). It should be noted that the NK1.1+TCRβ+ NKT population identifies the mature cells within all three NKT subsets while CD1d tetramer identifies NK1.1+/− type I iNKT cells and a portion of the less abundant type II subset. Chimeras reconstituted with Id2KO donor cells showed a striking reduction in the percentage the iNKT subset in the liver and bone marrow compared to controls receiving Id2+ donor cells, while a decreased percentage for total NKT cells (NK1.1+TCRβ+) was observed in all tissues (Fig. 1 B and C). We observed the most severe defect in the liver, where the percentage of total NKT cells recovered from mice reconstituted with Id2KO cells was approximately 10-fold lower than in chimeras that received Id2+ cells (Fig. 1C) and where the percentage of the iNKT cell population was reduced approximately 5-fold (Fig. 1C). Similarly, analysis of absolute NKT cell numbers revealed an approximately 20-fold reduction of NK1.1+TCRβ+ NKT cells and an approximately 6-fold reduction in CD1d tetramer+ iNKT cells recovered from livers of chimeras that received Id2KO compared to Id2+ donor cells (Fig. 1D). In bone marrow, we found a significantly lower frequency and total number of CD1d tetramer+ iNKT cells in Id2KO compared to Id2WT reconstituted recipients; a similar but not statistically significant trend was observed for NK1.1+TCRβ+ NKT cells (Fig. 1 C and D). While significant differences in frequency of NK1.1+TCRβ+ NKT cells were observed in the thymus and spleen, this was not maintained the analysis of absolute cell numbers (Fig. 1 C and D). As previously reported (16–18, 23), the percentage of TCRβ+ NK1.1− T cells (Fig. 1B) or CD4+ and CD8+ T cells (Fig. S2) was not altered by Id2-deficiency in any of the tissues examined, but the NK cell population (NK1.1+TCRβ−) was absent (Fig. 1B). Given the consistent and dramatic effect resulting from the loss of Id2 expression on hepatic NKT cells, we focused primarily on this population.
The final maturation step for many NKT cells is indicated by up-regulation of NK1.1 expression and can be completed after cells leave the thymus (1–3). To investigate the possibility of an Id2-mediated defect in NKT cell development, the percentages of mature and immature NKT cells, assessed by CD44 and NK1.1 expression, among hepatic and thymic iNKT cells from Id2KO and Id2+ chimeras were analyzed. There were no significant differences in the percentages or absolute numbers of thymic iNKT cells at any stage of development between Id2KO and Id2WT cells, indicating that early development of iNKT cells in the thymus was not affected by loss of Id2 (Fig. 1E). In contrast, examination of the iNKT cell subsets in the liver revealed a lower percentage of mature CD44hi NK1.1+ iNKT cells and a corresponding 2-fold increase in the percentage of immature CD44hi NK1.1− iNKT cells in the Id2KO chimera (Fig. 1E). However, a dramatic loss in the absolute number of cells was apparent for both CD44hi NK1.1− and CD44hi NK1.1+ iNKT populations, indicating that a failure to accumulate iNKT cells was not likely due simply to impaired up-regulation/expression of NK1.1 as the NK1.1− precursors were also diminished (Fig. 1E).
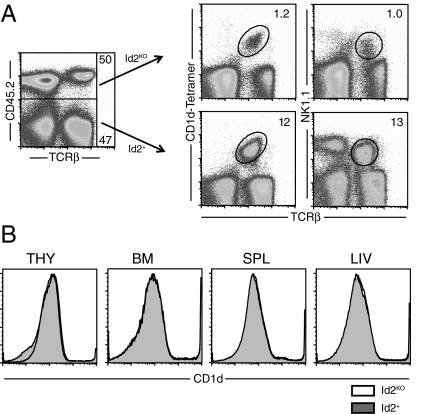
Importantly, we also found that the defective accumulation of Id2KO iNKT cells was not rescued in mixed chimeras where Id2+ donor cells were also present (Fig. 2A). Here, even when progenitors were mixed and allowed to reconstitute congenically distinct recipients, the Id2KO donor cells gave rise to approximately 10- to 12-fold fewer iNKT cells compared to the Id2+ cells within the same recipient. Thus, the presence of Id2+ cells did not restore the NKT cell population, highlighting an intrinsic role for Id2 in supporting the formation of an iNKT cell population. Furthermore, we did not observe any differences in the expression of CD1d, a molecule important for NKT cell maturation, between the Id2KO and Id2+ donor cells in thymus, bone marrow, spleen, or liver (Fig. 2B).
Fig. 2.
Id2KO NKT cell defect is intrinsic. Analysis of Id2KO/Id2WT mixed chimeras. (A) Id2KO (CD45.2) and Id2WT (CD45.1) NKT cells were isolated from livers of mixed chimeras and identified by expression of congenic marker, NK1.1, TCRβ, or CD1d tetramer. (B) CD1d surface staining of lymphocytes from thymus, bone marrow, spleen or liver from Id2KO or Id2WT mixed chimeras; Id2KO unfilled and Id2WT shaded. Data are representative of at least two independent experiments with n = 3–4 Id2KO or Id2WT pairs.
Id2KO NKT Cells Produce Cytokines upon Activation and Display an Activated Phenotype.
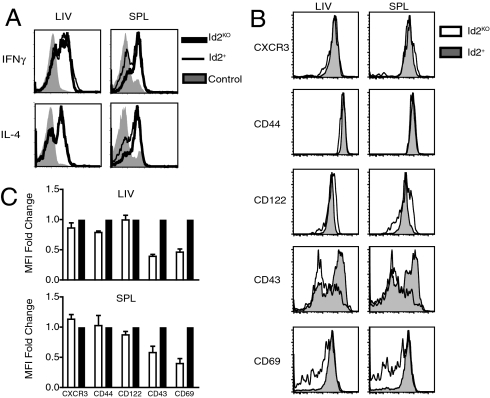
To determine if the few Id2KO NKT cells that were generated could function upon activation, we injected Id2KO or Id2+ reconstituted chimeras with 2 μg αGalCer. Mice were killed 1.5 h after injection, at which point IFNγ and IL-4 production by hepatic and splenic iNKT cells was assessed using intracellular cytokine staining and FACS analysis. Id2KO iNKT cells produced similar levels of cytokines after activation compared to their Id2+ counterparts (Fig. 3A). iNKT cells also express many cell-surface markers characteristic of activated/memory T cells (CD69+, CD44hi, and CD122+) (1–3). To characterize Id2-deficient iNKT cells further, expression of indicated phenotypic markers by mature iNKT cells was assessed. Id2KO iNKT cells and their wild-type counterparts expressed similar levels of CD122 (IL-2Rβ), CD44, and CXCR3 (Fig. 3B). However, we observed lower levels of CD69 and CD43, which may indicate partially impaired activation and/or maturation (Fig. 3B). Thus, the small numbers of mature Id2KO iNKT cells that can be identified in the liver have some of the phenotypic markers of maturation and can respond to stimulation with αGalCer in vivo.
Fig. 3.
Phenotype and function of Id2-deficient NKT cells. (A) Intracellular cytokine staining was used to detect IFN-γ and IL-4 expression by NKT cells harvested from Id2KO or Id2+ chimeras that were given 2 μg αGalCer i.p. 1.5 h before sacrifice. NKT cells from Id2KO chimeras (thick black line), Id2+ chimeras (thin black line), or unstimulated control B6 mice (gray shaded) were identified by CD45.2+ TCRβ+ NK1.1+ expression. Data are representative of three independent experiments with n = 2 Id2KO and Id2+ pairs. (B) Representative flow cytometry plots displaying expression of indicated cell surface molecules by lymphocytes recovered from spleen and liver of Id2KO (unfilled) or Id2+ (shaded) donor cells obtained from chimeras. Mature NKT cells were identified by CD45.2, CD1d tetramer and NK1.1 expression. (C) Average (± SEM) fold change of geometric mean fluorescence intensity between Id2KO and Id2WT iNKT cells for indicated activation marker. Data are representative of four pairs of Id2KO and Id2+ chimeras analyzed in two independent experiments.
Id2KO NKT Cells Undergo Increased Apoptosis in Liver but Show Normal Migration.
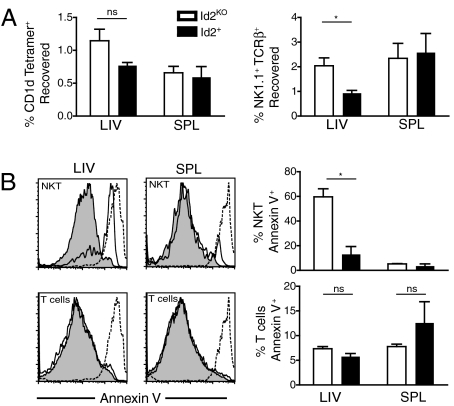
To assess whether Id2KO iNKT cells failed to accumulate in the liver due to impaired migration to this tissue, we transferred CD45.2+ B220−CD8− splenocytes from Id2KO or Id2+ chimeras into CD45.1+ host mice and measured the percentage of donor NKT cells recovered in the spleen and liver after 24 h. Whether identifying the NKT population by expression of NK1.1 and TCRβ or by CD1d tetramer, comparable to increased percentages Id2KO and Id2+ NKT cells were recovered from spleens and livers of host mice, indicating that trafficking to the peripheral organs by the Id2KO iNKT cells was not impaired and was perhaps enhanced (Fig. 4A).
Fig. 4.
Absence of Id2 does not affect NKT cells migration, but results in increased apoptosis. (A) Id2KO and Id2+ NKT cells were isolated from livers and spleens of host mice and identified by expression of congenic marker (CD45.2+), NK1.1, and TCRβ, or CD1d tetramer. Data shown is average (± SEM), (n = 6–7 for NK1.1+ TCRβ+ migration assay, n = 3 for CD1d tetramer+ migration assay). (B) Lymphocytes were isolated from Id2KO and Id2+ chimeras and were stained with Annexin V and analyzed by flow cytometry. (Left) Representative plots for donor gated Id2KO (unfilled) or Id2+ (shaded) NK1.1+TCRβ+ NKT cells or NK1.1−TCRβ+ T cells or dead cells (dotted line). (Right) Average (± SEM) of % Annexin V positive cells (n = 3). Data are representative of three independent experiments each with n = 2–3 Id2KO and Id2+ pairs. Statistical significance was determined using unpaired two-tailed t-test where *, P < 0.05.
The level of apoptosis among NKT cells was next evaluated; lymphocytes from liver and spleen were harvested from Id2KO or Id2+ chimeras and stained for Annexin V. More Id2KO than Id2+ hepatic NK1.1+TCRβ+ cells stained Annexin V+ (≈60% vs. ≈15%), while no differences were observed between the splenic populations or between conventional T cells in liver or spleen, indicating that many of the hepatic Id2KO NKT cells were undergoing apoptosis (Fig. 4B). Normal cytokine production by Id2KO NKT cells (Fig. 3) in the context of dramatically higher cell death is perhaps surprising; however, activation with αGalCer may rescue or accelerate cell death, leaving only functional cells to assay. It was not possible to measure Annexin V staining for iNKT cells as CD1d tetramer staining was lost with the conditions required for Annexin V binding. These results indicated that the lower percentage of iNKT cells observed in the livers of Id2KO chimeras was due to impaired survival.
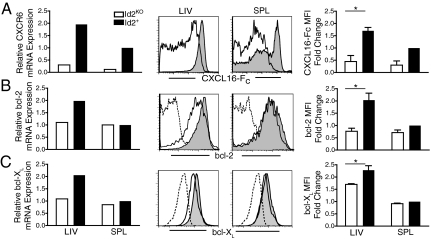
Id2KO NKT Cells Have Diminished Expression of CXCR6, bcl-2, and bcl-XL.
CXCR6, a chemokine receptor, and its ligand, CXCL16, are essential for iNKT cell accumulation in the liver (13, 14). CXCR6 is expressed at high levels by resting iNKT cells and at intermediate levels by effector and memory T cells (22). To determine if Id2 deficiency affected CXCR6 expression by NKT cells, we first analyzed CXCR6 mRNA by qPCR, comparing expression by NKT cells from Id2KO or Id2+ reconstituted chimeras. Both hepatic and splenic Id2KO iNKT cells showed an approximately 8- to 10-fold reduction in CXCR6 mRNA expression compared to Id2+ iNKT cells (Fig. 5A, left), suggesting that Id2 influences CXCR6 expression in this cell type. We next examined surface expression of the CXCR6 protein using a Fc fusion protein of its ligand, CXCL16-Fc, as described previously (12). NKT cells from Id2KO chimeras showed dramatically reduced levels of CXCL16-Fc binding in both spleen and liver (Fig. 5A, right), while the total conventional T cell population expressed low levels of CXCR6 that were slightly lower among Id2KO cells (Fig. S3A). As CXCR6 has been suggested to promote survival of hepatic iNKT cells, Id2KO NKT cells may fail to receive an essential survival signal due to lower CXCR6 expression (10, 11).
Fig. 5.
Hepatic NKT cells in Id2KO chimeras have diminished expression of CXCR6, bcl-2 and bcl-XL. Relative mRNA and protein expression of (A) CXCR6, (B) bcl-2, and (C) bcl-XL. qPCR was performed on sorted NKT cells from Id2KO or Id2+ chimeras based on expression of CD45.2, NK1.1, and TCRβ. CXCR6, bcl-2, and bcl-XL mRNA expression for each sample was normalized to splenic Id2+ NKT cells. (Left) qPCR data are representative of two independent sorts with n = 3–4 chimeras. (Center) For flow cytometry NKT cells were identified by expression of congenic marker, NK1.1, and TCRβ. Isotype controls dashed lines; Id2KO unfilled; Id2+ shaded. Flow cytometry data are representative of at least four pairs of Id2KO and Id2+ chimeras. (Right) Average (± SEM) fold change of geometric mean fluorescence intensity between Id2KO and Id2WT NKT cells. Statistical significance was determined using unpaired two-tailed t-test where *, P < 0.05.
However, reduced CXCR6 expression does not provide a complete explanation for the Id2KO iNKT cell defect as CXCR6-and CXCL16-deficient mice show a less severe loss of NKT cells (10, 11) (Fig. S4). In our study of Id2KO CD8+ effector cells, we also observed a defect in survival that correlated with diminished bcl-2 expression and enhanced bim expression (17). Further, bim has been implicated in negatively regulating survival of activated iNKT cells (25). To determine if Id2 deficiency affected expression of bcl-2 family members in iNKT cells, we used qPCR to compare the relative mRNA levels of a panel of pro- and anti-survival molecules (17). We found that bcl-2 and bcl-XL mRNA levels within sorted splenic and hepatic Id2KO and Id2+ NKT cells were consistently 2- to 3-fold decreased by hepatic Id2KO but not splenic Id2KO NKT cells when compared to Id2WT cells (Fig. 5 B and C, left). We further confirmed, by intracellular staining, that down-regulation of bcl-2 and bcl-XL protein expression by Id2KO cells was liver-specific (Fig. 5 B and C, right) and NKT cell specific, as conventional Id2KO T cells showed similar expression to Id2WT T cells (Fig. S3). Interestingly, CXCR6-deficient hepatic NKT cells expressed lower levels of intracellular bcl-2 protein, but displayed normal levels of bcl-XL (Fig. S4), further suggesting that Id2 regulates factors in addition to CXCR6 to support NKT cell survival. The preferential down-regulation of these two anti-apoptotic molecules in liver provide a mechanistic explanation for the liver-specific defect found for Id2KO NKT cells.
Bim Deficiency Rescues Id2KO NKT Cell Defect.
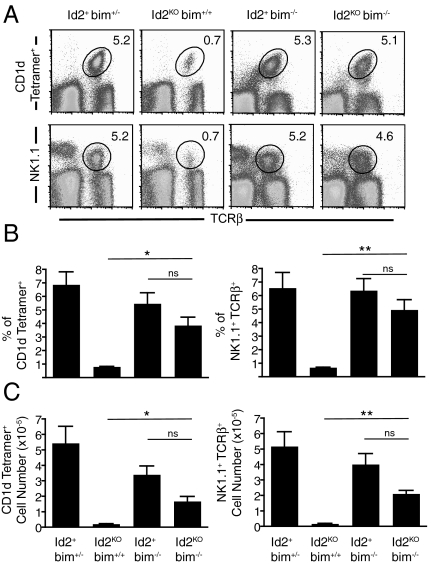
To determine if the lower levels of anti-apoptotic molecules was the cause of the Id2KO iNKT cell defect, we tested if inhibition of the mitochondrial pathway of apoptosis through loss of bim could rescue the phenotype. We generated chimeras reconstituted with Id2WTbim+/−, Id2KObim+/+, Id2+bim−/−, or Id2KObim−/− fetal liver cells. Chimeras reconstituted with Id2KObim−/− donor cells showed a dramatic recovery of the iNKT cell population in the liver compared to chimeras receiving Id2KObim+/+ donor cells (Fig. 6). The percentage of CD1d tetramer+ iNKT and NK1.1+TCRβ+ NKT cells recovered from mice reconstituted with Id2KObim−/− cells was approximately 4- to 5-fold higher than in chimeras that received Id2KObim+/+ cells (Fig. 6 A and B). Similarly, analysis of absolute NKT cell numbers revealed an approximately 8-fold increase in both NK1.1+TCRβ+ NKT cells and CD1d tetramer+ iNKT cells recovered from livers of chimeras that received Id2KObim−/− donor cells compared to Id2KObim+/+ donor cells (Fig. 6C). However, the number of donor iNKT cells in the thymus and spleen of recipients that received bim-deficient donor cells was not significantly altered compared recipients that received Id2WT donor cells, suggesting the rescue is not due to increased thymic output in the absence of bim (Fig. S5). The minor difference in percentage and number between Id2KObim−/− and Id2+bim−/− hepatic NKT cell populations was not significant. Thus, the removal of bim-mediated cell death overcomes the survival defect of Id2KO NKT cells.
Fig. 6.
Bim deficiency rescues hepatic Id2KO NKT cells. (A) Representative flow cytometry plots identifying NKT cells by congenic marker, NK1.1, and TCRβ, or CD1d tetramer staining of hepatic lymphocytes recovered from Id2+bim+/−, Id2KObim+/+, Id2+bim−/−, or Id2KO bim−/− fetal liver chimeras. (B) Average percentage (± SEM) of indicated populations from (A). (C) Absolute cell number for indicated populations from (A). Data are representative of three independent experiments with n = 3–5 per group. Statistical significance determined using unpaired two-tailed t-test where P < 0.01, *, P < 0.005, ***, ns >0.05.
We have shown that Id2 deficiency results in perturbations of NKT cell homeostasis including a dramatic reduction in accumulation of both Vα14i and total NKT cells in the liver (Fig. 1). Id2-deficient NKT cells displayed increased apoptosis compared to their Id2-sufficient counterparts in the liver, suggesting that Id2 expression promotes their survival (Fig. 4). These results highlight a role for Id2 in NKT cell homeostasis and are reminiscent of defects observed in NK and CD8+ T cells where a requirement for Id2 occurs at late stages of maturation upon acquisition of effector function, coincident with down-regulation of Id3 (16, 17, 22, 26).
Much like the defect we observe in the absence of Id2, CXCR6KO (10, 11) and CXCL16KO mice (13), have a significant loss of hepatic but relatively normal iNKT cell populations in other tissues. Interestingly, we found that Id2KO NKT cells showed substantially reduced expression of CXCR6 in all tissues tested (Fig. 5). The liver-specific iNKT cell defect in CXCR6KO mice has been proposed to result from increased apoptosis (10) and impaired maturation (11). We observed increased apoptosis (Fig. 4) and diminished numbers of both NK1.1− and NK1.1+ Id2KO NKT cells (Fig. 1) compared to their Id2+ counterparts in the liver. This observation suggests lower CXCR6 expression deprives Id2-deficient hepatic iNKT cells of an essential survival signal, preventing accumulation of mature cells (Fig. 4). Thus, splenic and thymic iNKT cells may depend on different survival signals, which do not require CXCR6 expression. Finally, we did not observe any defects in cytokine production by the Id2-deficient iNKT cells, while CXCR6KO and CXCL16KO NKT cells were found to have decreased cytokine production, indicating that the residual levels of CXCR6 expressed by Id2KO NKT cells were sufficient to mediate more normal maturation and activation.
Interestingly, we observed a more severe hepatic phenotype than the CXCR6KO mice and find fewer iNKT cells in the bone marrow unlike CXCR6KO mice, suggesting that reduced CXCR6 expression may not fully explain the severity and extent of the iNKT cell defect in Id2-deficient mice. We found that bcl-2 and bcl-XL were both expressed at lower levels in the Id2KO iNKT cells that localized to the liver, supporting a role for Id2 in promoting cell survival (Fig. 5). Loss of expression of pro-survival molecules could not be accounted for solely by down-regulation of CXCR6 expression as CXCR6KO iNKT cells had less bcl-2 but normal bcl-XL expression (Fig. S4). Of note, the Id2WT hepatic iNKT cells expressed higher levels of these anti-apoptotic molecules compared to their splenic counterparts, perhaps indicating that iNKT cells require greater protection from apoptosis when localized to the liver. Importantly, the survival and accumulation defect of Id2KO iNKT cells was largely rescued by eliminating bim, a key molecule promoting apoptosis of lymphocytes, including iNKT cells, emphasizing the role of Id2 in regulating survival by influencing components of the intrinsic apoptotic pathway (Fig. 6).
Studies addressing Id2 function in NK cell development demonstrate that Id proteins are required to control E protein activity, which is accomplished cooperatively by Id2 and Id3 during development but by Id2 alone at later stages as Id3 is down-regulated (16, 26). Interestingly, Id2 mRNA levels were equivalent between NKT cells recovered from spleen and liver, while Id3 levels were approximately 5-fold higher in splenic compared to hepatic NKT cells in wild-type mice, perhaps explaining the normal NKT population in the Id2KO spleen and why Id2 is crucial for hepatic the NKT population (Fig. 1B). Future experiments will be required to establish if the sole function of Id2 in NKT cell homeostasis is to diminish E protein activity and whether activity of E protein transcription factors may influence NKT cell development or function. It is well established that high levels of E protein expression can lead to programmed cell death and that developmental progression and survival require modulation of E protein activity by Id proteins (20, 27, 28). Furthermore, bim is known to be target directly induced by E proteins (29). In agreement, the apoptotic death of Id2KO CD8+ T cells resulting from a failure to attenuate E protein activity has been correlated with up-regulation of pro-apoptotic bim expression and diminished anti-apoptotic bcl-2 expression (17, 29). Thus, we hypothesize that the regulation of E proteins, which is mediated, at least in part, by Id2 expression controls NKT cell survival by balancing expression of bcl-2 family members.
Materials and Methods
Mice.
Mice were bred and housed in specific pathogen-free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of California San Diego. Id2KO mice were generated as previously described (12) and maintained on the C57/BL6 background. For the generation of fetal liver chimeras, 5–10 × 106 Id2KO, Id2+/−, or Id2+/+ E14-E15.5 fetal liver cells and 5–10 × 105 RAGKO cells were injected i.v. into lethally irradiated (900 RAD) recipient mice of a distinct CD45 congenic marker. Bone marrow chimeras were generated by transferring 5–10 × 106 B220−CD4−CD8− bone marrow cells obtained from Id2KO or Id2+ fetal liver chimeras into lethally irradiated recipient mice of a distinct congenic marker. For the generation of mixed fetal liver chimeras, 5 × 106 Id2KO or Id2WT E14.5 CD45.2+ fetal liver cells and 5 × 106 Id2WT CD45.1+ B220−CD4−CD8− bone marrow cells were injected i.v. into lethally irradiated (900 RAD) recipient CD45.1+ mice. All chimeras were rested for at least 8 weeks to allow reconstitution of the host. Mixed bone marrow chimeras were generated by transferring 6 × 106 B220−CD4−CD8− bone marrow cells obtained from CD45.2+ Id2KO or Id2WT fetal liver chimeras and 4 × 106 Id2WT CD45.1+ B220-CD4-CD8- bone marrow cells into lethally irradiated recipient CD45.1+ mice. All mixed chimeras were rested for at least 10 weeks to allow reconstitution of the host. Lymphocytes from CXCR6-deficient (CXCR6KO) mice (10) were kindly provided by Dr. Matloubian.
Flow Cytometry.
Single cell suspensions were prepared from liver, thymus, spleen, and bonemarrow of chimeras. Hepatic NKT cells were isolated as previously described (15). Fc receptors were blocked with anti-FcγRII/III (2.4G2) and then stained with the indicated monoclonal antibodies. The following fluorochrome-conjugated monoclonal antibodies were used: TCRβ FITC (clone H57–597), CD45.1 FITC (A20), CD45.2 FITC (104), CD43 FITC (1B11) (Biolegend), CD69 PE (H1.2F3), ICOS PE (7E.17G9), CD1d Tetramer PE [NIH Tetramer Core Facility (mCD1d/PBS57)] or generated as previously described (30), TCRβ PE (H57–597), CD122 PE (TM-b1), IFNγ PE (XMG1.2), IL-4 PE (11B11), CXCR3 PE (1C6/CXCR3) (BD Biosciences), goat anti-human IgG Fcγ PE (Jackson Immunoresearch), CD45.1 PeCy7 (A20), NK1.1 PerCP Cy5.5 (PK136), CD45.2 PerCP Cy5.5 (104), NK1.1 APC (PK136), TCRβ APC (H57–597), CD45.2 APC (104), CD44 APC, Klrg1 APC (2F1), Annexin V APC (Invitrogen), and CD45.2 Alexa 750 (104). CXCL16-Fc fusion protein was generously provided by Dr. M. Matloubian and used as previously described (6). Intracellular staining of bcl-2 and bcl-XL were performed with BD Cytofix/Cytoperm Plus Kit. PE hamster Anti-mouse bcl-2 (3F11) monoclonal antibodies and isotope control PE hamster IgG (A19–3) were purchased from BD Pharmigen; and bcl-XL (54H6) Rabbit mAb (Alexa Fluor 488 Conjugate) and isotype control Rabbit IgG (DA1E) mAb (Alexa Fluor 488 Conjugate) were purchased from Cell Signaling Technology. Samples were collected on FACSCalibur or FACSAria (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Quantitative PCR.
Relative levels of mRNA among indicated populations of sorted CD45.2+NK1.1+TCRβ+ NKT cells were compared by qPCR. Id2, Id3, Bcl-2, and Bcl-XL mRNA levels were assessed using non-specific product detection (SYBR Green, Stratagene). CXCR6 mRNA expression was detected using a TaqMan probe and primer sequences specific for CXCR6 (TaqMan Gene Expression Assay, Applied Biosystems). Id3 Forward: GACTCTGGGACCCTCTCTC, Id3 Reverse: ACCCAAGTTCAGTCCTTCTC, Id2 Forward: ACCAGAGACCTGGACAGAAC, Id2 Reverse: AAGCTCAGAAGGGAATTCAG. Primers for bcl-2 and bcl-XL have been previously described (17). Samples were normalized to HPRT or GAPDH expression.
Migration Assay.
Spleens were harvested from Id2KO or Id2+ chimeras and B220+ CD8+ splenocytes were depleted using MACS columns (Miltenyi Biotec). Thirteen to 18 × 106 Id2KO or Id2+ B220− CD8− splenocytes were injected i.v. into CD45.1 hosts. Recipient mice were killed 24 h after adoptive transfer and lymphocytes recovered from liver and spleen were analyzed by flow cytometry to determine the percentage of donor NKT cells (CD45.2) recovered.
Supplementary Material
Acknowledgments.
We thank Dr. Murre for generously providing the Id2-deficient mice and constructive discussions; Dr. Matloubian and Joyce Hu (University of California, San Francisco) for the generous gifts of the CXCL16-Fc fusion protein, CXCR6-deficient cells, and advice; and Drs. Johnson, Hamerman, David, and Werneck for critical review of the manuscript. This work was supported by an Investigator award from the Cancer Research Institute; Pew Scholar Award; National Institutes of Health Grants (A.G.) and R37 AI71922 (to M.K.); University of California San Diego, Center for Molecular Genetics training grant (Y.Y.); Deutsche Forschungsgemeinschaft Grant CA 715/1-1 (to M.A.C.); and a Leukemia Lymphoma Society fellowship (L.M.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908249106/DCSupplemental.
References
- 1.Godfrey DI, Berzins SP. Control points in NKT cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda JL, Gapin L. Developmental program of mouse Valpha14i NKT cells. Curr Opin Immunol. 2005;17:122–130. doi: 10.1016/j.coi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seino K, Taniguchi M. Functionally distinct NKT cell subsets and subtypes. J Exp Med. 2005;202:1623–1626. doi: 10.1084/jem.20051600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronenberg M. Toward an understanding of NKT cell biology: Progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 6.Tupin E, Kinjo Y, Kronenberg M. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol. 2007;5:405–417. doi: 10.1038/nrmicro1657. [DOI] [PubMed] [Google Scholar]
- 7.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston B, Kim CH, Soler D, Emoto M, Butcher EC. Differential chemokine responses and homing patterns of murine TCR alpha beta NKT cell subsets. J Immunol. 2003;171:2960–2969. doi: 10.4049/jimmunol.171.6.2960. [DOI] [PubMed] [Google Scholar]
- 10.Geissmann F, et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Germanov E, Veinotte L, Cullen R, Chamberlain E, Butcher EC, Johnston B. Critical role for the chemokine receptor CXCR6 in homeostasis and activation of CD1d-restricted NKT cells. J Immunol. 2008;181:81–91. doi: 10.4049/jimmunol.181.1.81. [DOI] [PubMed] [Google Scholar]
- 12.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 13.Shimaoka T, et al. Critical role for CXC chemokine ligand 16 (SR-PSOX) in Th1 response mediated by NKT cells. J Immunol. 2007;179:8172–8179. doi: 10.4049/jimmunol.179.12.8172. [DOI] [PubMed] [Google Scholar]
- 14.Emoto M, Mittrucker HW, Schmits R, Mak TW, Kaufmann SH. Critical role of leukocyte function-associated antigen-1 in liver accumulation of CD4+NKT cells. J Immunol. 1999;162:5094–5098. [PubMed] [Google Scholar]
- 15.Ohteki T, Maki C, Koyasu S, Mak TW, Ohashi PS. Cutting edge: LFA-1 is required for liver NK1.1+TCR alpha beta+ cell development: Evidence that liver NK1.1+TCR alpha beta+ cells originate from multiple pathways. J Immunol. 1999;162:3753–3756. [PubMed] [Google Scholar]
- 16.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannarile MA, et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 18.Yokota Y, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 19.Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nat Rev Immunol. 2001;1:193–199. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- 20.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 21.Rivera R, Murre C. The regulation and function of the Id proteins in lymphocyte development. Oncogene. 2001;20:8308–8316. doi: 10.1038/sj.onc.1205091. [DOI] [PubMed] [Google Scholar]
- 22.Rolf J, et al. Molecular profiling reveals distinct functional attributes of CD1d-restricted natural killer (NK) T cell subsets. Mol Immunol. 2008;45:2607–2620. doi: 10.1016/j.molimm.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Lian RH, Kumar V. Murine natural killer cell progenitors and their requirements for development. Semin Immunol. 2002;14:453–460. doi: 10.1016/s1044532302000805. [DOI] [PubMed] [Google Scholar]
- 24.Park KW, et al. Inhibitor of DNA binding 2 is a small molecule-inducible modulator of peroxisome proliferator-activated receptor-gamma expression and adipocyte differentiation. Mol Endocrinol. 2008;22:2038–2048. doi: 10.1210/me.2007-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uldrich AP, et al. NKT cell stimulation with glycolipid antigen in vivo: Costimulation-dependent expansion, Bim-dependent contraction, and hyporesponsiveness to further antigenic challenge. J Immunol. 2005;175:3092–3101. doi: 10.4049/jimmunol.175.5.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, Gapin L. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood. 2006;107:2797–2805. doi: 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel I, Murre C. Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc Natl Acad Sci USA. 1999;96:996–1001. doi: 10.1073/pnas.96.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci USA. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.