Abstract
Historically, dwarfism was the major genetic defect in U.S. beef cattle. Aggressive culling and sire testing were used to minimize its prevalence; however, neither of these practices can eliminate a recessive genetic defect. We assembled a 4-generation pedigree to identify the mutation underlying dwarfism in American Angus cattle. An adaptation of the Elston-Steward algorithm was used to overcome small pedigree size and missing genotypes. The dwarfism locus was fine-mapped to BTA6 between markers AFR227 and BM4311. Four candidate genes were sequenced, revealing a nonsense mutation in exon 15 of cGMP-dependant type II protein kinase (PRKG2). This C/T transition introduced a stop codon (R678X) that truncated 85 C-terminal amino acids, including a large portion of the kinase domain. Of the 75 mutations discovered in this region, only this mutation was 100% concordant with the recessive pattern of inheritance in affected and carrier individuals (log of odds score = 6.63). Previous research has shown that PRKG2 regulates SRY (sex-determining region Y) box 9 (SOX9)-mediated transcription of collagen 2 (COL2). We evaluated the ability of wild-type (WT) or R678X PRKG2 to regulate COL2 expression in cell culture. Real-time PCR results confirmed that COL2 is overexpressed in cells that overexpressed R678X PRKG2 as compared with WT PRKG2. Furthermore, COL2 and COL10 mRNA expression was increased in dwarf cattle compared with unaffected cattle. These experiments indicate that the R678X mutation is functional, resulting in a loss of PRKG2 regulation of COL2 and COL10 mRNA expression. Therefore, we present PRKG2 R678X as a causative mutation for dwarfism cattle.
Keywords: achondroplasic dwarfism, BTA6, cGKII, recessive genetic disease
Bovine dwarfism was widely observed from the late 1940s through the 1960s, making it one of the greatest problems in the history of the U.S. beef cattle industry (1). Even President Eisenhower showed great interest in the disease, because he raised Angus cattle on his ranch. Over the years, the disease has cost beef producers millions of dollars. Affected animals represent an economic loss, because of slow growth and lack of market value. Breeders have attempted to eliminate the disease by using test matings or removing parents in a herd known to produce affected offspring. These methods are extremely costly, and did not remove all of the carriers in the past. Yet by the 1970s, producers thought that dwarfism had been eliminated. Unfortunately, that was not the case, as 6 dwarf calves were discovered on several ranches across the United States in the spring of 2002. Today, there is no good estimate of the number of affected individuals in the United States. Rumors indicate that affected individuals are often not reported, and are removed from herds at birth to prevent negative publicity for the cattle breeder. Cattle breeders with suspect carriers have lost their livelihood due to the negative connotations associated with the damaging history of dwarfism in the beef industry. Development of a definitive genetic test will finally allow producers to eliminate dwarfism while preserving genetically superior carrier animals with desirable genetic backgrounds in the Angus breed.
A multitude of dwarfism phenotypes occur in nature. Dwarfism occurs in many species, and several animal models exist. Notably, transgenic models in the mouse have been used to understand the implications of mutations within the fibroblast growth factor receptor 3 (FGFR3) gene. In humans, achondroplasia is caused primarily by a dominant mutation in FGFR3 (2). Angus dwarfism is also characterized as a form of achondroplasia. Affected individuals have diminished endochondral ossification of growth plates, protrusions of the alar wing of the basisphenoid bone into the cranial cavity, abnormalities of the ventral vertebral bodies, and curvature of the vertebral processes (3). In American Angus, dwarfism appears to follow a recessive pattern of inheritance.
In cattle, few forms of dwarfism have been characterized at the molecular level (i.e., Dexter and Japanese Brown cattle) (4, 5). Disproportionate dwarfism in Japanese brown cattle is a recessive phenotype caused by insufficient endochondral ossification and abnormal arrangement of growth plate chondrocytes. Affected individuals have shortened stature, joint abnormalities, and ateliosis (5). Our group previously investigated the possibility that Angus dwarfism was caused by either of the 2 mutations in the LIMBIN gene that cause dwarfism in Japanese brown cattle. Analysis of the 2 mutations showed that Angus dwarf calves were wild-type for the LIMBIN mutations (3). We also investigated the possibility that dwarfism in the Angus and Dexter breeds could share a common genetic cause. Dexter dwarfism is a form of chondrodysplasia also caused by defective endochondral ossification due to 2 different mutations in aggrecan 1 (ACAN) (6). Homozygous mutants display a “bulldog” phenotype, including a retruded muzzle, cleft palate, abdominal hernia, shorted and misshaped long bones, and lethality at approximately 7 months of gestation. Individuals heterozygous for mutations in the ACAN gene exhibit disproportionate dwarfism (4, 6). Angus dwarf samples were wild-type for Dexter dwarfism markers. Therefore, we concluded that Angus dwarfism was likely caused by a novel mutation.
Herein, we describe the fine mapping and analysis of positional candidate genes for dwarfism in American Angus cattle using a unique adaptation of the Elston-Steward algorithm to refine the position of the disease locus. We present strong evidence that a nonsense mutation within the kinase domain of PRKG2 is the causative mutation for Angus dwarfism based on linkage mapping, cell culture, in vivo expression data, and similarities with existing mutant rodent models.
Results
Linkage Analysis Reveals a High Signal on BTA6.
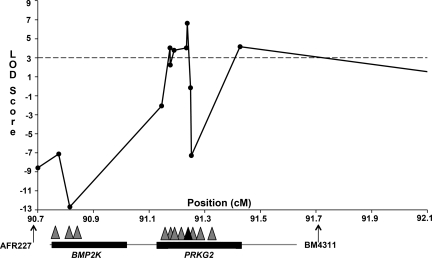
To find the Angus dwarfism locus, we began by genotyping an Angus pedigree consisting of 26 individuals, including 6 affected animals (Fig. S1). We prioritized genotyping of chromosomes that contained genes that have been shown to be associated with dwarfism in humans. After investigating several chromosomes with no statistical association, a significant log of odds (LOD) score of 4.53 was detected on BTA6 between 2 microsatellite markers, AFR227 and BM4311. Because no other statistically significant linkage signals were detected, we focused on fine mapping BTA6. We added an additional 19 microsatellite marker genotypes within or closely flanking the critical region to verify the position of the dwarfism locus. Analysis of these new markers increased the size of the critical region from 0.8 to 2.8 cM, flanked by AFR227 and BMS511. The size of the critical region expanded, because increased marker information was provided in the affected individuals by the new marker genotypes distal to BM4311 (Fig. S2). However, the new critical region still encompassed the original region with approximately the same LOD score.
Positional Candidate Gene Analysis: Discovery of a Nonsense Mutation in PRKG2.
Four positional candidate genes were prioritized for resequencing based on their known function in bone development or relationship to genes known to be involved in bone development. They were bone morphogenetic protein 2 kinase (BMP2K), bone morphogenetic protein 3 (BMP3), fibroblast growth factor 5 (FGF5), and cGMP-dependant type II protein kinase (PRKG2). We sequenced the exons and some flanking intron sequence of each candidate gene in a WT, dwarf carrier, and dwarf animal. Additionally, we sequenced one Hereford and one Brahman DNA sample as negative controls, which resulted in the identification of 75 single nucleotide polymorphisms (SNPs). We used the Brahman, Hereford, and unaffected Angus as control samples to determine if a given SNP followed the recessive pattern of inheritance for Angus dwarfism. Few heterozygous SNPs were found to be informative in BMP3 or FGF5 in Angus, but many heterozygous informative SNPs were detected in BMP2K and PRKG2 (Tables S1–S4). Seven SNP were detected within exons; however, only one was nonsynonymous. This mutation resulted in a C to T transition [Nt2032 (C-T)] that caused a nonsense mutation in exon 15 of PRKG2, herein referred to as the R678X marker. The T mutation was homozygous in the dwarf, heterozygous in carriers, and not present in the WT Angus, Hereford, or Brahman controls that we sequenced. Upon genotyping our pedigree, we found 100% concordance of the mutation with dwarf carrier and affected status. We genotyped 11 SNPs that surrounded the R678X PRKG2 mutation and found a maximal LOD score of 6.63 for the marker interval, including the nonsense mutation (Fig. 1). The marker genotypes for the 6 affected individuals and 2 unaffected full siblings (sibs) show a relatively large segment of homozygosity in the dwarfs, but a much smaller one in the unaffected full sibs (Fig. S2). We genotyped additional breeds, including 8–10 randomly selected samples from Brahman, Hereford, Holstein, Limousin, and South Devon breeds, as well as crossbred cattle composed of Maine Anjou, Charolais, Simmental, and Angus (n = 54). However, we were unable to detect the R678X PRKG2 mutation in additional breeds, indicating that this mutation may be Angus breed specific.
Fig. 1.
The maximum LOD score for Angus dwarfism on BTA6 occurs at the C/T nonsense mutation, indicated by a black triangle, in exon 15 of PRKG2. Microsatellite markers are indicated by arrows; SNP markers are indicated by triangles.
The PRKG2 R678X Mutation Causes a Loss of COL2 Regulation in Cell Culture.
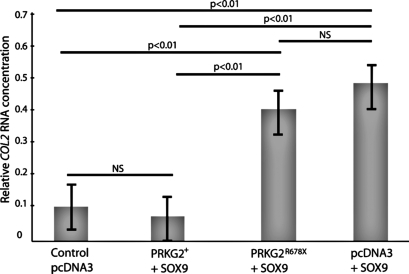
Based on previous studies in the rat (7), we designed a cell culture experiment to determine the functionality of the PRKG2 R678X mutation found in bovine dwarfs. Human hepatoma (HUH-7) cells were transiently transfected with one of 4 treatments: (i) wild-type bovine (bt) PRKG2 + human (h) SOX9; (ii) R678X btPRKG22 + hSOX9; (iii) hSOX9 + empty vector; and (iv) empty vector (negative control). The PRKG2 protein is known to regulate SOX9 and downstream COL2 mRNA levels. PRKG2 downregulates COL2 by activation of SOX9 (7). Therefore, we hypothesized that the truncated dwarf PRKG2 kinase domain would not be able to downregulate COL2. Cells transfected with the R678X btPRKG2 vector had a 38% increase in COL2 mRNA compared with cells transfected with WT btPRKG2 (Bonferroni corrected P < 0.01; Fig. 2). There was no difference in COL2 levels between the control (empty vector only) and WT PRKG2 +hSOX9 treatment (not significant, P > 0.70). The hSOX9 + empty vector control treatment exhibited the same COL2 expression as the R678X btPRKG2 + hSOX9 treatment (not significant, P > 0.30) and increased COL2 expression compared with btPRKG2 + hSOX9 (Bonferroni corrected P < 0.01) and the empty vector alone (Bonferroni corrected P < 0.01). These findings indicate that the R678X PRKG2 mutation severely reduced the ability of PRKG2 to regulate SOX9 to levels comparable to a complete loss of function of PRKG2 (vector + hSOX9 treatment alone).
Fig. 2.
Comparison of COL2 mRNA expression levels between WT btPRKG2 and R678X btPRKG2 transfected HUH-7 cells. The lines above the bar graph denote the accompanying statistical difference for the 6 treatment comparisons. The P value directly above each line is associated with the difference between only the outermost 2 bars underneath the line. All P values are Bonferroni corrected to account for multiple testing. NS, not significant.
Individuals Homozygous for the R678X Mutation Show Reduced Stature.
More than 20 embryos were produced by carrier × carrier and dwarf × carrier matings; however, only 6 embryos survived to parturition. We blindly genotyped these 6 individuals using the R678X PRKG2 marker within 2 weeks of birth and successfully predicted dwarf status. Animals predicted to be dwarfs by the R678X PRKG2 marker had shorter metatarsals (reduced 4.5 cm; P < 0.01) and fused ulna/radius (reduced 7.5 cm; P < 0.01) bones. Although not significant, there were also trends toward significant differences between normal and dwarf metacarpals (reduced 2.75 cm; P < 0.09), femur (reduced 3.9 cm; P < 0.17), and tibia (reduced 4.9 cm; P < 0.11). No significant differences were observed between these animals for head length, calvarium (skull) width, or bone circumference for each of the major long bones previously listed (P > 0.25). Estimates of differences in stature and body length were also calculated for live dwarf and normal animals at ≈210 days of age. Differences in height of 15.8 cm (P < 0.0001) and 20.7 cm of spine length (P < 0.0001) were observed between animals declared normal versus those declared dwarf at birth by the R678X genotype. Based on height differences, gross differences in long bone length, and changes in vertebral length, we feel confidant that both unaffected and dwarf animals were produced in these matings.
Individuals Homozygous for the R678X Mutation Have Altered COL2 and COL10 Gene Expression in Growth Plate Cartilage.
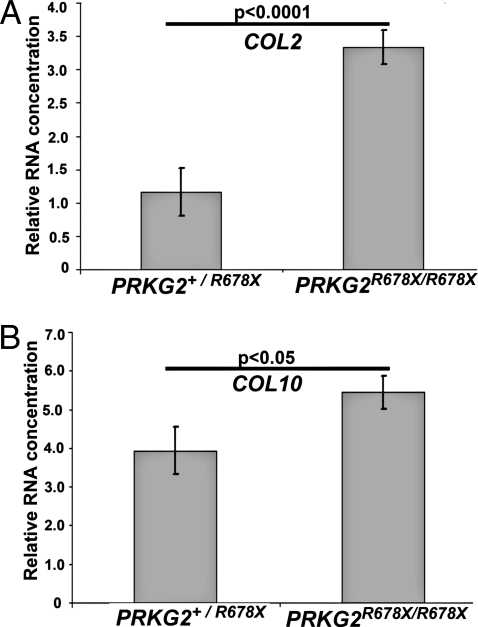
Expression of COL2 and COL10 in growth plate cartilage was compared between 4 calves predicted to be dwarfs (PRKG2R678X/R678X) and 2 calves predicted to be unaffected (PRKG2R678/+) at approximately 7 months of age. Real-time PCR analysis confirmed a considerable increase in COL2 mRNA in dwarf cattle compared with normal cattle (P < 0.0001; Fig. 3A). Real-time PCR of COL10 indicated an increase in expression in dwarfs compared with normal individuals (P < 0.05; Fig. 3B). Expression levels of COL2 and COL10 were increased in dwarf (PRKG2R678X/R678X) cattle, indicating expression levels comparable to PRKG2 null dwarf rats (7).
Fig. 3.
In vivo gene expression of COL2 and COL10 was measured in bovine PRGK2R678X/R678X (dwarf) and PRKG2+/R678X (unaffected) tibia growth plates. (A) Difference in COL2 expression in dwarf R678X PRKG2 mutants versus normal cattle growth plate. (B) Difference in COL10 expression in dwarf R678X PRKG2 mutants versus normal cattle growth plate.
There Is No Evidence of Nonsense-Mediated Decay Due to the R678X Mutation.
The R678X mutation occurs in the 15th exon of 18 exons, leading us to test the possibility of nonsense-mediated RNA decay. We tested PRKG2 mRNA expression by quantitative real-time PCR. No difference in PRKG2 expression was detected between R678X homozygous (dwarf) and R678X heterozygous (unaffected) individuals (P > 0.20). Because the R678X mutation is in the kinase domain, we would expect truncation of this domain to cause a loss of function even if the mRNA was produced. We wondered if the truncation could somehow occur near enough the end of the protein to not alter the structure of the protein. To predict the effect of the R678X mutation on PRKG2 protein structure, we used the SwissModel homology protein-modeling program to compare WT and R678X PRKG2 (8, 9). The modeling results indicate that a conformational change likely occurs (Fig. S3). Furthermore, a comparison of the primary amino acid sequence across 7 species suggests that the mutation resides in a conserved block of 11-aa residues (Fig. S4).
Discussion
We believe the use of the Elston-Steward algorithm—specifically, the ability to accommodate all possible genotypes for common ancestors in our small pedigree—was critical to our ability to quickly find the Angus dwarfism locus. The linkage mapping results strongly indicated that the R678X mutation in PRKG2 was the causative mutation for Angus dwarfism. Of the 76 mutations detected in the dwarfism-critical region, the R678X mutation was one of only 7 coding mutations, and the only nonsynonymous coding mutation. Evaluation of dwarf and unaffected full-sib genotypes indicates a small region of homozygosity unique to affected individuals. Homozygosity in unaffected full sibs at markers flanking the R678X mutation caused the reduced LOD scores near the causative mutation as a result of analyzing 2-marker haplotypes (Fig. 1). Analysis of a larger haplotype does not exclude these flanking marker intervals; however, this limitation did not deter our ability to conclude which SNP was the causative mutation.
To evaluate the physiological effects in Angus dwarfs, we produced 6 dwarf calves by embryo transfer. Because only 6 of 20 individuals survived full term in pregnancy, this may indicate that embryo death may be an additional consequence in PRKG2 mutants. This observation is consistent with research that indicates PRKG2 may function in preovulatory follicles as a response to luteinizing hormone and progesterone (10). Analysis of bone lengths indicates that both dwarf and normal individuals were produced from our mating experiment. Analysis of such small data is difficult because of the limited degrees of freedom available. This experiment does not prove causality of the mutation, because there was likely considerable sharing of large chromosomal segments between these individuals and those in our linkage mapping pedigree due to shared common ancestors.
Rodent models show that PRKG2 is a logical candidate gene for dwarfism in Angus cattle. Pfeifer et al. (11) created a knockout of PRKG2 in the mouse, which resulted in mice that exhibited a dwarfism phenotype. These mice exhibited an unorganized growth plate with abnormal stacking of chondrocytes. More recently, Chikuda et al. (7) discovered a deletion in the coding region of PRKG2 that caused dwarfism in the KMI rat. The loss of growth plate organization observed in these dwarf rats is very similar to what is observed in the Angus dwarf growth plate (Fig. S5).
To explain the role of PRKG2 in growth plate organization, Chikuda et al. (7) showed that PRKG2 regulates SOX9 via phosphorylation. Phosphorylation of SOX9 is required for nuclear translocation and transcriptional regulation of COL2 to COL10 within the growth plate. This is a key developmental checkpoint that allows proper transition of chondrocytes from a proliferative to hypertrophic growth phase. As a consequence, KMI mini-rats show increased COL2 and decreased COL10 mRNA expression levels, consistent with constitutive proliferative growth. Angus dwarfs also show a lack of regulation of COL2 and COL10 gene expression, as indicated by real-time PCR experiments. Functional changes in COL2 mRNA levels in R678X PRKG2 mutants are consistent with those observed in the KMI mini-rat. Results from cell culture indicate that the R678X PRKG2 mutation was sufficient to change COL2 mRNA expression. Control transfections using only SOX9 did not significantly differ in COL2 expression levels from R678X mutants, indicating that dwarf cattle would have COL2 gene expression similar to a null PRKG2 individual. Interestingly, the change in COL10 expression in dwarf cattle was opposite of what was expected. Dwarf cattle show increased expression of COL10, unlike the rat PRKG2 mutants. This could be due to differences in tissue collection, small sample size, temporal changes in expression, or species differences. Alternatively, differences in COL10 expression could be explained, because bovine expression was measured in vivo, whereas expression measurements in the rat were carried out in cell culture. Because the R678X mutation occurred before the last exon, we considered nonsense-mediated decay as a cause of PRKG2 loss of function. However, we observed no change in PRKG2 expression in bovine growth plate cartilage. This would suggest that the change in PRKG2 was caused at the protein level and not due to reduced PRKG2 gene expression. It is possible that we were unable to detect a difference in expression due to small sample size or the lack of homozygous WT individuals in our assay. Because the mutation truncates a kinase domain, the functional change in likely at the level of the PRKG2 protein. Because severe coding mutations cause dwarfism in both transgenic and naturally occurring dwarf animals, it is logical that a similar mutation in cattle would cause dwarfism as well.
The function of PRKG2 remains widely unknown, but it is clearly an important signal transduction pathway in numerous tissues (e.g., brain, growth plate cartilage, jejunum, kidney, lung, and pancreas). Its potential roles are diverse, ranging from kidney disease to cancer and chondrodysplasia (12, 13). Interestingly, the knockout of PRKG2 shows no symptoms of infection when treated with STa toxin of E. coli (11). This may suggest that genetic variation in PRKG2 is beneficial to circumvent bacterial infection. The role of PRKG2 in transport of Na+, H+, HCO3#x2212;, Cl−, and water have been well documented in the small intestine (11, 14, 15). Additionally, PRKG2 has been shown to be highly expressed in the brain with a largely unknown function (http://www.brain-map.org/). Study of PRKG2 knockouts indicated that PRKG2 might regulate the circadian clock, behavior, and alcoholism (16–19). The use of PRKG2 as a therapeutic treatment in medicine has considerable potential. It was previously suggested by Pfeifer et al. (11) that PRKG2 could be used to enhance the process of bone healing. More recently, it has been suggested that the protein kinase G pathway may be important in developing treatments for diabetes (20). The role of PRKG2 to regulate the cystic fibrosis transmembrane conductance regulator (CFTR) is particularly interesting. Analysis of the PRKG2 pathway along with cAMP pathways may yield valuable information needed to create new treatments for cystic fibrosis patients. Angus dwarfs do exhibit labored, “raspy” breathing, but it is unclear if this is due to a lung phenotype or skeletal malformation. They do not exhibit additional phenotypes; however, many of referenced phenotypes are difficult to measure without examination at the molecular level.
The mutation for dwarfism in Angus is located on chromosome 6, along with numerous quantitative trait loci (QTLs) for production traits. Of particular interest are QTLs for growth rate, milk yield, and milk fat and protein that would be selected for optimal calf growth and maternal ability (see http://bovineqtl.tamu.edu/; http://www.vetsci.usyd.edu.au/reprogen/QTL_Map/; and http://www.animalgenome.org/QTLdb/cattle.html). Potential selection pressure for the QTL regions could have increased homozygosity of the region, which led to increased observance of the dwarfism allele. Additionally, the Angus breed went through a transition period where animals were selected to be short and stocky. It is possible that the occurrence of the mutation is a holdover from earlier years when smaller frame size was desired.
We present a strong argument that a nonsense mutation at R678X of PRKG2 causes dwarfism in Angus cattle. First, considering a recessive gene action model, the mutation is 100% concordant with the pedigree, including both dwarfs and dwarf carriers. Second, the mutation causes a premature stop codon in a kinase domain. Third, knockout mouse and naturally occurring rat PRKG2 mutants exhibit dwarfism with growth plate phenotypes similar to bovine dwarfs. Fourth, transfection of WT versus R678X PRKG2 demonstrates that the R678X mutation causes loss of PRKG2 function. Last, expression analysis from dwarf and normal cattle show that this mutation alters COL2 and COL10 expression levels. Because COL2 and COL10 expression level and timing are critical to proper growth plate development, it is likely that such a functional change will alter endochondral ossification. The immediate impact of this research is a genetic test for use by Angus producers to remove dwarfism. Continued research on knockouts and naturally occurring PRKG2 mutants may unlock the doors to medical treatments for many debilitating genetic diseases. Additional functional analysis is underway to characterize the impact of the R678X PRKG2 mutation on bovine physiology.
Materials and Methods
Primer sequences for PCR and descriptions of general laboratory techniques are provided in SI Text.
Population.
We constructed a 4-generation pedigree to determine the relationship between 6 individuals diagnosed as dwarfs (Fig. S1). This pedigree consists of 26 individuals that are related to 2 founders. Carrier and dwarf phenotypes were determined based on production of dwarf calves and diagnosis of dwarfism by veterinarians. Of the 26 individuals, DNA was available for only 23 individuals.
Identification and Analysis of Positional Candidate Genes.
Very few bovine scaffold sequences were available for the Angus dwarfism locus in the spring of 2005. Traditional comparative genomic approaches were used to overcome this problem. We used the human-bovine comparative radiation hybrid map (http://cagst.animal.uiuc.edu/RHmap2004/index.html) to identify positional candidate genes (21). Human gene sequence was used to BLAST query bovine gene sequence from the preliminary bovine genome draft. Because this process required repeated BLAST searches and manual assembly of segmented exon and UTR sequences, we created an automated pipeline called BEAP (BLAST Extension and Alignment Program) to expedite sequence assembly and reduce sequence handling errors. We used BEAP to query bovine sequences and perform an in silico chromosome walk using CAP3 to assemble retrieved sequences into contigs (22). We identified 20 genes in cattle by BLAST analysis within or closely surrounding the critical region.
Statistical Analysis/Elston Stewart Methodology.
We used an extension of the Elston-Stewart algorithm in a model-based linkage analysis to map the genomic location most likely to contain the dwarfism locus (23, 24). The method was modified to make it more computationally feasible when pedigrees contain multiple loops to common ancestors. The implementation is described in the following citations (24–26). The implementation of linkage mapping has been described previously (27). The model for linkage mapping assumes the disease allele follows a recessive pattern of inheritance that is present at a low frequency in the Angus population. Briefly, likelihood ratios were calculated for each marker interval assuming (i) the dwarf mutation is at the center of this interval (L1) and (ii) the dwarf mutation is in another linkage group (L2). The log base 10 of this likelihood ratio (L1/L2) resulted in the LOD score. Likelihood (L) can be expressed as
where Pr(y) is a vector of dwarfism phenotypes, and Pr(G) is a vector of genotypes at the markers flanking the interval and the dwarf mutation. A LOD score greater than 3 was deemed significant. We used multiple disease allele frequencies in the model for the founder population to determine the influence of disease allele frequency on LOD score.
Matings and Data Generated from a Breeding Experiment.
Affected and unaffected full-sib calves were produced using embryo transfer. Two cows, mother (carrier) and daughter (dwarf), were mated to a bull known to be a carrier of dwarfism. There were more than 20 embryos produced for implantation into surrogate mothers. Only 6 embryos survived to full term, 3 from each mating. The 6 calves were raised under the guidelines of the Iowa State University Institutional Animal Care and Use Committee protocols. These 6 calves were within the normal size range for newborn Angus calves and not distinguishable as dwarfs at birth. A DNA sample was collected from each calf and tested blindly to determine the genotype for the PRKG2 R678X marker. Calves were weighed, and height and bone measurements were taken monthly to approximate lengths for the head, femur, metacarpal, metatarsal, tibia, and spine. In addition, circumferences were measured for head, chest, metacarpal, knee, and pastern. All measurements were performed in triplicate to reduce measurement error. At approximately weaning age, the calves were killed to collect tissues for functional analysis of the PRKG2 R678X mutation and histological study. Growth plate cartilage was collected from the tibia of each individual. Length measurements were taken for the femur, head, tibia, fused ulna/radius, metacarpal, and metatarsal.
Statistical Analysis of Long Bone Length.
Analysis of long bones and measurements from the 6 calves produced in the breeding experiment, described previously, using PROC GLM (SAS Institute). For consistency and to avoid overfitting of the model, each model was fitted with the same factors, regardless of the independent variable (bone length). For long bone lengths, the model was
where gi denotes dwarfism genotype (yes or no), and e is the random error term. For live measurements (spine and height lengths), the model was
where s denotes sex, and Gr is the birth group (encompassing age, dam, and common environment).
Cloning, Cell Culture, Transfection, Real-Time PCR and Analysis.
We cloned the full-length bovine PRKG2 cDNA into pGEM T-easy vector (Promega). The insert was then shuttled to the pcDNA3 expression vector (Invitrogen) by digestion with XbaI and ApaI restriction enzymes (New England Biolabs) followed by ligation with T4 ligase (Promega). This bovine PRKG2 vector was then mutated to contain the PRKG2 exon 15 R678X mutation by site-directed mutagenesis using the QuikChange II kit (Stratagene). The resulting vectors were verified using restriction fragment length polymorphism (RFLP) and DNA sequence analysis. Cell culture was performed as described (7). Briefly, HUH-7 cells were transiently transfected using Lipofectamine 2000 (Invitrogen) and treated with 8Br-cGMP (Sigma) 4 h posttransfection. Cells were transfected with the following vector combinations (treatments): (1) WT bovine (bt) PRKG2 + human (h) SOX9; (2) R678X btPRKG22 +hSOX9; (3) hSOX9 + empty vector; and (4) empty vector (negative control). Treatments 1, 2, and 4 were replicated 4 times in 2 separate experiments, for a total of 8 replicates. Treatment 3 was replicated 4 times in a single experiment along with the other treatments. Cells were harvested 72 h posttransfection and total RNA was isolated with the Qiagen RNeasy Kit (Qiagen) according to manufacturer's protocol. RNA was reverse transcribed with SuperScript III (Invitrogen) for downstream amplification of cDNA in a real-time PCR that used Syber Green chemistry (Bio-Rad). COL2 mRNA levels were evaluated using the primers F- CTTAGGCCCGAGAGAGAAGG, R- ACTCAGGGTGGCAGAGTTTC, and the housekeeping gene GAPDH: F- TGATGACATCAAGAAGGTGGTGAAG, R- CCTTGGAGGCCATGTAGGCCAT. Real-time PCR was replicated 3 times for each treatment for both COL2 and GAPDH expression.
In Vivo Real-Time PCR Analysis of Bovine Growth Plate Cartilage.
Expression levels were measured in 4 R678X homozygote and 2 R678X carriers produced from the previously described breeding experiment. Total RNA from growth plate cartilage was analyzed by replicating each sample 3 times for the target and housekeeping gene in 2 separate plate replicates (i.e., 6 total replicates of both the target and housekeeping gene) using the same COL2 primers, COL10 primers F- AAT CTG AAA TGC AAG GTG CT, R- AAG ACT CAA ATA GTC ATT AAA GCA AA, PRKG2 primers F- TGGTTTTAATTGGGAGGGACT, R- AAGTCTTTGTCCCAGCCTGA, and β-actin as the housekeeping gene, F- AA GGA CTC GTA CGT GGG GGA TGA, R- AA GGA CTC GTA CGT GGG GGA TGA. All genes were amplified using the same thermocycling protocol: (1) 95 °C for 10 min, (2) 95 °C for 15 s, (3) 60 °C for 30 s, (4) repeat steps 2 and 3, 40×, (5) 60 °C + 0.5 °C/cycle for 70 cycles, (6) end.
Statistical Analysis of Real-Time PCR.
Standard curves for the reference gene were made and evaluated for all genes analyzed. Real-time PCR expression data were analyzed first using MyiQ software to normalize the data (Bio-Rad). The normalized expression data were then statistically analyzed using the natural log of the starting quantity of cDNA for transfection data and the ddCt method (28) for cattle expression data in PROC GLM (SAS Institute). The model used to analyze the cell culture transfection data were
where y is the ln starting value of COL2 mRNA adjusted for ln starting value of housekeeping gene mRNA (ln starting number COL2 − ln starting number GAPDH), p is the real-time plate replicate, t is treatment 1−4 as described in Results, d is the experiment date, and e represents error. The P values calculated from the cell culture data were Bonferroni corrected to adjust for multiple testing (29). The model used to analyze the expression data from cattle growth plate was
where y is the ddCt, target gene (either COL2 or COL10) − housekeeping gene (β-actin), for the dwarf subtracted from the target gene − housekeeping gene, for a unaffected individual; p is the real-time plate replicate; g is the genotype, and e represents error.
Supplementary Material
Acknowledgments.
The authors thank B. Schumann, C. Hines, J. Collins, J. Frerichs, B. Mote, J. R. Tait, C. Charlier, H. Takeda, N. Tama, J. Cavanagh, I. Tammen, and T. Aboellail for technical assistance and discussion. Benoit de Crombrugghe (Houston, TX) kindly provided the SOX9 expression vector. We acknowledge support from the Department of Biotechnology, Ministry of Science and Technology, Government of India (fellowships to B.P.M., D.K., and R.S.K.), and contributions from the American Angus Association and the USDA Cooperative State Research, Education, and Extension Service. Funding for this project of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa (Project No. NRSP-8), was also provided by the Hatch Act and the State of Iowa.
Footnotes
Conflict of interest statement: J.E.K. and J.M.R. have patented a genetic test to detect the bovine PRKG2 R678X mutation.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ386770, GQ386771, GQ386772, and GQ386773). Single nucleotide polymorphism data were submitted to the SNP database under NCBI nos. 159831023–159831097.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904513106/DCSupplemental.
References
- 1.McCann LP. The Battle of the Bull Runts. Columbus, OH: 1974. [Google Scholar]
- 2.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 3.Mishra BP, Reecy JM. Mutations in the limbin gene previously associated with dwarfism in Japanese brown cattle are not responsible for dwarfism in the American Angus breed. Anim Genet. 2003;34:311–312. doi: 10.1046/j.1365-2052.2003.01020.x. [DOI] [PubMed] [Google Scholar]
- 4.Harper PA, Latter MR, Nicholas FW, Cook RW, Gill PA. Chondrodysplasia in Australian Dexter cattle. Aust Vet J. 1998;76:199–202. doi: 10.1111/j.1751-0813.1998.tb10129.x. [DOI] [PubMed] [Google Scholar]
- 5.Takeda H, et al. Positional cloning of the gene LIMBIN responsible for bovine chondrodysplastic dwarfism. Proc Natl Acad Sci USA. 2002;99:10549–10554. doi: 10.1073/pnas.152337899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh JA, et al. Bulldog dwarfism in Dexter cattle is caused by mutations in ACAN. Mamm Genome. 2007;18:808–814. doi: 10.1007/s00335-007-9066-9. [DOI] [PubMed] [Google Scholar]
- 7.Chikuda H, et al. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004;18:2418–2429. doi: 10.1101/gad.1224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 9.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sriraman V, Rudd MD, Lohmann SM, Mulders SM, Richards JS. Cyclic guanosine 5′-monophosphate-dependent protein kinase II is induced by luteinizing hormone and progesterone receptor-dependent mechanisms in granulosa cells and cumulus oocyte complexes of ovulating follicles. Mol Endocrinol. 2006;20:348–361. doi: 10.1210/me.2005-0317. [DOI] [PubMed] [Google Scholar]
- 11.Pfeifer A, et al. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science. 1996;274:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann F, Feil R, Kleppisch T, Schlossmann J. Function of cGMP-dependent protein kinases as revealed by gene deletion. Physiol Rev. 2006;86:1–23. doi: 10.1152/physrev.00015.2005. [DOI] [PubMed] [Google Scholar]
- 13.Vaandrager AB, Hogema BM, de Jonge HR. Molecular properties and biological functions of cGMP-dependent protein kinase II. Front Biosci. 2005;10:2150–2164. doi: 10.2741/1687. [DOI] [PubMed] [Google Scholar]
- 14.Vaandrager AB, Bot AG, de Jonge HR. Guanosine 3′,5′-cyclic monophosphate-dependent protein kinase II mediates heat-stable enterotoxin-provoked chloride secretion in rat intestine. Gastroenterology. 1997;112:437–443. doi: 10.1053/gast.1997.v112.pm9024297. [DOI] [PubMed] [Google Scholar]
- 15.Vaandrager AB, et al. Differential role of cyclic GMP-dependent protein kinase II in ion transport in murine small intestine and colon. Gastroenterology. 2000;118:108–114. doi: 10.1016/s0016-5085(00)70419-7. [DOI] [PubMed] [Google Scholar]
- 16.El-Husseini AE, Bladen C, Williams JA, Reiner PB, Vincent SR. Nitric oxide regulates cyclic GMP-dependent protein kinase phosphorylation in rat brain. J Neurochem. 1998;71:676–683. doi: 10.1046/j.1471-4159.1998.71020676.x. [DOI] [PubMed] [Google Scholar]
- 17.Oster H, et al. cGMP-dependent protein kinase II modulates mPer1 and mPer2 gene induction and influences phase shifts of the circadian clock. Curr Biol. 2003;13:725–733. doi: 10.1016/s0960-9822(03)00252-5. [DOI] [PubMed] [Google Scholar]
- 18.Scherer-Oppliger T, Leimbacher W, Blau N, Thony B. Serine 19 of human 6-pyruvoyltetrahydropterin synthase is phosphorylated by cGMP protein kinase II. J Biol Chem. 1999;274:31341–31348. doi: 10.1074/jbc.274.44.31341. [DOI] [PubMed] [Google Scholar]
- 19.Werner C, et al. Importance of NO/cGMP signalling via cGMP-dependent protein kinase II for controlling emotionality and neurobehavioural effects of alcohol. Eur J Neurosci. 2004;20:3498–3506. doi: 10.1111/j.1460-9568.2004.03793.x. [DOI] [PubMed] [Google Scholar]
- 20.McCarty MF. cGMP may have trophic effects on beta cell function comparable to those of cAMP, implying a role for high-dose biotin in prevention/treatment of diabetes. Med Hypotheses. 2006;66:323–328. doi: 10.1016/j.mehy.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Everts-van der Wind A, et al. A high-resolution whole-genome cattle-human comparative map reveals details of mammalian chromosome evolution. Proc Natl Acad Sci USA. 2005;102:18526–18531. doi: 10.1073/pnas.0509285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koltes JE, Hu Z-L, Fritz E, Reecy JM. BEAP: The BLAST Extension and Alignment Program—a tool for contig construction and analysis of preliminary genome sequence. BMC Res Notes. 2009;2:11. doi: 10.1186/1756-0500-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannings C, Thompson EA, Skolnick MH. Probability functions on complex pedigrees. Adv Appl Prob. 1978;10:26–61. [Google Scholar]
- 24.Elston RC, Stewart J. A general model for the genetic analysis of pedigree data. Hum Hered. 1971;21:523–542. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez SA, Fernando RL, Guldbrandtsen B, Totir LR, Carriquiry AL. Sampling genotypes in large pedigrees with loops. Genet Sel Evol. 2001;33:337–367. doi: 10.1186/1297-9686-33-4-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez SA, et al. Irreducibility and efficiency of ESIP to sample marker genotypes in large pedigrees with loops. Genet Sel Evol. 2002;34:537–555. doi: 10.1186/1297-9686-34-5-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott J. Estimation of the recombination fraction in human pedigrees: Efficient computation of the likelihood for human linkage studies. Am J Hum Genet. 1974;26:588–597. [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates Inc; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.