Abstract
OBJECTIVE
SIRT1 has pleiotropic metabolic functions. We investigated whether SIRT1 genetic variation is associated with obesity.
RESEARCH DESIGN AND METHODS
In 6,251 elderly subjects from the prospective, population-based Rotterdam Study, three single nucleotide polymorphisms (SNPs) in the SIRT1 gene were studied in relation to BMI and risk of obesity (BMI ≥30 kg/m2) and prospectively with BMI change after 6.4 years of follow-up. We used cross-sectional data from 2,347 participants from the Erasmus Rucphen Family (ERF) study for replication.
RESULTS
Minor alleles of rs7895833 (G = 20.2%) and rs1467568 (A = 36.8%) were associated with lower BMI in the Rotterdam Study (P = 0.02 and 0.04) and in the replication cohort ERF study (P = 0.03 and 0.008) and in both studies combined (P = 0.002 for both SNPs), with a 0.2–0.4 kg/m2 decrease in BMI per allele copy. Carriers of these alleles had 13–18% decreased risk of obesity (for rs7895833 in the Rotterdam Study: odds ratio 0.79 [95% CI 0.67–0.94], P = 0.007; in the ERF study: 0.93 [0.73–1.19], P = 0.37; and in the studies combined 0.87 [0.77–0.97], P = 0.02; for rs1467568 in the Rotterdam Study: 0.80 [0.68–0.94], P = 0.007; in the ERF study: 0.85 [0.72–0.99], P = 0.04; and in the studies combined: 0.82 [0.73–0.92], P = 0.0009). In the Rotterdam Study, the two variants were also associated with a lower BMI increase during 6.4 years of follow-up (P = 0.01 and 0.08).
CONCLUSIONS
Two common variants in SIRT1 are associated with lower BMI in two independent Dutch populations. Carriers of these variants have 13–18% decreased risk of obesity and gain less weight over time. The availability of SIRT1 stimulators makes these findings relevant in light of the growing obesity epidemic.
SIRT1 belongs to the Sirtuin protein family of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetyleases conserved in evolution from bacteria to humans. In lower organisms such as yeast, flies, and worms, the silent information regulator Sir2 protein is related to longevity (1,2) and to lifespan extension after caloric restriction (3–5). Humans have seven sirtuins (SIRT1–7) (6), of which SIRT1 has the highest homology to yeast Sir2. SIRT1 controls numerous physiological processes and protects cells against stress (2,7–13). SIRT1 also has an important function in endocrine signaling, specifically in glucose and fat metabolism (14–18). Increased hepatic SIRT1 activity enhances gluconeogenesis and inhibits glycolysis (15,16). In the pancreas, SIRT1 stimulates insulin secretion in response to glucose (17,18). In adipose tissue, SIRT1 interacts with peroxisome proliferator–activated receptor (PPAR)-γ to repress its transcriptional activity, leading to inhibition of adipogenesis during fasting and activation of lipolysis (14). This results in fat loss, which is an important component of the effect of caloric restriction on longevity in mammals (14). It is not known whether caloric restriction can extend lifespan in humans, but clear beneficial effects on cardiovascular risk factors have been described (19,20). Fasting leads to upregulation of SIRT1 in adipose tissue of mice, pigs, and humans (21–23). Based on findings in lower organisms, it has been hypothesized that stimulation of SIRT1 by agonists may mimic the beneficial effects of caloric restriction in mammals. Resveratrol, a naturally occurring SIRT1 agonist present in red wine and grapes, was indeed recently shown to prevent diet-induced obesity and insulin resistance in mice and to improve their survival on a high-calorie diet (24,25). If activation of SIRT1 can result in loss of body fat without decreasing caloric intake, this could open the door for novel treatment and prevention strategies for obesity and related diseases. However, in humans, effects of SIRT1 stimulation have not been investigated in vivo, and there is concern that generalized SIRT1 activation may also have pro-ageing or adverse health effects, due to potential pleiotropic and tissue-dependent physiological functions (26–29).
Based on the above findings, we hypothesized that genetic variation in SIRT1 may influence BMI and the risk of obesity in humans. Previous smaller studies, which have examined the relation between variants in SIRT1 and obesity, have led to inconsistent findings. A recent case-control study of 1,068 obese patients and 313 normal-weight control subjects found a SIRT1 single nucleotide polymorphism (SNP) associated with obesity risk. Unexpectedly, male but not female carriers of the allele associated with lower obesity risk had increased visceral adiposity on computed tomography scans. This was observed only after adjustment for BMI (30). Another recent small study (31) investigating associations of SIRT1 SNPs with metabolic response to lifestyle intervention found no association of four SIRT1 SNPs in 917 overweight subjects with baseline BMI or with BMI change after 9 months follow-up. No population-based studies with large sample size and replication are available on the association between SIRT1 genetic variation, BMI, BMI change in time, and risk of obesity. We therefore assessed the association of variation in the SIRT1 gene with BMI and the risk of being overweight or obese in the large cohort of elderly subjects of the Rotterdam Study. For replication, we used subjects with a wide age range from a genetically isolated population in the Netherlands, participating in the Erasmus Rucphen Family (ERF) study. In the Rotterdam Study, we also assessed prospectively the relation of SIRT1 variants with BMI change on follow-up.
RESEARCH DESIGN AND METHODS
The Rotterdam Study.
The Rotterdam Study is a prospective, population-based cohort study among 7,983 subjects aged ≥55 years from the district of Rotterdam, the Netherlands. The study was designed to investigate the incidence and determinants of chronic disabling diseases. Rationale and design have been described previously (32,33). Informed consent was obtained from each participant, and the medical ethics committee of the Erasmus Medical Center Rotterdam approved the study. At baseline (1990–1993), all participants were interviewed and underwent extensive physical examination. At the second follow-up (1997–1999), these examinations were performed according to the same protocol.
The ERF study.
This study is a family-based cohort study that is embedded in the Genetic Research in Isolated Populations Program in the southwest of the Netherlands. The aim of this program was to identify genetic risk factors in the development of complex disorders (34–36). For the ERF study, 22 families that had at least five children baptized in the community church between 1850 and 1900 were identified with the help of genealogical records. All living descendants of these couples and their spouses were invited to take part in the study. Data collection started in June 2002 and was finished in February 2005. In this study, we focused on 2,347 participants for whom complete phenotypic, genotypic, and genealogical information was available. The medical ethics committee of Erasmus Medical Center Rotterdam approved of both studies, and informed consent was obtained from all participants.
Measurements.
For the two studies, identical protocols were used for assembling phenotypic and genotypic information. Height (cm) and weight (kg) were measured at the initial examination and, in the Rotterdam Study, also at follow-up examinations, in standing position wearing indoor clothes without shoes. BMI was computed as weight in kilograms divided by height in meters squared (kg/m2).
Genotyping.
Three tagging SNPs, rs7895833, rs1467568, and rs497849, were selected from the HapMap database (available at http://www.hapmap.org) that, together with constructed haplotypes, covered 100% of the common (minor allele frequency >10%) variation of the SIRT1 gene in Caucasians. Genotyping of the SIRT1 SNPs was performed by Taqman on genomic DNA isolated from peripheral leukocytes by standard salting-out procedures. Results were analyzed by the ABI Taqman 7900HT using the sequence detection system 2.22 software (Applied Biosystems, Foster City, CA). To confirm the accuracy of genotyping results, 332 (5%) randomly selected samples were regenotyped with the same method. No inconsistencies were observed. All used primers and probes are available on request.
Statistical analyses.
Hardy-Weinberg equilibrium of the three SIRT1 SNPs was tested with the GENEPOP package (37). Subjects were grouped according to genotype for individual SNP alleles and by allele copy number of haplotype alleles. We inferred multimarker haplotypes in the Rotterdam Study only from these SNPs using the program Phase (38). In the ERF study, haplotypes were not determined because they could not be inferred with high certainty due to the complex pedigree structure.
Haplotype alleles were numbered in order of decreasing frequency in the population (Fig. 1). Subjects were grouped according to genotype. Genotype groups were based on allele copy number (0, 1, and 2, corresponding to noncarriers, heterozygote carriers, and homozygote carriers, respectively, of the most common haplotype alleles).
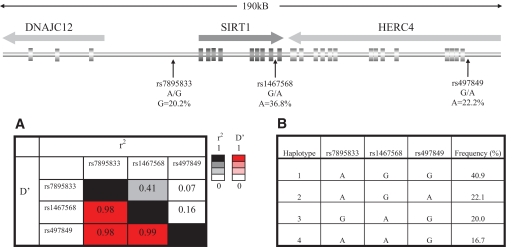
FIG. 1.
Schematic representation of the SIRT1 gene with the localization of the three tagging SNPs. A: LD plot of the SNPs with estimates of r2 in the upper right corner and D′ in the left lower corner. B: Haplotype construction and observed frequencies of the haplotypes in the Rotterdam Study.
The relation between BMI and SIRT1 genotypes was assessed using linear regression analysis, assuming an additive model. The odds ratio of being overweight (BMI ≥25 kg/m2) or obese (BMI ≥30 kg/m2) compared with normal weight (BMI 18.5–25 kg/m2) was assessed using logistic regression. The 95% CIs of the odds ratios (ORs) were calculated as the exponent of the regression coefficient and its standard error. For the assessment of an association between SIRT1 genotypes and BMI at follow-up in the Rotterdam Study, we used BMI data of the second follow-up measurement, which was (means ± SD) 6.4 ± 0.4 years after baseline. Linear regression analysis was also used for the association between SIRT1 genotypes and BMI change from baseline to second follow-up.
In the Rotterdam Study, genetic outliers as identified by the identity-by-state (IBS) clustering analysis clustering >3 SDs away from the population mean were removed prior to the analyses. Therefore, this population is ethnically homogenous. P values were corrected by the inflation factor using genomic control method (39). In the Rotterdam Study, λs (obtained after running genome-wide association [GWA] analysis for these traits) were small (1.049, 1.045, and 1.036 for BMI, the risk of overweight, and the risk of obesity, respectively).
Since genetic association may be influenced by family relationships, we performed the analyses in the ERF study with a polygenic model using a variance component approach with the pedigree information in SOLAR (Sequential Oligogenic Linkage Analysis Routine [available at http://solar.sfbrgenetics.org]). The program takes into account the familial relationships by estimating heritability from the pedigree data. Subsequently, analyses were corrected for residual genomic inflation, which was very small (λ = 1.02, values obtained from GWA studies).
We performed a meta-analysis using fixed effects on the results of the two cohorts using the software Review Manager (available at www.cc-ims.net/RevMan/RevMan5). We performed a search to add data from the literature to our meta-analysis using PubMed, with the search terms in humans “SIRT1” or “SIRT1 and polymorphisms” and “overweight” or “obesity” or “BMI” or “body weight” on 1 April 2009. Only two studies (30,31) were found. Because the study by Weirich et al. (31) was small and contained no information on the risk of obesity, we performed meta-analysis for the risk of obesity using results from the Rotterdam and ERF studies, together with the published data from the study by Peeters et al. (30). We used the rs1467568 and rs7069102 SNPs, which are in almost perfect linkage disequilibrium (LD) (D′ = 0.99 and r2 = 0.96) to meta-analyze results across studies. All analyses were adjusted for age and sex. The statistical analyses were performed using SPSS software (version 15.0) in the Rotterdam Study and with SOLAR software (package 2.1.2) in the ERF study.
RESULTS
General characteristics.
In Table 1, baseline characteristics of the two study populations are presented. In both studies, a slight majority of participants were females. BMI was 1.1% higher in subjects from the ERF study than in the Rotterdam Study at baseline.
TABLE 1.
General and body composition characteristics of two study populations
| Rotterdam Study | ERF study | |
|---|---|---|
| n | 6,251 | 2,347 |
| Women | 3,665 (59) | 1,385 (55) |
| Age (years) | 69.1 ± 8.8 | 47.5 ± 13.8 |
| Range | 55.1–99.0 | 17.6–85.3 |
| Height (m) | 1.67 ± 0.09 | 1.68 ± 0.09 |
| Weight (kg) | 73.2 ± 12.0 | 75.3 ± 15.4 |
| BMI (kg/m2) baseline | 26.3 ± 3.7 | 26.6 ± 4.4 |
| BMI (kg/m2) at follow-up 2 (n = 3,630) | 26.8 ± 3.9 | |
| BMI change (kg/m2)* | 0.55 ± 1.8 | |
| Range | −10.0 to +11.0 |
Data are means ± SD or n (%), unless otherwise indicated.
*Change in BMI from baseline to follow-up 2 examination.
SIRT1 genotype.
Figure 1 shows the schematic representation of the SIRT1 locus with the localization of the three tagging SNPs and an LD plot of the SNPs (D′ and r2) as well as haplotype construction and observed frequencies of the haplotypes in the Rotterdam Study. In the Rotterdam Study, allele and genotype distributions of the three tagging SNPs of SIRT1 follow Hardy-Weinberg equilibrium proportions (P > 0.10). LD between the SNPs was high (D′ > 0.8), which enabled us to infer multimarker haplotypes in the Rotterdam Study with high confidence. The haplotype frequencies of the four most common haplotypes were 40.9, 22.1, 20.0, and 16.7%. As shown in Fig. 1, rs497849 fully tags haplotype 2 and rs7895833 fully tags haplotype 3. In the ERF study, we genotyped the two SNPs showing association with obesity in the Rotterdam Study. These SNPs showed slightly different frequencies between the two studies but were also in Hardy-Weinberg equilibrium proportions in the ERF study.
Relation of SIRT1 variants with BMI.
Table 2 shows the relationship of SIRT1 SNPs with BMI in the Rotterdam Study at baseline and at the second follow-up examination and the replication data of two SIRT1 SNPs in the ERF study. In the Rotterdam Study, two of three SNPs, rs7895833 and rs1467568, had a significant association with BMI at baseline (P = 0.02 and 0.04 for two SNPs at baseline, respectively, and P = 0.02 for both SNPs at the follow-up examination) with an allele-dose effect. Carriers of the minor alleles of the two SNPs had, on average, 0.2–0.3 kg/m2 decreased BMI per allele copy. The third SNP (rs497849) that tags haplotype 2 showed no association with BMI. Haplotype analysis offered little additional information. For haplotype 1, which contains the major alleles for two SNPs associated with BMI (rs7895833 and rs1467568), we observed a significant positive association with BMI that was stronger at the second follow-up (β = 0.273, P = 0.008) than at baseline (β = 0.130, P = 0.07). Haplotype 4 showed no association with BMI (data not shown).
TABLE 2.
BMI in the Rotterdam Study and the ERF study by SIRT1 genotype and longitudinal BMI change in the Rotterdam Study
| Genotype | Rotterdam Study baseline |
Rotterdam Study follow-up |
Rotterdam Study BMI change |
ERF study |
P combined‡ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF* | Effect size per allele copy† | P trend | MAF | Effect size per allele copy | P trend | MAF | Effect size per allele copy | P trend | MAF | Effect size per allele copy | P trend | ||
| rs7895833 | 0.20 | −0.190 (0.08) | 0.02 | 0.20 | −0.291 (0.12) | 0.02 | 0.20 | −0.126 (0.05) | 0.01 | 0.23 | −0.350 (0.16) | 0.03 | 0.002 |
| rs1467568 | 0.37 | −0.149 (0.07) | 0.04 | 0.37 | −0.238 (0.10) | 0.02 | 0.37 | −0.075 (0.04) | 0.08 | 0.41 | −0.372 (0.14) | 0.008 | 0.002 |
| rs497849 | 0.22 | +0.030 (0.08) | 0.70 | 0.22 | −0.054 (0.11) | 0.62 | 0.22 | −0.020 (0.05) | 0.69 | — | |||
| Haplotype 1 | 0.41 | +0.130 (0.07) | 0.07 | 0.41 | +0.273 (0.10) | 0.008 | 0.41 | +0.070 (0.05) | 0.11 | — | |||
Analyses adjusted for age, sex, and genomic control and, in ERF, also for family structure.
*Minor allele rs7895833; G, rs1467568; A, rs497849.
†Effect size is difference in BMI (kg/m2) (SE) or difference in BMI change per allele.
‡P combined analysis for Rotterdam Study baseline and the ERF study. MAF, minor allele frequency.
Similar to the findings in the Rotterdam Study, carriers of the minor alleles of rs7895833 and rs1467568 had a lower BMI in the ERF study (P = 0.03 and 0.008 for the two SNPs, respectively). To exclude that associations between the SNPs and BMI were explained by a difference in body height, we also analyzed the relation between the SNPs and height. No differences in height were seen between the genotypes (data not shown).
Combined analysis of relation of SIRT1 variants with BMI in two studies.
The combined analysis (Table 2) on the outcomes of the two studies, the Rotterdam Study baseline and the ERF study, showed a highly significant association with BMI (P = 0.002 for both SNPs).
Relation of SIRT1 variants with BMI change during follow-up in the Rotterdam Study.
Table 2 also presents data from linear regression analysis of change in BMI or body weight from baseline to second follow-up for three SNPs in the SIRT1 gene in the Rotterdam Study over a period of 6.4 years on average. The two SNPs, rs7895833 and rs1467568, that were associated with BMI at baseline and follow-up and with the risk of overweight and obesity were also associated with BMI change at the second follow-up examination. After adjustment for age and sex, the P value for trend was 0.01 for rs7895833 and P = 0.08 for rs1467568. Adjustment for BMI at baseline did not change these results, and findings were similar when change of body weight instead of BMI was assessed (data not shown).
SIRT1 variants and risk of overweight/obesity.
Table 3A presents the odds ORs and 95% CIs for the risk of being overweight (BMI ≥25 kg/m2) compared with being normal weight (BMI 18.5–25 kg/m2) for the two SNPs in the SIRT1 gene in the Rotterdam Study at baseline and in the replication cohort of the ERF study. In the Rotterdam Study, carriers of the minor alleles of rs7895833 had 12% decreased OR (95% CI) of being overweight compared with noncarriers (0.88 [0.79–0.99], P = 0.03). For rs1467568, a similar, yet not significant, trend was seen with an OR of 0.91 (0.82–1.02). Similar results were observed in the ERF study, with the strongest and most significant effect for rs1467568. At the second follow-up measurement of the Rotterdam Study (n = 3,630), the risk of overweight in relation to SIRT1 was similar to the risk at baseline but did not reach statistical significance (for rs7895833: 0.91 [0.79–1.05], P = 0.21; for rs1467568: 0.90 [0.78–1.04], P = 0.14, data not shown).
TABLE 3.
Risk of being overweight (A) or obese (B) in the Rotterdam Study at baseline, the ERF study, and studies combined by SIRT1 genotype
| Genotype | n case/control subjects | OR (95% CI) | P value dominant model | P value additive model | P value recessive model | Genotype | n case/control subjects | OR (95% CI) | P value dominant model | P value additive model | P value recessive model | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A: Overweight (BMI >25 kg/m2) | rs7895833 | rs1467568 | ||||||||||
| Rotterdam Study | AA | 2,389/1,390 | 1 (reference) | GG | 1,497/863 | 1 (reference) | ||||||
| AG + GG | 1,311/869 | 0.88 (0.79–0.99) | 0.03 | 0.03 | 0.42 | GA + AA | 2,207/1,398 | 0.91 (0.82–1.02) | 0.09 | 0.27 | 0.85 | |
| ERF study | AA | 854/487 | 1 (reference) | GG | 509/259 | 1 (reference) | ||||||
| AG + GG | 559/359 | 0.95 (0.84–1.06) | 0.35 | 0.08 | 0.01 | GA + AA | 910/593 | 0.88 (0.84–0.91) | 0.02 | 1.9 × 10−6 | 0.02 | |
| Studies combined | 0.91 (0.84–0.99) | 0.02 | 0.006 | 0.02 | 0.89 (0.83–0.97) | 0.005 | 3.5 × 10−6 | 0.06 | ||||
| B: Obese (BMI >30 kg/m2) | rs7895833 | rs1467568 | ||||||||||
| Rotterdam Study | AA | 577/1,390 | 1 (reference) | GG | 377/863 | 1 (reference) | ||||||
| AG + GG | 282/869 | 0.79 (0.67–0.94) | 0.007 | 0.008 | 0.20 | GA + AA | 483/1,398 | 0.80 (0.68–0.94) | 0.007 | 0.01 | 0.34 | |
| ERF study | AA | 276/487 | 1 (reference) | GG | 164/259 | 1 (reference) | ||||||
| AG + GG | 161/359 | 0.93 (0.73–1.19) | 0.37 | 0.23 | 0.25 | GA + AA | 277/593 | 0.85 (0.72–0.99) | 0.04 | 0.05 | 0.35 | |
| Studies combined | 0.87 (0.77–0.97) | 0.02 | 0.009 | 0.10 | 0.82 (0.73–0.92) | 0.0009 | 0.002 | 0.20 |
Analyses adjusted for age, sex, and genomic control and, in the ERF study, also for family structure. BMI of control subjects 18.5–25 kg/m2.
In Table 3B, the ORs and 95% CIs for the risk of being obese (BMI ≥30 kg/m2) compared with being normal weight (BMI 18.5–25 kg/m2) are presented for two SNPs in the SIRT1 gene in the two studies. In the Rotterdam Study, both SNPs were associated with the risk of being obese. Carriers of minor alleles of the two SNPs had a 20–21% decreased risk (for rs7895833: 0.79 [0.67–0.94], P = 0.007; for rs1467568: 0.80 [0.68–0.94], P = 0.007). Similar trends were seen in the ERF study population, only statistically significant for rs1467568. At the second follow-up measurement of the Rotterdam Study, the association with risk of obesity was similar for carriers of the two SNPs (for rs7895833: 0.78 [0.64–095], P = 0.01; for rs1467568: 0.69 [0.57–0.84], P = 0.0002, data not shown).
Combined analysis of the risk of overweight/obesity in two studies.
Table 3 also presents the data from the meta-analysis of the risk of overweight or obesity for the two SNPs in the SIRT1 gene in the Rotterdam Study at baseline and the ERF study. Carriers of the minor alleles of two SNPs had 9–11% decreased risk of being overweight compared with noncarriers (for rs7895833: OR 0.91 [95% CI 0.84–0.99], P = 0.02; for rs1467568: 0.89 [0.83–0.97], P = 0.005), with evidence for allele dose effects (P value additive model = 0.006 and 3.5 × 10−6 for rs7895833 and rs1467568, respectively). The risk for obesity was decreased by 13–18% (for rs7895833: 0.87 [0.77–0.97], P = 0.02; for rs1467568: 0.82 [0.73–0.92], P = 0.0009), with allele dose effects (P value additive models 0.009 and 0.002 for rs7895833 and rs1467568, respectively). The P values for the recessive model were always higher than those for the dominant or additive models.
Meta analysis with published data.
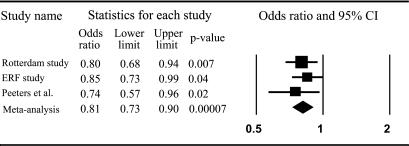
Figure 2 shows a forest plot from the meta-analysis of risk of obesity for carriers compared with noncarriers of the A allele of rs1467568 in the Rotterdam and ERF studies and the C allele of rs7069102 from the study by Peeters et al. (30). The A and C alleles of these SNPs are in high LD (r2 = 0.96). The combined OR for the three studies was 0.81 (95% CI 0.73–0.90) (P = 0.00007).
FIG. 2.
Meta-analysis on the risk of obesity from three studies by SIRT1 genotype for carriers compared with noncarriers of the A allele of rs1467568 in the Rotterdam Study and ERF study and the C allele of rs7069102 from the study by Peeters et al. (30).
DISCUSSION
In this study, we show in two large and independent Dutch Caucasian populations that the minor alleles of two common SIRT1 variants are associated with a decreased BMI. Carriers of these two common genetic variants had 9–11% decreased risk of being overweight and 13–18% decreased risk of being obese compared with noncarriers. In line with these findings, we also observed in the Rotterdam Study that carriers of these SIRT1 variants had a smaller increase of BMI over a 6.4-year follow-up period.
Our study is hypothesis driven and based on the known functions of SIRT1 in cell cultures and animal studies, such as an inhibitory effect of SIRT1 on PPAR-γ in adipose tissue (14) and a stimulatory effect on PPAR coactivator-1α (PGC-1α) (24,25). This powerful transcriptional coactivator is regarded as a key mediator of many of the known beneficial effects of physical activity on skeletal muscle physiology (40). Treatment of mice with resveratrol leads to increased mitochondrial biogenesis and increased energy expenditure in mice, possibly by SIRT1-mediated increase in PGC-1α activity (25). It is also possible that the effects on BMI are caused by an influence of SIRT1 on appetite and energy intake, since SIRT1 is highly expressed in brain (41).
Interestingly, SIRT1 has recently been shown to modulate CLOCK-gene expression (42–44), and it may thus form an intriguing link between sensing of cellular metabolism and the circadian clock, which merits further study.
Inhibitory effects of SIRT1 on differentiation of skeletal myoblasts have been shown under glucose-restricted conditions in relation to activation of the AMP-activated protein kinase (45). Therefore, we cannot exclude that effects on lean mass as well as on fat mass explain the relation with BMI that we observed. Further studies are needed to investigate the relation between SIRT1 genetic variants and human body composition traits and muscle strength. Yet, the strong association with the risk of obesity that we found makes a predominant effect on lean mass unlikely since obesity is associated more strongly with excess adipose tissue than with excess muscle mass.
This is the first large, population-based study reporting an association between SIRT1 genetic variation and BMI and obesity in humans with validation in an independent, population-based cohort. Two recently published genetic studies in humans corroborate our findings. In a Belgian case-control study with 1,068 obese patients and 313 normal-weight control subjects, carriers of the minor allele of the SNP rs7069102 (which is in high LD [r2 = 0.96] with our SNP rs1467568) had a reduced obesity risk (OR 0.74 [95% CI 0.57–0.96], P = 0.025) (30). After including their results in a meta-analysis with our results, the association became more significant and the combined P value decreased from 9 × 10−4 to 7 × 10−5. In their study, the variant that was protective against obesity was associated with increased visceral obesity as measured by computed tomography scanning after adjusting for BMI in obese male, but not female, subjects. This unexpected finding may be caused by overadjustment when the decrease in BMI is caused by a decrease in lean as well as in fat mass or represent a real sex-specific effect, which needs further investigation. A second recent study in 917 overweight German Caucasian subjects found no significant change in BMI by SIRT1 genotype, yet we discovered in their data a similar decrease in BMI as we found for two SNPs in high LD with ours (31). For carriers of the minor variants of rs7069102 (r2 = 0.96 with rs1467568), BMI was 2.4% lower, and for rs730821 (r2 = 1.0 with rs7895833), BMI was 6.1% lower, compared with noncarriers. These differences were not significant, possibly because of low power. Recent GWA studies have shown that common SNPs contributing to common complex diseases have modest effects and require large sample sizes to be discovered (46). The absence of SIRT1 among the genome-wide significant new findings in recent GWA studies for BMI may be explained by the stringent criteria used to correct for multiple testing in GWA studies and shows the added value of candidate gene analyses.
The strength of our study is the use of two large and independent population-based cohorts with consistency of findings between studies and between cross-sectional and longitudinal analyses. The findings robustly show that SIRT1 variants influence human obesity. A limitation of our study is the use of tagging SNPs. The SNPs we selected are noncoding and we therefore assume these two variants to be linked with one or more functional variants within the SIRT1 gene or its regulatory regions. This will require more in-depth molecular studies. Future fine mapping and resequencing of the SIRT1 gene may detect such functional variants. Further studies into underlying mechanisms and body composition are also needed as well as prospective studies on a relation with obesity-associated diseases. Because we have no information on functionality of the genetic variants, we cannot conclude from our data whether the lower BMI that we observed in carriers of the minor alleles of the two SIRT1 SNPs is associated with increased or decreased SIRT1 activity. However, based on the known functions of SIRT1, one could speculate that the minor alleles of rs7895833 and rs1467568 are linked with functional variants leading to increased SIRT1 activity.
The results of our study may have important clinical implications. Obesity has become a global epidemic and represents an important risk factor for type 2 diabetes, hypertension, cardiovascular disease, stroke, some types of cancer, and disability. Few, if any, effective options for treatment and prevention are available. The findings of our study are in line with speculations that modulators of SIRT1 activity may decrease BMI and the risk of becoming overweight and obese and may thus provide a valuable new strategy for treatment and prevention of obesity and its related diseases. An attractive aspect of our findings is that, in contrast to novel genetic findings from GWA studies, many data exist on SIRT1 biological functions, while in addition several SIRT1 modifiers have already been identified. The protective effects of the SIRT1-stimulating flavenoid resveratrol against diet-induced obesity in mice may potentially apply to humans as well (25), but its effects are not only SIRT1 mediated (47,48) and low bioavailability is a concern (49). More research on pharmacology and possible side effects in humans is necessary for this compound as well as for recently developed more potent SIRT1 activators (50). Alternatively, SIRT1 inhibitors might be of use in cachexia.
In summary, we found that carriers of two common variants in the SIRT1 gene have lower BMI, a 13–18% decreased risk of being obese, and less BMI gain in time. Together with results from recent studies, these consistent findings warrant research into a potential role for SIRT1 modulators in prevention and treatment of human obesity.
Acknowledgments
The Rotterdam study was funded by the Netherlands Organization for Scientific Research under the Research Institute for Diseases in the Elderly (grant 014-90-001) and the European Commission (QL46-CT-2002-02629, GENOMOS, Health-F2-2008-201865, GEFOS) and is supported by the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (project no. 050-060-810). The ERF study was supported by a joint grant from the Center of Medical Systems Biology.
No potential conflicts of interest relevant to this article were reported.
The contributions of the participants, general practitioners, and pharmacists of the Rotterdam Study and the ERF study are gratefully acknowledged. We thank Mila Jhamai for genotyping and Slavica Pecioska for assistance with statistical analyses.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Sinclair DA, Guarente L: Extrachromosomal rDNA circles: a cause of aging in yeast. Cell 1997; 91: 1033– 1042 [DOI] [PubMed] [Google Scholar]
- 2.Blander G, Guarente L: The Sir2 family of protein deacetylases. Annu Rev Biochem 2004; 73: 417– 435 [DOI] [PubMed] [Google Scholar]
- 3.Lin SJ, Defossez PA, Guarente L: Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000; 289: 2126– 2128 [DOI] [PubMed] [Google Scholar]
- 4.Rogina B, Helfand SL: Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 2004; 101: 15998– 16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Tissenbaum HA: Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev 2006; 127: 48– 56 [DOI] [PubMed] [Google Scholar]
- 6.Frye RA: Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 2000; 273: 793– 798 [DOI] [PubMed] [Google Scholar]
- 7.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W: Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001; 107: 137– 148 [DOI] [PubMed] [Google Scholar]
- 8.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME: Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004; 303: 2011– 2015 [DOI] [PubMed] [Google Scholar]
- 9.Araki T, Sasaki Y, Milbrandt J: Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 2004; 305: 1010– 1013 [DOI] [PubMed] [Google Scholar]
- 10.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J: Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res 2004; 95: 971– 980 [DOI] [PubMed] [Google Scholar]
- 11.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Neri C: Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet 2005; 37: 349– 350 [DOI] [PubMed] [Google Scholar]
- 12.Pillai JB, Isbatan A, Imai S, Gupta MP: Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem 2005; 280: 43121– 43130 [DOI] [PubMed] [Google Scholar]
- 13.Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isono M, Isshiki K, Uzu T, Kashiwagi A, Koya D: Silent information regulator 2 (SIRT1) attenuates oxidative stress-induced mesangial cell apoptosis via p53 deacetylation. Free Radic Biol Med 2006; 40: 2175– 2182 [DOI] [PubMed] [Google Scholar]
- 14.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L: Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 2004; 429: 771– 776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P: Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005; 434: 113– 118 [DOI] [PubMed] [Google Scholar]
- 16.Frescas D, Valenti L, Accili D: Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem 2005; 280: 20589– 20595 [DOI] [PubMed] [Google Scholar]
- 17.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S: Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab 2005; 2: 105– 117 [DOI] [PubMed] [Google Scholar]
- 18.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L: Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol 2006; 4: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson-Meyer DE, Heilbronn LK, Redman LM, Newcomer BR, Frisard MI, Anton S, Smith SR, Alfonso A, Ravussin E: Effect of calorie restriction with or without exercise on insulin sensitivity, β-cell function, fat cell size, and ectopic lipid in overweight subjects. Diabetes Care 2006; 29: 1337– 1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefevre M, Redman LM, Heilbronn LK, Smith JV, Martin CK, Rood JC, Greenway FL, Williamson DA, Smith SR, Ravussin E: Caloric restriction alone and with exercise improves CVD risk in healthy non-obese individuals. Atherosclerosis 2008; 203: 206– 213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA: Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004; 305: 390– 392 [DOI] [PubMed] [Google Scholar]
- 22.Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY: Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett 2008; 582: 2417– 2423 [DOI] [PubMed] [Google Scholar]
- 23.Pedersen SB, Olholm J, Paulsen SK, Bennetzen MF, Richelsen B: Low Sirt1 expression, which is upregulated by fasting, in human adipose tissue from obese women. Int J Obes (Lond) 2008; 32: 1250– 1255 [DOI] [PubMed] [Google Scholar]
- 24.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA: Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006; 444: 337– 342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J: Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 2006; 127: 1109– 1122 [DOI] [PubMed] [Google Scholar]
- 26.Yang T, Fu M, Pestell R, Sauve AA: SIRT1 and endocrine signaling. Trends Endocrinol Metab 2006; 17: 186– 191 [DOI] [PubMed] [Google Scholar]
- 27.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, Bedalov A: Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res 2006; 66: 4368– 4377 [DOI] [PubMed] [Google Scholar]
- 28.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L: Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev 2008; 22: 1753– 1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Xu W, McBurney MW, Longo VD: SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab 2008; 8: 38– 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peeters AV, Beckers S, Verrijken A, Mertens I, Roevens P, Peeters PJ, Van Hul W, Van Gaal LF: Association of SIRT1 gene variation with visceral obesity. Hum Genet 2008; 124: 431– 436 [DOI] [PubMed] [Google Scholar]
- 31.Weyrich P, Machicao F, Reinhardt J, Machann J, Schick F, Tschritter O, Stefan N, Fritsche A, Haring HU: SIRT1 genetic variants associate with the metabolic response of Caucasians to a controlled lifestyle intervention: the TULIP study. BMC Med Genet 2008; 9: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA: Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol 1991; 7: 403– 422 [DOI] [PubMed] [Google Scholar]
- 33.Hofman A, Breteler MM, van Duijn CM, Krestin GP, Pols HA, Stricker BH, Tiemeier H, Uitterlinden AG, Vingerling JR, Witteman JC: The Rotterdam Study: objectives and design update. Eur J Epidemiol 2007; 22: 819– 829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaessen N, Heutink P, Houwing-Duistermaat JJ, Snijders PJ, Rademaker T, Testers L, Batstra MR, Sandkuijl LA, van Duijn CM, Oostra BA: A genome-wide search for linkage-disequilibrium with type 1 diabetes in a recent genetically isolated population from the Netherlands. Diabetes 2002; 51: 856– 859 [DOI] [PubMed] [Google Scholar]
- 35.Sayed-Tabatabaei FA, van Rijn MJ, Schut AF, Aulchenko YS, Croes EA, Zillikens MC, Pols HA, Witteman JC, Oostra BA, van Duijn CM: Heritability of the function and structure of the arterial wall: findings of the Erasmus Rucphen Family (ERF) study. Stroke 2005; 36: 2351– 2356 [DOI] [PubMed] [Google Scholar]
- 36.Santos RL, Zillikens MC, Rivadeneira FR, Pols HA, Oostra BA, van Duijn CM, Aulchenko YS: Heritability of fasting glucose levels in a young genetically isolated population. Diabetologia 2006; 49: 667– 672 [DOI] [PubMed] [Google Scholar]
- 37.Raymond M, Rousset F: GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 1995; 86: 248– 249 [Google Scholar]
- 38.Stephens M, Smith NJ, Donnelly P: A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978– 989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bacanu SA, Devlin B, Roeder K: The power of genomic control. Am J Hum Genet 2000; 66: 1933– 1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handschin C, Spiegelman BM: The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 2008; 454: 463– 469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I: Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 2005; 16: 4623– 4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P: The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008; 134: 329– 340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U: SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008; 134: 317– 328 [DOI] [PubMed] [Google Scholar]
- 44.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai SI, Bass J: Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009; 324: 651– 654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V: Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 2008; 14: 661– 673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN: Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008; 9: 356– 369 [DOI] [PubMed] [Google Scholar]
- 47.Grubisha O, Smith BC, Denu JM: Small molecule regulation of Sir2 protein deacetylases. FEBS J 2005; 272: 4607– 4616 [DOI] [PubMed] [Google Scholar]
- 48.Zhang J: Resveratrol inhibits insulin responses in a SirT1-independent pathway. Biochem J 2006; 397: 519– 527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Lastra CA, Villegas I: Resveratrol as an anti-inflammatory and anti-aging agent: mechanisms and clinical implications. Mol Nutr Food Res 2005; 49: 405– 430 [DOI] [PubMed] [Google Scholar]
- 50.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH: Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 2007; 450: 712– 716 [DOI] [PMC free article] [PubMed] [Google Scholar]