Abstract
OBJECTIVE
Digenic causes of human disease are rarely reported. Insulin via its receptor, which is encoded by INSR, plays a key role in both metabolic and growth signaling pathways. Heterozygous INSR mutations are the most common cause of monogenic insulin resistance. However, growth retardation is only reported with homozygous or compound heterozygous mutations. We describe a novel translocation [t(7,19)(p15.2;p13.2)] cosegregating with insulin resistance and pre- and postnatal growth deficiency. Chromosome translocations present a unique opportunity to identify modifying loci; therefore, our objective was to determine the mutational mechanism resulting in this complex phenotype.
RESEARCH DESIGN AND METHODS
Breakpoint mapping was performed by fluorescence in situ hybridization (FISH) on patient chromosomes. Sequencing and gene expression studies of disrupted and adjacent genes were performed on patient-derived tissues.
RESULTS
Affected individuals had increased insulin, C-peptide, insulin–to–C-peptide ratio, and adiponectin levels consistent with an insulin receptoropathy. FISH mapping established that the translocation breakpoints disrupt INSR on chromosome 19p15.2 and CHN2 on chromosome 7p13.2. Sequencing demonstrated INSR haploinsufficiency accounting for elevated insulin levels and dysglycemia. CHN2 encoding β-2 chimerin was shown to be expressed in insulin-sensitive tissues, and its disruption was shown to result in decreased gene expression in patient-derived adipose tissue.
CONCLUSIONS
We present a likely digenic cause of insulin resistance and growth deficiency resulting from the combined heterozygous disruption of INSR and CHN2, implicating CHN2 for the first time as a key element of proximal insulin signaling in vivo.
The genetic susceptibility to insulin resistance can involve the disruption of a single gene (e.g., INSR) or may involve the interplay of many genetic loci (including PPARG, FTO, HNF1B, etc.). However, there is currently only one known digenic disorder of insulin resistance resulting from mutations in peroxisome proliferator–activated receptor gamma (PPARG) and protein phosphatase 1 regulatory subunit 3 (PPPTR3A) (1). Compound heterozygous mutations in these genes, which are primarily involved in carbohydrate or lipid metabolism, respectively, can combine to produce a phenotype of extreme insulin resistance and lipodystrophy (1). Interestingly, individuals who possess only one mutation have normal insulin levels demonstrating that disruption of both genes and therefore pathways is necessary to result in disease (1).
INSR encodes the insulin receptor with a key role in both major arms of the insulin signaling pathways, specifically the metabolic pathway mainly via IRS-1/Akt2/AS160 signaling and the growth pathway mainly via IRS-2/extracellular signal–related kinase (ERK) signaling (2). INSR mutations are the most common cause of monogenic insulin resistance and cause a clinical spectrum of disease ranging from type A insulin resistance to the most severe form of insulin receptoropathy, leprechaunism (also known as Donohue's syndrome) (3–6).
Growth is a complex biological process with multiple interacting pathways. The key pathways involved in growth include the insulin signaling pathway and the growth hormone/insulin-like growth factor pathway (7–9). Intrauterine growth is also regulated by multiple fetal and maternal factors including genetic and epigenetic factors and various environmental factors (10).
We describe a family with a reciprocal translocation [t(7,19)(p15.2;p13.2)] cosegregating with insulin resistance and pre- and postnatal growth deficiency. We demonstrate that the breakpoint on chromosome 19 disrupts INSR, causing monoallelic expression. Haploinsufficent INSR individuals have type A insulin resistance with no apparent severe growth deficiency (5). Given the short stature and intrauterine growth retardation also seen in this family, we hypothesized this could be due to either complete loss of the functional INSR protein (due to the second INSR allele harboring a mutation or due to a dominant negative effect of a mutant INSR protein) or a digenic syndrome due to disruption of a second gene involved in growth by the chromosome 7 breakpoint. The first two hypotheses were excluded by the demonstration of a normal INSR DNA sequence and monoallelic INSR expression, while further cytogenetic analysis established that the second breakpoint on chromosome 7p15.2 disrupted CHN2 encoding β-2 chimerin. The demonstration that disruption of CHN2 affects both pre- and postnatal growth suggests that chimerins may play an important role in early growth and development.
RESEARCH DESIGN AND METHODS
We report a family with pre- and postnatal growth deficiency, insulin resistance, and early-onset diabetes (Table 1). The female proband was born at term weighing 1.85 kg (<5th centile), and all other intrauterine growth parameters were <5th centile (Table 1). Her growth was maintained below the 5th centile. She was hypoglycemic at birth and treated with intravenous 10% glucose infusion for 36 h. She has mild dysmorphic features with a triangular face, irregular teeth, hypertrichosis, a masculine appearance, and no history of miscarriage. The proband was diagnosed with diabetes requiring insulin treatment at age 15 years and since then has been on insulin therapy with variable dosage, notably requiring less insulin throughout pregnancy and dietary treatment alone for 2 months postnatally. She is currently managed with gradually increasing insulin doses (see supplementary methods Table 1, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0787/DC1). Her fasting insulin and C-peptide levels are greatly elevated (Table 1). Karyotype analysis identified the following karyotype: [46, XX, t(7,19)(p15.2;p13.2)].
TABLE 1.
Clinical and biochemical characteristics of patients with the novel translocation
| Proband | Son (10th centile) | |
|---|---|---|
| Current age (years) | 24 | 2 |
| Gestational age (weeks) | 40 | 35 |
| Birth weight (kg) (5th–95th centile) | 1.85 (2.69–4.03) | 1.9 (1.84–3.2) |
| Length at birth (cm) (5th–95th centile) | 43 (43.47–52.69) | 39 (44.08–49.7) |
| Head circumference at birth (cm) | 33 (10th centile) | 32 |
| Current weight (kg) | 38 | 10.3 (3rd centile) |
| Current height (cm) (mid-parental height) | 145 (161) | 80 (< 3rd centile) |
| Current BMI (kg/m2) (normal range) | 18 (19–25) | 16 (15–18) |
| Acanthosis nigricans | No | No |
| Hypertrichosis (facial, limbs, and trunk) | Yes (aged 11 years) | No |
| Menarche (years) | 11 | — |
| Age at diagnosis of hyperglycemia (years) | 15.9 (symptoms from age 10 years) | 0.25 |
| Current treatment | Insulin | Diet |
| Glucose in the neonatal period (mmol/l) (normal >2.6 mmol/l) | 3.2 | 1.4 |
| Fasting glucose at diagnosis (mmol/l) (normal <7 mmol/l) | 11.2 | 2.3–8 (preprandially) |
| Ketonuria | Trace | Negative |
| Autoantibodies (GAD, IA2A) | Negative | Negative |
| Insulin (pmol/l) (5th–95th centile) | 3,724 (15.2–159) | 6,025 (18–46.8) |
| C-peptide (pmol/l) (normal 160–1,100 pmol/l) | 2,931 | 8,081 |
| Insulin–to–C-peptide ratio (normal <0.1) | 1.27 | 0.74 |
| Adiponectin (ng/ml)* | 22.3 | 26.1 |
| Triglycerides (mmol/l) | 0.62 (0.55–1.9) | 1.0 |
| Cholesterol (mmol/l) | 3.9 (3.5–5.5) | 4.3 (3.8–4.5) |
| HDL (mmol/l) | 1.62 (0.8–1.8) | 1.61 (0.82–0.94) |
| LDL (mmol/l) | 1.68 (0.8–2.2) | 2.05 |
| IGF-1 (normal range for age) | 873 ng/ml (90–500) | 203.7 ng/ml (70–380) |
| Proteinuria (normal range) (mg/24 h) | 353.4 (<30) (pre-pregnancy) | NA |
Data are control means (95% CI) unless otherwise indicated.
*Range for adults (9.8 +/− 1.26 ng/ml). NA, not available.
The proband's son was the first-born child of a nonconsanguineous marriage delivered by caesarean section at 35 weeks' gestation. Aminocentesis at 16 weeks' gestation identified the following karyotype [46, XY, t(7,19)(p15.2;p13.2)]. The son weighed 1.9 kg (<10th centile), and his length was <5th centile (Table 1). He has no dysmorphic features. He had recurrent neonatal hypoglycemia and was noted to have preprandial hypoglycemia and postprandial hyperglycemia at age 3 months. His fasting insulin and C-peptide levels were greatly elevated (Table 1). He is currently age 22 months and continues on dietary management; his growth trajectory remains below the 3rd centile.
The proband's parents are nonconsanguineous and have no evidence of diabetes, growth retardation, or dysmorphic features. The height of the proband's mother, father, and older sister is 162, 174, and 173 cm, respectively. Chromosomal analysis in both parents revealed a normal karyotype.
Biochemical analysis.
Insulin and C-peptide levels were assayed using a radioimmunoassay (Immunotech, Prague, Czech Republic). Adiponectin was also assayed using a radioimmunoassay (Linc, Millipore, U.K.), and normative data were acquired from 27 healthy control subjects.
Bioinformatic tools.
The University of California Santa Cruz Web site (http://genome.ucsc.edu/), Online Mendelian Inheritance in Man (OMIM) (www.ncbi.nlm.nih.gov), and GeneSniffer (www.genesniffer.org) were used to identify and prioritize biological candidate genes within the region of both breakpoints.
Chromosome preparation, DNA isolation, and establishment of patient cell lines.
Peripheral blood samples were obtained from the proband and her son, and conventional methods were used to prepare metaphase spreads for FISH analysis, extract genomic DNA, and establish an Epstein-Barr virus (EBV)-transformed lymphoblastoid cell line.
Fluorescence in situ hybridization analysis.
Fluorescence in situ hybridization (FISH) analysis was performed using standard techniques (see online supplementary methods).
Gene dosage investigations.
Multiplex ligation-dependent probe amplification (MLPA) and SYBR green analysis were used to quantify gene dosage of INSR and CHN2, respectively (see online supplementary methods).
Sequencing.
All 22 exons and exon-intron boundaries of INSR were amplified and sequenced on an ABI 3700 genetic analyzer (Applied Biosystems, Warrington, U.K.). Sequences were compared with the published sequence (NM_000208) using Mutation Surveyor version 3.4 (Softgenetics, Cambridge, U.K.). The entire cDNA sequence of INSR was amplified in nine overlapping fragments and sequenced as above. Coding sequences with known single nucleotide polymorphisms (SNPs) of CHN2 and adjacent biologically plausible genes (GHRHR, JAZF1, GRB10) were amplified and sequenced in patient genomic DNA. All primer sequences are available in the online supplementary methods.
RNA extraction, retrotranscription, and gene expression studies.
RNA was extracted and retrotranscribed from patient EBV–transformed lymphoblastoid cell lines, and the proband's subcutaneous adipose tissue was obtained by needle biopsy using standard methods (see online supplementary methods). A commercially available RNA library from a standard panel of tissues (Human Total RNA Master Panel II) was purchased from Clontech (Saint-Germain-en-Laye, France). Gene expression analysis was performed by quantitative real-time PCR (qRT-PCR) on an ABI 7900HT analyzer using inventoried and designed assays (Applied Biosystems). Data were normalized to the mean of three housekeeping genes (11). Details of assays and housekeeping genes are provided in the online supplementary methods.
RESULTS
Karyotype analysis revealed an apparently balanced translocation [t(7,19)(p15.2;p13.2)] in both the proband and her affected son. The proband's unaffected parents had normal karyotypes, suggesting the translocation arose de novo in the proband. We hypothesized that the breakpoint of this translocation disrupted one or more genes involved in the etiology of the insulin resistance and growth deficiency in both subjects. Bioinformatics identified a total of 67 genes within the breakpoint regions on chromosomes 19p13.2 and 7p15.2. The most plausible biological candidate was INSR on chromosome 19. Biochemical analysis showed raised insulin, C-peptide, insulin–to–C-peptide ratio, and adiponectin levels in both individuals, suggesting disruption of INSR (Table 1).
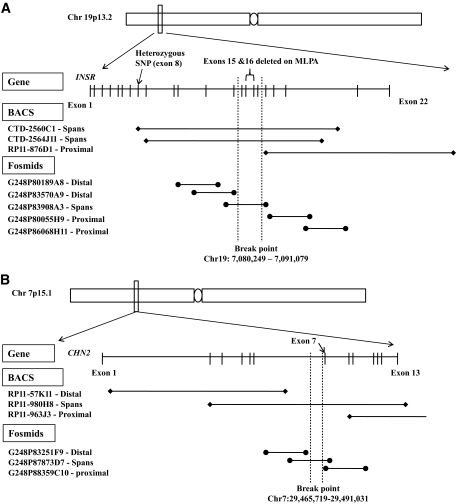
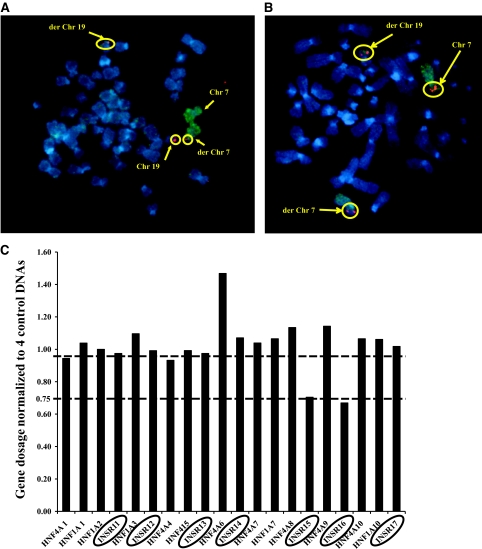
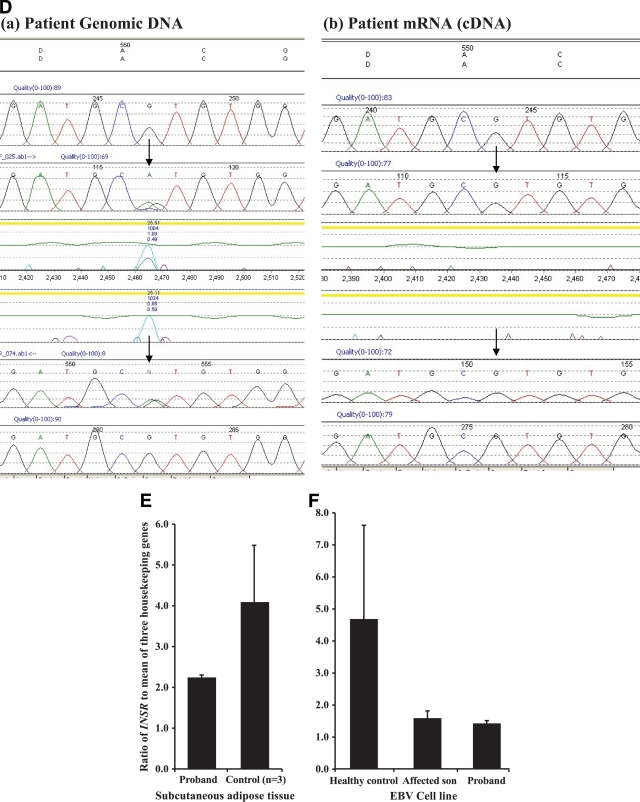
FISH analysis of proband chromosomes using bacterial artificial chromosomes (BACs) and fosmids narrowed down the region of the breakpoint on chromosome 19 to an ∼10-Kb region entirely within the genomic sequence of INSR (Figs. 1A and 2A), predicted to be between exons 13 and 15. To investigate possible cryptic microdeletions within the INSR sequence, MLPA analysis was performed in both patients. Taking into account the resolution limits of FISH, we decided to perform MLPA analysis on a broader DNA region spanning exons 11–17. A microdeletion between 2.4–6.6 Kb in size including exons 15 and 16 was detected (Fig. 2C). Direct sequencing of the entire coding region of INSR in patient genomic DNA excluded an INSR mutation. An informative heterozygous SNP (c.1650G>A;p.A550A) identified in patient genomic DNA was shown to be monoallelic in patient RNA, establishing INSR haploinsufficiency and excluding a potential dominant negative mutational mechanism (Fig. 2D). Reduced INSR expression in both patient-derived EBV cell line and adipose tissue cDNA was also demonstrated (Fig. 2E and F).
FIG. 1.
A: Map of the INSR region on chromosome 19p13.2 illustrating the BACs and fosmids selected for FISH analysis. BACs (∼150–200 Kb in size) and fosmids (∼40 Kb in size) overlapping over the entire genomic sequence of INSR were selected and obtained for FISH analysis. Also shown are the results of FISH analysis, which demonstrated that the minimal breakpoint region is 10.8 Kb and all within the genomic sequence of INSR. B: Map of the CHN2 region on chromosome 7p15.1 illustrating the BACs and fosmids selected for FISH analysis. BACs (∼150–200 Kb in size) and fosmids (∼40 Kb in size) overlapping the entire genomic sequence of CHN2 were selected and obtained for FISH analysis. Also shown are the results of FISH analysis, which demonstrated that the minimal breakpoint region is 25.3 Kb and all within the genomic sequence of CHN2.
FIG. 2.
A: FISH analysis showing disruption of the BAC CTD-2560C1 containing the genomic sequence of INSR. As shown, the digoxigenein-labeled BAC CTD−2560C1 (red) spans the breakpoint as there are three signals apparent where it hybridizes to chromosome 19, derivative chromosome 19, and derivative chromosome 7. The two chromosome 7 homologs are identified by a fluoroisothiocyanate (FITC)-labeled chromosome-specific paint (green). Chromosomes are counterstained with DAPI (blue). B: FISH analysis showing disruption of the BAC RP11-980H8 containing the genomic sequence of CHN2. As shown, the digoxigenein-labeled BAC RP11-980H8 (red) spans the breakpoint with three signals apparent where it hybridizes to chromosome 7, derivative chromosome 7, and derivative chromosome 19. The two chromosome 7 homologs are identified by a FITC-labeled chromosome-specific paint (green). Chromosomes are counterstained with DAPI (blue). C: Identification of a cryptic microdeletion (exons 15–16) within INSR using MLPA. Graph of normalized gene dosage of INSR exons 11–17 and control HNF1A and HNF4A exons run at the same time. Dosage quotients were calculated from average crossing points of triplicate samples using the comparative Ct (ΔΔCt) method. A ratio of 1 implies normal gene dosage, and a ratio below 0.75 suggests a deletion. Exons 15 and 16 of INSR are deleted with a ratio of less than 0.75. D: Demonstration of monoallelic expression of INSR in a patient-derived lymphoblastoid cell line. The exon 8 silent variant, c.1650G>A, p.A550A, is heterozygous in genomic DNA, but sequencing of patient cDNA reveals monoallelic expression due to the heterozygous SNP only showing one allele at c.1650G. E: INSR gene expression studies in the proband's subcutaneous adipose tissue–derived cDNA showed reduced expression compared with that from three BMI-matched samples. F: INSR gene expression studies in patient EBV cell line–derived cDNA showed reduced expression in both the proband and her affected son compared with a healthy control sample. (A high-quality color digital representation of this figure is available in the online issue.)
To explain the clinical phenotype observed in this family and determine the mutational mechanism for the growth deficiency, we also mapped the translocation breakpoint on chromosome 7. A number of strong biological candidate genes for growth mapped to the region including JAZF1 (12–14) and GHRHR (15). Disruption of both genes was excluded by FISH analysis (data not shown), while gene expression studies on patient EBV–transformed lymphocyte cell line derived–cDNA, compared with healthy control subjects, demonstrated that JAZF1 expression was not altered (see online supplementary results Fig. 1). GHRHR is only expressed in the pituitary so it was not possible to investigate patient gene expression levels. To exclude a growth hormone–releasing hormone (GHRH) receptor defect, the proband underwent a GHRH-arginine stimulation test that illustrated a normal response (see online supplementary results Table 1).
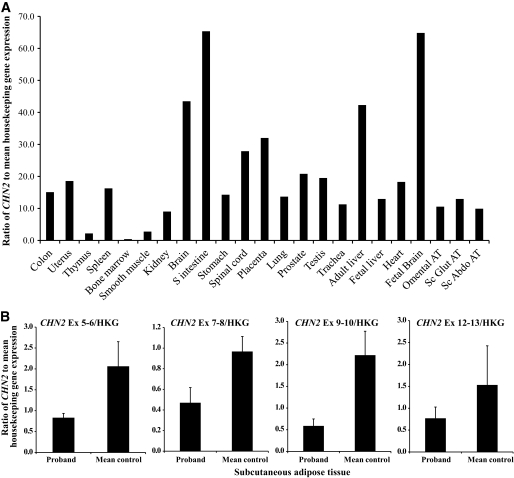
Further mapping on chromosome 7 restricted the breakpoint to 25.3 Kb entirely within the genomic sequence of CHN2 (Figs. 1B and 2B). Gene dosage studies established that there was no loss of the coding region of CHN2 (data not shown). Monoallelic CHN2 expression could not be demonstrated as no informative coding SNPs were identified in patient genomic DNA. Gene expression studies established that CHN2 is expressed in human brain and insulin-sensitive tissues including liver, adipose tissue (subcutaneous and omental), and muscle (Fig. 3A). Expression analysis in patient adipose tissue–derived cDNA compared with healthy control subjects demonstrated reduced CHN2 expression (Fig. 3B).
FIG. 3.
A: Expression profile of CHN2 in a panel of healthy human tissues to identify tissue distribution, shown is the expression in brain and insulin-sensitive tissues. The ratio of CHN2 expression relative to the mean of three housekeeping genes (PPIA, GAPDH, and 18s) was analyzed in a panel of human tissues. B: CHN2 expression is reduced in patient adipose tissue–derived cDNA compared with that of three matched healthy control samples. Gene expression studies to identify the expression levels of CHN2 (relative to the mean of three housekeeping genes) were performed using a combination of inventoried and designed assays to cover all known transcripts of CHN2. All transcripts of CHN2 were reduced in patient subcutaneous (SC) adipose tissue (AT) compared with those of three healthy control subjects (mean control).
Given the proximity of the translocation breakpoint to an imprinted region on the chromosome implicated in Silver-Russell syndrome (SRS) and the possibility of a positional effect of the translocation on gene expression, we excluded involvement of GRB10—the SRS candidate gene in this region—by establishing that GRB10 gene expression levels were normal in patient adipose tissue (see online supplementary results Fig. 2) (16). Monoallelic GRB10 expression could not be confirmed due to a lack of informative coding SNPs in the GRB10 coding sequence.
DISCUSSION
Digenic causes of human disease are rare in the literature but present an opportunity to model possible gene-gene interactions, which may provide insights into common metabolic disorders including type 2 diabetes. We report a family with a novel reciprocal translocation [t(7,19)(p15.2;p13.2)] resulting in the first reported case of severe insulin resistance, diabetes, and growth deficiency resulting from the synergistic disruption of INSR and CHN2.
It is well established that INSR mutations are the most common cause of monogenic insulin resistance. Most mutations are missense mutations in the β subunit that have dominant negative activity. However, truncating mutations have a similar effect. Biochemical features supporting the diagnosis of an INSR defect in the family described include markedly elevated insulin and C-peptide levels as a response to severe insulin resistance and due to reduced insulin clearance (17), a raised insulin–to–C-peptide ratio (17), and raised adiponectin levels relative to the degree of insulin resistance in these subjects (18). This is confirmed by our genetic investigations, which demonstrated disruption of INSR by a reciprocal translocation and monoallelic INSR expression. However, our genetic studies at the INSR locus do not explain the severe pre- and postnatal growth deficiency observed in both subjects. Variable penetrance of INSR haploinsufficiency may result in phenotypic heterogeneity. Published evidence from the parents of children with Donohue's syndrome shows that haploinsufficient parents have a mildly deranged or normal metabolic phenotype, although some may have marked insulin resistance. However, this is usually in the presence of obesity or other risk factors (5,19).
The family described shares some features with the more severe autosomal recessive insulin receptoropathies (Donohue's syndrome and Rabson-Mendenhall syndrome). Both affected individuals have an intermediate phenotype with neonatal hypoglycemia, severe insulin resistance, hyperglycemia in childhood, and intrauterine and postnatal growth deficiency; however, they have no evidence of premature mortality. The proband also has mild dysmorphic features with minimal subcutaneous fat, overdeveloped musculature (masculinization), and abnormal dentition—all features seen in severe autosomal recessive insulin receptoropathies.
The genotype-phenotype correlation in patients with INSR mutations is likely to be affected by the mutation site and possible modifier loci (20). Moreover, evidence from mice that are double heterozygous for null alleles in insr and irs-1 (insr+/−irs-1+/−) genes display a synergistic effect on insulin resistance with a 5- to 50-fold rise in insulin levels despite the expected ∼50% reduction in the protein levels of INSR and IRS-1 (21), suggesting that gene-gene interactions may play a significant role in severe insulin resistance. In this family, we have the rare opportunity to define a possible modifier locus by mapping the second breakpoint of the translocation. Mapping of the breakpoint on chromosome 7 demonstrated disruption of CHN2, which encodes β-2 chimerin. Chimerins are ligand-activated Rac-specific GTPase-activating proteins (22), which are expressed in many human tissues, especially brain, pancreas, and insulin-sensitive tissues. Chimerins are downstream effectors of tyrosine kinase receptors and have been shown to regulate growth in an inhibitory manner via suppression of Rac and extracellular signal–related kinase phosphorylation (23). Decreased expression of CHN2 is associated with high-grade malignant gliomas, duodenal adenocarcinomas, and breast tumors (23), suggesting that chimerins are tumor suppressors; however, increased expression of CHN2 is reportedly associated with lymphomas (24). We therefore propose that reduced CHN2 expression, a gene with a known role in growth pathways, contributes to a novel digenic syndrome of insulin resistance and dysglycemia due to disruption of INSR combined with marked pre- and postnatal growth deficiency due to disruption of CHN2.
There is a possible role for phenotypic modification caused by disruption of regulation of other biologically plausible genes close to the chromosome 7p breakpoint as there are several strong biological contenders. Although these genes are a long way from the breakpoint, disruption of the spatial relationship between these genes and unknown regulatory elements, as seen with other translocations, cannot be excluded (25). GHRHR maps to chromosome 7p15.2, and mutations in GHRHR are a known cause of dwarfism of Sindh (15). However, a GHRH test performed on the proband confirmed a normal response, thereby excluding a defect in the GHRH receptor. JAZF1 on chromosome 7p15.1–15.2 is a recently described type 2 diabetes susceptibility gene with a known role in growth (12–14). However, JAZF1 gene expression levels are not altered in either the proband or her son. SRS has also been linked to chromosome 7p, and the minimal overlap region has been delineated to chromosome 7p11.2, ∼25 Mb away from the breakpoint (26). This region contains multiple biological candidates with the strongest evidence supporting GRB10 (16). GRB10 encodes growth factor receptor binding protein 10, which is also known as insulin receptor binding protein and is maternally imprinted (16). Mice carrying maternally inherited targeted deletions of grb10 display a growth phenotype that recapitulates the SRS phenotype described in patients (16). However, our investigations have shown that there are no differences in GRB10 gene expression levels in the proband and her son compared with normal healthy control subjects.
In summary, we describe a novel cytogenetic defect resulting in a syndrome of severe insulin resistance, dysglycemia, and pre- and postnatal growth deficiency. The simultaneous heterozygous disruption of INSR and CHN2 without loss of other candidate growth-related genes in the region suggests this represents a novel digenic disorder and implicates CHN2 for the first time in insulin's metabolic and growth-promoting actions in vivo.
Supplementary Material
Acknowledgments
This study was funded in Oxford by Diabetes U.K. (BDA:RD05/0003131) and the Medical Research Council (MRC) (81696) and in Slovakia by the Slovakian Research and Development Support Agency (51-014205) and the Slovak Ministry of Health (2005/15-NEDU-01). E.V.V., N.W., and K.S.E. are supported by the Wellcome Trust. S.G.I.S. is a Diabetes U.K. Clinical Training Fellow, and A.L.G. is an MRC New Investigator.
No potential conflicts of interest relevant to this article were reported.
We are grateful to Pauline Sutton and Marjorie Gilbert for adiponectin assays and to Adrian Edwards for establishing the EBV-transformed cell lines. We thank Dr. Sue Price and Dr. Helen Stewart for valuable discussions on SRS and Alica Mitkova for her technical assistance.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Savage DB, Agostini M, Barroso I, Gurnell M, Luan J, Meirhaeghe A, Harding AH, Ihrke G, Rajanayagam O, Soos MA, George S, Berger D, Thomas EL, Bell JD, Meeran K, Ross RJ, Vidal-Puig A, Wareham NJ, O'Rahilly S, Chatterjee VK, Schafer AJ: Digenic inheritance of severe insulin resistance in a human pedigree. Nat Genet 2002; 31: 379– 384 [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi CM, Emanuelli B, Kahn CR: Critical nodes in signaling pathways: insights into insulin action. Nat Rev Mol Cell Biol 2006; 7: 85– 96 [DOI] [PubMed] [Google Scholar]
- 3.Kahn CR, Flier JS, Bar RS, Archer JA, Gorden P, Martin MM, Roth J: The syndromes of insulin resistance and acanthosis nigricans: insulin-receptor disorders in man. N Engl J Med 1976; 294: 739– 745 [DOI] [PubMed] [Google Scholar]
- 4.Krook A, Brueton L, O'Rahilly S: Homozygous nonsense mutation in the insulin receptor gene in infant with leprechaunism. Lancet 1993; 342: 277– 278 [DOI] [PubMed] [Google Scholar]
- 5.Longo N, Wang Y, Smith SA, Langley SD, DiMeglio LA, Giannella-Neto D: Genotype-phenotype correlation in inherited severe insulin resistance. Hum Mol Genet 2002; 11: 1465– 1475 [DOI] [PubMed] [Google Scholar]
- 6.O'Rahilly S, Moller DE: Mutant insulin receptors in syndromes of insulin resistance. Clin Endocrinol (Oxf) 1992; 36: 121– 132 [DOI] [PubMed] [Google Scholar]
- 7.Rother KI, Accili D: Role of insulin receptors and IGF receptors in growth and development. Pediatr Nephrol 2000; 14: 558– 561 [DOI] [PubMed] [Google Scholar]
- 8.Accili D, Drago J, Lee EJ, Johnson MD, Cool MH, Salvatore P, Asico LD, Jose PA, Taylor SI, Westphal H: Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet 1996; 12: 106– 109 [DOI] [PubMed] [Google Scholar]
- 9.Baker J, Liu JP, Robertson EJ, Efstratiadis A: Role of insulin-like growth factors in embryonic and postnatal growth. Cell 1993; 75: 73– 82 [PubMed] [Google Scholar]
- 10.Woo M, Patti ME: Diabetes risk begins in utero. Cell Metab 2008; 8: 5– 7 [DOI] [PubMed] [Google Scholar]
- 11.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40: 638– 645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson A, Marroni F, Hayward C, Franklin CS, Kirichenko AV, Jonasson I, Hicks AA, Vitart V, Isaacs A, Axenovich T, Campbell S, Dunlop MG, Floyd J, Hastie N, Hofman A, Knott S, Kolcic I, Pichler I, Polasek O, Rivadeneira F, Tenesa A, Uitterlinden AG, Wild SH, Zorkoltseva IV, Meitinger T, Wilson JF, Rudan I, Campbell H, Pattaro C, Pramstaller P, Oostra BA, Wright AF, van Duijn CM, Aulchenko YS, Gyllensten U: Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum Mol Genet 2009; 18: 373– 380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins LL, Lee YF, Heinlein CA, Liu NC, Chen YT, Shyr CR, Meshul CK, Uno H, Platt KA, Chang C: Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proc Natl Acad Sci U S A 2004; 101: 15058– 15063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wajnrajch MP, Gertner JM, Harbison MD, Chua SC, Jr, Leibel RL: Nonsense mutation in the human growth hormone-releasing hormone receptor causes growth failure analogous to the little (lit) mouse. Nat Genet 1996; 12: 88– 90 [DOI] [PubMed] [Google Scholar]
- 16.Abu-Amero S, Monk D, Frost J, Preece M, Stanier P, Moore GE: The genetic aetiology of Silver-Russell syndrome. J Med Genet 2008; 45: 193– 199 [DOI] [PubMed] [Google Scholar]
- 17.Hojlund K, Hansen T, Lajer M, Henriksen JE, Levin K, Lindholm J, Pedersen O, Beck-Nielsen H: A novel syndrome of autosomal-dominant hyperinsulinemic hypoglycemia linked to a mutation in the human insulin receptor gene. Diabetes 2004; 53: 1592– 1598 [DOI] [PubMed] [Google Scholar]
- 18.Semple RK, Soos MA, Luan J, Mitchell CS, Wilson JC, Gurnell M, Cochran EK, Gorden P, Chatterjee VK, Wareham NJ, O'Rahilly S: Elevated plasma adiponectin in humans with genetically defective insulin receptors. J Clin Endocrinol Metab 2006; 91: 3219– 3223 [DOI] [PubMed] [Google Scholar]
- 19.Elsas LJ, 2nd, Longo N, Langley S, Griffin LD, Shuster RC: Molecular genetics of severe insulin resistance. Yale J Biol Med 1989; 62: 533– 547 [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor SI: Prenatal screening for mutations in the insulin receptor gene: how reliably does genotype predict phenotype? J Clin Endocrinol Metab 1995; 80: 1493– 1495 [DOI] [PubMed] [Google Scholar]
- 21.Bruning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR: Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 1997; 88: 561– 572 [DOI] [PubMed] [Google Scholar]
- 22.Caloca MJ, Delgado P, Alarcon B, Bustelo XR: Role of chimaerins, a group of Rac-specific GTPase activating proteins, in T-cell receptor signaling. Cell Signal 2008; 20: 758– 770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruinsma SP, Baranski TJ: Beta2-chimaerin in cancer signaling: connecting cell adhesion and MAP kinase activation. Cell Cycle 2007; 6: 2440– 2444 [DOI] [PubMed] [Google Scholar]
- 24.Nishiu M, Yanagawa R, Nakatsuka S, Yao M, Tsunoda T, Nakamura Y, Aozasa K: Microarray analysis of gene-expression profiles in diffuse large β-cell lymphoma: identification of genes related to disease progression. Jpn J Cancer Res 2002; 93: 894– 901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gloyn AL, Ellard S, Shepherd M, Howell RT, Parry EM, Jefferson A, Levy ER, Hattersley AT: Maturity-onset diabetes of the young caused by a balanced translocation where the 20q12 break point results in disruption upstream of the coding region of hepatocyte nuclear factor-4alpha (HNF4A) gene. Diabetes 2002; 51: 2329– 2333 [DOI] [PubMed] [Google Scholar]
- 26.Monk D, Bentley L, Hitchins M, Myler RA, Clayton-Smith J, Ismail S, Price SM, Preece MA, Stanier P, Moore GE: Chromosome 7p disruptions in Silver Russell syndrome: delineating an imprinted candidate gene region. Hum Genet 2002; 111: 376– 387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.