Abstract
Sequence-specific DNA-binding transcription factors (TFs) are often termed as ‘master regulators’ which bind to DNA and either activate or repress gene transcription. We have computationally analysed the soybean genome sequence data and constructed a proper set of TFs based on the Hidden Markov Model profiles of DNA-binding domain families. Within the soybean genome, we identified 4342 loci encoding 5035 TF models which grouped into 61 families. We constructed a database named SoybeanTFDB (http://soybeantfdb.psc.riken.jp) containing the full compilation of soybean TFs and significant information such as: functional motifs, full-length cDNAs, domain alignments, promoter regions, genomic organization and putative regulatory functions based on annotations of gene ontology (GO) inferred by comparative analysis with Arabidopsis. With particular interest in abiotic stress signalling, we analysed the promoter regions for all of the TF encoding genes as a means to identify abiotic stress responsive cis-elements as well as all types of cis-motifs provided by the PLACE database. SoybeanTFDB enables scientists to easily access cis-element and GO annotations to aid in the prediction of TF function and selection of TFs with functions of interest. This study provides a basic framework and an important user-friendly public information resource which enables analyses of transcriptional regulation in soybean.
Keywords: soybean, transcription factors, abiotic stress, database
1. Introduction
Sequence-specific DNA-binding transcription factors (TFs) are the key molecular switches that control or influence many of the biological processes such as development, growth, cell division and responses to environmental stimuli in a cell or organism. By being capable of activating or repressing transcription of multiple target genes, they affect the metabolism, physiological balance and progression in cells and the responses of cells to the environment.1–3 TFs form complex regulatory networks at the transcriptional level and through protein–protein interactions among themselves or with proteins of other classes. Protein–protein interactions may also form with other transcriptional regulators such as chromatin remodelling/modifying proteins to recruit or block access of RNA polymerases to the DNA template. The specific interactions between TFs and a family of cis-regulatory sequences described by a consensus motif play a central part in how genetic regulatory proteins affect spatial and temporal gene expression.4 Additionally, alterations in the activity and regulatory specificity of TFs are emerging as a major source of diversity and evolutionary adaptation.5,6
In the past decade, the availability of complete genome sequences and the development of high-throughput experimental techniques have enabled scientists to compile complementary information describing the function and organization of TF regulatory systems in a number of organisms. The identification, characterization and classification of TFs at the genome-wide level will provide an important resource for researchers who are interested in studying the regulation of gene expression. Similar to other proteins, TFs are comprised evolutionarily conserved units called ‘domains’, which belong to families that can occur in many different proteins. The majority of TFs can be grouped into a number of different families according to the specific type of DNA-binding domain (DBD) that is present within their sequence.7–10 Using bioinformatics approaches, computational studies have documented valuable TF repertoires by searching for genes containing DBDs within individual organisms ranging from prokaryotes to eukaryotes or by searching across all completely sequenced genomes.7,8,10–18
In plants, ∼7% of the genome encodes putative TFs.19 Despite their importance as a fundamental component of biological systems, the TF repertoires for many plant genomes remain largely unknown and understudied. Analyses of expressed sequence tag (EST) and genome sequence databases have indicated that legumes encode more than 2000 TFs per genome. At the present time, less than 1% of these putative TFs have been genetically and functionally characterized.19 Our basic knowledge of TFs and their role in transcriptional regulation is derived from molecular biological and genetic investigations. Proper characterization of particular TFs often requires a detailed study in the biological context of a whole TF family, since functional redundancy is a common occurrence within TF families.20–24 Furthermore, since TFs control the expression of the genome, it is not possible to completely understand their function without performing detailed functional studies at a genome-wide level.7,25–27
Soybean (Glycine max L.) is a nutritionally important crop which provides an abundant source of oil and protein for worldwide human consumption.28–31 In addition, soybean is also viewed as an attractive crop for the production of renewable fuels such as biodiesel. Due to its symbiosis with nitrogen fixing bacteria, soybean can fix atmospheric nitrogen and therefore requires minimal input of nitrogen fertilizer. Agricultural dependence on nitrogen fertilizer often accounts for the single largest energy input in agronomic practices.32 With the recent completion of the soybean genomic sequence (http://www.phytozome.net/soybean#C Soybean Genome Project, DOE Joint Genome Institute), the identification, isolation and functional analysis of important genes will be accelerated. From a biotechnology perspective, this resource will be especially important for studying regulatory genes involved in plant productivity, seed quality, nitrogen fixation and the sensing/response and adaptation to the environment. Within the soy genome model, ∼975 Mb has been captured in 20 chromosomes and 66 153 protein-coding loci have been predicted (http://www.phytozome.net/soybean#C). With the completion of the soybean genome sequence, the full complement of TF-encoding genes from this important crop can be characterized and functionally analysed.
In this report, we searched for sequence-specific DNA-binding TFs using a prediction method which uses 51 Hidden Markov Models (HMMs) from the Pfam database. We also used 11 models, which were originally created by HMMbuild of HMMER2 package, to identify the domains within the putative TF proteins. The computational results predict that the soybean genome contains 5035 TF protein models coded from 4342 loci in 61 families. We created a database named ‘SoybeanTFDB’. This database provides open access for researchers to all relevant and basic information on functional motifs, full-length cDNAs, promoter regions, genomic distribution, gene duplication and multiple sequence alignment of the DBDs for each TF family. Since most of these TFs have not been experimentally characterized for regulatory function as indicated by assessment in PubMed, we searched for their putative regulatory function by assessing annotations of the gene ontology (GO) using comparative analysis with their Arabidopsis counterparts. As a complement to this functional prediction using GO annotations, we also mapped all putative cis-regulatory elements that were documented within the PLACE database on all TF encoding genes. In this analysis, we placed a particular emphasis on abiotic stress responsive cis-elements. Knowledge gained from identifying the presence of stress responsive cis-elements, in addition to GO annotation, enables effective prediction of stress responsive TFs. Taken together, in this study, we demonstrate a comprehensive and high-quality census of TFs encoded within the soybean genome. These results provide a solid foundation for further systematic characterization of soybean TFs using traditional molecular approaches and/or genomic techniques at either the single-gene level or family-wide scale.
2. Materials and methods
2.1. Identification of TF repertoire in soybean
To identify TF encoding genes from the annotations of Glyma1 in the soybean genome, 51 HMMs of Pfam33 and those of 11 originally created using HMMbuild of the HMMER2 package (http://hmmer.janelia.org/) were applied, which corresponded to a total of 61 TF families (Supplementary Table S1). The modelled proteome data of annotated genes in Glyma1, which were downloaded from Phytozome (http://www.phytozome.net/), were subjected to a profile search for HMM dataset using Pfam-HMM with set thresholds of E-value, E < 1e−5 (Supplementary Table S1). The search results for each of the TF families were then applied to retrieve discovered regions as conserved DBDs and related annotations. To further classify genes with a conserved MYB domain into three subgroups: (R1)R2R3_MYB, MYB_related and atypical_MYB, the MYB soybean protein sequences were searched against previously classified Arabidopsis MYB genes34 using blastp (E < 1e−5) and each top hit combination was applied to the classification. To avoid possible contaminations of pseudo response regulator or histidine kinase sequences into the GARP_ARRB family, genes containing CCT, CHASE, HATPase_c and HisKA together with Response_reg of Pfam domains were searched by InterProScan. Genes, which hit in this search, were subsequently removed from the GARP_ARRB family.
The putative TF encoding genes discovered in the soybean genome were classified into the following four categories based on their potential functionality as TFs. The first group of TFs (Category A) consists of TF encoding genes showing sequence identity ≥95% and a blastn E ≤ 1e−100 with GenBank soybean sequences having a functional description as TFs. Category A genes were classified with the highest confidence level after assessment with the PubMed database. The second group of TFs (Category B) is comprised TFs which have an equivalent protein domain arrangement (blastp E ≤ 1e−30) for regulatory function in well-annotated plants, such as Arabidopsis and/or rice. The third group of TFs (Category C) combines possible TFs which show a significant hit with each of the HMM models used for DBD prediction (Pfam-HMM E ≤ 1e−20). The last group contains TFs which have promiscuous HMM models with a threshold of settled E-values.
2.2. Structural and functional annotations for putative soybean TFs
For annotating TF encoding genes in soybean,35 we used protein and cDNA sequences of soybean TFs as queries against the following protein and nucleotide datasets using the BLAST algorithm:36 the nr protein DB of NCBI (ftp://ftp.ncbi.nih.gov/blast/db); the protein data presented in TAIR release 8 (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets); the protein data from UniProt (http://www.uniprot.org/); the TIGR/MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) release 6; the soybean representative cDNA sequences in UniGene (ftp://ftp.ncbi.nih.gov/repository/UniGene/); the TIGR Transcript Assemblies (http://plantta.jcvi.org/); the Plant GDB (http://www.plantgdb.org/); the sequence sets of ESTs and high throughput cDNAs (HTCs) of RIKEN soybean full-length cDNA clones (http://rsoy.psc.riken.jp/);37 the cDNAs of the previous version of the soybean genome annotation (Glyma0, Phytozome) and the target sequences of the Affymetrix soybean GeneChip (GPL4592 of NCBI GEO platform accession). All of the similarity searches using blastn were performed with threshold E < 1e−100, and the top scoring hit for each query was applied. All similarity searches with blastp against protein datasets were performed with a threshold E < 1e−5 to find possible functional descriptions for TF encoding genes. The top scoring hit for each query was applied.
Conserved domains in the protein sequence of putative TF encoding genes were identified with InterProScan and the InterPro DB (http://www.ebi.ac.uk/interpro/) to predict structures of DBD of TFs together with other functional domains and associated GO terms. All domains and those positions predicted by the search were retrieved and implemented them into our database. To determine the global characteristic features of functional categories of TF encoding genes of soybean, the TFs were assigned to possible GO terms based on a blastp similarity search to find Arabidopsis counterparts together with those annotated GOs of TAIR8. Particular emphasis was placed on sequences serving under the ‘biological process’ functional category.
TFs have been widely reported in plant TF databases such as DATF, AtTFDB, RARTF and PlnTFDB for Arabidopsis and DRTF, GRASIUS and PlnTFDB for rice. To annotate all putative soybean TFs in relation to Arabidopsis and rice counterparts, soybean TF sequences were assigned to annotation data related to TF families provided from each of the aforementioned databases based on sequence similarity searches between soybean proteome data and those of Arabidopsis and rice.8,14,38–42 The interrelated dataset of soybean genes, in combination with related Arabidopsis and rice TF annotations, were implemented into the SoybeanTFDB to provide cross references with other plant TFDBs.
2.3. Gene duplications and gene clusters in soybean TF families
Gene duplications and gene clusterings in soybean TF families were estimated by analysing the amino acid sequences of TF genes found on soybean chromosomes. Specifically, the presence of gene pairs or gene clusters of closely homologous genes based on global sequence similarity with threshold of more than 60% amino acid sequence identity using cd-hit program of CD-HIT package were investigated.43 Gene clusters are defined as genetic loci containing three or more closely homologous genes. Once identified, the pairs or gene clusters were used to assess the chromosomal allocation of highly homologous genes. Genes in tandem duplication are arbitrarily defined as those occurring within a sequence distance of 50 kb. On the other hand, genes that are duplicated in the same chromosome but reside >50 kb from each other are referred to as ‘Duplications in same chromosome’. ‘Duplications in different chromosomes’ indicate pairs of highly homologous genes which reside on different chromosomes.
2.4. Discovery of cis-regulatory motifs in promoter regions of TF genes
To discover cis-regulatory motifs located in the promoter regions of each putative soybean TF gene and to investigate the enriched representation of cis-motifs in each TF family, cis-motif sequences from the PLACE database (version 30, 469 entries) (http://www.dna.affrc.go.jp/PLACE/)44 and the stress responsive cis-motifs previously reported45 were used as queries to search against the Glyma1 genome scaffold sequence using the fuzznuc program of EMBOSS package (http://emboss.sourceforge.net/). The results of pattern matches were subsequently assessed to identify matched sequences located on the −500, −1000 and −3000 bp upstream sequences from the putative transcription start site for each TF encoding gene defined in the Glyma1 annotation. The cis-element search results were implemented into the SoybeanTFDB as a searchable property. In addition, these search results were also incorporated as an annotation track of the genome browser (Gbrowse).
To assess the enrichment for the representative allocation of each cis-element identified on upstream sequences of each TF family, we analysed cis-element representations in the −1000 bp promoter region of TF members for TF families containing more than 50 gene loci to compare cis-element representations of randomly sampled gene loci of Glyma1. The computation of the overrepresentation test and its significance were performed by a Z-test as previously described.46
2.5. Construction of a web accessible database
The database is implemented in MySQL and the web interface of Perl CGI and Java script run on the Apache Web server. The definition strings used for sequence similarity searches for each database, the domain searches by InterProScan, cis-motif names from the PLACE database and the assigned GO terms have been assembled as a keyword database enabling users to specify queries on any keyword and to retrieve relevant information for genes from the SoybeanTFDB. A BLAST server was implemented to provide a similarity search interface for queried sequences using NCBI BLAST together with soybean Glyma1-related sequences, as well as those from Arabidopsis and rice. Generic Genome Browser (Gbrowse)47 was also implemented in SoybeanTFDB with Glyma1 genome annotations released by Phytozome to visualize the gene annotations of the putative TF encoding genes together with cis-motifs found on the upstream sequence of the TF genes. All of the data in the SoybeanTFDB are accessible not only through a web interface but also as downloadable files from the website. The cross references of corresponding data for each of the entries were also implemented into the SoybeanTFDB together with the URLs for each of the original referenced data to provide hyperlinks on the web interface with seamless navigations.
3. Results and discussion
3.1. Identification of the soybean TF repertoire
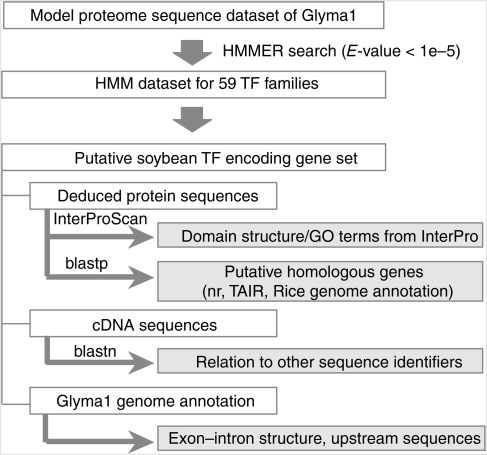
For the purpose of identifying the repertoire of TFs within the soybean genome, we first define a class of proteins which bind DNA in a sequence-specific fashion. A protein is classified as a TF if it has a significant match to a model that we annotated as being a DBD, with the significance thresholds for HMM matches. Supplementary Table S1 summarized the HMMs used in TF predictions. For each HMM, we examined the description and associated literature to assess their sequence-specific DNA-binding capabilities. The pipeline that we used to predict soybean TFs began with retrieving the complete set of predicted proteins from the completely sequenced soybean genome. This approach was then followed by a HMMER search with all HMMs taken from the Pfam database (Fig. 1). In total, 4342 putative TF encoding loci which showed a significant match with these selected DBDs were extracted from the soybean genome sequence Glyma1 model (http://www.phytozome.net/soybean#C). These putative TFs represent 6.56% of the total number of predicted genes in soybean (Table 1 and Supplementary Table S2). In soy, this percentage of TFs to total gene number was similar to what has been observed for Arabidopsis. In the Arabidopsis genome, there are at least 1968 TFs which account for 7.23% of the total number of genes. Although the number of TFs generally increase with the number of genes in a genome, interestingly the percentage of TF genes described in rice (3.68%) is less than expected (Table 1). The identified soybean TFs were classified into 61 families based on the presence of domains that were specific for the family (Table 2). Among the identified TFs, a significant proportion of the soybean TF repertoire has not been annotated with full-length open reading frames in the Glyma1 model. As a means to address this deficiency, we took advantage of our recently released soybean FL-cDNA collection of 4712 complete sequences and 68 661 ESTs to assess the Glyma1 annotation of the identified TFs (http://www.legumebase.agr.miyazaki-u.ac.jp).37 Table 2 summarized the full-length information of the soybean TF encoding genes annotated by Glyma1 and the FL-cDNA collection. Detailed information for each gene is available on Supplementary Table S2. Next, we then grouped the TFs into four categories according to our confidence in their structure and functionality by assessing PubMed and relevant databases as described in Materials and methods (Fig. 2, and Supplementary Table S3). Relevant information of the soybean TF repertoire can be easily accessed at our website SoybeanTFDB (http://soybeantfdb.psc.riken.jp). Information that is readily available for the TF repertoire includes nucleotide and amino acid sequences, promoter regions and domain alignments within the family as well as multiple alignments with putative Arabidopsis and homologous rice genes.
Figure 1.
Schematic workflow of the computational pipeline used to discover and annotate genes encoding putative transcription factors in soybean.
Table 1.
Numbers of TFs in Arabidopsis, rice and soybean
| Species | No. of non-redundant TF gene locia | No. of non-redundant gene locib | Referenced annotation | Percentage of TFsc | Database | URL |
|---|---|---|---|---|---|---|
| A. thaliana | 1922 | 27 235 | TAIR8 | 7.06 | DATF | http://datf.cbi.pku.edu.cn/ |
| 1968 | 7.23 | RARTF | http://rarge.g.sc.riken.jp/rartf/ | |||
| 1961 | 7.20 | PlnTFDB | http://plntfdb.bio.uni-potsdam.de/v2.0/index.php?sp_id=ATH | |||
| 1737 | 6.38 | AtTFDB | http://arabidopsis.med.ohio-state.edu/AtTFDB/ | |||
| 1358 | 4.99 | DBD | http://dbd.mrc-lmb.cam.ac.uk/DBD/index.cgi?About | |||
| O. sativa | 1928 | 56 797 | Rice Pseudomole-cule Release 6 | 3.39 | DRTF | http://drtf.cbi.pku.edu.cn/ |
| 2095 | 3.69 | PlnTFDB | http://plntfdb.bio.uni-potsdam.de/v2.0/index.php?sp_id=OSAJ | |||
| 2141 | 3.77 | GRASSIUS | http://grassius.org/summary.html | |||
| 1629 | 2.87 | DBD | http://dbd.mrc-lmb.cam.ac.uk/DBD/index.cgi?About | |||
| G. max | 4342 | 66 210 | Glyma1.0 | 6.56 | SoybeanTFDB | http://soybeantfdb.psc.riken.jp |
aNumber of predicted non-redundant TF gene loci in each genome.
bNumber of predicted non-redundant gene loci in each genome.
cPercentage of TFs per genome.
Table 2.
Characteristics of soybean transcription factors
| TF gene families | No. of modelsa | No. of gene locib | No. of models (FL)c | No. of models (not FL)d | No. of models assigned with RIKEN FL ESTe | No. of models assigned with RIKEN FL-HTCf | |
|---|---|---|---|---|---|---|---|
| 1 | (R1)R2R3_MYB | 333 | 319 | 246 | 87 | 37 | 6 |
| 2 | ABI3VP1 | 163 | 139 | 121 | 42 | 35 | 7 |
| 3 | Alfin-like | 27 | 18 | 26 | 1 | 7 | 2 |
| 4 | AP2_EREBP | 405 | 382 | 306 | 99 | 78 | 31 |
| 5 | ARF | 75 | 58 | 69 | 6 | 29 | 6 |
| 6 | ARID | 22 | 20 | 17 | 5 | 7 | 0 |
| 7 | atypical_MYB | 89 | 78 | 67 | 22 | 22 | 2 |
| 8 | Aux_IAA | 126 | 85 | 120 | 6 | 41 | 14 |
| 9 | BBR-BPC | 21 | 10 | 21 | 0 | 1 | 1 |
| 10 | BES1 | 21 | 18 | 17 | 4 | 8 | 2 |
| 11 | bHLH | 390 | 325 | 317 | 73 | 72 | 18 |
| 12 | bZIP | 205 | 148 | 194 | 11 | 49 | 8 |
| 13 | C2C2_Zn-CO-like | 101 | 84 | 87 | 14 | 31 | 13 |
| 14 | C2C2_Zn-Dof | 87 | 81 | 78 | 9 | 13 | 5 |
| 15 | C2C2_Zn-GATA | 65 | 63 | 53 | 12 | 14 | 1 |
| 16 | C2C2_Zn-YABBY | 28 | 18 | 27 | 1 | 8 | 1 |
| 17 | C2H2_Zn | 270 | 258 | 211 | 59 | 42 | 5 |
| 18 | C3H-TypeI | 178 | 151 | 154 | 24 | 50 | 14 |
| 19 | CAMTA | 14 | 14 | 12 | 2 | 4 | 0 |
| 20 | CCAAT_Dr1 | 23 | 16 | 20 | 3 | 4 | 2 |
| 21 | CCAAT_HAP2 | 42 | 23 | 40 | 2 | 9 | 2 |
| 22 | CCAAT_HAP3 | 47 | 39 | 34 | 13 | 5 | 2 |
| 23 | CCAAT_HAP5 | 26 | 23 | 20 | 6 | 6 | 0 |
| 24 | CPP | 22 | 17 | 17 | 5 | 1 | 0 |
| 25 | E2F_DP | 23 | 14 | 21 | 2 | 3 | 0 |
| 26 | EIL | 14 | 13 | 11 | 3 | 6 | 2 |
| 27 | GARP_ARRB | 21 | 21 | 16 | 5 | 4 | 2 |
| 28 | GARP_G2-like | 104 | 82 | 95 | 9 | 20 | 6 |
| 29 | GeBP | 17 | 17 | 6 | 11 | 4 | 1 |
| 30 | GRAS | 127 | 117 | 101 | 26 | 29 | 7 |
| 31 | GRF | 10 | 8 | 9 | 1 | 3 | 0 |
| 32 | HB | 283 | 242 | 240 | 43 | 67 | 20 |
| 33 | HMG-box | 50 | 26 | 43 | 7 | 18 | 3 |
| 34 | HRT | 1 | 1 | 1 | 0 | 1 | 1 |
| 35 | HSF | 65 | 59 | 54 | 11 | 11 | 6 |
| 36 | JUMONJI | 54 | 51 | 37 | 17 | 8 | 0 |
| 37 | LFY | 3 | 3 | 2 | 1 | 0 | 0 |
| 38 | LIM | 41 | 32 | 38 | 3 | 5 | 0 |
| 39 | LUG | 10 | 9 | 10 | 0 | 2 | 0 |
| 40 | MADS | 220 | 186 | 141 | 79 | 7 | 0 |
| 41 | MBF1 | 3 | 3 | 3 | 0 | 2 | 1 |
| 42 | MYB_related | 168 | 135 | 138 | 30 | 35 | 8 |
| 43 | NAC | 205 | 187 | 159 | 46 | 37 | 11 |
| 44 | Nin-like | 23 | 23 | 17 | 6 | 2 | 0 |
| 45 | PcG | 94 | 86 | 76 | 18 | 11 | 1 |
| 46 | PHD | 333 | 285 | 287 | 46 | 84 | 16 |
| 47 | PLATZ | 40 | 33 | 34 | 6 | 8 | 2 |
| 48 | S1Fa-like | 4 | 4 | 4 | 0 | 2 | 0 |
| 49 | SAP | 2 | 2 | 1 | 1 | 0 | 0 |
| 50 | SBP | 58 | 48 | 46 | 12 | 15 | 2 |
| 51 | SRS | 24 | 22 | 18 | 6 | 0 | 0 |
| 52 | TCP | 61 | 61 | 39 | 22 | 13 | 3 |
| 53 | Trihelix | 34 | 33 | 31 | 3 | 9 | 4 |
| 54 | TUB | 37 | 24 | 33 | 4 | 18 | 2 |
| 55 | ULT | 32 | 32 | 5 | 27 | 0 | 0 |
| 56 | VOZ | 8 | 7 | 7 | 1 | 2 | 1 |
| 57 | Whirly | 11 | 7 | 11 | 0 | 2 | 0 |
| 58 | WRKY_Zn | 219 | 198 | 167 | 52 | 47 | 12 |
| 59 | zf-HD | 57 | 56 | 37 | 20 | 4 | 1 |
| 60 | zf-TAZ | 8 | 8 | 6 | 2 | 1 | 0 |
| 61 | ZIM | 57 | 34 | 55 | 2 | 28 | 13 |
| Total | 5035 | 4342 | 4032 | 1003 | 1003 | 249 |
aNumber of predicted TF models in Glyma1 model.
bNumber of predicted TF loci in Glyma1 model.
cNumber of predicted full-length TF models in Glyma1 model.
dNumber of predicted not full-length TF models in Glyma1 model.
eNumber of predicted full-length TF models in soybean assigned with RIKEN full-length ESTs.
fNumber of predicted full-length TF models in soybean assigned with RIKEN full-length high throughput cDNAs.
Figure 2.
The distributions of soybean TF encoding genes are classified into four categories of annotation levels. Category A includes soybean gene models showing sequence identity ≥95% and a blastn E ≤ 1e−100 with GenBank soybean sequences having a functional description as TFs. Category B includes gene models which have an equivalent protein domain arrangement (blastp E ≤ 1e−30) for regulatory function in well-annotated plants, such as Arabidopsis and/or rice. Category C includes gene models which show a significant hit with each of the HMMs used for DBD prediction (Pfam-HMM E ≤ 1e−20). Category D includes TF genes which have promiscuous HMMs with a threshold of settled E-values.
Our prediction method depends heavily on the content of the Pfam database and the ability of the search algorithms to detect the DBDs in protein sequences, thus there are a few possible sources of inaccuracies in this prediction method. In addition, although the Glyma1 model contains more than 98% of known soybean protein-coding genes in its assembly, part of the TF repertoire may be clarified in the future by fine-tuning of the annotation. Finally, our literature analysis depends on the existing available published information pertaining to each gene, which will need to be updated as new findings are reported. The availability of updated HMM libraries or refinements of existing ones and better fine-tuned annotation and continuous searches for newly reported literature will enable us to improve the TF prediction coverage. We will continue to update the website with new information when it becomes available.
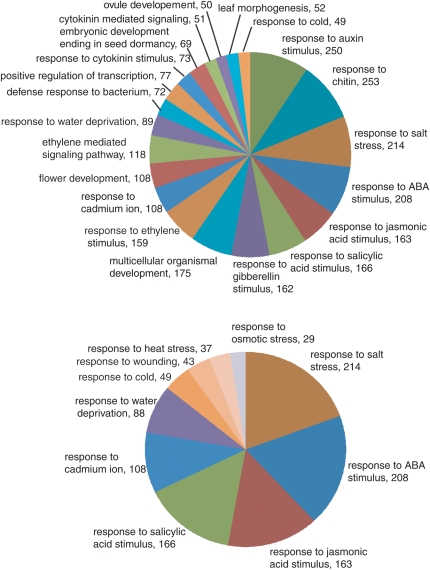
Literature analysis, which is achieved by assessing corresponding genes deposited as soybean TF genes in the GenBank core nucleotide division together with associated identifiers of PubMed, has revealed that the majority of soybean TFs remains experimentally uncharacterized. Thus, we attempted to further extend our current knowledge base regarding their regulatory function by assessing the putative functions of soybean TFs via comparative analyses with relevant GO annotations of Arabidopsis in TAIR. First, we analysed the profile of GO terms at the biological process level which could be assigned to soybean TFs based on sequence similarity searches against Arabidopsis counterparts having GO terms in TAIR. In order to grasp the overall representation of GO terms in applied entries of soybean TFs, all of the assigned terms were counted after the similarity searches were completed. With the exception of ‘regulation of transcription’, ‘DNA binding’ and ‘biological process’, the top 21 most abundant terms were subsequently used to classify the TFs. The contig results of these total 21 GO terms for each soybean TF, which was based on similarity with Arabidopsis TFs, are provided in Supplementary Table S4. Figure 3A illustrates the distribution of soybean TFs in the 21 most abundant GO terms. A significant proportion of soybean TFs are related to stress and hormone responses (Fig. 3B), indicating the important role of TFs in controlling these biological processes. Of these assigned regulatory functions, responses to auxin, chitin and salt stress are the most highly represented. It is acknowledged that these annotations are the first steps in functional prediction, and researchers must use original publications as a source for a higher level of detailed information. In addition, it is ideal if functional analyses can be performed in order to gain a detailed understanding of gene function. Overall, these analyses emphasize the limited amount of functional information that we know regarding the biological processes that most of the TFs mediate, even for model plants such as Arabidopsis. Directing research efforts into uncharacterized TFs—for example, using high-throughput genomic surveys to describe the key features combined with a detailed examination using traditional molecular approaches—will undoubtedly accelerate our functional understanding of these important regulatory genes. The NAC TF family, which is widely distributed in plants but so far has not been found in other eukaryotes, is an excellent example of how research interests can suddenly arise the following key findings. The acceleration in functional studies has revealed their diverse functions in different biological aspects and future follow-up studies will rapidly improve our understanding of the regulatory function of NAC members. A greater understanding of how TFs operate will be subsequently translated into their potential applications to enhance plant productivity.4,48,49
Figure 3.
The representative distributions of the GO terms for biological processes associated with soybean TF encoding genes. The top 21 abundantly found GO terms were assigned based on homology searches against annotated Arabidopsis genes (A). Abundant distribution of TFs in GO terms related to the response to various types of abiotic stresses in the soybean TF dataset (B). Gene numbers are displayed next to the terms.
3.2. Structural feature of TFs
As mentioned above, the most common classification of TFs is based on the structure of their DBD.7,14 Grouping TFs by their structural domains has been extremely useful in gaining insights into how they recognize and bind specific DNA sequence. This strategy has also been proven successfully for characterizing their evolutionary histories as well. Moreover, the DBD may provide clues to their biological function. For example, ABI3/VP1 TFs are often associated with the regulation of abscisic acid (ABA)-responsive genes during seed development.50
Since structural features of TF families have been extensively characterized in other reports, we do not cover this in detail within this report.8 However, it is worthy to note that soybean contains a number of large families which consist of more than 100 members (Supplementary Table S5). For example, the large AP2_EREBP family alone contains 405 TF models and accounts for a total of 8.04% of the TF repertoire. The bHLH and (R1)R2R3_MYB TFs also represent major families with 390 and 333 members, respectively, which together occupy 14.36% of the TF repertoire. These observations agree with the previous studies in Arabidopsis and rice, which confirmed that the same three families contain the highest numbers of TFs in these model systems (Supplementary Table S5). In addition, the plant-specific NAC family, which comprises 205 models in soybean, represents a similar ratio in Arabidposis and rice (Supplementary Table S5). Taken together, these results suggest a similar tendency in the evolution of major TF families in plants. Furthermore, given that the size of TF families is influenced in part by the number of different DNA sequences that they are able to recognize, the DBDs of AP2_EREBP, bHLH and (R1)R2R3_MYB TF families may be able to diversify their collection of target sequences. As a result, they occur in the greater numbers in a genome.4,51,52
3.3. Chromosomal distribution and gene duplications of TFs
Our analysis has indicated that the soybean TF families are scattered throughout the genome. The larger families, such as AP2_EREBP, (R1)R2R3_MYB, have members that are distributed on every chromosome in soybean (Supplementary Table S2). The local distribution of TF genes relative to each other is also of interest. Previous studies have described duplications and clusters of highly homologous genes. In Arabidopsis, tandem gene duplications and large-scale duplications on different chromosomes may account for >60% of the genome.7 In soybean, we were able to distinguish between two types of duplications and clusters based on the evolutionary history of the TF-coding genes that they contain. The first type of duplications and clusters consists of a series of paralogous genes, suggesting that they arose through repeated tandem duplications which originated from a founding locus. In contrast, the second type of duplications and clusters is not comprised paralogous genes. We anticipate that the TF-coding genes in these duplications and clusters arose independently of each other at diverse locations within the genome. Over time, it is likely that they relocated to form these duplications and clusters. Table 3 summarizes gene duplications and gene clusters in soybean TF families. Closely related genes, which are defined by >60% amino acid sequence identity, account for ∼77.75% of the total number in the TF families (Table 3). Pairs of duplicated genes on different chromosomes are most common and gene clusters of three or more highly related genes are also widely found (Table 3). On the basis of the distance of their occurrence, a few of the duplicated genes could be classified arbitrarily as either genes that were duplicated on same chromosome or genes that were tandemly duplicated. Evolutionary studies and haploid genome analysis have suggested that the soybean genome experienced a tetraploidization event which occurred an estimated 10–15 million years ago. Since then, the soybean genome has gone through extensive gene rearrangements and deletions to become diploidized.53 Therefore, we can observe in soybean that multigene families, including TF families, contain highly related genes.24,54
Table 3.
Classification of homologous soybean TF genes
| TF family | No. of gene locia | No. of genes with close homologb | Percentage of genes with close homolog | No. of individual genes | Duplications in different chromosomesc | Duplications in same chromosomed | Tandem duplicationse | No. of gene clusters/no. of genes in cluster (no. of chromosmes)f |
|---|---|---|---|---|---|---|---|---|
| (R1)R2R3_MYB | 318 | 261 | 82.08 | 57 | 50 | 2 | 1 | 43/155 (20) |
| ABI3VP1 | 139 | 90 | 64.75 | 49 | 20 | 1 | 2 | 12/44 (17) |
| Alfin-like | 18 | 18 | 100.00 | 0 | 1 | 0 | 0 | 1/16 (11) |
| AP2_EREBP | 381 | 309 | 81.10 | 72 | 74 | 1 | 0 | 42/159 (20) |
| ARF | 58 | 45 | 77.59 | 13 | 8 | 1 | 0 | 7/27 (16) |
| ARID | 20 | 9 | 45.00 | 11 | 1 | 0 | 0 | 2/7 (6) |
| atypical_MYB | 78 | 42 | 53.85 | 36 | 14 | 1 | 1 | 3/10 (6) |
| Aux_IAA | 85 | 72 | 84.71 | 13 | 9 | 0 | 0 | 13/54 (15) |
| BBR-BPC | 10 | 10 | 100.00 | 0 | 2 | 0 | 0 | 1/6 (4) |
| BES1 | 18 | 17 | 94.44 | 1 | 3 | 0 | 0 | 2/11 (9) |
| bHLH | 323 | 269 | 83.28 | 54 | 63 | 1 | 2 | 38/137 (20) |
| bZIP | 148 | 124 | 83.78 | 24 | 26 | 1 | 0 | 18/70 (20) |
| C2C2_Zn-CO-like | 84 | 67 | 79.76 | 17 | 19 | 0 | 0 | 7/29 (13) |
| C2C2_Zn-Dof | 81 | 60 | 74.07 | 21 | 17 | 1 | 0 | 7/24 (13) |
| C2C2_Zn-GATA | 63 | 53 | 84.13 | 10 | 11 | 0 | 0 | 9/31 (15) |
| C2C2_Zn-YABBY | 18 | 15 | 83.33 | 3 | 2 | 0 | 0 | 3/11 (8) |
| C2H2_Zn | 257 | 187 | 72.76 | 70 | 61 | 2 | 2 | 15/57 (18) |
| C3H-TypeI | 151 | 123 | 81.46 | 28 | 25 | 0 | 1 | 15/71 (17) |
| CAMTA | 14 | 12 | 85.71 | 2 | 6 | 0 | 0 | 0/0 (0) |
| CCAAT_Dr1 | 16 | 14 | 87.50 | 2 | 2 | 0 | 0 | 3/10 (9) |
| CCAAT_HAP2 | 23 | 16 | 69.57 | 7 | 2 | 0 | 0 | 3/12 (10) |
| CCAAT_HAP3 | 39 | 32 | 82.05 | 7 | 4 | 0 | 0 | 4/24 (14) |
| CCAAT_HAP5 | 23 | 19 | 82.61 | 4 | 3 | 0 | 1 | 3/11 (9) |
| CPP | 17 | 10 | 58.82 | 7 | 3 | 0 | 0 | 1/4 (4) |
| E2F_DP | 14 | 11 | 78.57 | 3 | 2 | 0 | 0 | 2/7 (5) |
| EIL | 13 | 12 | 92.31 | 1 | 2 | 0 | 0 | 2/8 (6) |
| GARP_ARRB | 20 | 13 | 65.00 | 7 | 3 | 0 | 0 | 2/7 (6) |
| GARP_G2-like | 82 | 59 | 71.95 | 23 | 18 | 0 | 0 | 7/23 (13) |
| GeBP | 17 | 13 | 76.47 | 4 | 2 | 0 | 0 | 2/9 (8) |
| GRAS | 117 | 102 | 87.18 | 15 | 23 | 0 | 0 | 14/56 (20) |
| GRF | 8 | 2 | 25.00 | 6 | 1 | 0 | 0 | 0/0 (0) |
| HB | 240 | 197 | 82.08 | 43 | 31 | 0 | 0 | 33/135 (20) |
| HMG-box | 26 | 22 | 84.62 | 4 | 5 | 0 | 0 | 2/12 (10) |
| HRT | 1 | 0 | 0.00 | 1 | 0 | 0 | 0 | 0/0 (0) |
| HSF | 59 | 48 | 81.36 | 11 | 12 | 0 | 0 | 7/24 (17) |
| JUMONJI | 51 | 28 | 54.90 | 23 | 8 | 0 | 1 | 3/10 (9) |
| LFY | 3 | 2 | 66.67 | 1 | 1 | 0 | 0 | 0/0 (0) |
| LIM | 32 | 28 | 87.50 | 4 | 1 | 0 | 0 | 5/26 (15) |
| LUG | 9 | 8 | 88.89 | 1 | 2 | 0 | 0 | 1/4 (4) |
| MADS | 186 | 154 | 82.80 | 32 | 25 | 2 | 1 | 18/98 (19) |
| MBF1 | 3 | 3 | 100.00 | 0 | 0 | 0 | 0 | 1/3 (2) |
| MYB_related | 135 | 112 | 82.96 | 23 | 17 | 1 | 1 | 16/74 (19) |
| NAC | 187 | 173 | 92.51 | 14 | 26 | 0 | 1 | 30/119 (20) |
| Nin-like | 23 | 15 | 65.22 | 8 | 4 | 0 | 0 | 2/7 (4) |
| PcG | 86 | 56 | 65.12 | 30 | 21 | 0 | 0 | 4/14 (9) |
| PHD | 285 | 188 | 65.96 | 97 | 51 | 1 | 0 | 18/84 (20) |
| PLATZ | 33 | 27 | 81.82 | 6 | 2 | 0 | 1 | 4/21 (13) |
| S1Fa-like | 4 | 4 | 100.00 | 0 | 0 | 0 | 0 | 1/4 (4) |
| SAP | 2 | 2 | 100.00 | 0 | 1 | 0 | 0 | 0/0 (0) |
| SBP | 48 | 39 | 81.25 | 9 | 9 | 0 | 0 | 6/21 (13) |
| SRS | 22 | 18 | 81.82 | 4 | 4 | 0 | 0 | 3/10 (8) |
| TCP | 61 | 44 | 72.13 | 17 | 19 | 0 | 0 | 2/6 (3) |
| Trihelix | 33 | 25 | 75.76 | 8 | 11 | 0 | 0 | 1/3 (3) |
| TUB | 24 | 20 | 83.33 | 4 | 2 | 0 | 0 | 3/16 (12) |
| ULT | 24 | 20 | 83.33 | 4 | 1 | 1 | 0 | 2/16 (1) |
| VOZ | 7 | 7 | 100.00 | 0 | 1 | 0 | 0 | 1/5 (4) |
| Whirly | 7 | 6 | 85.71 | 1 | 3 | 0 | 0 | 0/0 (0) |
| WRKY_Zn | 198 | 166 | 83.84 | 32 | 49 | 0 | 2 | 17/64 (20) |
| zf-HD | 56 | 46 | 82.14 | 10 | 7 | 0 | 1 | 6/30 (15) |
| zf-TAZ | 8 | 5 | 62.50 | 3 | 1 | 0 | 0 | 1/3 (3) |
| ZIM | 34 | 27 | 79.41 | 7 | 8 | 0 | 0 | 3/11 (9) |
aNumber of predicted TF loci found in soybean chromosomes (Glyma1 model).
bGenes were considered closely homologs if they showed >60% amino acid sequence identity.
cPairs of closely homologous genes which are duplicated in different chromosomes.
dPairs of closely homologous genes which are duplicated in same chromosome but resided >50 kb apart from each other.
ePairs of closely homologous genes which are duplicated in same chromosome but resided <50 kb apart from each other.
fClusters of three or more closely homologous genes.
3.4. Promoter regions of the TFs and the discovery of cis-elements in the TF promoter regions
Cis-regulatory elements, which are the binding sites for TFs located in the promoter regions of genes, are the functional elements that determine the timing and location of transcriptional activity. Over the years, extensive promoter analyses have identified a large number of cis-elements, which are important molecular switches involved in the transcriptional regulation of a dynamic network of gene activities controlling various biological processes such as abiotic stress responses, hormone responses and developmental processes.45,55 The PLACE database (http://www.dna.affrc.go.jp/PLACE/) has consolidated all of the published cis-motifs that have been identified to date. In addition, a number of stress responsive cis-motifs are also reported, which are of great interest to our area of research.45 To facilitate the functional characterization of soybean TFs, we retrieved the promoter regions for all of the TF genes from soybean genomic sequence database. Specifically, we retrieved 500, 1000 and 3000 bp of sequence upstream from putative transcription start sites. We provided this data on our website in addition to other relevant information on the TFs for convenient downloading. The −500, −1000 and −3000 bp promoter regions were subjected to an extensive in silico analyses to search for the existence of all putative known cis-regulatory motifs. In addition, we also analysed the enrichment of all of the cis-motifs in each TF family using −1000 bp promoter regions as described in Materials and methods. Information on the cis-elements located in the promoter region of each TF is accessible on the detailed page of each TF gene under ‘cis-motif prediction’ function (Fig. 4C). In addition, our website provides the ‘cis-motif (PLACE)’ search function, which enables the search for all types of cis-motifs provided by the PLACE database in promoter region of any TF and/or the search for those TFs which contains the cis-motif(s) of interest (Fig. 4A). In combination with GO annotations (Fig. 4H), these data can facilitate the systematic functional predictions of soybean TFs.
Figure 4.
The web-based user interface of SoybeanTFDB and a demonstration of a typical example of related annotations for a putative soybean TF encoding gene. The interface of SoybeanTFDB provides search queries for the names of TF families, keywords, sequence identifiers, identifiers of domains supported by InterProScan, GO terms and available cis-motifs (A). The search results are listed for each of the TF families with a description of corresponding genes based on similarity searches (B). Users are able to navigate to the detailed annotation pages to browse the related annotations. The detailed annotation pages provide summarized basic information on each of the gene models annotated in Glyma1 with gene structure. The figure for a gene structure is accessible via a hyperlink to a genome browser which is browsed together with other sequences allocated onto the soybean genome (C). The sequences of cDNAs and proteins are provided and all clickable buttons navigate users to the blast search interface directory (D). The similarity search results for each of the entries against NCBI nr, gene models of Arabidopsis and rice with detailed search results and hyperlinks to the original data (E). Resultant hierarchical clustering of homologous soybean TF genes can be browsed with multiple alignment of each cluster (F). The identifiers assigned provide hyperlinks to the annotation web pages of Arabidopsis and rice TF databases. Information of other sequence identifiers for representative transcript sequence databases, including PlantGDB, UniGene and TIGR GI, as well as the probe ID of target sequences on the soybean Affymetrix GeneChip, are also accessible. Furthermore, the interface also provides corresponding hyperlinks to the FL-cDNA provided from RIKEN (http://rsoy.psc.riken.jp/) (G). The GO terms assigned to each of the entries based on InterProScan and sequence similarity search against the annotated genes of Arabidopsis of TAIR8 (H). The domain structure predicted by InterProScan is provided (I). The result of a cis-motif sequence pattern search of promoter regions for each gene is shown together with genomic gene structure (J).
Numerous cis-elements have been reported for their essential roles in determining the tissue-specific or stress-induced expression patterns of genes.45,55 Recently, a systematic combinatorial in silico analysis of cis-motifs and expression patterns in Arabidopsis indicated a positive correlation between multi-stimuli response genes and cis-element density in upstream regions.56 Inspection of the relationship of the existence of cis-regulatory elements and the expression patterns of the TF genes can therefore help predict the function of the respective TFs during development, in different organs, cell types or in response to various endo- or exogenous stimuli. Additionally, quantitative models that describe how combinations of cis-elements dictate changes in expression will play an important role for enriching our understanding of the transcriptional response of individual genes to environmental perturbations.26
3.5. Cis-element- and comparative sequence analysis-based prediction of abiotic stress responsive TFs in soybean
Plants respond to environmental changes by altering large-scale transcriptional responses. The exquisite sensitivity and specificity of these responses are controlled in large part by the cis-regulatory elements. The molecular mechanisms regulating gene expression in response to abiotic stresses have been studied by analysing the cis- and trans-acting elements, i.e. the sequence-specific binding TFs.4,45
Genes induced by stresses are classified into two groups: functional genes and regulatory genes. The regulatory group includes genes encoding various TFs which can regulate various stress-inducible genes cooperatively or separately and may constitute gene networks. Identification and functional analysis of these stress-inducible TFs should provide more information on the complex regulatory gene networks that are involved in stress responses. At the present time, the functions for most of stress responsive TF encoding genes are not fully understood. Some of the stress-inducible TFs have been overexpressed in transgenic plants and result in stress-tolerant phenotypes.4,45,49
Recent studies have substantiated that sequence similarity-based clustering of the members of several TF families correlates with their function. Phylogenetic analysis of the AP2_EREBP and NAC families of soybean and the rice NAC family with orthologs from other plant species whose stress responsive expression pattern and/or function are known, resulted in a nearly perfect match between sequence conservation and function or expression patterns. These similarities clearly demonstrate that this can serve as a reliable approach to rationalize systematic functional predictions of different TF families.21,24,54 Moreover, increasing evidence indicates that the cis-motifs are highly conserved among orthologous or paralogous genes and coregulated genes and defined cis-elements can effectively aid in the genome-wide screening of ABA and abiotic stress responsive genes.57–59 These observations together prompted us to investigate in a comprehensive fashion the relationship between TFs and abiotic stress with the integration of cis-element annotation and comparative sequence analysis using stress responsive GO terms which aimed to identifying soybean TFs which may function in abiotic stress response. We, therefore, carried out comparative sequence analysis with stress-responsive Arabidopsis TFs to predict the soybean TFs with stress responsive GO terms (Fig. 3B). We also characterized information on stress-responsive cis-element distributions in promoter regions of each soybean TF gene on our webpage for querying and searching for putative stress responsive TFs in each family using ‘cis-motif (stress responsive)’ search function. With the help of our soybean TF database, we can use, for example, the ‘cis-motif (stress responsive)’ search function to identify TF genes which harbour major known stress responsive cis-motif(s) in their promoter regions (Fig. 4A). Next, we screen the identified TFs using GO annotation provided for each TF on detailed annotation page (Fig. 4H). Thus, we will be able to identify the putative stress responsive TFs based on both the existence of stress responsive cis-motif(s) and the associated stress responsive GO terms. The predicted stress responsive function of the identified TFs shall be then confirmed by experiments. The existence of major stress responsive cis-motifs enriched in −1000 bp promoter regions for a number of TF families was summarized in Table 4.
Table 4.
Enriched cis-regulatory motifs found in promoter region (1000 bp upstream from transcription start site of each gene) of genes encoding each of the TF families
| Cis-motif namea | Cis-motif patterna | TF family | No. of gene loci hit | No. of gene loci | Mean observedb | Mean expectedc | Z-score | P-value (<0.001) |
|---|---|---|---|---|---|---|---|---|
| ABRE1 | [TC]ACGTGGC | C2C2_Zn-CO-like | 4 | 84 | 47.385 | 14.074 | 8.81 | 0 |
| C2C2_Zn-GATA | 3 | 63 | 47.784 | 14.074 | 8.92 | 0 | ||
| C3H-TypeI | 6 | 151 | 39.851 | 14.074 | 6.82 | 4.577E−12 | ||
| JUMONJI | 2 | 51 | 39.435 | 14.074 | 6.71 | 9.7873E−12 | ||
| NAC | 5 | 187 | 26.593 | 14.074 | 3.31 | 0.00046339 | ||
| TCP | 5 | 61 | 82.433 | 14.074 | 18.08 | 0 | ||
| WRKY_Zn | 6 | 198 | 29.952 | 14.074 | 4.20 | 0.000013318 | ||
| ABRE2 | ACGTG[GT]C | (R1)R2R3_Myb | 26 | 319 | 81.996 | 55.79 | 3.59 | 0.00016627 |
| AP2_EREBP | 31 | 382 | 81.157 | 55.79 | 3.47 | 0.00025671 | ||
| Aux_IAA | 7 | 85 | 82.312 | 55.79 | 3.63 | 0.00014072 | ||
| C2C2_Zn-CO-like | 13 | 84 | 154.522 | 55.79 | 13.52 | 0 | ||
| C2C2_Zn-Dof | 10 | 81 | 123.231 | 55.79 | 9.24 | 0 | ||
| C2C2_Zn-GATA | 5 | 63 | 79.277 | 55.79 | 3.22 | 0.00064947 | ||
| C3H-TypeI | 12 | 151 | 79.117 | 55.79 | 3.19 | 0.00070084 | ||
| JUMONJI | 7 | 51 | 137.741 | 55.79 | 11.22 | 0 | ||
| MADS | 18 | 186 | 96.524 | 55.79 | 5.58 | 1.2169E−08 | ||
| NAC | 27 | 187 | 144.379 | 55.79 | 12.13 | 0 | ||
| TCP | 8 | 61 | 131.2 | 55.79 | 10.33 | 0 | ||
| Atypical_MYB | 10 | 78 | 128.071 | 55.79 | 9.90 | 0 | ||
| CE1 | TGCCACCGG | C2H2_Zn | 1 | 258 | 3.952 | 0.571 | 4.39 | 5.7069E−06 |
| JUMONJI | 1 | 51 | 19.698 | 0.571 | 24.83 | 0 | ||
| WRKY_Zn | 4 | 198 | 20.174 | 0.571 | 25.44 | 0 | ||
| CRT | GGCCGACAT | AP2_EREBP | 1 | 382 | 2.538 | 0.324 | 3.88 | 0.000051901 |
| C2H2_Zn | 1 | 258 | 3.86 | 0.324 | 6.20 | 2.8371E−10 | ||
| DRE | TACCGACAT | ABI3VP1 | 1 | 139 | 7.255 | 0.77 | 7.36 | 9.0372E−14 |
| AP2_EREBP | 3 | 382 | 7.931 | 0.77 | 8.13 | 2.2204E−16 | ||
| NAC | 1 | 187 | 5.351 | 0.77 | 5.20 | 9.9255E−08 | ||
| ICEr2 | ACTCCG | AP2_EREBP | 22 | 382 | 57.453 | 37.698 | 3.25 | 0.00057027 |
| C2C2_Zn-GATA | 4 | 63 | 63.223 | 37.698 | 4.20 | 0.000013136 | ||
| JUMONJI | 4 | 51 | 78.537 | 37.698 | 6.73 | 8.7457E−12 | ||
| PHD | 20 | 285 | 70.006 | 37.698 | 5.32 | 5.1702E−08 | ||
| TCP | 4 | 61 | 65.403 | 37.698 | 4.56 | 2.5263E−06 | ||
| MYBR | TGGTTAG | C2H2_Zn | 19 | 258 | 73.778 | 48.032 | 3.90 | 0.000047443 |
| MADS | 13 | 186 | 69.508 | 48.032 | 3.26 | 0.00056509 | ||
| TCP | 5 | 61 | 82.123 | 48.032 | 5.17 | 0.000000118 | ||
| MYCR | CACATG | (R1)R2R3_Myb | 98 | 319 | 307.51 | 230.595 | 5.93 | 1.5169E−09 |
| ABI3VP1 | 41 | 139 | 295.074 | 230.595 | 4.97 | 3.3303E−07 | ||
| AP2_EREBP | 112 | 382 | 293.307 | 230.595 | 4.83 | 6.6647E−07 | ||
| ARF | 19 | 58 | 327.992 | 230.595 | 7.51 | 2.9865E−14 | ||
| Aux_IAA | 23 | 85 | 270.693 | 230.595 | 3.09 | 0.00099623 | ||
| C2C2_Zn-CO-like | 25 | 84 | 298.315 | 230.595 | 5.22 | 8.9042E−08 | ||
| C2C2_Zn-Dof | 29 | 81 | 358.556 | 230.595 | 9.87 | 0 | ||
| C2H2_Zn | 70 | 258 | 271.297 | 230.595 | 3.14 | 0.00085076 | ||
| Myb_related | 38 | 135 | 281.165 | 230.595 | 3.90 | 0.000048357 | ||
| bZIP | 42 | 148 | 283.077 | 230.595 | 4.05 | 0.000026039 | ||
| NACR | ACACGCATGT | C2H2_Zn | 1 | 258 | 3.823 | 0.494 | 4.93 | 4.1633E−07 |
| WRKY_Zn | 1 | 198 | 4.944 | 0.494 | 6.59 | 2.2463E−11 | ||
| bZIP | 1 | 148 | 6.804 | 0.494 | 9.34 | 0 |
aAccording to Yamaguchi-Shinozaki et al.45
bThe mean values observed were calculated by counting motif pattern hit in 1000 random samplings in each 1000 trials for promoter pools of each TFs.
cThe mean values expected were calculated by counting motif patterns hit in 1000 random samplings in each 1000 trials for promoter pools of all genes annotated in soybean genome.
3.6. RIKEN soybean TF database
We constructed a TF database named SoybeanTFDB which is based on the identified soybean TF repertoire. Access to our database is available via the following link: http://soybeantfdb.psc.riken.jp, and all of the data described above are available for viewing and immediate downloading. The scientific community can browse predictions for a total of 5035 TF models and receive classifications for submitted nucleotide and protein sequences. Multiple alignments of amino acid sequences within TF families are also available for downloading and can be used for the construction of phylogenetic trees. We also provided clustered results showing amino acid similarity with different levels of amino acid identity (30, 60 and 90%), search functions for functional motif information of InterProScan, cis-motifs in promoter regions of TFs and GO annotations. Furthermore, cross-references and links to other databases such as Arabidopsis TAIR8, TIGR rice, UniProt, SoyBase, soybean FL-cDNA and other TF databases such as AtTFDB, DATF, RARTF, DRTF, Grassius, PlnTFDB are available. On the first page of SoybeanTFDB, we provide four types of search keywords to find an entry: ‘TF search’, ‘Similarity search’, ‘Genome browser’ and ‘Quick search’. Similarity search allows search using either nucleotide or amino acid sequence of any TF gene. Genome browser enables search using gene IDs and Quick search allows search function via any essential keywords such as ‘NAC’. Within the TF search keyword, we provide seven search functions (Fig. 4A). Figure 4 illustrates the web-based user interface of SoybeanTFDB with a detailed description. One can easily carry out functionality predictions for any TF of interest based on GO annotations and cis-motif search results. For instance, putative abiotic stress responsive TFs can be searched based on the existence of stress responsive cis-motifs and GO annotations. Thus, the database that we have developed consolidates comprehensive information for all of the members of soybean TF repertoire. This database is a very user-friendly interface which aims to meet the broad demands of researchers who strive to perform research with soybean TFs with the goal of gaining greater understanding of their putative roles in plant development, differentiation and environmental responses. Taken together, SoybeanTFDB will serve as an in silico analysis-based basic platform for the elucidation of regulatory mechanisms underlying different developmental and physiological processes and stress responses. We strongly feel that this database will rapidly accelerate the progress in ‘transcription factoromics’ of soybean, comparative genomics of TF repertoires both within legume species and between legumes and other species, as well as facilitate genetic engineering programs to improve the productivity of soybean grown in adverse conditions.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
Funding support from Grants-in-Aid (Start-up) for Scientific Research, Ministry of Education, Culture, Sports, Science, and Technology of Japan (No. 21870046) is gratefully appreciated.
Supplementary Material
Acknowledgements
The soybean sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.g.ov/ in collaboration with the scientific user community.
References
- 1.Riechmann J.L., Ratcliffe O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000;3:423–34. doi: 10.1016/s1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- 2.Czechowski T., Bari R.P., Stitt M., Scheible W.R., Udvardi M.K. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 2004;38:366–79. doi: 10.1111/j.1365-313X.2004.02051.x. [DOI] [PubMed] [Google Scholar]
- 3.Tran L.-S.P., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods Enzymol. 2007;428:109–28. doi: 10.1016/S0076-6879(07)28006-1. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 5.Bustamante C.D., Fledel-Alon A., Williamson S., et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–7. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 6.Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 7.Riechmann J.L., Heard J., Martin G., et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–10. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 8.Riaño-Pachón D.M., Ruzicic S., Dreyer I., Mueller-Roeber B. PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo A.Y., Chen X., Gao G., et al. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res. 2008;36:D966–9. doi: 10.1093/nar/gkm841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson D., Charoensawan V., Kummerfeld S.K., Teichmann S.A. DBD-taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherry J.M., Adler C., Ball C., et al. SGD: Saccharomyces genome database. Nucleic Acids Res. 1998;26:73–9. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Rueda E., Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–47. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray P.A., Fu H., Luo P., et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–7. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 14.Iida K., Seki M., Sakurai T., et al. RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res. 2005;12:247–56. doi: 10.1093/dnares/dsi011. [DOI] [PubMed] [Google Scholar]
- 15.Reece-Hoyes J.S., Deplancke B., Shingles J., Grove C.A., Hope I.A., Walhout A.J. A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adryan B., Teichmann S.A. FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–3. doi: 10.1093/bioinformatics/btl143. [DOI] [PubMed] [Google Scholar]
- 17.Moreno-Campuzano S., Janga S.C., Perez-Rueda E. Identification and analysis of DNA-binding transcription factors in Bacillus subtilis and other Firmicutes—a genomic approach. BMC Genomics. 2006;7:147. doi: 10.1186/1471-2164-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J., Park J., Jang S., et al. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors, Bioinformatics. 2008;24:1024–5. doi: 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- 19.Udvardi M.K., Kakar K., Wandrey M., et al. Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 2007;144:538–49. doi: 10.1104/pp.107.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran L.-S.P., Nakashima K., Sakuma Y., et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–98. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y., You J., Xie K., Xie W., Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics. 2008;280:535–46. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 22.Liao Y., Zou H.F., Wei W., et al. Soybean GmbZIP144, GmbZIP162 and GmbZIP178 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta. 2008;228:225–40. doi: 10.1007/s00425-008-0731-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q.Y., Tian A.G., Zou H.F., et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 24.Tran L.-S.P., Quach T.N., Guttikonda S.K., et al. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genomics. 2009;281:647–64. doi: 10.1007/s00438-009-0436-8. [DOI] [PubMed] [Google Scholar]
- 25.Gong W., Shen Y.P., Ma L.G., et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004;135:773–82. doi: 10.1104/pp.104.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gertz J., Cohen B.A. Environment-specific combinatorial cis-regulation in synthetic promoters. Mol. Syst. Biol. 2009;5:244. doi: 10.1038/msb.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 28.Ososki A.L., Kennelly E.J. Phytoestrogens: a review of the present state of research. Phytotherapy Res. 2003;17:845–69. doi: 10.1002/ptr.1364. [DOI] [PubMed] [Google Scholar]
- 29.Shinozaki K. Acceleration of soybean genomics using large collections of DNA markers for gene discovery. DNA Res. 2007;14:235. doi: 10.1093/dnares/dsm031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai T., Kogiso M. Soyisoflavones and immunity. J. Med. Invest. 2008;55:167–73. doi: 10.2152/jmi.55.167. [DOI] [PubMed] [Google Scholar]
- 31.Tran L.-S.P., Nguyen H.T. Future Biotechnology of Legumes. In: Emerich W.D., Krishnan H., editors. Nitrogen Fixation in Crop Production. Madison, WI, USA: The American Society of Agronomy, Crop Science Society of America and Soil Science Society of America; 2009. pp. 265–308. [Google Scholar]
- 32.Graham P.H., Vance C.P. Legumes: importance and constraints to greater use. Plant Physiol. 2003;131:872–7. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sammut S.J., Finn R.D., Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinform. 2008;9:210–9. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- 34.Yanhui C., Xiaoyuan Y., Kun H., et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006;60:107–24. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang S., Thibaud-Nissen F., Childs K.L., Zhu W., Buell C.R. Plant genome annotation methods. Methods Mol. Biol. 2009;513:263–82. doi: 10.1007/978-1-59745-427-8_14. [DOI] [PubMed] [Google Scholar]
- 36.Altschul S.F., Madden T.L., Schaffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umezawa T., Sakurai T., Totoki Y., et al. Sequencing and analysis of approximately 40000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 2008;15:333–46. doi: 10.1093/dnares/dsn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davuluri R.V., Sun H., Palaniswamy S.K., et al. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo A., He K., Liu D., et al. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–9. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 40.Gao G., Zhong Y., Guo A., et al. DRTF: a database of rice transcription factors. Bioinformatics. 2006;22:1286–7. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- 41.Palaniswamy S.K., James S., Sun H., Lamb R.S., Davuluri R.V., Grotewold E. AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–29. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yilmaz A., Nishiyama M.Y., Jr, Fuentes B.G., et al. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 2009;149:171–80. doi: 10.1104/pp.108.128579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 44.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi-Shinozaki K., Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Nemhauser J.L., Mockler T.C., Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donlin M.J. Using the generic genome browser (GBrowse) Curr. Protoc. Bioinform. 2007 doi: 10.1002/0471250953.bi0909s17. Chapter 9, Unit 9.9. [DOI] [PubMed] [Google Scholar]
- 48.Valliyodan B., Nguyen H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006;9:189–95. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 49.Nakashima K., Ito Y., Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazarova G., Zeng Y., Kermode A.R. Cloning and expression of an ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene homologue of yellow-cedar (Chamaecyparis nootkatensis) J. Exp. Bot. 2002;53:1219–21. doi: 10.1093/jexbot/53.371.1219. [DOI] [PubMed] [Google Scholar]
- 51.Luscombe N.M., Thornton J.M. Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J. Mol. Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 52.Itzkovitz S., Tlusty T., Alon U. Coding limits on the number of transcription factors. BMC Genomics. 2006;7:239. doi: 10.1186/1471-2164-7-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlueter J.A., Lin J.Y., Schlueter S.D., et al. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics. 2007;8:330. doi: 10.1186/1471-2164-8-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang G., Chen M., Chen X., et al. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) J. Exp. Bot. 2008;59:4095–107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandepoele K., Quimbaya M., Casneuf T., De Veylder L., Van de Peer Y. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;50:535–46. doi: 10.1104/pp.109.136028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walther D., Brunnemann R., Selbig J. The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet. 2007;3:e11. doi: 10.1371/journal.pgen.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W., Ruan J., Ho T.H., You Y., Yu T., Quatrano R.S. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in. Arabidopsis thaliana. Bioinformatics. 2005;21:3074–81. doi: 10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
- 58.Kim D.W., Lee S.H., Choi S.B., et al. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell. 2006;18:2958–70. doi: 10.1105/tpc.106.045229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Won S.K., Lee Y.J., Lee H.Y., Heo Y.K., Cho M., Cho H.T. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009;150:1459–73. doi: 10.1104/pp.109.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.