Abstract
Organ morphogenesis requires cooperation between cells, which determine their course of action based upon location within a tissue. Just as important, cells must synchronize their activities, which requires awareness of developmental time. To understand how cells coordinate behaviors in time and space, we analyzed Drosophila egg chamber development. We found that the transcription factor Tramtrack69 (TTK69) controls the fates and shapes of all columnar follicle cells by integrating temporal and spatial information, restricting characteristic changes in morphology and expression that occur at stage 10B to appropriate domains. TTK69 is required again later in oogenesis: it controls the volume of the dorsal-appendage (DA) tubes by promoting apical re-expansion and lateral shortening of DA-forming follicle cells. We show that TTK69 and Notch compete to repress each other's expression and that a local Ecdysone signal is required to shift the balance in favor of TTK69. We hypothesize that TTK69 then cooperates with spatially restricted co-factors to define appropriate responses to a globally available (but as yet unidentified) temporal signal that initiates the S10B transformations.
Keywords: Drosophila, Notch, Tramtrack69 (TTK69; TTK), Ecdysone, Dorsal appendage, Epithelial morphogenesis, Patterning, Oogenesis, Apical constriction
INTRODUCTION
Pattern formation is generally thought of as the process by which cells determine their identity within a larger structure through the use of mechanisms such as morphogen gradients (Wolpert, 1989; Moussian and Roth, 2005; Ashe and Briscoe, 2006). Equally important is the more poorly understood process by which cells determine identity as a function of developmental time. Steroid hormones can induce global transitions by transforming tissues throughout the body (Riddiford, 1976; Hiruma et al., 1999; Banerjee and Clayton, 2007). At the other extreme, microRNAs can regulate single-cell decisions that control temporal aspects of the worm lineage (Frasch, 2008), and a defined sequence of transcription factors regulates neuroblast identity based on time of cell birth during Drosophila embryogenesis (Grosskortenhaus et al., 2005). On the tissue level, the addition of somites to the vertebrate trunk and the addition of segments to the posterior of short-germband insect embryos require synchronized gene expression and cell movements (Davis and Patel, 2002; Kalcheim and Ben-Yair, 2005). In general, however, the mechanisms that cells use to coordinate their activities remain largely unexplored.
We study these questions in the context of the morphological changes required to build the Drosophila eggshell, focusing on the dorsal appendages (DAs: eggshell protrusions that facilitate gas exchange). DA tube formation resembles gastrulation or neurulation but occurs within egg chambers, which mature through 14 stages (S) (King, 1970; Spradling, 1993). Egg chambers contain 16 germ cells — 15 nurse cells and a single oocyte — surrounded by a somatic epithelium that, by mid-oogenesis, consists of the columnar follicle cells over the oocyte and the squamous stretch cells overlying the nurse cells. During S10B-S14, while the nurse cells transfer their contents into the oocyte and undergo cell death, the DAs develop from two patterned subsets of dorsal anterior columnar follicle cells.
By S10B, Epidermal growth factor (EGF), Decapentaplegic (DPP), and Notch (N) have patterned the columnar follicle cells into four subtypes (Dobens and Raftery, 2000) (Fig. 1I), each expressing unique markers (Dorman et al., 2004; Ward and Berg, 2005). The main body follicle cells (forming no specialized structures) (Margaritis, 1985), occupy the majority of the egg chamber and express low levels of the transcription factor Broad (BR). The roof cells, which form the roof and sides of each DA, express high BR. Remaining columnar follicle cells form a ‘T’ shape (‘The T’) lacking BR. Those T cells that contact the roof cells become DA floor cells; the rest form the operculum. During S10B—S14, the roof and floor cells form, then elongate, the DAs (Dorman et al., 2004). Tube formation involves apical constriction of roof cells while floor cells elongate beneath the roof cells and zipper together. Elongation involves anterior migration of the tube, coordinated with roof- and floor-cell shape changes.
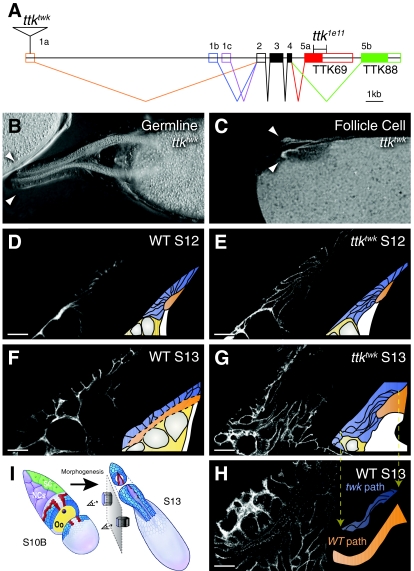
Fig. 1.
tramtrack69 is required in the follicle cells for proper morphogenesis. (A) The ttk locus. The twk P element disrupts 1a-containing transcripts while the 1e11 deletion removes most of the TTK69 zinc finger (red box) but not the BTB domain. (B) Germline ttktwk clone at S14 with normal appendages (arrowheads). (C) S14 whole follicular epithelium clone of ttktwk marked by absence of nuclear GFP. Chorion autofluorescence reveals short appendages (arrowheads). (D-H) DA-forming cell shapes before tube elongation at S12 (D,E) and after at S13 (F,G,H) in wild-type (D,F,H) and ttktwk (E,G) egg chambers. Images are single confocal slices; lateral cell membranes are stained with antibody against alpha-spectrin. D,E,G,H are lateral views. Wild-type DA-forming cells rotate ventrally as they migrate anteriorly. To maintain comparable view of lateral cell surfaces, the image in F is rotated to a ventrolateral perspective. H shows an unrotated (lateral) view of a wild-type S13 DA. Most DA-forming cells lie above the image plane. A higher section would show their basal surfaces, obscuring the lumen. Insets diagram the cell and DA shapes: DA-forming follicle cells (blue), lumen (orange) and nurse cell nuclei (gray). DA cells normally shorten while expanding their apices (F), but fail to do so in ttktwk (G). Nevertheless, ttktwk basal surfaces migrate anteriorly, resulting in thin, elongated cells. (H) Wild-type S13 from a lateral view, showing the bend in the DA. Inset in H compares the shape of elongated ttktwk cells from G (blue, pulled down from the inset in G) to the shape of the wild-type lumen from H (orange). Other features are omitted. The lateral surfaces of ttktwk cells trace the same sigmoidal path as the wild-type DA lumen. (I) Schematic representation of S10B and S13 egg chambers (not to scale) showing the morphogenesis between these stages. Note that the DA-forming cells rotate 90° as they migrate anteriorly. Thus, a lateral section reveals lateral cell surfaces at S10B. At S13, however, lateral cell surfaces are only visible at the base of the appendages. In the anterior a lateral section reveals basal cell surfaces. Adapted with permission (Dorman et al., 2004). Scale bars: 20 μm.
Tramtrack69 (TTK69; TTK — FlyBase) is a zinc-finger transcription factor that contains a Bric-a-brac-Tramtrack-Broad (BTB) dimerization domain (Fig. 1A). TTK69 functions in numerous tissues as a transcriptional repressor, e.g. downstream of N in asymmetric cell divisions to repress neural cell fates (Okabe et al., 2001). Previous work from our lab studying the twin peaks (ttktwk) allele, which disrupts expression of the gene late in oogenesis, demonstrates that TTK69 is required to elongate the DA tubes (French et al., 2003).
Here we describe a novel mechanism by which TTK69 promotes DA elongation, and we uncover a broader role for the gene in integrating temporal and spatial cues at mid-oogenesis. ttktwk mutants fail to re-expand the apices of DA-forming cells. Mosaic analyses with a null allele, ttk1e11, demonstrate that TTK69 is required in all columnar follicle cells to prevent location-inappropriate behaviors at the transition from S10A to S10B. Furthermore, we show that TTK69 and N regulate each other's expression and in turn are controlled by Ecdysone signaling during this key developmental switch.
MATERIALS AND METHODS
Drosophila strains
We used the following alleles and transgenic constructs: ttktwk (French et al., 2003), FRT82B ttk1e11 (Baonza et al., 2002), Su(H)SF8 FRT40A (Schweisguth and Posakony, 1994), N55e11 FRT101 (Kidd et al., 1983; Couso and Martinez Arias, 1994; Tweedie et al., 2009), UAS-TTK69 ttk1e11 (French et al., 2003), UAS-N-DeltaEN B2a-3/TM3 (constitutively activated N, UAS-N-CA) (Fuerstenberg and Giniger, 1998), UAS-EcR-B1W650A (dominant negative EcR, UAS-EcR-DN) (Hu et al., 2003), rhomboid-lacZ.8.3 (rho-lacZ) (Ip et al., 1992), and the N reporter E(Spl)mβ-CD2 (gift of Wu-Min Deng).
Strains used to generate mosaics: hsFLP; FRT82B ovoD (Chou and Perrimon, 1996), UAS-FLP e22c-GAL4; FRT82B Ubi-GFP.nls (Duffy et al., 1998), hsFLP UAS-GFP.nls tub-GAL4/FM7; +; FRT82B tub-GAL80/TM6B (gift of Bruce Edgar), hsFLP tub-GAL4 UAS-GFP.nls; tub-GAL80 FRT40A/CyO (gift of Bruce Edgar), hsFLP; Act-FRT-CD2-FRT-GAL4, UAS-GFP/TM6B (gift of Bruce Edgar), and Ubi-GFP FRT101; hsFLP/CyO (gift of Hannele Ruohola-Baker).
Mosaic analysis
We used FLP/FRT (Chou and Perrimon, 1992; Xu and Rubin, 1993), ovoD (Chou et al., 1993) for germline clones, and e22c-GAL4 (Duffy et al., 1998) for large ttktwk clones. For MARCM mosaics (Lee and Luo, 2001), we heat shocked for 1 hour and dissected after 2-4 days. N55e11 clones were generated by heat shocking twice for 1 hour a day apart, dissecting ovaries 5-7 days later. Mosaic overexpression employed FLP-out GAL4 (Pignoni and Zipursky, 1997), heat shocking for 10 minutes and dissecting after 2 days.
Immunostaining
Ovary immunostaining was as described (Ward and Berg, 2005), using mouse anti-alpha-spectrin [1:50, 3A9-concentrate, Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA] (Dubreuil et al., 1987), mouse anti-BR-core (1:500, 25E9.D7-concentrate, DSHB) (Emery et al., 1994), rat anti-DE-cadherin (1:50, DCAD2) (Oda et al., 1994), rabbit anti-TTK69 (1:200) (Badenhorst, 2001), rat anti-TTK69 (1:100) (Read and Manley, 1992), mouse anti-N-ICD (1:500, C17.9C6-concentrate, DSHB) (Fehon et al., 1990), mouse anti-GFP (1:200, Molecular Probes/Invitrogen, Eugene, OR) rabbit anti-GFP (1:200, Molecular Probes/Invitrogen), rabbit anti-β-galactosidase (1:3000, Cappel, West Chester, PA), rabbit anti-CD2 (1:50, Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-CD2 (1:50, AbD Serotec, Oxford, UK). Secondary detection was by goat anti-mouse, rabbit and rat Alexa fluor 488-, 555-, 568- and 647-conjugated antibodies (1:200, Molecular Probes/Invitrogen).
RESULTS
TTK69 is required in the follicle cells for apical re-expansion and cell shortening
ttktwk mutants produce correctly patterned but short DAs (French et al., 2003). TTK69 is required in the follicle cells themselves: ttktwk whole-follicle cell clones recapitulated the ttktwk phenotype, whereas germline clones produced wild-type eggs (Fig. 1B,C). Although ttktwk also disrupts chorion production, DA elongation is independent of this process, as tube elongation occurs normally in defective chorion 2 (dec2; Cp36 — FlyBase) mutants (French et al., 2003).
To determine which morphogenetic processes fail in ttktwk, we examined cell shapes during DA elongation (Fig. 1D-H). ttktwk starts with a correctly formed tube. Tube elongation requires both anterior/lateral migration and apical re-expansion. As the surface area of the lumen is the sum of these apices, re-expansion determines lumen size. This process failed in ttktwk; at S13, mutant cells remained apically constricted (Fig. 1G) whereas wild-type cells became cuboidal (Fig. 1F). Note that the follicle cells rotate 90 degrees as they migrate (Fig. 1I). We therefore rotated the perspective in Fig. 1F to maintain the view of lateral cell surfaces.
Remarkably, anterior/lateral migration of the ttktwk basal surfaces occurred correctly, delivering the basal surfaces to wild-type locations while the apical surfaces were left behind. The resulting highly elongated lateral surfaces trace a sigmoidal path (Fig. 1G) identical to the shape of a wild-type DA (Fig. 1H). The inset in Fig. 1H compares this shape (orange) to the shape of ttktwk mutant cells (blue). Thus, the ttktwk defect is a failure to re-expand apical surfaces and shorten lateral surfaces, not in migration or path finding. These results implicate TTK69 in regulating apical and lateral domain size.
ttk1e11 mutant cells block DA formation and constrict apically regardless of location
ttktwk is a hypomorph that disrupts TTK69 expression only late during oogenesis (French et al., 2003). To expand our understanding of the role of ttk69 in DA formation, we made mosaics of ttk1e11, which deletes a portion of the TTK69 zinc-finger domain (Fig. 1A). Although ttktwk cells formed a tube, ttk1e11 mutant cells did not participate in DA morphogenesis (Fig. 2A). Patches of mutant cells clumped together and blocked surrounding cells, occasionally splitting the DA.
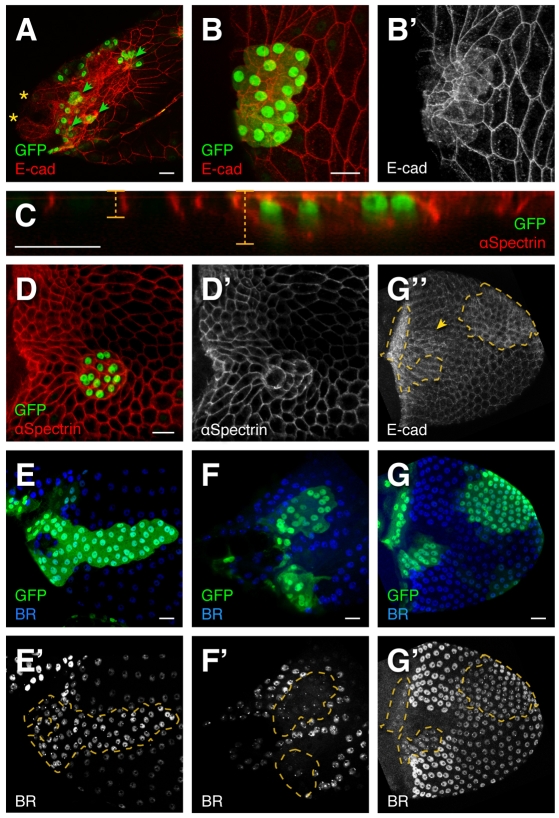
Fig. 2.
ttk1e11 disrupts cell shape and patterning. (A) Small clones of ttk1e11 (marked by GFP) result in elevated E-cadherin and extremely constricted apices. ttk1e11 clones (arrowheads) blocked the elongation of the single DA shown in A, causing it to split into two lobes (asterisks). The right appendage, not pictured, lies deep on the far side of the egg chamber. (B,B′) Magnified view of a ttk1e11 clone with severely constricted apices. E-cadherin expression is increased apically and cytoplasmically (B′), resembling expression in the T. Compare cytoplasmic E-cadherin in ttk1e11 clones (B′,G″, outline) to the wild-type T (G″, arrowhead). E-cadherin and GFP expression also reveal basal surfaces that are much larger than apical surfaces. (C) A confocal re-slice through a ttk1e11 clone with alpha-spectrin staining lateral surfaces. ttk1e11 cells are taller than their neighbors. (D,D′) Basal surfaces of ttk1e11 cells are rounded. (E-G″) ttk1e11 affects BR expression. (E) Lateral view at S13. ttk1e11 cells in the main body exhibit elevated BR. Appearance of more tightly packed nuclei within the clone is not due to extra rounds of division but to the smaller size of ttk1e11 cells; this difference is pronounced at this stage because wild-type follicle cells have enlarged to accommodate oocyte growth. (F) Dorsal view at S13. ttk1e11 cells in the roof express reduced BR. (G,G′) Dorsal view at S10B. ttk1e11 cells in the T maintain BR. At this stage, wild-type BR expression in the main body is still as high as BR in ttk1e11 cells but decreases over time (E′). Scale bars: 20 μm.
ttk1e11 also produced dramatic defects outside the DAs (Fig. 2, 141/141 clones) resulting in apical constriction of normally cuboidal cells. Egg chambers grow rapidly and vary in size, so we compared apical area between wild-type and ttk1e11 cells within individual egg chambers. In a typical example (Fig. 2B), ttk1e11 apices measured 7.9% of wild type (31.5 μm2 vs 398.8 μm2). Combined with the ttktwk phenotype, this result suggests that TTK69 regulates apical surface size.
Adjacent wild-type cells stretched to maintain epithelial integrity (Fig. 2B′). ttk1e11 basal surfaces were slightly shrunken (Fig. 2B-D) and rounded (Fig. 2D), but far larger than their apices (apically constricted, not just smaller). These phenotypes were not secondary to the role of TTK69 in endocycle/amplification (Jordan et al., 2006; Sun et al., 2008): reduction in volume as a result of lower DNA content would not explain apices smaller than basal surfaces. Although we did note that ttk1e11 nuclei appeared more closely packed at late stages (Fig. 2E′), this effect was not pronounced at earlier stages (Fig. 2G′, Fig. 6). Extra cell divisions at the S6 endocycle transition would affect all subsequent stages. Instead, tight packing of the ttk1e11 cells was caused by apical constriction; wild-type cells, by contrast, expanded to accommodate the growing oocyte. ttk1e11 cells elevated E-cadherin (encoded by shotgun — FlyBase) expression apically (Fig. 2A,B) and basally/cytoplasmically, resembling normal T cells (Fig. 2B′,G″) (James et al., 2002). Mutant cells were also taller than wild type (Fig. 2C). In conclusion, TTK69 regulates morphogenesis in all columnar follicle cells, preventing adoption of features normally specific to epithelial subregions (apical constriction in roof cells and high basal E-cadherin in T cells).
Fig. 6.
The ttk1e11 phenotypes are specific to S10B and later. Compare mutant cells (green arrowheads) with wild-type cells (blue arrowheads). (A-B′) ttk1e11 cells do not constrict their apices at S10A (average apical surface area: 52.2 μm2) (A) but do so at S10B (average apical surface area 34.4 μm2) (B). (C-D″) BR is normal in ttk1e11 cells at S10A (C,C′). The yellow arrowhead indicates a cluster of ttk1e11 mutant cells displaying an endocycle defect (resulting in condensed, brighter nuclei). At the beginning of S10B (D), BR is reduced in ttk1e11 clones, concurrent with the wild-type reduction of BR in the T (D′, yellow arrowhead). Neither roof nor ttk1e11 cells have begun to constrict their apices (D″). (E,E′) At S10A, N is uniformly high and unaffected by ttk1e11. (F,F′) UAS-EcR-DN does not affect cell shape at S10A. (G-G″) At S9, UAS-N-CA expression does not affect cell shape (G′) or TTK69 expression (G″). Scale bars: 20 μm.
TTK69 regulates Broad expression
As ttk1e11 cells exhibit features of multiple cell types, we asked whether ttk1e11, unlike ttktwk, affects cell fate. When DA morphogenesis begins at S10B, changes in BR expression mark the various cell types: BR is elevated in the roof, absent in the T and reduced elsewhere (Dorman et al., 2004). Therefore we examined BR expression in ttk1e11 mosaics.
Clones of ttk1e11 outside the high-BR-expressing roof region exhibited elevated BR compared with neighbors in the main body or T (Fig. 2E,G). Adjacent cells did not become floor: they continued expressing BR and failed to induce rhomboid-lacZ or elevate FAS3 (not shown). By contrast, ttk1e11 clones within the roof reduced BR (Fig. 2F′). Thus, removal of TTK69 resulted in expression of BR at a level between that of main body and high-BR cells (47/47 clones).
Note that the level of BR expressed by wild-type cells is dynamic over time. Main body follicle cells cease expression of br RNA and allow degradation of BR protein over time (Yakoby et al., 2008b). As a result, early S10B BR expression in ttk1e11 clones was very close to that of neighboring wild-type main body cells (Fig. 2G′), but expression remained high (or increased) in the clone as BR levels decreased over time elsewhere (Fig. 2E′). Although ttk1e11 cells express BR at a level similar to that of pre-S10B BR expression, these cells are not arrested at this earlier stage: they constricted their apices and modified E-cadherin expression, activities that occur only in subsequent stages. These data demonstrate that TTK69 is required for follicle-cell patterning, as well as morphogenesis.
TTK69 is required for the downregulation of N at S10B
We asked whether we could place ttk69 within the known follicle-cell patterning pathways. First we examined TTK69 expression throughout the course of oogenesis to determine whether its pattern suggested regulation by a particular pathway (Fig. 3A′-D′). Levels were low throughout mid-oogenesis (Fig. 3A′) then increased slightly by the beginning of S10B (Fig. 3B′). During the S10A-S10B transition, TTK69 became highly variable, with TTK69 missing, present at very low levels, or elevated in patches of cells, but with no clear pattern to this variation (Fig. 3E-G). Occasionally, a large patch of dorsal anterior cells showed reduced TTK69, but such cells were not reliably centered on the dorsal anterior and were not co-localized with any other marker (including N). After this period, TTK69 expression increased via transcription of the 1a isoform, as ttktwk clones (affecting only that isoform) first reduce TTK69 in early S10B (Fig. 3H). This result places an upper bound on when TTK69 acts to regulate main-body cell shape and BR expression, as ttktwk does not affect these processes. After S10B, TTK69 expression became elevated in the anterior (Fig. 3D′).
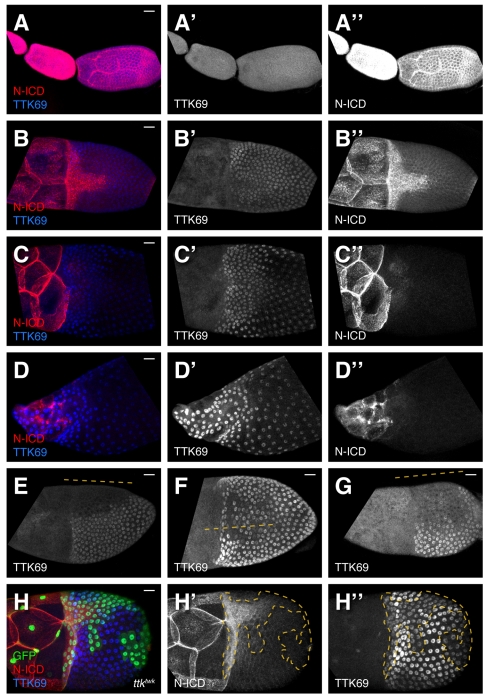
Fig. 3.
Notch intracellular domain and TTK69 expression during late oogenesis. (A-G) Confocal projections of wild-type egg chambers stained for N intracellular domain (N-ICD) and TTK69 collected using uniform exposure conditions. (A) High N and low TTK69 in S6-S9. (B) N clears to the T at early S10B and TTK69 is elevated. (C) No N expression remains in follicle cells at S11 except for a small region in the posterior (not shown). TTK69 levels become higher in the anterior. (D) At S13, N remains off and TTK69 is expressed yet more strongly in the anterior. At all stages, N is expressed in the nurse cells. (E-G) Three examples of early S10B egg chambers showing variable TTK69 expression. Lines indicate approximate dorsal midline position. (H-H″) A ttktwk clone at early S10B (T stage). At this stage, TTK69 is reduced but not eliminated in ttktwk cells. Scale bars: 20 μm.
A good candidate for an upstream regulator of TTK69 is N, a well-studied signaling protein that acts upstream of ttk69 in other tissues (Guo et al., 1996) and interacts with BR and TTK69 during oogenesis (Jordan et al., 2006; Ward et al., 2006; Sun et al., 2008). Early in oogenesis, N is expressed in all follicle cells (Fig. 3A″) (Xu et al., 1992). At the S10A-S10B transition, N disappears except in cells of the T (Fig. 3B″) (Ward et al., 2006) and some posterior cells (not shown). Shortly thereafter, expression in the T also stops (Fig. 3C″). Although the downregulation of N overlaps with the period of TTK69 variegation, the two patterns are not complementary.
N function at this time is key to establishing the four columnar cell types (Ward et al., 2006). Based on its role in other tissues, TTK69 could act downstream in this pathway to regulate BR. Analysis of cell cycle regulation, however, reveals that the relationship between TTK69 and N is not consistent during oogenesis. At S6, N promotes TTK69 expression to facilitate entry into the endocycle (Jordan et al., 2006). Later, during the endocycle/amplification transition at S10, N represses TTK69, preventing exit from the endocycle (Sun et al., 2008). If TTK69 acts downstream of N to control BR expression at S10B, the N pattern (expression only in the T) should be normal in ttk1e11 cells.
Instead, ttk1e11 cells maintained high N through the end of oogenesis (Fig. 4A,B; 62/62 clones). Distinct from the earlier membrane localization, ttk1e11 cells expressed N throughout the cytoplasm, resembling N expression in the T. Cytoplasmic localization of N may indicate elevated or reduced signaling (Vaccari et al., 2008); whether N actually signals in the T, however, is unknown. Although a N reporter is not elevated in ttk1e11 cells late in oogenesis (Sun et al., 2008), the last moment when TTK69 may function in patterning is at the S10A-S10B transition. We therefore examined a N reporter, E(Spl)mβ-CD2, near the T stage (see Fig. S1 in the supplementary material). Staining was low, punctate and variable, consistent with downregulation of signaling at this time. Reporter expression was not elevated in the T (see Fig. S1A in the supplementary material) or in most ttk1e11 clones (not shown). Occasionally, however, staining was elevated in ttk1e11 clones (see Fig. S1B in the supplementary material). This variability suggested that a dynamic interaction occurs at this time. We therefore investigated the relationship between TTK69 and N more fully.
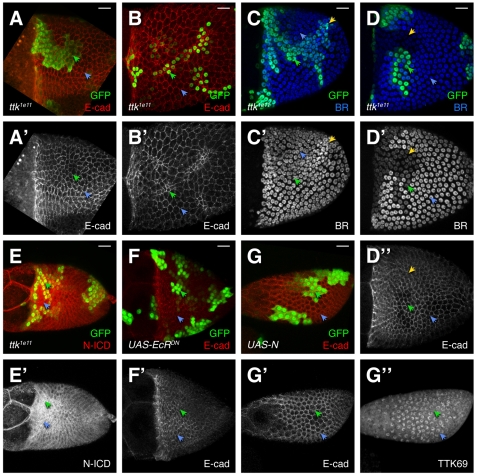
Fig. 4.
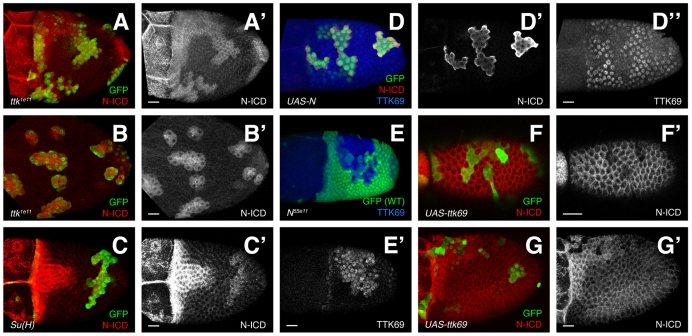
TTK69 and N are mutually repressive. (A-B′) Notch intracellular domain (N-ICD) expression is elevated in ttk1e11 clones. (A) Early S10B. N is maintained in the T and ttk1e11 cells. (B) At S13, (after expression in the T has ceased), N expression continues in ttk1e11 clones. (C,C′) At early S10B, Su(H)SF8 clones maintain N, albeit at a level lower than in the T. (D-D″) Overexpression of constitutively active N (red, D′) via FLP-out GAL4 (green) causes reduced TTK69 expression (blue, D″). (E,E′) N55e11 (null) cells upregulate TTK69 (E′) prematurely just before the natural upregulation of TTK69 during S10B. (F-G′) Overexpression of TTK69 reduces N at S9 (F) and in the T (G). Scale bars: 20 μm.
N and TTK69 form a mutually repressive feedback loop
In other developmental contexts, N signaling regulates TTK69 (Guo et al., 1996; Okabe et al., 2001; Jordan et al., 2006; Sun et al., 2008). Thus, we were surprised to find elevated N protein in ttk1e11 cells. This expression could result from loss of negative feedback regulation. To test this hypothesis, we blocked N signaling downstream of N by removing Suppressor of Hairless (Su(H)) (de Celis et al., 1996). Indeed, early in S10B, Su(H)SF8 cells outside the T maintained N expression (Fig. 4C). Unlike ttk1e11, Su(H)SF8 clones in later stages lacked N (not shown), indicating that while delayed, N downregulation can proceed without SU(H)-mediated N signaling. Alternatively residual function or perdurance in Su(H)SF8 cells could suffice for delayed clearance. This result supports the idea that N is involved in a negative feedback loop but does not explain how TTK69 is involved.
To test the interaction between N and TTK69 more directly, we made gain-of-function, FLP-out clones. We expressed constitutively active N (UAS-N-CA) or wild-type TTK69 and observed expression of the other protein. Activated N reduced TTK69 at S10 and later (Fig. 4D; 13/13 clones). This result contrasts with observations of earlier stages, when loss of N reduces TTK69 (Jordan et al., 2006), but is consistent with N acting upstream to repress TTK69 at S10 (Sun et al., 2008). Is the opposite true? Does removal of N induce upregulation of TTK69? The answer is ‘No’ as late as S9 (Sun et al., 2008). After S10B, however, TTK69 is naturally upregulated, leaving a very narrow but critical window during the S10A-S10B transition. While N clearance to the T had begun, but not yet finished (directly before normal TTK69 upregulation), TTK69 was upregulated prematurely in N null cells (N55e11; Fig. 4E; 4/4 clones).
The interaction between TTK69 and N works in both ways: overexpression of TTK69 resulted in severe reduction of N (20/20 clones) at S9 (Fig. 4F) and the T of early S10B (Fig. 4G). Later in oogenesis, N expression is naturally absent and cannot be reduced further by TTK69. Taken together, these data are inconsistent with a linear pathway that places TTK69 downstream of N; instead, N and TTK69 are mutually repressive. Although their patterns are not directly opposed (Fig. 3), this relationship is consistent with the transition to high TTK69 expression shortly after N reduction. These data suggest that TTK69 and N exist in a bistable, mutually repressive feedback loop during mid-to-late oogenesis, which ensures a rapid, reliable clearance of N and upregulation of TTK69.
Expression of activated N and overexpression of TTK69 cause ttk1e11-like phenotypes
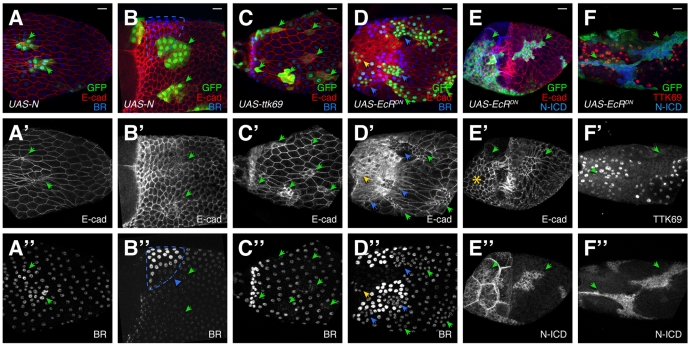
As multiple mechanisms exist to ensure rapid downregulation of N and upregulation of TTK69, we investigated the consequences of preventing or accelerating this switch. Trapping cells in an early, high-N/low-TTK69 state using activated N resulted in ttk1e11-like phenotypes (Fig. 5A,B). Columnar follicle cells constricted their apices (Fig. 5A′,B′; 11/11 clones) and expressed altered BR (12/12 clones). BR was elevated in the main body (Fig. 5A″), whereas cells in the roof failed to induce high-BR expression (Fig. 5B″ arrowhead).
Fig. 5.
Expression of activated N, TTK69 and dominant-negative Ecdysone receptor causes defects in follicle cell shape and patterning. FLP-out clones (green/arrowheads) of UAS-N-CA, UAS-ttk69 or UAS-EcR-DN. (A-B″) N-CA cells constrict their apices (A′,B′) and alter BR levels. BR is elevated in the main body late in oogenesis (A″) and reduced in the roof (B″, blue triangle). Expected roof cell region is outlined in blue. (C-C″) UAS-ttk69 causes elevated E-cadherin, reduced cell size (C′), and reduced BR (C″). (D-D″) UAS-EcR-DN results in small cells (D′); BR (D″) is reduced in the roof (blue arrowheads) and maintained in the T (yellow arrowhead). (E-E″) UAS-EcR-DN blocks stretch cell formation (E′, asterisk). (F-F″) UAS-EcR-DN blocks N downregulation (E″,F″) and TTK69 upregulation (F′). Note the nuclear exclusion of TTK69 in the clone (F′). Scale bars: 20 μm.
Accelerating the transition to a high-TTK69/low-N state with UAS-ttk69 resulted in defects as well. Cells expressed elevated, membrane-bound E-cadherin and altered BR expression. Unlike ttk1e11 cells, they were smaller overall, not apically constricted (Fig. 5C′; 38/38 clones), and BR was downregulated in all regions (Fig. 5C″; 24/26 clones contained cells with downregulated BR). These phenotypes were highly variable, even within a single egg chamber, possibly in part from use of FLP-out GAL4 in endoreplicated tissue (each cell can activate different numbers of cassettes). These data show that the balance between TTK69 and N is critical for establishing proper fate and shape in columnar follicle cells.
Ecdysone signaling promotes TTK69 and represses N expression
The mutually repressive relationship between TTK69 and N will resist change without outside influence, but the dominant factor switches during S10B. What signal(s) might induce this transition? Ecdysone is a promising candidate. Usually organism-wide, at S10, the germ cells within a single egg chamber produce a local Ecdysone signal (Buszczak et al., 1999). Blocking Ecdysone at this time disrupts TTK69 expression, whereas activated N reduces that signaling. These observations have led to the conclusion that relief of N signaling allows Ecdysone to activate TTK69 (Sun et al., 2008). Furthermore expression of dominant-negative Ecdysone receptor (EcR-DN) induces cell shape phenotypes similar to that of ttk1e11 (Hackney et al., 2007). In light of our expanded understanding of the N-TTK69 relationship at this stage, we sought to clarify the role of Ecdysone using dominant-negative Ecdysone receptor, EcR-DN (Cherbas et al., 2003).
EcR-DN resulted in high N when wild-type cells lacked N (Fig. 5E″,F″; 39/39 clones). TTK69 was reduced in or excluded from nuclei (Fig. 5F′), indicating Ecdysone may regulate TTK69 by promoting nuclear localization. EcR-DN also prevented adoption of correct fates and shapes (Fig. 5D,F). Unlike ttk1e11 and N-CA cells, EcR-DN-expressing cells were smaller but not apically constricted (compare the GFP-filled cell bodies to the apical E-cadherin in Fig. 2B and Fig. 5A vs Fig. 5D,E), and nuclei were also slightly smaller. Like ttk1e11, EcR-DN cells failed to adopt post-S10B BR levels, failing to upregulate BR in the roof or clear BR from the T (Fig. 5D″; 18/18 clones). Unlike ttk1e11, however, EcR-DN expression prevented the earlier process of stretch cell formation, leaving mutant cells over the nurse cells with apical surface sizes similar to the columnar follicle cells overlying the oocyte (Fig. 5E′). Together, these data show that EcR activity defines multiple cell types during oogenesis in part by promoting downregulation of N and upregulation of TTK69.
TTK69 is required for proper response to the onset of S10B of oogenesis
A common thread connecting the mutants discussed in this paper is the adoption of aspects of other follicle cell fates regardless of position within the epithelium, such as apical constriction, which normally occurs only in roof cells, and elevated basal E-cadherin, normally restricted to the T. This result is remarkable because in wild type these behaviors occur only in cells that receive well-defined, spatially localized signals (EGF and DPP). How does removal of TTK69, a transcriptional repressor, lead to these phenotypes in cells far from the sources of these ligands? We envision two possibilities. (1) Once EGF and DPP are activated at appropriate levels by their autoregulatory cascades (Yakoby et al., 2008b), they themselves induce the behaviors of the dorsal anterior cells (providing both spatial and temporal cues). Removal of TTK69, then, induces constitutive, ligand-independent activation of these downstream responses. (2) Alternatively, EGF and DPP provide only spatial information. At the S10A-S10B transition, all follicle cells receive another, temporal signal, and TTK69 integrates these signals to restrict each cell's response to one appropriate to the region. As ttk1e11 cells show behaviors characteristic of cells receiving widely varying levels of EGF and DPP activity, we favor this latter hypothesis.
If ttk1e11 phenotypes reflect wild-type S10B behaviors and are triggered by the same signals, we should observe defects only at stages after initiation of the wild-type processes. Indeed, removal of TTK69 or expression of activated N or EcR-DN produced effects in S10B and later (Fig. 6B, Fig. 5B,E) but not in S9 or S10A (Fig. 6A,F,G). Furthermore, N-CA was insufficient to cause TTK69 downregulation before S10 (probably because N is naturally high and TTK69 low at this stage already; Fig. 6G″). Similarly, N and BR expression were not affected at S10A in ttk1e11 clones (Fig. 6C,E), but were altered at S10B and onward (Fig. 6D, Fig. 4A,B).
Apical constriction occurred simultaneously in ttk1e11 and wild-type roof cells, whereas apices of wild-type cells outside the roof greatly expanded at this time to accommodate oocyte growth. This expansion could create the appearance of active apical constriction if the apices of the mutant cells simply did not grow. To determine whether apical constriction of ttk1e11 cells was active like roof-cell apical constriction, we measured the area of ttk1e11 (and wild-type) apices before (n=3) and after (n=4) the onset of roof-cell apical constriction. Before roof-cell apical constriction, wild type and ttk1e11 mutant cells were not significantly different in size (44.98 μm2 vs 42.73 μm2, respectively; P=0.6). Later, ttk1e11 apices shrank significantly to 30.86 μm2 (P=0.0009). By contrast, the average wild-type cell had grown to 256.19 μm2. Therefore, roof and ttk1e11 cells act concurrently, and ttk1e11 cells, like their roof-cell counterparts, actively constrict.
Finally, wild-type and mutant cells behaved in a coordinated fashion (Table 1). Fig. 6D captures an egg chamber midway through the S10A-S10B transition, during reduction of BR in the T but before BR elevation or apical constriction in the roof; here, ttk1e11-mutant cells express BR at a reduced level, below that of main-body follicle cells and similar to that of the cells in the T (Fig. 6D′). This low level of BR is surprising as very shortly thereafter (<4 hours later, in mid-S10B), when BR is elevated in the roof, ttk1e11 cells expressed BR at a higher level, similar to levels in main-body follicle cells (Fig. 2G′). These data suggest that ttk1e11 cells first downregulate BR concurrently with the T, then upregulate BR concurrently with the high-BR cells, resulting in BR expression at a level in between that of the two populations. Likewise, apical constriction and elevation of basal E-cadherin are synchronized between ttk1e11 cells, roof cells and T cells. In Fig. 6D″, neither wild-type nor mutant cells have begun apical constriction; both begin this process later at the end of S10B. Taken together, these results suggest that a temporal signal, distinct from the well-characterized EGF and DPP signals and available to all follicle cells, induces a sequence of changes at S10B. The level of EGF and DPP signaling that each cell experiences restricts its response to this temporal signal to one that is appropriate for its location, and it does so in a TTK69-dependent manner.
Table 1.
Sequence of events during the formation of the dorsal-anterior follicle-cell subtypes of late oogenesis
| Roof cells | T | Mainbody | ttk1e11 cells | |
| S10A-S10B transition | N expression cleared | N expression cleared | ||
| Early S10B | BR eliminated | BR reduced | ||
| Mid S10B | BR upregulated | E-cadherin upregulated on basal surfaces; N expression cleared | BR increased; E-cadherin upregulated on basal surfaces | |
| Late S10B | Apical constriction starts | BR reduced | Apical constriction starts | |
| S12-S13 | Apical re-expansion |
Although rapid, the S10A-S10B transition does not occur all at once, but in a sequence. ttk1e11 cells produce phenotypes in synchrony with the related wild-type events (e.g. apical constriction is simultaneous in ttk1e11 and roof cells).
DISCUSSION
We have demonstrated that TTK69 plays a central role in regulating the behavior of follicle cells during mid-to-late oogenesis. At S10 and again at S12, TTK69 integrates temporal and spatial information to coordinate the behaviors of subpopulations of follicle cells.
Elsewhere in development, TTK69 acts as a determinant of cell fate. Best studied is its role in binary cell-fate decisions downstream of N-mediated lateral inhibition, such as in asymmetric sensory organ precursor (SOP) cell division, where N becomes active in one of two daughter cells, leading to TTK69 expression and repression of neural determinants (Guo et al., 1996; Okabe et al., 2001). TTK69 also acts downstream of N to promote endocycle entry at S6 (Jordan et al., 2006), and in the transition from endocycle to chorion gene amplification at S10 (Sun et al., 2008). Recent studies demonstrate a role for TTK69 in morphogenesis. ttktwk, an allele of ttk69 that does not disrupt patterning, prevents DA elongation (French et al., 2003), and TTK69 is required for proper cell shape and fate during tracheal morphogenesis (Araújo et al., 2007).
In this paper, we advance our understanding of TTK69 significantly by reporting four novel mechanisms of TTK69 action. First, we show that TTK69 controls DA tube volume by inducing apical expansion and lateral shortening, independent of basal extension. Second, we find that loss of TTK69 in mid-oogenesis causes cells to adopt inappropriate fates without inducing a binary cell-fate switch. Third, we reveal a surprising, mutually repressive relationship between TTK69 and N that is modulated by Ecdysone receptor activity. Finally, we demonstrate that TTK69 coordinates the response to temporal and spatial signals and thereby ensures the fidelity of egg chamber maturation.
Apical re-expansion of ttktwk roof cells fails
Although apical constriction has been well studied (Pilot and Lecuit, 2005), less is known about apical re-expansion and the mechanisms that determine final apical size (Wu and Beitel, 2004; Laprise et al., 2009). Surprisingly, ttktwk reveals independent control of apical and basal surfaces. Although DA-forming cells elongate, the tube extends only when roof-cell apices expand.
ttk1e11 causes dramatic shape and fate alterations
The ttk1e11 allele deletes a portion of the TTK69 zinc finger. Although long regarded as null (Xiong and Montell, 1993), we considered the possibility that some truncated protein, expressed even at undetectable levels, might cause gain-of-function effects. Several lines of evidence support the argument against this possibility. First, heterozygous cells, the majority in the ttk1e11 mosaics, show no phenotype; thus, any gain-of-function effect would be recessive, a very rare occurrence. Furthermore, N-CA phenocopies ttk1e11 and causes TTK69 downregulation, independently demonstrating that TTK69 reduction produces the ttk1e11 phenotypes.
ttk1e11 caused dramatic shape and fate transformations in all columnar follicle cells, leading to cells with highly constricted apices, elevated E-cadherin, and altered BR levels. These changes did not result from constitutive activation of BR or cell-shape determinants, as S10A and earlier egg chambers did not exhibit any of these phenotypes. Thus, TTK69 regulates diverse processes at S10B. Unlike its role in SOP cell division, TTK69 does not switch cells from one type to another. Rather, ttk1e11 cells take on aspects of many S10B follicle cell subtypes, not all of which are normally exhibited by a single cell (e.g. high basal E-cadherin and apical constriction).
Ecdysone receptor function is required for TTK69 upregulation and N downregulation
Ecdysone signaling was required to flip the bistable relationship between N and TTK69 toward TTK69 at S10. Inactivating Ecdysone receptor via a dominant-negative construct largely mirrored ttk1e11. Additional phenotypes, such as failure to generate stretch cells, were probably due to earlier requirements for EcR function, processes that do not involve TTK69. Interestingly, TTK69 levels were sometimes elevated in the cytoplasm and excluded from the nuclei of EcR-DN-expressing cells, indicating that Ecdysone may promote nuclear localization of TTK69. This mechanism could provide more rapid and reversible control over TTK69 activity than transcriptional regulation. The mutually repressive relationship between TTK69 and N could accelerate this change, once triggered.
How does Ecdysone signaling produce this change? Autocrine signaling within the germline could induce downregulation of Delta, thereby reducing N activity. At the same time, Ecdysone signaling to the follicle cells could stabilize TTK69 in the nucleus, modulating expression of target genes that control N degradation. Alternatively, as activated N can block Ecdysone signaling at S10B (Sun et al., 2008), decreased Delta activity could allow Ecdysone receptor function in the follicle cells, indirectly activating TTK69. Regardless of the exact mechanism, these relationships would act as a positive feedback loop to ensure a rapid and reliable switch between the two states.
TTK69 is required for proper response to the onset of S10B
The transition from S10A to S10B is marked by the adoption of specific follicle-cell fates in positions specified by EGF and DPP signaling. Strikingly, many of the phenotypes we observed appeared at S10B but not S10A, revealing roles for TTK69, N and EcR in regulating temporal maturation to S10B. Thus, fundamental differences exist between S10A and S10B follicle cells. We hypothesize that a signal experienced by all follicle cells induces this change. Importantly this signal cannot be a combination of EGF and DPP, as these signals are spatially restricted to the dorsal anterior at this stage, and ttk1e11, UAS-N-CA and UAS-EcR-DN have equivalent effects in all columnar follicle cells.
What is this signal? Probably, several factors contribute. Ecdysone itself is an interesting candidate. While ttk1e11 mutant and N-CA-expressing cells were apically constricted after S10B, EcR-DN-expressing cells more closely resembled S10A cells (smaller overall with smaller nuclei). This distinction could indicate that Ecdysone is required to progress beyond S10A. EcR-DN-expressing cells, however, do not perfectly resemble S10A cells. Differences could be due to dominant effects of EcR-DN complexes, which could strongly repress EcR targets rather than simply failing to activate them (Cherbas et al., 2003). Another potential input at this time could be prostaglandin signaling (Tootle and Spradling, 2008), which induces nurse-cell dumping. Finally, the egg chamber grows during this period and overall egg chamber nutrition could determine the timing of the S10A-S10B transition. Insulin signaling is required earlier for vitellogenesis (Drummond-Barbosa and Spradling, 2001; Richard et al., 2005) and may continue to monitor growth during this period (Cavaliere et al., 2005).
How does TTK69 integrate this temporal signal with the spatial pattern to achieve appropriate responses at the correct time and place? ttk1e11 cells adopt aspects of multiple cell types. In wild type, each cell performs a subset of behaviors, repressing inappropriate responses in a ttk69-dependent fashion. For example, roof cells need TTK69 to prevent BR downregulation, whereas main body cells need TTK69 to prevent apical constriction. How does TTK69 repress different processes in different cells? Spatial information from DPP and EGF must contribute to TTK69 activity. TTK69 expression, however, although dynamic and variable, has no consistent spatial pattern and is required in all columnar follicle cells.
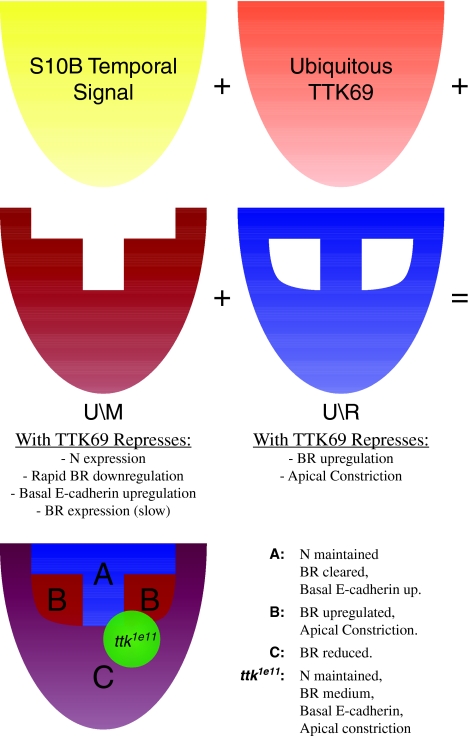
A conceptual solution (Fig. 7) is to imagine TTK69 working in concert with two co-factors that are spatially restricted by EGF and DPP signaling: one co-factor is expressed in a U\M pattern, all follicle cells except the T using the system described previously (Yakoby et al., 2008a), and one is present in a U\R pattern (all follicle cells except the roof). When combined with TTK69, the U\M factor would repress basal E-cadherin, N expression and the early clearance of BR that normally occurs in the T. In combination with TTK69, the U\R factor would repress apical constriction and BR upregulation, which normally occur in the roof. Removal of TTK69 causes a failure to repress all of these behaviors, leading to the observed phenotypes, with moderate BR levels as a result of both up- and downregulation. These co-factors could take the form of BTB transcription factors that dimerize with TTK69. Alternatively, they could modify TTK69 by phosphorylation or mono-ubiquitylation (Lehembre et al., 2000) or affect expression of other genes that alter the response to TTK69 function.
Fig. 7.
Conceptual model of spatiotemporal regulation by TTK69. One possible mechanism of TTK69 action. Spatially restricted co-factors (red and blue) work with TTK69 (orange) to prevent cells from responding inappropriately to a globally available temporal signal (yellow). The combination of each of these patterns results in the differential reaction of each subpopulation at the onset of S10B. Loss of TTK69 (green) disrupts all regulation.
In conclusion, we have identified several novel functions for TTK69. At S10, presumably in collaboration with region-specific interacting proteins, it facilitates spatially correct responses to a uniform temporal signal; its function at S12 coordinates a return to a cuboidal cell shape. The regulatory network by which TTK69, N and Ecdysone receptor control the progression of egg chambers from mid- to late oogenesis could serve as a simple model to explain how a bistable system flips its state to regulate the temporal progression of development.
Supplementary Material
Acknowledgements
We thank Alan Rubin for initial observations of ttk1e11 chorions; Hannele Ruohola-Baker, Bruce Edgar, Wu-Min Deng and the Bloomington Stock Center for strains; the Developmental Studies Hybridoma Bank for monoclonal antibodies; the National Science Foundation (0417129), UW Provost Bridge Fund (65-3460) and the National Institutes of Health (R01-GM079433) for funding to C.A.B.; M.J.B. was supported by a Developmental Biology Predoctoral Training Grant T32 HD007183 from the National Institutes of Child Health and Human Development. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/24/4187/DC1
References
- Araújo S. J., Cela C., Llimargas M. (2007). Tramtrack regulates different morphogenetic events during Drosophila tracheal development. Development 134, 3665-3676 [DOI] [PubMed] [Google Scholar]
- Ashe H. L., Briscoe J. (2006). The interpretation of morphogen gradients. Development 133, 385-394 [DOI] [PubMed] [Google Scholar]
- Badenhorst P. (2001). Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development 128, 4093-4101 [DOI] [PubMed] [Google Scholar]
- Banerjee I., Clayton P. (2007). The genetic basis for the timing of human puberty. J. Neuroendocrinol. 19, 831-838 [DOI] [PubMed] [Google Scholar]
- Baonza A., Murawsky C. M., Travers A. A., Freeman M. (2002). Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 4, 976-980 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Freeman M. R., Carlson J. R., Bender M., Cooley L., Segraves W. A. (1999). Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Development 126, 4581-4589 [DOI] [PubMed] [Google Scholar]
- Cavaliere V., Donati A., Hsouna A., Hsu T., Gargiulo G. (2005). dAkt kinase controls follicle cell size during Drosophila oogenesis. Dev. Dyn. 232, 845-854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L., Hu X., Zhimulev I., Belyaeva E., Cherbas P. (2003). EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development 130, 271-284 [DOI] [PubMed] [Google Scholar]
- Chou T. B., Perrimon N. (1992). Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131, 643-653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. B., Perrimon N. (1996). The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144, 1673-1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. B., Noll E., Perrimon N. (1993). Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119, 1359-1369 [DOI] [PubMed] [Google Scholar]
- Couso J. P., Martinez Arias A. (1994). Notch is required for wingless signaling in the epidermis of Drosophila. Cell 79, 259-272 [DOI] [PubMed] [Google Scholar]
- Davis G. K., Patel N. H. (2002). Short, long, and beyond: molecular and embryological approaches to insect segmentation. Annu. Rev. Entomol. 47, 669-699 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., de Celis J., Ligoxygakis P., Preiss A., Delidakis C., Bray S. (1996). Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122, 2719-2728 [DOI] [PubMed] [Google Scholar]
- Dobens L. L., Raftery L. A. (2000). Integration of epithelial patterning and morphogenesis in Drosophila ovarian follicle cells. Dev. Dyn. 218, 80-93 [DOI] [PubMed] [Google Scholar]
- Dorman J. B., James K. E., Fraser S. E., Kiehart D. P., Berg C. A. (2004). bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev. Biol. 267, 320-341 [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D., Spradling A. C. (2001). Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev. Biol. 231, 265-278 [DOI] [PubMed] [Google Scholar]
- Dubreuil R., Byers T. J., Branton D., Goldstein L. S., Kiehart D. P. (1987). Drosophilia spectrin. I. Characterization of the purified protein. J. Cell Biol. 105, 2095-2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B., Harrison D. A., Perrimon N. (1998). Identifying loci required for follicular patterning using directed mosaics. Development 125, 2263-2271 [DOI] [PubMed] [Google Scholar]
- Emery I. F., Bedian V., Guild G. M. (1994). Differential expression of Broad-Complex transcription factors may forecast tissue-specific developmental fates during Drosophila metamorphosis. Development 120, 3275-3287 [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., Muskavitch M. A., Artavanis-Tsakonas S. (1990). Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61, 523-534 [DOI] [PubMed] [Google Scholar]
- Frasch M. (2008). A matter of timing: microRNA-controlled temporal identities in worms and flies. Genes Dev. 22, 1572-1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R. L., Cosand K. A., Berg C. A. (2003). The Drosophila female sterile mutation twin peaks is a novel allele of tramtrack and reveals a requirement for Ttk69 in epithelial morphogenesis. Dev. Biol. 253, 18-35 [DOI] [PubMed] [Google Scholar]
- Fuerstenberg S., Giniger E. (1998). Multiple roles for notch in Drosophila myogenesis. Dev. Biol. 201, 66-77 [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R., Pearson B. J., Marusich A., Doe C. Q. (2005). Regulation of temporal identity transitions in Drosophila neuroblasts. Dev. Cell 8, 193-202 [DOI] [PubMed] [Google Scholar]
- Guo M., Jan L. Y., Jan Y. N. (1996). Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27-41 [DOI] [PubMed] [Google Scholar]
- Hackney J. F., Pucci C., Naes E., Dobens L. (2007). Ras signaling modulates activity of the ecdysone receptor EcR during cell migration in the Drosophila ovary. Dev. Dyn. 236, 1213-1226 [DOI] [PubMed] [Google Scholar]
- Hiruma K., Shinoda T., Malone F., Riddiford L. M. (1999). Juvenile hormone modulates 20-hydroxyecdysone-inducible ecdysone receptor and ultraspiracle gene expression in the tobacco hornworm, Manduca sexta. Dev. Genes. Evol. 209, 18-30 [DOI] [PubMed] [Google Scholar]
- Hu X., Cherbas L., Cherbas P. (2003). Transcription activation by the ecdysone receptor (EcR/USP): identification of activation functions. Mol. Endocrinol. 17, 716-731 [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Bier E., Levine M. (1992). The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 6, 1728-1739 [DOI] [PubMed] [Google Scholar]
- James K. E., Dorman J. B., Berg C. A. (2002). Mosaic analyses reveal the function of Drosophila Ras in embryonic dorsoventral patterning and dorsal follicle cell morphogenesis. Development 129, 2209-2222 [DOI] [PubMed] [Google Scholar]
- Jordan K. C., Schaeffer V., Fischer K. A., Gray E. E., Ruohola-Baker H. (2006). Notch signaling through tramtrack bypasses the mitosis promoting activity of the JNK pathway in the mitotic-to-endocycle transition of Drosophila follicle cells. BMC Dev. Biol. 6, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalcheim C., Ben-Yair R. (2005). Cell rearrangements during development of the somite and its derivatives. Curr. Opin. Genet. Dev. 15, 371-380 [DOI] [PubMed] [Google Scholar]
- Kidd S., Lockett T. J., Young M. W. (1983). The Notch locus of Drosophila melanogaster. Cell 34, 421-433 [DOI] [PubMed] [Google Scholar]
- King R. C. (1970). Ovarian Development in Drosophila Melanogaster New York: Academic Press; [Google Scholar]
- Laprise P., Lau K. M., Harris K. P., Silva-Gagliardi N. F., Paul S. M., Beronja S., Beitel G. J., McGlade C. J., Tepass U. (2009). Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase form a novel group of epithelial polarity proteins. Nature 459, 1141-1145 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251-254 [DOI] [PubMed] [Google Scholar]
- Lehembre F., Badenhorst P., Müller S., Travers A., Schweisguth F., Dejean A. (2000). Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol. Cell. Biol. 20, 1072-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis L. H. (1985). Structure and physiology of the eggshell. In Comprehensive Insect Physiology, Biochemistry and Pharmacology (ed. Gilbert L. L., Kerkut G. A.), pp. 153-230 Oxford: Pergamon Press; [Google Scholar]
- Moussian B., Roth S. (2005). Dorsoventral axis formation in the Drosophila embryo-shaping and transducing a morphogen gradient. Curr. Biol. 15, R887-R899 [DOI] [PubMed] [Google Scholar]
- Oda H., Uemura T., Harada Y., Iwai Y., Takeichi M. (1994). A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev. Biol. 165, 716-726 [DOI] [PubMed] [Google Scholar]
- Okabe M., Imai T., Kurusu M., Hiromi Y., Okano H. (2001). Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature 411, 94-98 [DOI] [PubMed] [Google Scholar]
- Pignoni F., Zipursky S. L. (1997). Induction of Drosophila eye development by decapentaplegic. Development 124, 271-278 [DOI] [PubMed] [Google Scholar]
- Pilot F., Lecuit T. (2005). Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev. Dyn. 232, 685-694 [DOI] [PubMed] [Google Scholar]
- Read D., Manley J. L. (1992). Alternatively spliced transcripts of the Drosophila tramtrack gene encode zinc finger proteins with distinct DNA binding specificities. EMBO J. 11, 1035-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard D. S., Rybczynski R., Wilson T. G., Wang Y., Wayne M. L., Zhou Y., Partridge L., Harshman L. G. (2005). Insulin signaling is necessary for vitellogenesis in Drosophila melanogaster independent of the roles of juvenile hormone and ecdysteroids: female sterility of the chico1 insulin signaling mutation is autonomous to the ovary. J. Insect. Physiol. 51, 455-464 [DOI] [PubMed] [Google Scholar]
- Riddiford L. M. (1976). Hormonal control of insect epidermal cell commitment in vitro. Nature 259, 115-117 [DOI] [PubMed] [Google Scholar]
- Schweisguth F., Posakony J. W. (1994). Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development 120, 1433-1441 [DOI] [PubMed] [Google Scholar]
- Spradling A. C. (1993). Developmental genetics of oogenesis. In The Development of Drosophila melanogaster (ed. Bates M., Arias A. M.). Plainview, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sun J., Smith L., Armento A., Deng W. M. (2008). Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J. Cell Biol. 182, 885-896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle T. L., Spradling A. C. (2008). Drosophila Pxt: a cyclooxygenase-like facilitator of follicle maturation. Development 135, 839-847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., Marygold S., Millburn G., Osumi-Sutherland D., Schroeder A., Seal R., Zhang H., Flybase Consortium (2009). FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 37, D555-D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008). Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180, 755-762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E. J., Berg C. A. (2005). Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech. Dev. 122, 241-255 [DOI] [PubMed] [Google Scholar]
- Ward E. J., Zhou X., Riddiford L. M., Berg C. A., Ruohola-Baker H. (2006). Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types. Dev. Biol. 297, 461-470 [DOI] [PubMed] [Google Scholar]
- Wolpert L. (1989). Positional information revisited. Development 107Suppl, 3-12 [DOI] [PubMed] [Google Scholar]
- Wu V. M., Beitel G. J. (2004). A junctional problem of apical proportions: epithelial tube-size control by septate junctions in the Drosophila tracheal system. Curr. Opin. Cell Biol. 16, 493-499 [DOI] [PubMed] [Google Scholar]
- Xiong W. C., Montell C. (1993). tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 7, 1085-1096 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223-1237 [DOI] [PubMed] [Google Scholar]
- Xu T., Caron L. A., Fehon R. G., Artavanis-Tsakonas S. (1992). The involvement of the Notch locus in Drosophila oogenesis. Development 115, 913-922 [DOI] [PubMed] [Google Scholar]
- Yakoby N., Bristow C. A., Gong D., Schafer X., Lembong J., Zartman J. J., Halfon M. S., Schüpbach T., Shvartsman S. Y. (2008a). A combinatorial code for pattern formation in Drosophila oogenesis. Dev. Cell 15, 725-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoby N., Lembong J., Schüpbach T., Shvartsman S. Y. (2008b). Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development 135, 343-351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.