Abstract
BACKGROUND: Aberrant DNA methylation has been recognized in human breast carcinogenesis as a common molecular alteration associated with the loss of expression of a number of key regulatory genes. The present study was undertaken to determine whether methylation and expression of p16 and FHIT genes would correlate with the estrogen receptor (ER) and progesterone receptor (PR) status. METHODS: Methylation-specific polymerase chain reaction, messenger RNA (mRNA) expression analysis, immunohistochemistry, and Western blot analysis were performed to study the methylation of p16 and FHIT genes in 351 pairs of malignant/normal breast tissues. We examined the expression of ER and PR in those specimens by immunohistochemistry. Mutations of p16 and FHIT genes in tumors were detected by direct sequencing. RESULTS: The frequency of hypermethylation was 31.9% and 36.8% in p16 and FHIT genes, respectively, and showed significant harmony in concordant hypermethylation (P < .0001). In postmenopausal patients, methylation frequency in both genes is significantly higher in poorly and moderately differentiated tumors. Loss of protein expression of p16 and FHIT in 77 and 74 tumors, respectively, is associated with their methylation status in premenopausal women. CONCLUSION: We did not find any significant differences in tumor-related gene methylation patterns relevant to both ER and PR status of breast tumors.
Introduction
Breast cancer is the single most common cause of cancer-related mortality in women worldwide, accounting for an annual prevalence of more than 1 million new cases, representing nearly one fifth of all malignancies among females [1]. The incidence of breast cancer is increasing in many countries particularly in Asia, with more than 3% increase every year compared with just more than half a percent in developed countries [2]. Breast cancer among younger women is appearing as the leading cancer in India. These tumors are often estrogen receptor (ER)- and/or progesterone receptor (PR)-negative; however, their mechanistic basis remains elusive [3]. Aberrant DNA methylation has been increasingly recognized as a frequent molecular alteration in breast cancers. Hypermethylation of the CpG islands is associated with delayed replication, condensed chromatin, inhibition of transcription initiation, and silencing of genes [4]. Genes involved in cell cycle regulation (p16), cell adhesion (CDH1), DNA repair (BRCA1), and cell signaling pathway (ER, RARβ2) have been reported to undergo hypermethylation [5,6]. The product of INK4A locus, p16, encodes a cyclin-dependent kinase (CDK) inhibitor that functions as a negative regulator of cyclin/CDK complexes. It binds preferentially to CDK4/6 and prevents their association with D-type cyclins, thus inhibiting pRB phosphorylation and progression through the cell cycle [7,8]. It plays an important role in maintaining normal cellular properties, preventing both centrosome dysfunction and genomic instability [9]. Inactivation of p16INK4A is an early event in carcinogenesis [7,8], and loss of p16/Rb activity occurs through different mechanism such as deletion, mutation, and/or hypermethylation of p16INK4A [8]. Inactivation of p16 seems a crucial event in the development of several tumors [10]. The relevance of this defect in mammary carcinogenesis remains unclear. However, homozygous deletions of this gene are seen in half of the breast cancer cell lines and neither homozygous deletions nor point mutations are frequently observed in primary breast cancers, suggesting that these alterations might have been acquired in the cultures [11,12]. There are suggestions that aberrant hypermethylation may be a useful biomarker, with implications for breast cancer etiology, diagnosis, and management. FHIT is also found frequently inactivated in many tumor types, including those of breast, cervix, esophagus, digestive tract, lung, and bladder [13–17].
Prognostic factors considered negative for survival such as advanced diagnosis stage (enlarged tumor size, node involvement, and sometimes more distant metastasis), early onset disease (<50 years age), ER and PR status, poor differentiation of tumors and obesity might play an important role in the etiology of this disease. The aim of the present study was to evaluate the association between the major clinicopathologic features of breast cancer and methylation status of p16 and FHIT genes among primary breast cancer cases. We also tried to delineate more precisely the association of p16 and FHIT methylation and their expression using Northern and Western blot analyses along with immunohistochemistry profile among breast cancer patients from northern India.
Materials and Methods
Sample Size
A total of 379 breast tumor and adjacent normal tissue biopsies not infiltrated by tumors as confirmed by a pathologist were collected directly from patients attending the Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi. The samples were from breast tumors and corresponding adjacent normal-appearing tissues (from tissues located at least 3 cm away from the site at which the tumor was sampled). However, data from only 351 (299 ductal and 52 lobular) breast tumor biopsies have been analyzed owing to either insufficient quantity and/or quality of DNA and RNA of the other 28 tumor and/or adjacent tissue specimens. Biopsies were collected directly into sterile collection vials containing chilled PBS (pH 7.2) and were stored in liquid nitrogen or -80°C until further analysis. The tissue materials were used for DNA and RNA extraction. The present study was approved by the university and institutional review boards. The histologic type and grade of the tumors were classified according to the World Health Organization's criteria. Prior consent from the patients was also taken before starting the study. All breast specimens including tumor and normal adjacent tissue were reviewed by two experienced pathologists at the hospital. Slides prepared from these were subject to immunoperoxidase staining for ER (clone 6F11) and PR (clone 1A6) as per the manufacturer's (Novocastra Laboratories Ltd, Benton Lane, UK) recommendations. Cancers were considered receptor-positive if more than 10% of malignant cells showed nuclear staining. Cancers were classified as ER- and/or PR-positive.
Methylation-Specific Polymerase Chain Reaction
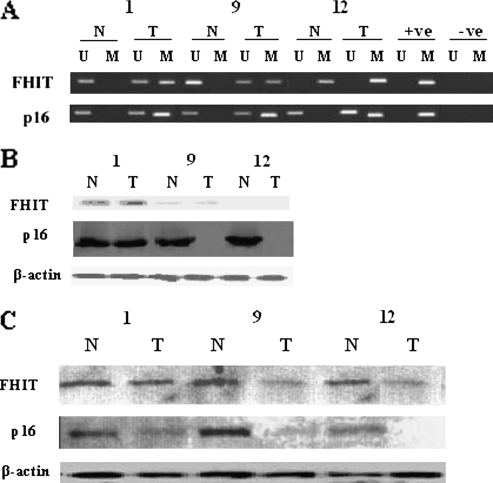
Methylation status of the promoter regions of p16 and FHIT genes was determined by methylation-specific polymerase chain reaction (MSP) as described previously [18]. Primer sequences and annealing temperatures for the MSP were as described previously in the literature [18,19]. Each reaction mixture contained 1x polymerase chain reaction (PCR) buffer, 1.5 mM MgCl2, 1.25 mM each of the dNTPs, primers (300 ng each/reaction), 1.5 U of Taq polymerase, and bisulfite-modified DNA (50 ng). Reactions were hot-started at 95°C and carried out for 35 cycles, and each cycle consisted of 45 seconds at 95°C, 75 seconds at 65°C and 60°C for p16 and at 69°C and 67°C for FHIT, respectively, for methylated and unmethylated specific PCR and 60 seconds at 72°C, followed by 7 minutes of final extension at 72°C. Each PCR product (15 µl each) was run on 2.5% agarose gel, stained with ethidium bromide, and visualized under UV illumination. DNA from peripheral blood lymphocytes of a healthy individual was treated with SssI methyltransferase (New England Biolabs, Inc, Ipswich, MA) was used as a positive control. To confirm the methylation status by bisulfite sequencing, a subset of the patients who show unmethylation and methylation in p16 and FHIT was chosen, and we found that these results match with the results obtained by MSP (data not shown). Appropriate controls were set up in each reaction. Representative examples are shown in Figure 1A.
Figure 1.
(A) Methylation status of p16 and FHIT genes in normal and tumor tissues from the same patient. (B) Northern blot analysis showing mRNA expression profile of FHIT, p16, and β-actin in different breast cancer patients. Lane marked N: normal breast tissue, T: tumor tissue. (C) Western blot analysis in the same patients analyzed for Northern blot.
Mutation Analysis in p16 and FHIT Genes
p16 (exon 2) and FHIT (exon 5–8) gene mutations analysis was performed by single-strand conformation polymorphism. Mutations were detected by direct sequencing of relevant genomic DNA fragments showing an aberrant profile in the single-strand conformation polymorphism analysis.
RNA Extraction and Northern Blot
RNA was extracted from the breast tumor tissues using guanidium-isothiocyante method was electrophoresed in 3-[N-morpholino]-propanesulfonic acid gel to ascertain the quantities of extracted RNA. From each sample, 1 µg of RNA was resolved on 1% agarose-4-morpholinepropanesulfonicacid/formaldehyde gel, transferred to a nylon membrane, and hybridized for 16 to 18 hours to a radiolabeled FHIT or p16 complementary DNA probe. After 16 to 18 hours of hybridization, the membrane was washed three times with a washing solution (2x SSC, 0.1% SDS) for 15 minutes each at 65°C, and the membrane was exposed to a Kodak X-Omat X-ray film (Carestream Health, Inc, Rochester, NY) in x-ray cassette with intensifying screen at -20°C for 24 hours, and the autoradiogram was developed. Representative examples are shown in Figure 1B.
Western Blot Analysis
Frozen tissue samples were homogenized in lysis buffer (20 mM Tris-HCl at pH 7.5, 1 mM EDTA, 1 mM EGTA, 150 mM sodium chloride, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mg/ml leupeptin, 1 M phenylmethylsulfonyl fluoride), and protein content was quantified. A total of 25 µg of protein was run on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel, transferred on to nitrocellulose membranes, probed either with an antihuman p16 (1:500; BD Pharmingen, San Jose, CA) or FHIT (1:2000; Zymed Laboratories, San Francisco, CA) and electrotransferred to a nitrocellulose membrane. The membrane was incubated in a blocking buffer (1 g/L Tween-20 with 50 g/L nonfat dry milk in Tris-buffered saline) for 2 hours. The membrane strip was then incubated with FHIT antibody overnight at 4°C. The expressions of p16 and FHIT protein were detected using routine methods. The same blots were reprobed with antiactin monoclonal antibodies to assure equal loading in each lane. Representative examples are shown in Figure 1C.
Immunohistochemistry for p16 and FHIT
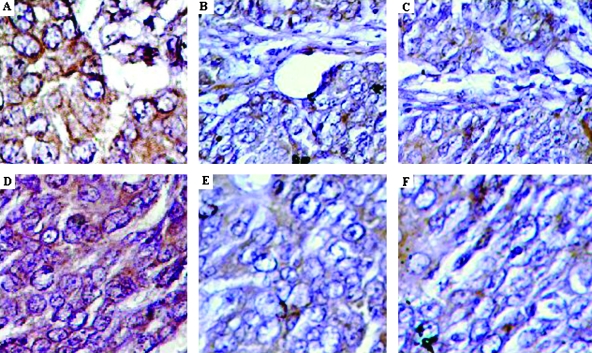
Deparaffinized tissue sections in citrate buffer were heated in microwave for 15 minutes for antigen retrieval for p16 and FHIT genes. Tissue sections were incubated with 3% H2O2 (15 minutes) to block endogenous peroxidase with 10% normal horse serum in phosphate-buffered saline (10 minutes). Primary antibodies against p16 and FHIT (1:200 dilutions; Zymed Laboratories) were applied at room temperature for 1 hour followed by binding to secondary antibody and streptavidin horseradish peroxidase conjugate (20 minutes; Dako, Carpinteria, CA). Normal colonic epithelial cells served as internal positive controls with membrane staining. Cytoplasmic, nuclear, and membrane expressions were recorded separately as no expression, weak expression, or moderate/strong expression. Positivity in each compartment (cytoplasm, nucleus, or membrane) was defined as moderate/strong expression in that compartment. We calculated activation score as the sum of nuclear score (+2 = positive expression; +1 = weak expression; 0 = no expression), cytoplasmic score (+2 = positive expression; +1 = weak expression; 0 = no expression), and membrane score (0 = positive membrane expression; +1 = negative membrane expression). Appropriate positive and negative controls were included in each experiment. Coded slides were scored by one of the investigators (M.R.). At least 25% slides were checked independently by other investigator(s) to rule out any bias. Representative examples are shown in Figure 2 (A to F).
Figure 2.
Immunohistochemical detection of p16 and FHIT protein in breast cancer patients. (A–C) Progressive loss of p16 in different patients: (A) normal epithelium shows consistent and strong staining for p16, (B) low or reduced p16 expression, and (C) no expression of p16. (D–E) Progressive loss of FHIT in different patients: (D) normal epithelium of breast tissue stains consistently and strongly for FHIT protein; (E) low or reduced expression of FHIT gene; and (F) no expression of FHIT protein in breast cancer patients.
Statistical Analysis
The χ2 test was used to compare the distributions of the methylation status of individual genes and various features of breast cancer. Unconditional logistic regression was used to assess odds ratios (ORs) and 95% confidence intervals (CIs). All models were adjusted for age at diagnosis, smoking, and education level. Potential confounding effects from other demographic factors and known breast cancer risk factors, including age at menarche, age at menopause, parity, family history of breast cancer, and body mass index, were also examined, and no appreciable confounding was observed. All statistical tests were based on two-sided probability. Statistical analyses were conducted using SPSS Version 14.0 (SAS Institute, Cary, NC). Differences were considered statistically significant for P < .05.
Results
Primary clinical and pathologic characteristics of patients are shown in Table 1. The frequency of hypermethylation was 31.9% and 36.8% for p16 and FHIT genes, respectively. Promoter region CpG hypermethylation for any of these two genes was identified in 165 (47%) of 351 primary tumors. Seventy-six patients (21.6%) showed hypermethylation for both genes (Table 2). Methylation profile of these genes in normal adjacent tissues from the same patient was also investigated, and we found very low methylation (1.4% and 1.6% of p16 and FHIT, respectively). Patients who showed methylation in either of these genes in normal tissue were found to be methylated in tumor tissue as well. Among premenopausal women, hypermethylation of both p16 and FHIT is significantly higher (OR = 1.71, 95% CI = 1.1–2.89, P = .039 and OR = 1.79, 95% CI = 1.2–2.98, P = .025, respectively, for p16 and FHIT; Table 3) compared with postmenopausal women. The relationship between individual gene hypermethylation status with clinical and pathologic features of breast cancer was evaluated stratifying by the menopausal status (Table 4). Associations varied somewhat by menopausal status. For premenopausal women, there was a significant association of p16 hypermethylation with ER status, whereas FHIT hypermethylation status is significantly associated with TNM stage (Table 4). In postmenopausal women, p16 hypermethylation is significantly associated with histologic grade of the tumors, whereas FHIT methylation is associated with age at diagnosis of the disease. In postmenopausal women, methylation of both p16 and FHIT is significantly associated with the differentiation of the tumors (OR = 3.18, 95% CI = 1.53–9.61 and OR = 3.09, 95% CI = 1.27–7.96 for p16; OR = 2.51, 95% CI = 1.15–6.65 and OR = 2.86, 95% CI = 1.07–7.42 for FHIT gene, respectively; Table 5). Methylation of FHIT gene in premenopausal women is significantly associated with stage of tumors (TNM; OR = 6.1, 95% CI = 1.68–21.52 and OR= 6.2, 95% CI = 1.7–22.6; Table 5). No associations were observed for tumors exhibiting negativity with ER and PR status (Table 5). We found concordant hypermethylation (P < .001) in p16 and FHIT genes.
Table 1.
Clinicopathologic Parameters in Breast Cancer Patients.
| Clinicopathologic Variables | Patients | Percentage |
| Age (years) | ||
| <50 | 156/351 | 44.5 |
| >50 | 195/351 | 55.5 |
| Nodal association | ||
| Positive | 231/351 | 65.8 |
| Negative | 120/351 | 34.2 |
| Histologic grading | ||
| PD | 156/351 | 44.4 |
| MD | 138/351 | 39.3 |
| WD | 57/351 | 16.4 |
| Histologic status | ||
| Infiltrating ductal carcinoma | 299/351 | 85.4 |
| Infiltrating lobular carcinoma | 52/351 | 14.8 |
| ER | ||
| Positive | 293/351 | 83.5 |
| Negative | 58/351 | 16.5 |
| PR | ||
| Positive | 271/351 | 77.2 |
| Negative | 80/351 | 22.8 |
| Methylation | ||
| p16 methylation | 112/351 | 31.9 |
| FHIT methylation | 129/351 | 36.8 |
| p16 gene expression (mRNA) | ||
| Normal | 284/351 | 80.9 |
| Low | 45/351 | 12.8 |
| Nil | 22/351 | 6.3 |
| FHIT gene expression (mRNA) | ||
| Normal | 277/351 | 78.9 |
| Low | 43/351 | 12.3 |
| Nil | 31/351 | 8.8 |
| p16 gene expression (immunohistochemistry) | ||
| Normal | 284/351 | 80.9 |
| Low | 45/351 | 12.8 |
| Nil | 22/351 | 6.3 |
| FHIT gene expression (immunohistochemistry) | ||
| Normal | 277/351 | 78.9 |
| Low | 43/351 | 12.3 |
| Nil | 31/351 | 8.8 |
| TNM clinical stage | ||
| I | 43/351 | 12.3 |
| II | 138/351 | 39.3 |
| III and IV | 170/351 | 48.4 |
| Premenopausal status | ||
| p16 methylation | 46/117 | 39.3 |
| FHIT methylation | 53/117 | 45.3 |
| Postmenopausal status | ||
| p16 methylation | 66/234 | 28.2 |
| FHIT methylation | 76/234 | 32.5 |
Table 2.
Correlation between p16 and FHIT Methylation in Breast Cancer Patients.
| Promoters | FHIT Methylated | FHIT Unmethylated | Total | OR (95% CI) |
| p16 methylated | 76 | 36 | 112 | 7.68* (4.5–12.6) |
| p16 unmethylated | 53 | 186 | 239 | 1.0 (reference) |
| Total | 129 | 222 | 351 |
Odds ratios and 95% CI were estimated with unconditional logistic model adjusted for various confounding factors such as age at the time of diagnosis and smoking habits
P = .05.
Table 3.
Association of Hypermethylation in p16 and FHIT Genes in Association with Premenopausal and Postmenopausal Status in Breast Cancer Patients.
| Promoters | Premenopausal | Postmenopausal | P | OR (95% CI) |
| p16 unmethylated | 71 | 168 | 1.0 (reference) | |
| p16 methylated | 46 | 66 | .039 | 1.71* (1.1–2.89) |
| FHIT unmethylated | 64 | 158 | 1.0 (reference) | |
| FHIT methylated | 53 | 76 | .025 | 1.79* (1.2–2.98) |
Odds ratios and 95% CI were estimated with unconditional logistic model adjusted for various confounding factors such as age at the time of diagnosis and smoking habits
P < .05.
Table 4.
Association of p16 and FHIT Hypermethylation with Selected Clinicopathologic Factors.
| Clinicopathological Factors | Premenopausal Women | Postmenopausal Women | ||||||
| p16 | FHIT | p16 | FHIT | |||||
| M | UM | M | UM | M | UM | M | UM | |
| Age at diagnosis (years) | ||||||||
| <50 | 41 | 56 | 42 | 49 | 2 | 11 | 2 | 11 |
| 50–59 | 5 | 15 | 11 | 15 | 58 | 137 | 59 | 136 |
| ≥59 | - | - | - | - | 6 | 20 | 14 | 12 |
| P | .21 | .82 | .44 | .02* | ||||
| Histologic grade | ||||||||
| Well | 2 | 10 | 3 | 9 | 6 | 39 | 8 | 37 |
| Moderate | 19 | 29 | 24 | 24 | 29 | 61 | 31 | 59 |
| Poor | 25 | 32 | 26 | 31 | 31 | 68 | 37 | 62 |
| P | .21 | .29 | .047* | .058 | ||||
| TNM stage | ||||||||
| I | 5 | 15 | 3 | 17 | 3 | 20 | 4 | 19 |
| II | 22 | 30 | 27 | 25 | 28 | 58 | 32 | 54 |
| III and IV | 19 | 26 | 23 | 22 | 35 | 90 | 40 | 85 |
| P | .35 | .01* | .18 | .19 | ||||
| ER status | ||||||||
| Positive | 43 | 53 | 47 | 49 | 57 | 140 | 63 | 134 |
| Negative | 3 | 18 | 6 | 15 | 9 | 28 | 13 | 24 |
| P | .009* | .09 | .58 | .71 | ||||
| PR status | ||||||||
| Positive | 39 | 50 | 41 | 48 | 54 | 128 | 57 | 125 |
| Negative | 7 | 21 | 12 | 16 | 12 | 40 | 19 | 33 |
| P | .08 | .77 | .35 | .49 | ||||
| ER/PR | ||||||||
| Both positive | 19 | 33 | 23 | 29 | 26 | 68 | 28 | 66 |
| Either positive | 12 | 28 | 16 | 24 | 24 | 70 | 31 | 63 |
| Both negative | 15 | 10 | 14 | 11 | 16 | 30 | 17 | 29 |
| P | .047* | .44 | .51 | .69 | ||||
Significant at *P < .05.
Table 5.
Estimated OR (95% CI) for These Two Genes in Premenopausal and Postmenopausal Women with Selected Factors.
| Clinicopathologic Factors | Premenopausal Women, OR (95% CI) | Postmenopausal Women, OR (95% CI) | |||
| p16 | FHIT | p16 | FHIT | ||
| Histologic grade | |||||
| Well | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | |
| Moderate | 3.37 (0.65–17.25) | 3.11 (0.85–10.16) | 3.18* (1.53–9.61) | 2.51* (1.15–5.65) | |
| Poor | 3.96 (0.82–18.79) | 2.61 (0.92–9.72) | 3.09* (1.27–7.96) | 2.86* (1.07–7.42) | |
| TNM stage | |||||
| I | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | |
| II | 2.4 (1.1–2.9) | 6.1* (1.68–21.52) | 3.38 (0.97–11.36) | 2.71 (1.21–8.2) | |
| III and IV | 2.3 (0.7–6.9) | 6.2* (1.7–22.6) | 2.43 (1.14–8.94) | 2.35 (1.08–8.9) | |
| ER status | |||||
| Positive | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | |
| Negative | 0.22 (0.06–1.54) | 0.59 (0.1–4.23) | 0.81 (0.46–2.15) | 1.25 (0.63–2.76) | |
| PR status | |||||
| Positive | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | |
| Negative | 0.49 (0.17–1.2) | 0.92 (0.56–2.31) | 0.82 (0.41–1.53) | 1.36 (0.71–2.97) | |
| ER/PR status | |||||
| Both positive | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | |
| Either positive | 0.81 (0.34–2.68) | 0.91 (0.36–2.07) | 1.04 (0.55–1.39) | 1.25 (0.73–2.75) | |
| Either negative | 2.71 (1.21–5.89) | 1.68 (0.74–3.89) | 1.51 (0.75–3.36) | 1.43 (0.81–3.52) | |
Odds ratios and 95% CI were estimated with unconditional logistic model adjusted for various confounding factors such as age at the time of diagnosis and smoking habits.
Significant at P < .05.
The MSP approach only allows the samples to be scored either positive or negative for the presence of methylated 5′CpG island sequences. Therefore, no discrimination can be made in the degree of 5′CpG island methylation in any of these samples. Hence, to establish a relationship between positive MSP results with reduced p16 and FHIT mRNA expressions, we compared p16 and FHIT expression levels between the samples displaying methylated and unmethylated p16 and FHIT genes, for those patients where both expression and MSP data were available. Methylation of p16 gene is significantly associated with low/loss of expression (OR = 1.87, 95% CI = 1.1–3.25; Table 6). Similarly, low/loss of FHIT expression is significantly correlated with methylation status (OR = 2.59, 95% CI = 1.54–4.38; Table 6). We hypothesized that mutations in p16 and FHIT genes can also lead to reduced/low expression irrespective of their methylation status. However, we did not find any mutations in p16 and FHIT genes that could result in the loss of their expression in tumors.
Table 6.
Association between Methylation of p16 and FHIT and Their Expression Analyzed by mRNA Analysis and Immunohistochemistry.
| Gene | Low/Nil Expression | Normal Expression | OR (95% CI) |
| p16 unmethylated | 19 | 220 | 1.0 (reference) |
| p16 methylated | 29 | 86 | 1.87* (1.1–3.25) |
| FHIT unmethylated | 32 | 184 | 1.0 (reference) |
| FHIT methylated | 42 | 93 | 2.59* (1.54–4.38) |
Significant at P < .05.
Discussion
We evaluated the association between gene hypermethylation and expression of p16 and FHIT genes with various clinicopathologic characteristics in primary breast tumors. The aim of the study was to further understand the role of the previously mentioned genes in the natural history of breast carcinogenesis as well as a molecular predictor of disease progression. Promoter hypermethylation for at least one gene was found in 47% of the breast tumors. Methylation frequencies of various genes associated with breast cancer varied in different populations. In this study, we found that methylation frequencies for p16 and FHIT genes in our cohort were slightly different to those reported previously [20–22]. This variation could be attributed to the sample tissue with a high proportion of cancerous cells or to ethnic differences.
The most common inactivated tumor suppressor gene in human cancer (p16) is a CDK inhibitor that form complexes with CDK4, CDK6, and D-type cyclins to arrest cell cycle's progression from G1 to S phase [23]. Hypermethylation of p16 gene is an early and critical step in breast cancer development [24,25]. In the present study, we found higher methylation frequency for p16 and FHIT genes if patients show positivity for both ER and PR status compared with ER-negative especially among premenopausal women. However, we did not observe any associations of p16 hypermethylation with other clinicopathologic features of breast cancer. The strongest association observed in this study of specific gene methylation in p16 and FHIT genes was found in postmenopausal women with poorly differentiated tumors. This finding is important because it links DNA hypermethylation with the histologic appearance of breast cancers. Previous studies have also reported associations between CpG island methylation and poor histologic differentiation of breast tumors [20,21,26,27]. It is possible that p16 might play an important role in the initiation and progression of certain premalignant lesions and carcinoma that can act as a crucial event in cell transformation. Loss of p16 protein expression resulting from its methylation played an important role in the evolution of this event. We found reduced FHIT expression in a subset of BRCA2 linked breast cancer (data not shown here). However, FHIT expression is either absent or significantly reduced in some patients (22%), and this loss of the expression is within the range reported elsewhere [14,28]. A significant proportion of breast cancer patients (78%) showing normal FHIT expression without any apparent methylation further indicate that routes independent of this gene may be involved in the pathogenesis of this disease, and these findings are in agreement with a previously published report [14]. We found a similar incidence of tumors, showing loss of expression of p16 genes that is comparable with previously published reports [29–31].
Methylation in breast cancer has long been linked to the hormone regulation, but this correlation is not intelligible yet. DNA methylation profiles in breast, endometrial, ovarian, and proximal colon cancers provide contradictory evidence for global hormone-specific DNA methylation signatures [32,33]. However, in this study, we did not find hormone biology to play any significant role in the methylation status of these genes. In postmenopausal women, FHIT gene hypermethylation is more frequent among ER- and PR-negative patients. However, p16 gene is less methylated in tumors that are negative for PR status both in premenopausal and postmenopausal women. In a recently published report, methylation of RASSF1A, CCND2, GSTP1, Twist, and APC is correlated well with ER status among breast cancer patients [34].
Major progress in controlling mortality and morbidity from cancer requires better understanding of molecular mechanisms underlying disease initiation. Analysis of early aberrant events is complex because by the time the tumor is detected, the cancer progenitor cells may have already undergone multiple changes both at genetic and epigenetic levels. In current clinical practice, both ER and PR status are very important to help determine patients who would benefit most from hormone therapy. Depending on the hormone receptor and menopausal status of the patient, different treatment regimens can be used. A recent retrospective clinical analysis of patients established that both ER-positive/PR-negative and ER-positive/PR-positive breast cancers are clinically and biologically distinct tumor subgroups. The presence of both PR and ER in breast cancer specimens suggests a likelihood of positive response to hormonal therapy, most probably because the presence of PR indicates an intact and functional ER pathway. However, in the current study, we found that tumors with both ER-negative-PR-positive phenotype (10%) and an ER-positive-PR-negative phenotype (9%) exist, and a previous category is another heterogenous group that has been found to be biochemically distinct from ER-PR-positive tumors [35]. When combined with the methylation status, these groups further have the potential to respond differently to the various treatment options currently available to patients. We observed that premenopausal patients who were negative for both ER and PR showed p16 and FHIT methylation more frequently compared with postmenopausal women. Therefore, a larger study is needed to confirm these findings. Both ER and PR expressions can be downregulated in a number of ways including the structural loss of ER and PR gene loci and/or hypermethylation in their promoter regions.
The strengths of the present study include its population-based study design, similar ethnic background, and relatively large sample size leading to relatively stable risk estimates. In summary, the present study suggests that p16 and FHIT hypermethylation is associated with histologic grade, although there is no indication that the hypermethylation of these genes is associated with ER and PR status. However, further research is needed to assess the association of these tumor characteristics with other breast cancer risk factors for better understanding the etiology of this cancer type.
Footnotes
The authors thank the Department of Science and Technology and University Grants Commission, New Delhi, for financial assistance for DST grant No-D.O.No.SP/SO/B-13/2001 and UGC grant No-F.3-94/2003 (S.R.).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Chopra R. The Indian scene. J Clin Oncol. 2001;19:106S–111S. [PubMed] [Google Scholar]
- 4.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Yan L, Davidson NE. DNA methylation in breast cancer. Endocr Relat Cancer. 2001;8:115–127. doi: 10.1677/erc.0.0080115. [DOI] [PubMed] [Google Scholar]
- 6.Szyf M, Pakneshan P, Rabbani PA. DNA methylation and breast cancer. Biochem Pharmacol. 2004;68:1187–1197. doi: 10.1016/j.bcp.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 9.McDermott KM, Zhang J, Holst CR, Kozakiewicz BK, Singla V, Tlsty TD. p16INK4a prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4:e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Sgroi D, Sterner CJ, Beauchamp RL, Pinney DM, Keel S, Ukei K, Rutter JL, Buckler AJ, Louis DN, et al. Mutational analysis of CDKN2 (MTS1/p16ink4) in human breast carcinomas. Cancer Res. 1994;54:5262–5264. [PubMed] [Google Scholar]
- 12.Silva J, Silva JM, Domínguez G, García JM, Cantos B, Rodríguez R, Larrondo FJ, Provencio M, España P, Bonilla F. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J Pathol. 2003;199:289–297. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- 13.Ohta M, Inoue H, Coticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–598. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 14.Gatalica Z, Lele SM, Rampy BA, Norris BA. The expression of Fhit protein is related inversely to disease progression in patients with breast carcinoma. Cancer. 2000;88:1378–1383. [PubMed] [Google Scholar]
- 15.Lee JI, Soria JC, Hassan K, Liu D, Tang X, El-Naggar A, Hong WK, Mao L. Loss of Fhit expression is a predictor of poor outcome in tongue cancer. Cancer Res. 2001;61:837–841. [PubMed] [Google Scholar]
- 16.Yang Q, Yoshimura G, Suzuma T, Tamaki T, Umemura T, Nakamura M, Nakamura Y, Wang X, Mori I, Sakurai T, et al. Clinicopathological significance of fragile histidine triad transcription protein expression in breast carcinoma. Clin Cancer Res. 2001;7:3869–3873. [PubMed] [Google Scholar]
- 17.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24:1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 18.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HJ, Kwon YM, Kim JS, Lee H, Park JH, Shim YM, Han J, Park J, Kim DH. Tumor-specific methylation in bronchial lavage for the early detection of non-small cell lung cancer. J Clin Oncol. 2004;22:2363–2370. doi: 10.1200/JCO.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Rong M, Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett. 2006;237:272–280. doi: 10.1016/j.canlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Tao MH, Shields PG, Nie J, Millen A, Ambrosone CB, Edge SB, Krishnan SS, Marian C, Xie B, Winston J, et al. DNA hypermethylation and clinicopathological features in breast cancer: the Western New York Exposures and Breast Cancer (WEB) Study. Breast Cancer Res Treat. 2009;114:559–568. doi: 10.1007/s10549-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallian S, Sedaghat M, Nassiri I, Frazmand A. Methylation status of p16INK4A tumour suppressor gene in Iranian patients with sporadic breast cancer. J Cancer Res Clin Oncol. 2009;135:991–996. doi: 10.1007/s00432-008-0534-8. [DOI] [PubMed] [Google Scholar]
- 23.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 24.Holst CR, Nuovo GJ, Esteller M, Chew K, Baylin SB, Herman JG, Tlsty TD. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–1601. [PubMed] [Google Scholar]
- 25.Tlsty TD, Crawford YG, Holst CR, Fordyce CA, Zhang J, McDermott K, Kozakiewicz K, Gauthier ML. Genetic and epigenetic changes in mammary epithelial cells may mimic early events in carcinogenesis. J Mammary Gland Biol Neoplasia. 2004;9:263–274. doi: 10.1023/B:JOMG.0000048773.95897.5f. [DOI] [PubMed] [Google Scholar]
- 26.Nass SJ, Herman JG, Gabrielson E, Iversen PW, Parl FF, Davidson NE, Graff JR. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60:4346–4348. [PubMed] [Google Scholar]
- 27.Yan PS, Perry MR, Laux DE, Asare AL, Caldwell CW, Huang TH. CpG island arrays: an application toward deciphering epigenetic signatures of breast cancer. Clin Cancer Res. 2000;6:1432–1438. [PubMed] [Google Scholar]
- 28.Ingvarsson S, Agnarsson BA, Sigbjornsdottir BI, Kononen J, Kallioniemi OP, Barkardottir RB, Kovatich AJ, Schwarting R, Hauck WW, Huebner K, et al. Reduced Fhit expression in sporadic and BRCA2-linked breast carcinomas. Cancer Res. 1999;59:2682–2689. [PubMed] [Google Scholar]
- 29.Dublin EA, Patel NK, Gillett CE, Smith P, Peters G, Barnes DM. Retinoblastoma and p16 proteins in mammary carcinoma: their relationship to cyclin D1 and histopathological parameters. Int J Cancer. 1998;79:71–75. doi: 10.1002/(sici)1097-0215(19980220)79:1<71::aid-ijc14>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen NH, Roos G, Emdin SO, Landberg G. Methylation of the p16 (Ink4a) tumor suppressor gene 5′-CpG island in breast cancer. Cancer Lett. 2001;163:59–69. doi: 10.1016/s0304-3835(00)00674-1. [DOI] [PubMed] [Google Scholar]
- 31.Munot K, Bell SM, Lane S, Horgan K, Hanby AM, Speirs V. Pattern of expression of genes linked to epigenetic silencing in human breast cancer. Hum Pathol. 2006;37:989–999. doi: 10.1016/j.humpath.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Campan M, Weisenberger DJ, Laird PW. DNA methylation profiles of female steroid hormone-driven human malignancies. Curr Top Microbiol Immunol. 2006;310:141–178. doi: 10.1007/3-540-31181-5_8. [DOI] [PubMed] [Google Scholar]
- 33.Widschwendter M, Siegmund KD, Muller HM, Fiegl H, Marth C, Muller-Holzner E, Jones PA, Laird PW. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 34.Sunami E, Shinozaki M, Sim MS, Nguyen SL, Vu AT, Giuliano AE, Hoon DS. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res. 2008;10:R46. doi: 10.1186/bcr2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiani J, Khan A, Khawar H, Shuaib F, Pervez S. Estrogen receptor α negative and progesterone receptor positive breast cancer: lab error or real entity. Pathol Oncol Res. 2006;12:223–227. doi: 10.1007/BF02893416. [DOI] [PubMed] [Google Scholar]