Abstract
Papillary thyroid carcinoma (PTC) is the most common endocrine malignancy and RET/PTC rearrangements represent key genetic events frequently associated to this cancer, enhancing proliferation and dedifferentiation by activation of the RET/PTC-RAS-BRAF-mitogen-activated protein kinase (MAPK) pathway. Recently, let-7 microRNA was found to reduce RAS levels in lung cancer, acting as a tumor suppressor gene. Here, we report that RET/PTC3 oncogenic activation in PCCL3 rat thyroid cells markedly reduces let-7f expression. Moreover, stable transfection of let-7 microRNA in TPC-1 cells, which harbor RET/PTC1 rearrangement, inhibits MAPK activation. As a result, let-7f was capable of reducing TPC-1 cell growth, and this might be explained, at least in part, by decreased messenger RNA (mRNA) expression of cell cycle stimulators such as MYC and CCND1 (cyclin D1) and increased P21 cell cycle inhibitor mRNA. In addition, let-7 enhanced transcriptional expression of molecular markers of thyroid differentiation such as TITF1 and TG. Thus, reduced expression of let-7f might be an essential molecular event in RET/PTC malignant transformation. Moreover, let-7f effects on thyroid growth and differentiation might attenuate neoplastic process of RET/PTC papillary thyroid oncogenesis through impairment of MAPK signaling pathway activation. This is the first functional demonstration of an association of let-7 with thyroid cancer cell growth and differentiation.
Introduction
MicroRNA (miRNA) are noncoding ∼22-nucleotide RNA that negatively regulate gene expression at the posttranscriptional level either by directly cleaving targeted messenger RNA (mRNA) or by repressing translation [1–3]. Originally discovered in Caenorhabditis elegans, these small RNA are now recognized as one of the major regulatory gene families in plants and animals [4]. Lethal-7 (let-7) is one of the most investigated miRNA, being required for timing of stem cell division and differentiation in C. elegans [5,6]. In the human genome, almost 700 miRNA genes have been discovered, and the estimated number of miRNA is as high as 1000 [7,8]. Although the biology of most miRNA is not well understood, countless studies have linked anomalous expression of these molecules to carcinogenesis [1]. In humans, let-7 was shown to contain multiple putative complementary sites in the 3′ untranslated region of all three RAS genes (HRAS, KRAS, and NRAS), and it functions to negatively regulate their protein levels [9]. Because RAS activation is associated with several cancers, let-7 downregulation or deletion could therefore play a role in tumorigenesis. In fact, let-7 overexpression was found to inhibit lung and colon cancer cell growth in vitro [9–11]. These findings underscore the importance of this small RNA as a tumor suppressor for cancers in which the RAS pathway is constitutively activated.
Papillary thyroid carcinoma (PTC) is the most prevalent endocrine malignancy in humans [12]. Along with others, this laboratory has shown that ∼70% of PTCs harbor genetic alterations in RET, RAS, or BRAF with practically no overlap, providing genetic evidence that constitutive linear signaling along the RET-RAS-BRAF-ERK pathway is key to their development [13,14]. In particular, RET gene rearrangements occur in up to 43% of PTCs [15]. These rearrangements result in the recombination of the intracellular kinase-encoding domain of RET with different activating genes, thereby generating RET/PTC fusion oncogenes. Biologic effects mediated by RET/PTC include enhanced proliferation and dedifferentiation, and they depend on the activation of the RET/PTC-RAS-BRAF-ERK axis [16]. This requirement for mitogen-activated protein kinase (MAPK) activation is supported by experimental evidence demonstrating that depletion of MAPK cascade components, such as RAS or BRAF, interferes with RET/PTC-induced ERK phosphorylation [17,18].
In this study, the potential involvement of let-7 miRNA in PTC development is investigated. Because let-7 was demonstrated to inhibit RAS expression, it might provide a tumor suppressor function in thyroid cancers with MAPK activation. Indeed, we show that let-7f induction in TPC-1 cells, a human PTC cell line that spontaneously harbors the RET/PTC1 oncogene, causes a marked reduction in cell proliferation and induces expression of molecular markers characteristic of thyroid differentiation. Our data suggest that let-7 miRNA is an essential regulator of thyroid carcinogenesis.
Materials and Methods
Cell Culture
PTC3–5 [19] and PCCL-BRAF cells were derived from PCCL3 rat thyroid cells to obtain doxycycline-inducible expression of RET/PTC3 and BRAFV600E oncogenes, respectively. These cells were maintained in Ham's F12 medium supplemented with 5% fetal bovine serum, 1 mIU/ml bovine thyroid-stimulating hormone (Sigma, St Louis, MO), 10 µg/ml insulin (Sigma), 5 µg/ml apotransferrin (Sigma), 10 nM hydrocortisone (Sigma), 100 U/ml penicillin/100 µg/ml streptomycin (Invitrogen Life Technologies, Carlsbad, CA), 1 µg/ml amphotericin (Invitrogen Life Technologies), and antibiotics at 37°C in a humidified 5% carbon dioxide incubator. RET/PTC3 and BRAFV600E expression was induced after treating with 1 µg/ml doxycycline (Calbiochem, San Diego, CA) for 72 hours. Cells cultivated in the absence of doxycycline were used as controls.
TPC-1, a human PTC cell line spontaneously harboring the RET/PTC1 rearrangement [20], was maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 100 U/ml penicillin/100 µg/ml streptomycin, and 1 µg/ml amphotericin at 37°C in a humidified 5% carbon dioxide incubator.
This study complied with the guidelines from the ethical committee of the Institute of Biomedical Sciences (no. 20/F43/L2), University of São Paulo.
Transfection
TPC-1 cells were cultured to 80% to 90% confluence and transfected with pH1-RNApuro-control or pH1-RNApuro-let-7f plasmids using Lipofectamine 2000 (Invitrogen Life Technologies). Stable cell lines were obtained by culturing the cells with 1 µg/ml puromycin (Calbiochem) for 2 to 3 weeks. Stable clones overexpressing let-7f were screened by quantitative polymerase chain reaction (PCR) and used for further experiments. pH1-RNApuro-control and pH1-RNApuro-let-7f plasmids were kindly donated by Dr Takashi Takahashi (Nagoya University Graduate School of Medicine, Nagoya, Japan).
MTT Assay
Cells were seeded into 96-well plates at a density of 8 x 103 cells per well. When the cells reached semiconfluence, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Molecular Probes, Eugene, OR) was added to the medium at a concentration of 0.125 mg/ml. After 3 hours, the medium was removed, and the cells were solubilized in 100 µl of 0.04 M HCl in isopropanol and the reaction product was measured by a spectrophotometer at 595 nm.
Western Blot Analysis
Cells were washed twice with ice-cold PBS and scraped in RIPA buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 0.1% SDS) containing 10% protease inhibitor cocktail (Sigma). Measurement of protein concentration was done using Bradford (Bio-Rad Laboratories, Hercules, CA), and 50 µg of each sample was fractionated by 10% SDS-PAGE and blotted onto a nitrocellulose membrane (Hybond-ECL; Amersham Biosciences, Little Chalfont, UK). Nonspecific binding sites were blocked by incubating with 5% nonfat dry milk in Tris-buffered saline-0.1% Tween-20. The following primary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were used: anti-ERK1 K23, anti-phospho-ERK E4, anti-H-RAS C20, anti-K-RAS F234, and anti-α-tubulin B7. The antigen-antibody complexes were visualized using horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence system (Amersham Biosciences). Expression was quantified using image densitometry with Scion Image Analysis Software (Scion Corporation, Frederick, MA).
Quantitative PCR
Total RNA was isolated using the phenol-chloroform method with the TRIzol reagent (Invitrogen Life Technologies). Complementary DNA (cDNA) was generated from 3 µg of total RNA in the presence of random hexamers and MMLV reverse transcription (RT; Invitrogen Life Technologies) as described previously [21]. Primers were designed using the program Primer Express (Perkin-Elmer Applied Biosystems, Foster City, CA) as follows: CCND1 (cyclin D1): 5′-CTGTGCATCTACACCGACAACTC and 5′-CCAGGTTCCACTTGAGCTTGTT; CDKN1A (P21): 5′-CTGGAGACTCTCAGGGTCGAA and 5′-GGCGTTTGGAGTGGTAGAAATCT; MYC: 5′-TTCGGGTAGTGGAAAACCAG and 5′-TCCTGTTGGTGAAGCTAACG; NKX2-1 (TITF1): 5′-AGCCTGTCCCACCTGAACT and 5′-ATAGCAAGGTGGAGCAGGACAT; TG: 5′-CCTGCTGGCTCCACCTTGTTT and 5′-CCTTGTTCTGAGCCTCCCATCGTT; RPL19: 5′-TCTCATGGAACACATCCACAA and 5′-TGGTCAGCCAGGAGCTTCTT. Each set of sense and antisense primers was chosen within distinct exons to avoid amplification of genomic DNA. PCR amplification was performed in triplicate using 50 ng of cDNA, 200 or 300 nM of each primer and SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK) in a GeneAmp 7300 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA). PCRs without cDNA resulted in no amplification of any target gene. The expression of each gene of interest was normalized with housekeeping gene RPL19. Data were analyzed using the Q-Gene program [22].
let-7 Mature miRNA Quantification
Quantitative analysis of miRNA was accomplished using a real-time PCR assay previously reported [23]. In the first step, a tailed miRNA-specific primer was used to convert the RNA template into the cDNA at 50°C for 30 minutes and at 85°C for 5 minutes (5′-CATGATCAGCTGGGCCAAGAAACTATA). Then, the full-length cDNA was quantified by real-time PCR using a combination of a locked nucleic acid (LNA)-containing miRNA primer (5′-T+GA+GGTAGGTAGATTG, LNA substitutions are preceded by a “+”), and a generic universal primer that is complementary to the tail (5′-CATGATCAGCTGGGCCAAGA). The LNA modification stabilizes the conformation of the sugar group and thereby increases the hybridization affinity of oligonucleotides that contain LNA bases [24]. Amplification of this chimeric cDNA was monitored by SYBR green real-time PCR under the following conditions: 50°C for 2 minutes, 95°C for 15 seconds and 40 cycles at 95°C for 15 seconds, 54°C for 30 seconds, and 72° for 30 seconds. Data were normalized with the expression of RNU6B (small nuclear RNA) for human cells or small nucleolar RNA (snoRNA) for rat cells using a specific amplification kit (Applied Biosystems, Warrington, UK).
Statistical Analysis
The results are presented as the mean ± SD and submitted to analysis of variance using the Bonferroni t-test. Differences were considered significant at P < .05.
Results
Doxycycline-Inducible Expression of RET/PTC3 in PCCL3 Rat Thyroid Cells
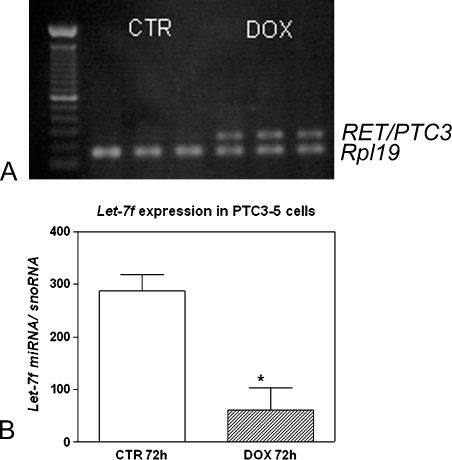
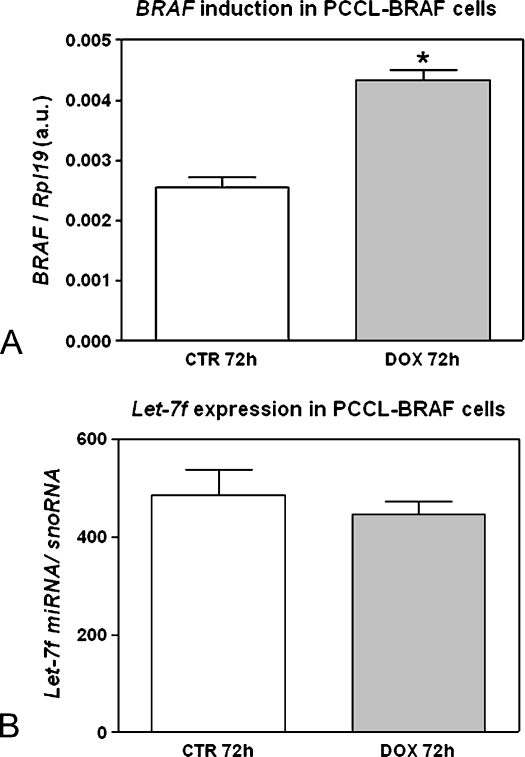
To determine whether the expression of mature let-7f miRNA is modulated by RET/PTC, PTC3–5 cells, derived from PCCL3 rat thyroid cells modified to contain doxycycline-inducible expression of RET/PTC3, were analyzed. RT-PCR was used to confirm the efficient induction of RET/PTC3 expression by treatment of the PTC3–5 cells with doxycycline for 72 hours (Figure 1A). The induced expression of the RET/PTC3 oncogene in PTC3–5 cells led to a strong reduction (76%) in let-7f miRNA expression (Figure 1B). However, induction of the BRAF oncogene in PCCL3 cells treated with doxycycline for 72 hours (Figure 2A) did not modulate let-7f mature miRNA expression (Figure 2B).
Figure 1.
Effect of the constitutive activation of the RET/PTC3 oncogene on let-7 miRNA expression. (A) Representative gel of the RT-PCR results showing RET/PTC3 mRNA induction after 72 hours of doxycycline (DOX) treatment. The reactions are normalized by comparison to the Rp/19 housekeeping gene. y-Axis represents 100-bp ladder. (B) RET/PTC3 induction leads to reduced let-7miRNA expression in PTC3–5 cells. y-Axis represents let-7f expression normalized by snoRNA. Columns in the graph represent mean ± SD of two independent reactions performed in triplicate. *P = .002 versus TPC-1 CTR.
Figure 2.
Effect of constitutive activation of the BRAF oncogene on let-7 miRNA expression. (A) BRAF induction in PCCL-BRAF cells after 72 hours of DOX treatment. y-Axis represents BRAF expression normalized by Rpl19. (B) let-7 expression does not fluctuate even when BRAF is induced for 72 hours. y-Axis represents let-7f expression normalized by snoRNA. Columns in the graph represent result as mean ± SD in arbitrary units (a.u.) of two independent quantitative RT-PCRs performed in triplicate. *P = .001 versus TPC-1 CTR.
Effects of let-7f Introduction on MAPK Signaling Pathway in TPC-1 Cells
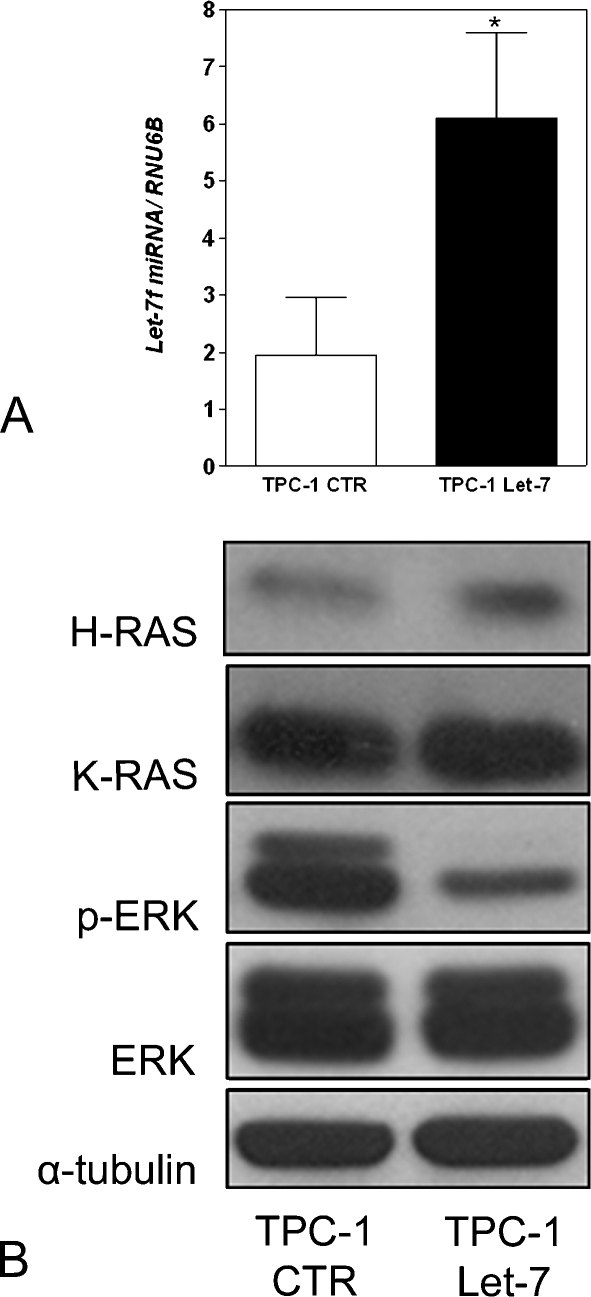
The observation that let-7f expression decreases in response to RET/PTC activation led us to explore the possibility that let-7f-reduced expression might contribute to papillary thyroid cancer development. To test this idea, let-7f was introduced into the TPC-1 cells, which spontaneously harbor the RET/PTC1 rearrangement, using expression constructs designed to synthesize mature miRNA of the let-7f-1 isomer under the control of the RNA polymerase-III H1-RNA gene promoter [11]. Real-time PCR was used to evaluate the expression of let-7f mature miRNA in TPC-1 control (CTR) and TPC-1 let-7 stable clones. let-7f was observed to have a basal expression level in TPC-1 cells, which was substantially increased after introducing the let-7f plasmid (Figure 3A). Because RET/PTC activation sequentially triggers stimulation of RAS, BRAF, and ERK to mediate its biologic effects, the potential influence of let-7f overexpression on ERK activation was evaluated. As expected, results of the Western blot analysis demonstrated that let-7f induction resulted in a decrease in phosphorylation of the ERK protein in TPC-1 cells (Figure 3B), even without altering H-RAS and K-RAS protein levels (expression quantified by densitometry and normalized by tubulin levels).
Figure 3.
Stable transfection of let-7 miRNA in TPC-1 cells. (A) Quantitative RT-PCR detection of the expression of let-7 miRNA in cells stably transfected with pH1-RNApuro-CTR and pH1-RNApuro-let-7f-1. y-Axis represents let-7f expression normalized by RNU6B. Columns in the graph represent mean ± SD of two independent reactions performed in triplicate. (B) Total cellular proteins were prepared and subjected to 10% SDS-PAGE electrophoresis. Immunoblot analyses were performed using specific antibodies against K-RAS, H-RAS, phosphorylated ERK (p-ERK), ERK1/2, and α-tubulin. *P = .017 versus TPC-1 CTR.
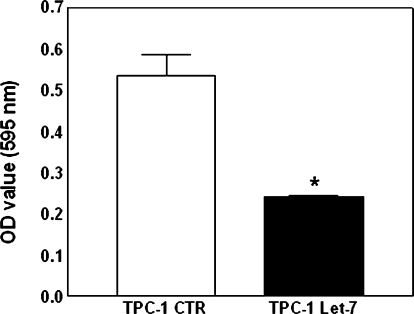
Growth Inhibitory Effect of let-7f Mature miRNA on TPC-1 Cells
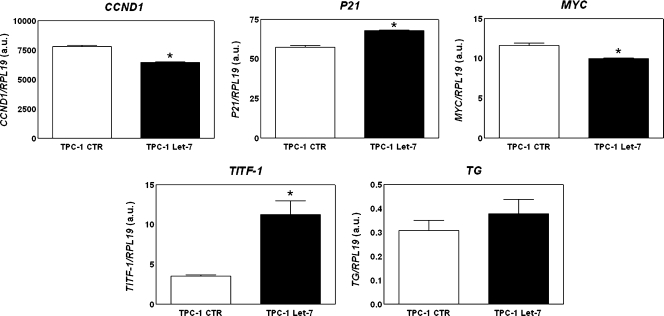
The MTT assay was used to examine whether let-7f expression is associated with PTC cell growth. let-7f overexpression markedly inhibited proliferation of TPC-1 cells by 55% (Figure 4). To identify genes involved in the inhibition of TPC-1 cell growth, the transcriptional expression of genes known for their action in the cell cycle, such as MYC, CCND1 (cyclin D1), and CDKN1A (p21), was examined. let-7f inhibited the mRNA expression of cell cycle stimulators such as MYC (17%) and CCND1 (14%). However, this small RNA also reduced the transcriptional expression of CDKN1A (18%), a gene that acts as a cell cycle inhibitor (Figure 5).
Figure 4.
Effect of let-7 miRNA on the proliferation of TPC-1 cells. Cells were seeded at equal density and cultured until they reached semiconfluence. They were then incubated with MTT for 3 hours. Data are presented as the mean ± SD of a single experiment performed in quintuplicate and are representative of two independent experiments. *P = .000 versus TPC-1 CTR.
Figure 5.
Effect of let-7 miRNA on gene expression. The graph shows the mRNA expression of the cell cycle-related genes MYC, CCND1, and CDKN1A, and the thyroid differentiation markers, TITF1 and TG, as determined by quantitative RT-PCR. y-Axis represents target gene expression normalized by RPL19. Columns in the graph represent mean ± SD in arbitrary units (a.u.) of two independent reactions performed in triplicate. *P = .000 (CCND1), .001 (P21), .006 (MYC), .001 (TITF-1) versus TPC-1 CTR.
Effects of let-7f on Transcriptional Expression of Thyroid Differentiation Genes
Because RET/PTC-induced thyroid cell dedifferentiation is mediated by activation of the RAS-BRAF-MAPK pathway, the possible biologic significance of let-7f in TPC-1 cell differentiation was explored. Transcription of specific molecular markers of thyroid differentiation such as thyroid transcription factor 1 (TITF1), thyroglobulin (TG), and sodium iodide symporter (NIS), was analyzed. let-7f induces a three-fold increase in TITF1 mRNA expression and also a slightly increase in TG transcription when compared with control cells (Figure 5). However, NIS mRNA expression was not detected in TPC-1 cells even with let-7f overexpression (data not shown).
Discussion
Alterations in RET, RAS, and BRAF genes are major contributors to the transformation of normal thyroid cells to PTC [16]. Recent studies have associated these genetic alterations with global changes in the expression of miRNA [25–28]. This information has led toward new understanding of the genetic mechanisms and pathways underlying the etiopathogenesis of PTCs. Despite all the information concerning changes in the expression of miRNA in thyroid cancer, the functions of these molecules in thyroid tumorigenesis are poorly understood. In this study, we have identified a functional association of let-7f miRNA with thyroid cancer for the first time. The let-7 family of miRNA includes 14 isomers; each isomer is typically located on a different chromosome [8]. This study focused on let-7f, which is located at 9q22.3, because it has been previously shown to inhibit proliferation of lung cancer cells [29]. We observed a reduced expression of let-7f mature miRNA in PCCL3 cells with activation of RET/PTC3, but not BRAF. This suggests that let-7f down-regulation is an important step in the biologic effects mediated by the RET/PTC oncogene during malignant transformation. Supporting our data, a previous study comparing miRNA expression profile in thyroid tissues from patients with papillary carcinoma and normal thyroid tissues showed that let-7f is downregulated in this cancer [28]. To clarify the function of let-7f in the thyroid cancer scenery, exogenous let-7f was overexpressed in TPC-1 papillary cancer cells that harbor the RET/PTC1 rearrangement. Although transfection of let-7f in TPC-1 cells repressed ERK phosphorylation, supporting the conclusion that RET/PTC-RAS-BRAF-ERK cascade activation is inhibited by this miRNA, significant modulation of K-RAS and H-RAS proteins was not observed after the induction of let-7f. Nevertheless, repression of MAPK signaling in concert with the modulation of genes related to cell cycle may explain, at least in part, the reduced cell growth of TPC-1 cells transfected with let-7f.
Previous studies from several groups have shown that constitutive activation of effectors along the RET/PTC-RAS-BRAF-ERK pathway impairs the expression of thyroid-specific genes, such as TG, TPO, NIS, and TSHR, and their corresponding transcription factors TITF1 and PAX8 [30–32]. During malignant progression, thyroid cancer cells undergo dedifferentiation, becoming more aggressive and refractory to treatment. We demonstrate here that let-7f is able to enhance the expression of some of these differentiation markers in TPC-1 cells, including TITF1, a major transcription factor in thyroid cells. TITF1 is also key gene in thyroid development; mice lacking this transcription factor are unable to develop a thyroid gland [33]. The role of let-7 during development is well established, and it may also play an important role in thyroid development through the regulation of TITF1 expression. Therefore, it is evident that let-7f is critical for proper regulation of thyroid cell growth and differentiation. It is known that let-7 has other targets in addition to RAS with respect to cancer, including the HMGA2 oncogene [34]. However, there is compelling evidence in the literature showing that activation of the MAPK signaling pathway is required for the development of RET/PTC-positive PTCs. Thus, the effects of let-7f in this specific cell (TPC-1) are likely due to its inhibitory effects on the MAPK pathway. Whether let-7f truly interacts with other mRNA that are important for its effects in TPC-1 cells is a question that remains to be answered. A few studies have previously associated alterations in miRNA with the development of PTCs, specifically using miRNA expression profiling to identify four miRNA (miR-146, miR-221, miR-222, and miR-181b) with high levels of expression in cancerous tissues [28,35]. In addition, it has been shown that the main genetic alterations observed in PTCs, the BRAF mutation and RET/PTC rearrangements, have global effects on the miRNA expression profile [26,27]. Along with these studies, our work demonstrates an association between this new class of small RNA and the development of thyroid carcinogenesis.
In summary, we show that reduced expression of let-7f is associated with RET/PTC malignant transformation. Furthermore, restoration of let-7f expression attenuates RET/PTC-mediated oncogenesis by preventing MAPK cascade activation and its resulting biologic effects, such as proliferation and dedifferentiation. Our data support a suppressor function of let-7f miRNA in thyroid cancer, revealing this molecule as a potential therapeutic target in patients with cancers harboring the RET/PTC fusion oncogene.
Acknowledgments
The authors thank J.A. Fagin (Memorial Sloan-Kettering Cancer Center, New York, NY) for kindly providing the PTC3–5 and TPC-1 cells and Takashi Takahashi (Nagoya University Graduate School of Medicine, Nagoya, Japan) for kindly donating pH1-RNApuro-control and pH1-RNApuro-let-7f plasmids.
Footnotes
This study was supported by research grants and a doctoral fellowship from the Foundation for Research Support of the State of São Paulo (FAPESP), Technological and Scientific Development National Council (CNPq), and the Coordination for the Improvement of Higher Education Personnel (CAPES).
Conflict of Interest Statement
The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Ricarte Filho JC, Kimura ET. MicroRNAs: novel class of gene regulators involved in endocrine function and cancer. Arq Bras Endocrinol Metabol. 2006;50:1102–1107. doi: 10.1590/s0004-27302006000600018. [DOI] [PubMed] [Google Scholar]
- 3.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 7.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 11.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 12.Howe HL, Wu X, Ries LA, Cokkinides V, Ahmed F, Jemal A, Miller B, Williams M, Ward E, Wingo PA, et al. Annual report to the nation on the status of cancer, 1975–2003, featuring cancer among U.S. Hispanic/Latino populations. Cancer. 2006;107:1711–1742. doi: 10.1002/cncr.22193. [DOI] [PubMed] [Google Scholar]
- 13.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 14.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev. 2007;28:742–762. doi: 10.1210/er.2007-0007. [DOI] [PubMed] [Google Scholar]
- 15.Fusco A, Santoro M. 20 years of RET/PTC in thyroid cancer: clinicopathological correlations. Arq Bras Endocrinol Metabol. 2007;51:731–735. doi: 10.1590/s0004-27302007000500010. [DOI] [PubMed] [Google Scholar]
- 16.Trovisco V, Soares P, Preto A, Castro P, Maximo V, Sobrinho-Simoes M. Molecular genetics of papillary thyroid carcinoma: great expectations. Arq Bras Endocrinol Metabol. 2007;51:643–653. doi: 10.1590/s0004-27302007000500002. [DOI] [PubMed] [Google Scholar]
- 17.Mitsutake N, Miyagishi M, Mitsutake S, Akeno N, Mesa C, Jr, Knauf JA, Zhang L, Taira K, Fagin JA. BRAF mediates RET/PTC-induced mitogenactivated protein kinase activation in thyroid cells: functional support for requirement of the RET/PTC-RAS-BRAF pathway in papillary thyroid carcinogenesis. Endocrinology. 2006;147:1014–1019. doi: 10.1210/en.2005-0280. [DOI] [PubMed] [Google Scholar]
- 18.Melillo RM, Castellone MD, Guarino V, De Falco V, Cirafici AM, Salvatore G, Caiazzo F, Basolo F, Giannini R, Kruhoffer M, et al. The RET/PTC-RAS-BRAF linear signaling cascade mediates the motile and mitogenic phenotype of thyroid cancer cells. J Clin Invest. 2005;115:1068–1081. doi: 10.1172/JCI22758. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Wang J, Knauf JA, Basu S, Puxeddu E, Kuroda H, Santoro M, Fusco A, Fagin JA. Conditional expression of RET/PTC induces a weak oncogenic drive in thyroid PCCL3 cells and inhibits thyrotropin action at multiple levels. Mol Endocrinol. 2003;17:1425–1436. doi: 10.1210/me.2003-0041. [DOI] [PubMed] [Google Scholar]
- 20.Ishizaka Y, Ushijima T, Sugimura T, Nagao M. cDNA cloning and characterization of ret activated in a human papillary thyroid carcinoma cell line. Biochem Biophys Res Commun. 1990;168:402–408. doi: 10.1016/0006-291x(90)92335-w. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo SE, Ebina KN, Kulcsar MA, Friguglietti CU, Kimura ET. Activin betaB expression in rat experimental goiter and human thyroid tumors. Thyroid. 2003;13:239–247. doi: 10.1089/105072503321582033. [DOI] [PubMed] [Google Scholar]
- 22.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 23.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 25.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cahill S, Smyth P, Denning K, Flavin R, Li J, Potratz A, Guenther SM, Henfrey R, O'Leary JJ, Sheils O. Effect of BRAFV600E mutation on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer. 2007;6:21. doi: 10.1186/1476-4598-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cahill S, Smyth P, Finn SP, Denning K, Flavin R, O'Regan EM, Li J, Potratz A, Guenther SM, Henfrey R, et al. Effect of ret/PTC 1 rearrangement on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer. 2006;5:70. doi: 10.1186/1476-4598-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 29.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 30.Knauf JA, Kuroda H, Basu S, Fagin JA. RET/PTC-induced dedifferentiation of thyroid cells is mediated through Y1062 signaling through SHC-RAS-MAP kinase. Oncogene. 2003;22:4406–4412. doi: 10.1038/sj.onc.1206602. [DOI] [PubMed] [Google Scholar]
- 31.Portella G, Vitagliano D, Borselli C, Melillo RM, Salvatore D, Rothstein JL, Vecchio G, Fusco A, Santoro M. Human N-ras, TRK-T1, and RET/PTC3 oncogenes, driven by a thyroglobulin promoter, differently affect the expression of differentiation markers and the proliferation of thyroid epithelial cells. Oncol Res. 1999;11:421–427. [PubMed] [Google Scholar]
- 32.De Vita G, Zannini M, Cirafici AM, Melillo RM, Di Lauro R, Fusco A, Santoro M. Expression of the RET/PTC1 oncogene impairs the activity of TTF-1 and Pax-8 thyroid transcription factors. Cell Growth Differ. 1998;9:97–103. [PubMed] [Google Scholar]
- 33.Kimura S, Ward JM, Minoo P. Thyroid-specific enhancer-binding protein/thyroid transcription factor 1 is not required for the initial specification of the thyroid and lung primordia. Biochimie. 1999;81:321–327. doi: 10.1016/s0300-9084(99)80077-7. [DOI] [PubMed] [Google Scholar]
- 34.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]