Abstract
Although anoikis resistance has been considered a hallmark of malignant phenotype, the causal relation between neoplastic transformation and anchorage-independent growth remains undefined. We developed an experimental model of murine melanocyte malignant transformation, where a melanocyte lineage (melan-a) was submitted to sequential cycles of anchorage blockade, resulting in progressive morphologic alterations, and malignant transformation. Throughout this process, cells corresponding to premalignant melanocytes and melanoma cell lines were established and show progressive anoikis resistance and increased expression of Timp1. In melan-a melanocytes, Timp1 expression is suppressed by DNA methylation as indicated by its reexpression after 5-aza-2′-deoxycytidine treatment. Methylation-sensitive single-nucleotide primer extension analysis showed increased demethylation in Timp1 in parallel with its expression along malignant transformation. Interestingly, TIMP1 expression has already been related with negative prognosis in some human cancers. Although described as a MMP inhibitor, this protein has been associated with apoptosis resistance in different cell types. Melan-a cells overexpressing Timp1 showed increased survival in suspension but were unable to form tumors in vivo, whereas Timp1-overexpressing melanoma cells showed reduced latency time for tumor appearance and increased metastatic potential. Here, we demonstrated for the first time an increment in Timp1 expression since the early phases of melanocyte malignant transformation, associated to a progressive gene demethylation, which confers anoikis resistance. In this way, Timp1 might be considered as a valued marker for melanocyte malignant transformation.
Introduction
Several studies have shown that cell interaction with extracellular matrix (ECM) is crucial for survival, proliferation, migration, and differentiation [1]. The physical link between them is mediated by integrins, α/β heterodimeric transmembrane glycoproteins that connect ECM proteins to actin cytoskeleton and play a critical role transducing bidirectional signals between extracellular and intracellular compartments [2]. Loss of integrin-matrix contacts results in a type of apoptosis, termed anoikis, even in the presence of all required growth factors and nutrients [3]. The acquired ability to survive independently of anchorage is known as a hallmark of malignant transformation, by increasing the survival in absence of attachment and facilitating migration and colonization at distant sites [4]. In animal models, anoikis-resistant tumors present increased incidence of metastatic lesions and number of circulating cells in blood [5,6]. Deletion of tumor suppressor genes, overexpression of oncogenes, alterations in adhesion molecules pattern, enhancement in survival signals, as well as inactivation of molecules involved in survival signaling render cells resistant to anoikis, which may be influenced by environment, genetic, and epigenetic factors, contributing to early steps of carcinogenesis and also tumor progression [7]. Although anoikis resistance has been described in many types of human malignancies, including melanoma [6], the causal relation between neoplastic transformation and anchorage-independent growth remains undefined.
The tissue inhibitor of metalloproteinase (Timp) family consists of four members (Timp1, 2, 3, and 4) that act as endogenous inhibitors of matrix metalloproteinases (MMPs) [8]. The balance between MMPs and Timps activities is involved both in normal and pathologic events such as wound healing, tissue remodeling, angiogenesis, invasion, and metastasis [9]. Almost all human cancers have been found to augment expression and activity of MMPs. Interestingly, Timps are often found upregulated in many tumors. TIMP1 expression has been related with negative prognosis in many human cancers including metastatic breast cancer, colorectal cancer, lymphoma, and non-small cell lung carcinoma, suggesting a promising value for TIMP1 as a prognostic marker for cancer [10,11]. Although well described as a MMP inhibitor, other cellular activities associated with Timp1 seem to be independent of MMP inhibition, such as proliferation promotion and apoptosis inhibition [12], including anoikis resistance [13]. In both cases, studies have been suggested that MMP-independent effects of TIMP1 on cell growth and survival may be mediated by their direct binding to a putative cell surface receptor [14].
Timp1 is expressed by a variety of cell types and tissues, and its expression is induced by external stimuli such as phorbol esters, serum, cytokines, and growth factors [9]. The mechanisms of Timp1 regulation have been intensively investigated but still remain largely unknown. Its induction occurs primarily at the transcription level involving activator protein 1, specificity protein 1, nuclear factor γB, and cyclic AMP response element transcription factors [12]. DNA methylation is an important mechanism of gene expression regulation [15], and some studies indicate that DNA methylation could be responsible for the regulation of Timp1 expression [16].
We studied the correlation between malignant transformation and anoikis resistance by using an experimental model of murine melanocyte malignant transformation [17], in which the nontumorigenic melanocyte lineage melan-a [18] was subjected to sequential cycles of anchorage blockade for 96 hours. Progressive morphologic alterations were observed along each deadhesion cycle, culminating in the establishment of nontumorigenic melan-a sublines corresponding to intermediate phases of malignant transformation found after two and four cycles of anchorage blockade (2C and 4C lineages, respectively). In addition, distinct tumorigenic lineages, both slow-growing (4C3-) and fast-growing melanoma lineages (4C3+ and Tm5), were established from spheroids formed after a new anchorage blockade cycle of 4C cells [17]. Previous gene expression analysis showed up-regulation of Timp1 gene expression in Tm5 melanoma cells (unpublished data). Here, we report an increase in Timp1 expression during melanoma genesis, related to progressive Timp1 gene demethylation, which is causally associated with the increase in anoikis resistance but not with the acquisition, by itself, of a fully transformed malignant phenotype.
Materials and Methods
Mice
Female C57Bl/6, 6 to 8 weeks old, were obtained from and housed in the animal facilities at the Instituto Nacional de Farmacologia, Universidade Federal de São Paulo (SP, Brazil). The animals were kept in a specific pathogen-free facility, under 12-hour daylight cycles, without food restriction, under the International Guiding Principles for Biomedical Research Involving Animals (CIOMS), Genebra (1985). All animal procedures were approved by our institutional animal care and ethics committee.
Cell Lines and Culture
Murine melanocyte lineage melan-a [18] and melan-a cell sublines 2C, 4C, 4C3-, 4C3+, and Tm5, established after submitting melan-a cells to sequential substrate adhesion impediment cycles [17], were maintained in RPMI 1640 (Gibco, Carlsbad, CA), pH 6.9, containing 5% fetal bovine serum (Gibco), 40 mg/L gentamicin, and 200 nM phorbol 12-myristate 13-acetate (Sigma, St Louis, MO) at 37°C in 5% CO2.
Substrate Adhesion Blockade Assay
To ensure an anchorage-independent growth condition, 105 cells were plated on 1% agarose-treated dishes and cultured for different times at 37°C in 5% CO2. Cells in suspension were collected from agarose-treated dishes, and cell viability was determined using the standard methyl thiazol tetrazolium assay (MTT; Amresco, Solon, OH) as described previously [19].
5-Aza-2′-Deoxycytidine Treatment
Melan-a cells were seeded at a density of 2 x 106 cells in a 10-mm dish. After 24 hours, cells were incubated with fresh culture medium with or without 5 µM of the demethylating agent 5-Aza-2′-deoxycytidine (5-Aza-CdR; Calbiochem, Darmstadt, Germany) for 48 hours. Every day, the medium was changed, and no significant cell death was observed. RNA was isolated from 5-Aza-CdR-treated and nontreated melan-a cells, and the expression of Timp1 was analyzed by reverse transcription-polymerase chain reaction (RT-PCR) using the housekeeping gene β-actin as an internal control. PCR products were visualized on an ethidium bromide-stained 1% agarose gel.
Reverse Transcription-Polymerase Chain Reaction
The cell lines were grown as a monolayer until 70% confluent, and total RNA was isolated with TRizol (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. One microgram of RNA was reverse-transcribed to complementary DNA (cDNA) with Superscript III (Invitrogen). The resulting single-strand cDNA were amplified by PCR in a reaction mixture containing 75 mM Tris-HCl, pH 9.0, 2 mM MgCl2, 50 mM KCl, 20 mM (NH4)2SO4, 0.4 mM of each deoxynucleotide triphosphate, 0.4 µM of each primer, 1 U of BioTools DNA Polymerase-Recombinant from Thermus thermophilus (BioTools, Madrid, Spain). The thermal cycling conditions were as follows: initial 5 minutes at 94°C, followed by cycles of denaturing at 94°C for 45 seconds, with combined annealing at 55°C for 45 seconds and extension at 72°C for 1 minute. To select the optimal PCR conditions, the number of PCR cycles was varied. The β-actin messenger RNA (mRNA) was used to normalize the amounts of RNA in the cell samples, and the intensities of the resulting PCR bands were used to calculate the ratio of Timp1/β-actin intensities for each cycle. Band intensities were quantified using ImageJ 1.38 software (Image Processing and Analysis in Java, Wayne Rasband, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/). Each PCR was done in triplicate. PCR fragment amplification was confirmed by agarose gel staining with ethidium bromide. The PCR primers were as follows: Timp1 forward 5′ GCTAAAAGGATTCAAGGC 3′; Timp1 reverse 5′ GCACAAGCCTAGATTCCG 3′; Actin forward 5′ CGAGGCCCAGAGCAAGAGAG 3′; Actin reverse 5′ AGGAAGAGGATGCGGCAGTGG 3′.
Western Blot Analysis
Cells of subconfluent cultures were trypsinized, rinsed with PBS, and lysed directly in cold lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 50 mM NaF, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 10 µg/ml aprotinin, and 10 µg/ml leupeptin) for 15 minutes on ice, followed by centrifugation at 14,000 rpm for 15 minutes at 4°C. The supernatant was collected and protein concentration was measure by Bio-Rad protein assay dye reagent concentrate (Bio-Rad, Hercules, CA). Equivalent amounts of protein (80 µg) were separated on 15% sodium dodecyl sulfate-polyacrylamide electrophoresis gel and transferred to polyvinylidene fluoride membranes (Amersham, Piscataway, NJ). Proteins were detected using rabbit polyclonal anti-Timp1 antibody (ABR, Golden, CA) and the signal was detected using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (KPL; Gaithersburg, MD) followed by development using chemiluminescence substrate (SuperSignal West Pico Chemiluminescent Substrate; Pierce Chemical, Rockford, IL).
Immunofluorescence Analysis
Cell lines (2 x 104 per well) were plated on glass coverslips, grown as a monolayer until 70%confluence, washed twice in PBS, fixed in 3.7% paraformaldehyde in PBS for 15 minutes at -20°C, and washed again in PBS. Coverslips were blocked with 1% BSA in PBS (1% BSA-PBS) for 30 minutes at room temperature. Cells were incubated for 1 hour with rabbit anti-Timp1 polyclonal antibody (ABR) at room temperature, washed three times with 1% BSA-PBS, and incubated with an Alexa 488-conjugated antirabbit immunoglobulin G antibody (Molecular Probes, Carlsbad, CA) for 45 minutes and for 5 minutes with 4′-6-diamidino-2-phenylindole. After five washes with PBS, slides were mounted with antifade solution and analyzed in a fluorescence microscope (Carl Zeiss, Göttingen, Germany).
Quantification of DNA Methylation by Methylation-Sensitive Single-Nucleotide Primer Extension
The average methylation at three different CpG sites present in Timp1 promoter was quantified using methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) as described previously [20]. Genomic DNA treated with sodium bisulfate as described by Frommer et al. [21]. Briefly, 6 µg of genomic DNA diluted in 20 µl of deionized water was denatured for 20 minutes at 95°C, and 5 µl of 3 M NaOH, pH 5.2, was added to the DNA and the solution was incubated for 20 minutes at 45°C. Denatured DNA was incubated in a total volume of 250 µl with freshly prepared 3.1 M sodium bisulfite/0.5 mM hydroquinone, pH 5.0, for 16 hours at 55°C under mineral oil. The bisulfite-treated DNA was purified by using a Wizard Mini Preparation Kit (Promega, Madison, WI), according to the manufacturer's instructions and resuspended in 50 µl of deionized water. NaOH was added to a final concentration of 0.3 M; DNA was incubated at room temperature for 10 minutes, followed by precipitation in 350 µl of a solution containing 20 mg/ml of glycogen, 0.3 M of NaOAc (pH 5.2), and 85% of ethanol for 50 minutes at -80°C. Precipitated DNA was resuspended in 40 µl of deionized water. Two microliters of bisulfite-treated DNA was amplified by PCR using the primers as follows: S1, 5′ TGGGTGGATGAGTAATG 3′, and Timp1 SNuPE reverse, 5′ CTAACTAAAAATCCTAATACCTACAA 3′. The PCR conditions were 94°C for 3 minutes, 44 cycles of 94°C for 1 minute, 52°C for 1 minute, and 72°C for 1 minute. The PCR products were separated by electrophoresis through a 2% agarose gel, purified from the gel using QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), and eluted in 40 µl of H2O. The single-nucleotide extensions were obtained adding 4 µl of the template to 6 µl of reaction mixture containing 1x PCR buffer, 1 µM SNuPE primer (S1, S2 5′ TTATTTTTTATTGTGTAGTTTTTGT 3′ or S3 5′ ATTAGAGGTAAGTGGGAGT 3′, one for each reaction), 1 µCi (32P) dCTP or (32P)dTTP, and 1 U of 1:1 Taq/TaqStart antibody (BD Biosciences Clontech, Palo Alto, CA). This mixture was incubated at 95°C for 30 seconds, at 51°C for 30 seconds, and finally at 72°C for 1 minute and then combined with 5 µl of stop solution (95% formamide, 20 mM EDTA [pH 8.0], 0.05% bromophenol blue, and 0.05% xylene cyanol) before being denatured at 94°C for 5 minutes and loaded onto a 15% denaturing polyacrylamide gel (Tris-borate-EDTA buffer, 14.25% acrylamide, 0.75% bis-acrylamide, 7 Murea). Methylation levels were quantified on a PhosphorImager Analysis System (Molecular Dynamics, Sunnyvale, CA). The methylation status of the Timp1 promoter and first exon represents the average methylation levels of three CpG sites.
Plasmid Construction and Transfection
Stable melan-a and Tm5 cell lines overexpressing Timp1 (ma T1S and Tm5 T1S, respectively) were generated by RT-PCR amplification of full-length coding sequence from Tm5 using the gene-specific EcoRI-flanked (underlined) forward primer 5′ TTGAATTCACCACCATGATGGCCCCCTTTGCA 3′ and XhoI-flanked (underlined) reverse primer 5′ TTCTCGAGTCATCGGGCCCCAAGGGATC 3′, digested with EcoRI and XhoI restriction enzymes and subcloning into the EcoRI/XhoI cut pcDNA3.1 vector (Invitrogen). The inserts were sequenced to confirm the desired DNA sequence. Stable transfectants were produced by transfecting with Lipofectamine 2000 Transfection Reagent (Invitrogen) and selected during 15 days in medium containing 2 mg/ml Geneticin (G418; Gibco). Transfection with pcDNA3.1 containing green fluorescence protein (GFP)-coding sequence (ma GFP and Tm5 GFP) was used as control for transfection efficiency. Transfectants were expanded and analyzed for transgene expression using semiquantitative RT-PCR, Western blot, and immunofluorescence analysis, as described above.
Flow Cytometry
Cell cycle analysis was performed to quantify sub-G1 population of cells in suspension. Percentage of cells presenting DNA fragmentation—sub-G1 population—were determined by the CellQuest software using a FACSCalibur cytometer (Becton Dickinson, San Juan, CA) as described previously [22]. Briefly, different cell lines were maintained in suspension (105 cells). After 96 hours, cells were washed, fixed with ice-cold ethanol (70% final concentration), resuspended in 0.5-ml solution containing 50 µg/ml propidium iodide and 20 µg/ml DNAse-free RNAse in PBS, and incubated for 30 minutes at 37°C. Ten thousand events were evaluated per assay.
Colony Formation in Soft Agar
Cells (103 cells per well) were suspended in 3 ml of 0.3% agarose in complete culture medium. This suspension was layered over 3 ml of 0.6% agarose medium base layer in 35-mm-diameter dishes. After 21 days, cells were stained with 2 ml of crystal violet solution and washed extensively with water, and colonies were counted using an inverted bright field microscope at a x2 magnification. This assay was performed in duplicate.
In Vivo Tumorigenesis Assay
Cells were harvested after trypsin treatment of subconfluent monolayers, counted, and then suspended in PBS. Cells (2 x 105) in 0.1 ml of PBS were injected subcutaneously into the flank of syngeneic C57Bl/6 female mice. Tumor growth was monitored by daily caliper measurements. Tumor volumes (mm3) were calculated as follows: (d2 x D) / 2. Each experimental group consisted of five animals.
Experimental Metastasis
Cells of subconfluent cultures were trypsinized, suspended in RPMI with 10%fetal calf serum, spun down, suspended in PBS, counted, and adjusted to the necessary concentration. Cells (2 x 105) in 0.1 ml of PBS were injected into the lateral tail vein of syngeneic C57Bl/6 mice with a 27-gauge needle. The mice were killed 21 days after injection, lungs were surgically removed, and metastatic foci in the lungs were macroscopically scored. Each experimental group consisted of five animals.
Databases
Oncomine Cancer Microarray database http://www.oncomine.org [23] was used to study gene expression of Timp1 in human melanoma, breast cancer, colorectal cancer, lymphoma, and non-small cell lung carcinoma. Only gene expression data obtained from a single study using the same methodology were compared. All data were log-transformed, median was centered per array, and the standard deviation was normalized to one per array [23]. Statistical comparisons in database were done with Student's t-tests and P values were calculated using adjustment for multiple testing and false discoveries as described at http://www.oncomine.org [23]. A gene was considered as overexpressed when its mean value in tumor samples was significantly higher to its mean value in the normal tissue counterpart (P ≤ .05).
Data Analysis
All experiments were repeated for at least two to three times with similar results. One-way analysis of variance (ANOVA) with Tukey multiple comparisons posttest and two-way repeated-measures ANOVA with Bonferroni posttest were performed using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA; www.graphpad.com). P values <.05 were considered statistically significant.
Results
Melanocyte Malignant Transformation Associated with Anoikis Resistance
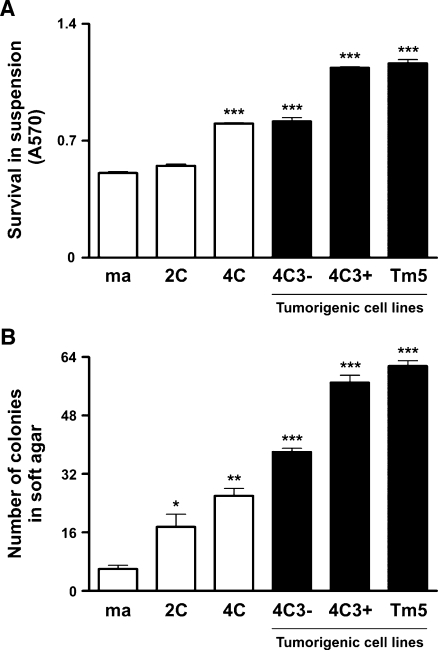
To study a possible correlation between neoplastic transformation and anchorage-independent growth, we analyzed the acquisition of anoikis resistance during murine melanocyte malignant transformation. Substrate adhesion blockade assay was performed by culturing cells in suspension for 96 hours in 1% agarose-coated dishes to prevent cell anchorage, and cell viability was determined by MTT assay. As shown in Figure 1A, melanoma cell lines became 1.6- to 2.5-fold more resistant to anoikis when compared with melan-a cells. In fact, the nontumorigenic 4C cell line also presented augmented anoikis resistance, similar to 4C3- slow-growing melanoma cells. The acquisition of this phenotype was also evaluated by culturing cells in soft agar during 21 days (Figure 1B) and revealed a similar result as the one found in substrate adhesion blockade assay (Figure 1A), showing that melanoma cells survive 6- to 10.3-fold more than the parental lineage. Together, these results show progressive increase in cell survival in anchorage-independent conditions during the first steps of malignant transformation along tumor progression, even after tumor establishment.
Figure 1.
Melanocyte malignant transformation associated with anoikis resistance. (A) Cell lines were maintained in suspension for 96 hours, and cell viability was determined by MTT assay. (B) Cell lines were cultured in soft agar for 21 days, and the number of colonies was counted. ma indicates melan-a cells; 2C and 4C, melan-a cells submitted to two and four substrate adhesion blockade cycles, respectively; 4C3-, slow-growing melanoma cell line; 4C3+, Tm5, fast-growing melanoma cell lines. Bars, SD of the mean of six samples. *P < .05, **P < .01, ***P < .001. P value was determined by one-way ANOVA.
Timp1 Expression Increases during Melanocyte Malignant Transformation and Neoplastic Progression
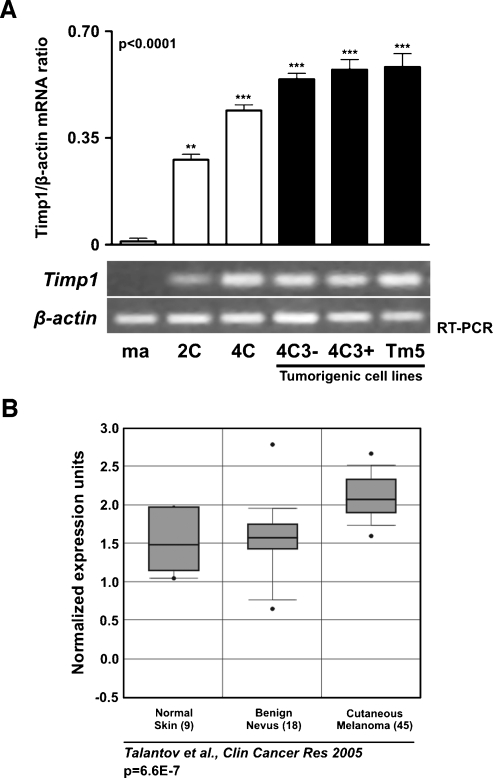
Some studies have shown increases in TIMP1 expression related to a poor prognosis in cancer [24,25]. Using our model, we wished to examine the expression pattern of Timp1 during melanocyte malignant transformation and neoplastic progression. Because Timp1 expression is induced by phorbol esters, and melan-a cells require it for growth, all cells in this study were cultured in the presence of phorbol 12-myristate13-acetate. The expression patterns of Timp1 in the different lineages representing distinct stages of neoplastic development (nontumorigenic 2C and 4C cell lines, tumorigenic slow-growing 4C3-melanoma lineage, and tumorigenic fast-growing 4C3+ and Tm5 melanoma lineages) were analyzed in comparison to those in parental melanocyte lineage melan-a. This analysis was performed in all lineages cultured in adhesion condition (melan-a, 2C, 4C, 4C3-, 4C3+, and Tm5) (Figure 2A) and also in melan-a cells cultured in suspension for 1, 3, 5, and 24 hours (data not shown). Semiquantitative RT-PCR revealed significant up-regulation of Timp1 expression in all six analyzed melan-a-derived cell lines cultured in adhesion conditions compared with that in melan-a cells (Figure 2A). Basal levels of Timp1 expression was only detected in melan-a cells cultured on plastic, whereas no expression was detectable by RT-PCR analysis in melan-a cultured in suspension for different times (data not shown). To compare our results with the findings in human melanoma, Timp1 expression was investigated in human cancer using the publicly available gene expression data, Oncomine Cancer Microarray database (http://www.oncomine.org) [23,26]. We found an augment in Timp1 expression in human melanoma compared with normal skin and benign nevus, their normal counterparts (Figure 2B). Together, these in silico data and our result suggest that Timp1 expression may be involved in melanoma genesis and/or progression (Figure 2A).
Figure 2.
Increased expression of Timp1 throughout melanocyte malignant transformation. (A) Timp1 expression was analyzed in melan-a cells and melan-a-derived cell lines by RT-PCR. The graph represents the Timp1/β-actin mRNA ratio for each sample obtained by semiquantitative RT-PCR for each cycle (P < .0001). The β-actin expression was used to normalize the amounts of mRNA in cell samples, and the intensities of resulting PCR bands were used to calculate the ratio of Timp1/β-actin intensities. Units on y-axis are arbitrary. (B) Box plots showing increased expression of TIMP1 in human melanoma compared with normal skin and benign nevus [24]. The y-axis represents normalized expression. Shaded boxes represent the interquartile range (25th–75th percentile). Whiskers represent the 10th to 90th percentile. The bars denote the median. Reference refers to microarray reference database. P values for correlation are shown below. ma indicates melan-a cells; 2C and 4C, melan-a cells submitted to two and four substrate adhesion blockade cycles, respectively; 4C3-, slow-growing melanoma cell line; 4C3+, Tm5, fast-growing melanoma cell lines. *P < .05, **P < .01, ***P < .001. P value was determined by one-way ANOVA.
Progressive Timp1 Promoter Demethylation Associated to Timp1 Expression along Neoplastic Progression
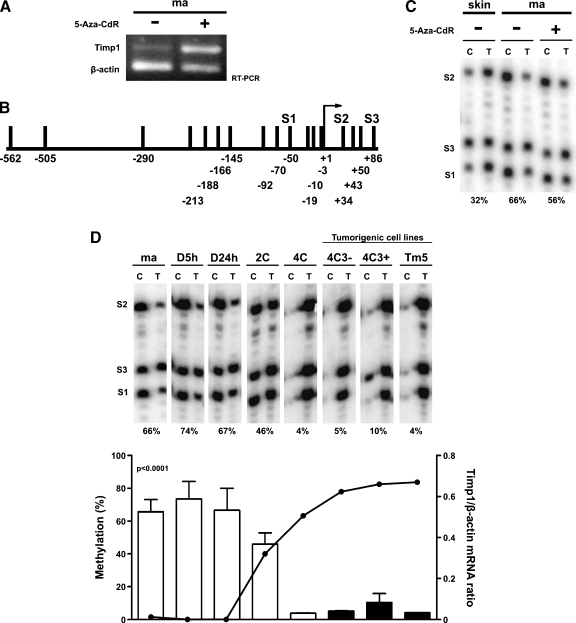
Given that previous data from our laboratory showed a significant alteration in the global DNA methylation level in this model along neoplastic transformation ([27] and data not shown), and DNA methylation was demonstrated as being a possible regulatory mechanism of Timp1 expression [16], we tested whether DNA demethylation induced by 5-Aza-CdR would induce Timp1 expression in melan-a cells. After treatment, Timp1 expression in melan-a cells was found to be highly elevated (Figure 3A). For that reason, we wished to confirm whether DNA methylation has a correlation with Timp1 expression in this model. Thus, using Ms-SNuPE assay, the average methylation at CpG dinucleotides within Timp1 promoter (S1) and first exon (S2 and S3; Figure 3B) was analyzed. The DNA methylation level in the promoter and exonic non-CpG island of Timp1 was quantified in all lineages, including ma-5-Aza-CdR cells, melan-a cells cultured in suspension for 5 and 24 hours, and nontreated melan-a-derived lineages cultured in adhesion (Figure 3, C and D). Whereas melan-a melanocytes presented the analyzed region methylated, nontumorigenic 4C cell line and all melanoma cell lines (4C3-, 4C3+, and Tm5) showed the same region demethylated. Curiously, nontumorigenic 2C cells showed an intermediate grade of demethylation in these CpG sites compared with parental cell line (melan-a) and melan-a-derived melanomas (4C3-, 4C3+, and Tm5). The Timp1 gene methylation level correlates strongly and inversely with its expression (Figure 3D).
Figure 3.
Timp1 promoter demethylation is associated with Timp1 expression along melanoma genesis. (A) Analysis of Timp1 expression by RT-PCR after in vitro treatment of melan-a cells with 5 µM5-Aza-CdR for 2 days. Every day, the medium was changed, and no significant cell death was observed between the two groups. ma- indicates nontreated melan-a cells (positive control); ma+, melan-a cells treated with 5-Aza-CdR. (B) Schematic representation of CpG sites location in Timp1 promoter and first exon. S1, S2, and S3 indicate CpG dinucleotide analyzed by Ms-SNuPE. Arrow indicates transcriptional start site. (C and D) Quantitative methylation analysis of three CpG sites in the promoter and first exon of Timp1 gene in various DNA samples using Ms-SNuPE. The percentage of methylation at each CpG site was determined by the C:T signal ratio (left axis). The average methylation of the three CpG sites is shown at the bottom of each panel. Methylation levels were quantified on a PhosphorImager Analysis System. In D, the curve in the graph represents Timp1/β-actin mRNA ratio (right axis). skin indicates DNA extracted from mouse normal skin; ma, melan-a cells; ma-5-Aza-CdR, melan-a cells treated with 5-Aza-CdR; D5h, D24h, melan-a cells cultured in suspension for 5 and 24 hours, respectively; 2C, 4C, melan-a cells submitted to two and four substrate adhesion, blockade cycles, respectively; 4C3-, slow-growing melanoma cell line; 4C3+, Tm5, fast-growing melanoma cell lines. P < .0001. P value was determined by one-way ANOVA.
Timp1 Overexpression Does Not Induce In Vitro Cell Proliferation
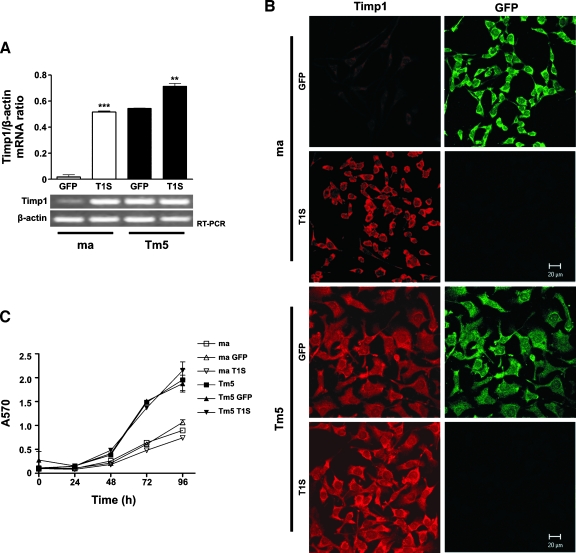
The progressive increase in Timp1 expression observed during melan-a malignant transformation suggests that its expression might be functionally related to neoplastic development. Considering the relationship between Timp1 expression and acquisition of anoikis-resistant phenotype [12], we tested the functional significance of increased Timp1 expression during neoplastic progression by transfecting Timp1 (T1S) in melan-a and Tm5 cells. Timp1 overexpression in transfected clones was confirmed by RT-PCR and immunofluorescence microscopy (Figure 4, A and B).
Figure 4.
Timp1 overexpression does not induce in vitro cell proliferation both in melan-a melanocytes and Tm5 melanoma cells. Timp1 expression was analyzed by RT-PCR (A) and indirect immunofluorescence (B) in melan-a and Tm5 cells transfected with Timp1. The graphics in A represent the Timp1/β-actin expression ratio for each sample obtained by RT-PCR. The β-actin expression was used to normalize the amounts of RNA in cell samples, and the intensities of the resulting PCR bands were used to calculate the ratio of Timp1/β-actin intensities. Units on y-axis are arbitrary. (B) Indirect immunofluorescence using Timp1-specific antibody (ABR). (C) In vitro cell proliferation was quantified by MTT assay. GFP-transfected cells were used as transfection control. ma indicates melan-a cells; Tm5, fast-growing melanoma cell line; GFP, GFP-transfected cells; T1S, Timp1-transfected cells. Bars, SD of the mean of five samples. **P < .01, ***P < .001. P value was determined by one-way ANOVA.
Homeostasis of cell number in tissues depends on the balance between proliferative and apoptotic indexes. Because Timp1 may regulate proliferation in some contexts [9,12], we asked whether increased Timp1 expression observed along malignant transformation was related to an altered proliferation rate. Accordingly, proliferation index was estimated by MTT assay in different times and revealed a very similar proliferation rate in Timp1-overexpressing cells and their controls, both in nontumorigenic and tumorigenic lineages (Figure 4C). These data showed that the augmented Timp1 expression observed during neoplastic progression (Figure 2) does not affect cell proliferation in this model.
Melan-a Melanocytes Overexpressing Timp1 Become Anoikis-Resistant
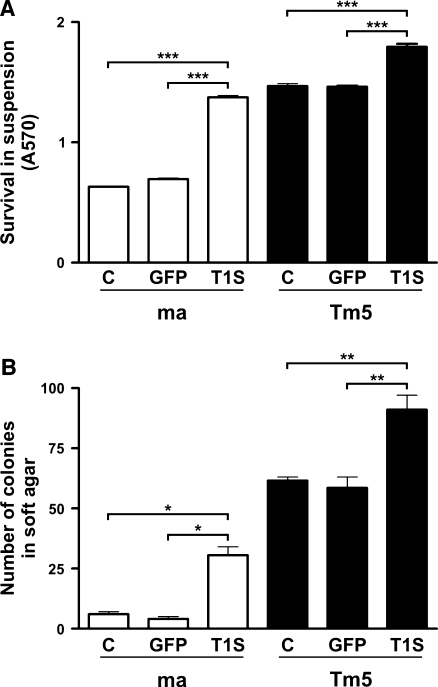
Apoptosis inhibition by Timp1 may result from its ability to stabilize cell-ECM interactions by inhibiting MMPs. In this context, Timp1 would not protect cells against apoptosis induced by loss of cell-substrate interactions. Then, melan-a, ma GFP, ma T1S, Tm5, Tm5 GFP, and Tm5 T1S cells were cultured in suspension conditions. Trypan blue staining showed 100%melan-a cell viability up to 24 hours in suspension (data not shown). After 96 hours, cell viability was determined by MTT assay, showing that Timp1-overexpressing cells remained more viable in suspension compared with their controls (Figure 5A). Ma T1S cells became resistant to anoikis when compared with their anoikis-sensitive controls (2.2-fold more than ma and 2-fold more than ma GFP). This resistance was also increased in Timp1-overexpressing melanoma cells (1.2-fold more than Tm5 and 1-fold more than Tm5 GFP), a lineage already resistant to anoikis (Figure 1). Anoikis-resistant phenotype was also evaluated by culturing cells in soft agar. As shown in Figure 5B, seven-fold ma T1S cells remained viable when compared with the control (ma and ma GFP cells), even after 21 days of being cultured in soft agar. In melanoma, the super expression of Timp1 was responsible for maintaining more cells viable in anchorage-independent condition (1.5-fold) when compared with its controls. Interestingly, maT1S cells were able to survive in soft agar as its tumorigenic counterpart. Thus, Timp1 expression can prevent anoikis in melan-a cells.
Figure 5.
Melan-a melanocytes overexpressing Timp1 become anoikis-resistant. Timp1-overexpressing melan-a and Tm5 cells were maintained in suspension for 96 hours. Cell viability (A) was determined by MTT assay in triplicate. In B, melan-a and Tm5 cells were cultured in soft agar medium for 21 days, and the number of colonies was counted. ma indicates melan-a cells; Tm5, fast-growing melanoma cell line; C, nontransfected cells; GFP, GFP-transfected cells; T1S, Timp1-transfected cells. Bars, SD of the mean of analyzed samples. *P < .05, **P < .01, ***P < .001. P value was determined by one-way ANOVA.
Timp1 Overexpression Is Related to In Vivo Aggressiveness Phenotype in Murine and Human Melanoma
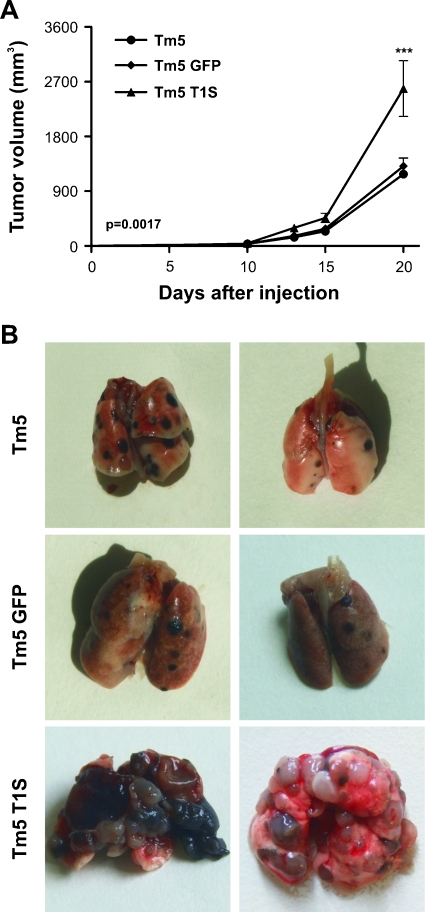
Because ma T1S cells were able to grow independent of anchorage in semisolid agar and this ability became a classic assay for in vitro evaluation of malignant transformation of various cultured cell lines [28], we examined whether Timp1 overexpression was capable to cause acquisition of a fully transformed malignant phenotype in these cells. Even after injecting 107 cells subcutaneously in the flank of syngeneic C57Bl/6 mice, no tumor development was observed in animals transplanted with melan-a, ma-5-aza-CdR, ma GFP, or ma T1S cells (data not shown). When melanoma cells overexpressing Timp1 (2 x 105 cells per animal) were injected subcutaneously or intravenously in mice (5 animals per group), the latency time for tumor appearance decreased, tumor volume increased (Figure 6A), and metastatic potential had a substantial augment (Figure 6B).
Figure 6.
Timp1 overexpression is related to in vivo aggressiveness phenotype in murine and human melanoma. (A) Tumorigenicity assay in vivo. Mice were subcutaneously injected with 2 x 105 Tm5 cells. Tumor growth was monitored by daily caliper measurements. (B) Experimental metastasis assay. Tm5 cells (2 x 105) were injected via caudal vein. Mice were killed 23 days later, and their lungs were removed. Each experimental group consisted of five animals. Tm5 indicates fast-growing melanoma cell line; Tm5 GFP, GFP-transfected Tm5 cells; Tm5 T1S, Timp1-transfected Tm5 cells. Bars, SD of the mean of five animals per group. ***P < .001. P value was determined by two-way ANOVA.
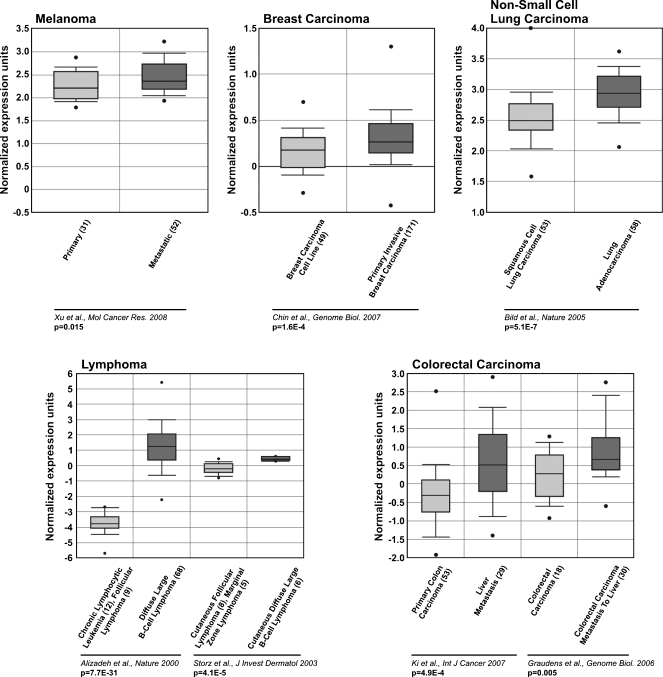
Because Timp1 overexpression in murine melanoma cells cause enhancement in tumor malignancy (Figure 6, A and B) and its increase is related with negative prognosis in many human cancers [10,11], we used Oncomine Cancer Microarray database [23] to investigate the expression of TIMP1 in human metastatic melanoma compared with primary melanoma [29]. Also, we evaluated TIMP1 expression in five other human malignant tumor types and their primary tumor counterparts [30–35]. Increased expression of TIMP1 was seen in all metastases analyzed when compared with primary tumors (Figure 7). It is important to note that the finding of increased expression levels of Timp1 in our direct analysis of tumor samples (Figure 6, A and B), associated with unfavorable clinical characteristics, is consistent with studies of microarray gene expression data in different human tumor types analyzed, including melanoma (Figure 7).
Figure 7.
Association of TIMP1 expression with prognosis. Representative data are shown across multiple independently published microarray studies as indicated. The y-axis represents normalized expression. Shaded boxes represent the interquartile range (25th–75th percentile). Whiskers represent the 10th to 90th percentile. Bars, median. P values for correlation are shown below.
Taken these data into account, TIMP1 might be a new useful marker for cancer progression because increased levels of its protein has been found in serum of patients with different aggressive malignancies [36,37], including melanoma [38]. Given that, we analyzed the level of Timp1 in supernatant of all cell lines studied in this work. Surprisingly, no detectable level of Timp1 protein was found by Western blot analysis (data not shown). This result may be explained by the fact that coimmunoprecipitation of Timp1 with β1-integrin were observed in all lineages studied here (data not shown), suggesting the involvement of Timp1 protein in intracellular signaling pathway, leading to anoikis-resistant phenotype.
Discussion
It is well known that both cell-cell and cell-ECM interactions play crucial roles in determining melanocyte appropriate functions in the epidermis, in humans, or in the dermis, in mice. Loss of these interactions is observed during melanoma progression, and human melanocytes are found ectopically in the dermis not only in melanoma lesions but also in dysplastic nevi, considered a premalignant lesion. The presence of normal cells in an inappropriate environment prevents their association with the ECM and then disrupts survival signals resulting in anoikis [3]. This kind of apoptosis is believed to contribute to the maintenance of proper cell number, geographic architecture, and position in epithelia organs. Apart from its relevance in physiological conditions, loss of anchorage dependency is believed to be one of the key mechanisms to tumor progression and malignant transformation because the cells are able to proliferate detached and spread from the primary tumor without undergoing anoikis [39,40]. The distinct ability of tumor cells to grow in suspension, semisolid media, or multicellular spheroid culture has been frequently linked to anchorage-independent growth [41] and, more recently, to anchorage independent survival [42]. Despite the strong correlation between the resistance of transformed cells to anoikis and their ability to form tumors in vivo, direct evidence for a causal relationship between them remains unknown.
To explain some of these interrelationships in a more direct manner, we established an in vitro murine carcinogenesis model [17], which specifically favors the acquisition of anoikis resistance of a nontumorigenic melanocyte lineage, melan-a [18]. In this model, the sequential exposure of melan-a cells to substrate anchorage impediment cycles leads to the establishment of cell lines with a markedly reduced susceptibility to anoikis (Figure 1). Melan-a cells sublines 2C and 4C became progressively more resistant to anoikis, although these lineages were not able to form tumors in vivo. Melanoma cell lines (4C3-, 4C3+, and Tm5), obtained by limiting dilution after a new deadhesion cycle of 4C cells, are also anoikis-resistant (Figure 1) and grow as tumors when injected subcutaneously in syngeneic mice, with different latency times for tumor appearance [17]. Previous cDNA microarray analysis revealed an up-regulation of Timp1 expression in Tm5 melanoma cells when compared with their parental cell line, melan-a. These results were initially unexpected considering the well-established function of Timp1 in the inhibition of MMPs-mediated ECM degradation, tumor cell invasion, and metastasis formation in different tumors [43–46] as well as in melanoma [47–52].
Besides the number of studies showing antitumor effects of Timp1, there are also different works reporting TIMP1 protumor effects in different tumor types [24,53–57], including melanoma [58]. Actually, TIMP1 is upregulated in many cancer types, and a high level of TIMP1 correlates with a poor prognosis [16,24,36,38,59–62]. In addition, studies have suggested that TIMP1 may also carry predictive information on response to treatment [63,64].
These contradictory events may be due to other important functions of Timp1. It has been shown that Timp1 induces proliferation and apoptosis in a wide range of cell types by mechanisms that are apparently independent of MMPs [12]. Previously, Li et al. [13] showed that TIMP1 expression protects human breast epithelial cells from different kinds of apoptosis, including anoikis. In the present study, we have shown during melanocyte malignant transformation a progressive upregulation of Timp1 even in melanocyte lineages corresponding to premalignant lesions (as 2C and 4C cell lines; Figure 2A), in parallel to a successive increase of anoikis-resistant phenotype (Figure 1). In fact, increase in TIMP1 expression is also seen during human melanoma genesis [26], as showed in Figure 2B.
It was shown that cancer genome is globally hypomethylated when compared with their normal counterparts [65], and changes in DNA methylation pattern, particularly in the promoter CpG islands, can also occur [66]. In our model, we observed a significant global hypermethylation when melanocytes are maintained in suspension and a progressive global DNA hypomethylation during tumor progression ([27], and data not shown). These results suggest that there are changes in the methylation pattern of melan-a cells during the substrate adhesion blockade cycles, which might be important for the expression of some genes and silencing of others, resulting to a new expression gene pattern related to the malignant phenotype. Taking these changes into account and considering the fact that Timp1 expression can be regulated by DNA methylation [52], Timp1 expression in melan-a cells was evaluated after treatment with 5-Aza-CdR (Figure 3A). The strong reexpression of Timp1 led us to use the Ms-SNuPE method to examine the methylation status in CpG sites in Timp1 promoter and in the first exon of melan-a cells treated and nontreated with 5-Aza-CdR (Figure 3C) and melan-a-derived lineages (Figure 3D). We demonstrated that Timp1, whose promoter and first exon present CpG-rich regions instead of CpG islands (Figure 3B), has its expression regulated by DNA methylation because our data show the augment of Timp1 expression in melan-a cells, occurring after 5-Aza-CdR treatment (Figure 3C). This result might explain a possible inverse correlation between promoter demethylation and Timp1 expression during melanocyte malignant transformation (Figure 3D). It is possible that other CpG sites are demethylated earlier and have more important transcription regulatory function than those studied. Whereas most studies report promoter CpG islands hypermethylation leading to gene silencing [67], some recent studies have shown that DNA methylation can also affect the transcription of genes whose 5′ untranslated region had low CpG density and, in case of these non-CpG islands genes, the correlation between methylation and transcription is inverse [68]. Aberrant expression of genes that do not contain bona fide CpG islands in their promoters, which is approximately 40% of human genes [69,70]. In addition, these works have shown correlations between methylation of non-CpG islands and tissue-specific expression. As the demethylation process seemed to be stable once initiated, its function would seem to be an enhancement of the amount of transcription of Timp1 in these cells to facilitate the anoikis resistance during the malignant transformation process.
To determine whether increased Timp1 expression would be associated with the increase of anoikis resistance and, consequently, with the acquisition of a fully transformed malignant phenotype as suggested by observations associating Timp1 with potentiation of carcinogenesis and with poor clinical outcome, we took a genetic approach using a transfection assay to overexpress Timp1 in two different lineages, the nontumorigenic melan-a melanocytes and the tumorigenic Tm5 melanoma cells (Figure 4). Results from the current study provide insight into Timp1 function as an inhibitor of cell death induced by anchorage-independent growth conditions and highlight the relevance of this inhibition once tumors have already formed. In melan-a cells, the overexpression of Timp1 resulted in an anoikis-resistant lineage capable not only of surviving but also of growing when they were cultured in suspension, even after 96 hours, and in soft agar medium for 21 days (Figure 5). This result is supported by observations suggesting that Timp1 could be an important contributor to epithelial carcinogenesis, especially during early stages of neoplastic progression [24]. In addition, these authors showed that Timp1 can inhibit tissue gelatinolytic activity in tumor stroma without inhibiting tumor progression or development of metastases.
Overexpression of Timp1 in malignant Tm5 melanoma augmented the anoikis resistance when cultured in suspension and increased the malignancy of these cells in vivo, suggesting that anoikis inhibition by Timp1 may be critical for anchorage-independent viability of disseminating cells during tumor cell metastasis (Figure 6). Taking into account that Tm5 T1S cells do not present an increase in the proliferation rate (Figure 4) or in the invasive potential in vitro (data not shown) compared with their control (Tm5 GFP), we believe that Timp1 has a role in tumor-promoting, ever since these cells showed a higher capability to survive in the absence of normal matrix components (Figure 5), resulting in the increase of tumor volume (Figure 6A). This fact also explains the increase in the number of metastatic foci in lungs when animals were inoculated with these cells (Figure 6B). This explanation was supported by Zhu et al. [6], who demonstrated that the selection in vitro of cells resistant to anoikis increases the metastatic potential of melanoma cells in a murine experimental model. In fact, many studies support the importance of anoikis resistance as a crucial phenotype for metastatic cells because tumor cells that intravasate into the circulation and extravasate into secondary tissue sites are either deprived of matrix or exposed to foreign matrix components [71]. Our data provide evidence that Timp1 expression is required for the acquisition of an anoikis-resistant phenotype in nontumorigenic melanocyte lineages and is implicated in a more aggressive phenotype of tumorigenic melanocyte cell lines. These data and ours are sustained by those reports showing that patients submitted to clinical trials with synthetic metalloproteinase inhibitors fail to show any alterations in overall survival or time to progression compared with placebo group [72,73]. By a meta-analysis approach, TIMP1 was found significantly overexpressed in metastatic melanoma compared with nonmetastatic melanoma [29], likewise in other human invasive tumor types, with TIMP1 overexpression correlated with unfavorable clinical characteristics [30–35].
In our studies, melanocyte malignant transformation was obtained after sequential cycles of substrate adhesion blockade. Increased levels of Timp1 seem to confer anoikis resistance in all cell lines derived from melanocytes anchorage blockade (premalignant and malignant cell lines). Considering that β1-integrins, the main receptor for ECM components, have already been described to take part of a cell surface complex with CD63 and TIMP1, activating integrin-mediated cell survival signaling pathways in human breast epithelial cells [14], it is reasonable suppose that alterations in cell-matrix interactions might interfere in Timp1 signaling pathways. Preliminary immunoprecipitation assay suggested Timp1 association with β1-integrin in all lineages studied in this work. Although the molecular mechanism by which the connection between Timp1 and β1-integrin induces cell survival in those cells remains to be defined, we have found that Timp1 is a potent inhibitor of anoikis, in which Timp1 expression is required for the acquisition of an anoikis-resistant phenotype in nontumorigenic melanocyte lineages, and is implicated in a more aggressive phenotype of tumorigenic melanocyte cell lines. Accordingly, in our model, the anoikis resistance is needed to melanoma malignancy but is insufficient for acquisition of tumorigenic competence by melanocytes cells in vivo.
All of these data support a critical function of TIMP1 and highlight the complex effects that this molecule can exert along tumorigenesis. As discussed elsewhere [9,12,74], there are many ways that could explain the TIMP1 protumor effects, and all of them should be considered and further investigated in our model.
In conclusion, these findings provide evidence that Timp1 protects murine melanoma cells from anoikis, and the increase in Timp1 expression along malignant transformation is associated with progressive gene demethylation. Consequently, the expression of Timp1 resulted in increased anoikis resistance, indicating that this molecule has protumor functions in this model, but alone is not capable of inducing a fully transformed malignant phenotype of nontumorigenic cells. Considering the elevated expression of Timp1 since the early phases of melanoma genesis associated with anoikis resistance, it is reasonable suppose that Timp1 expression might be a valued marker for melanocyte malignant transformation.
Acknowledgments
The authors thank Amanda Gonçalves dos Santos Silva for her help in some techniques.
Footnotes
This study was supported by Fundaàào de Amparo à Pesquisa do Estado de Sào Paulo (FAPESP), grant 06/61293-1 (M.G.J.).
References
- 1.Sepulveda JL, Gkretsi V, Wu C. Assembly and signaling of adhesion complexes. Curr Top Dev Biol. 2005;68:183–225. doi: 10.1016/S0070-2153(05)68007-6. [DOI] [PubMed] [Google Scholar]
- 2.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 3.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Díaz-Montero CM, McIntyre BW. Acquisition of anoikis resistance in human osteosarcoma cells. Eur J Cancer. 2003;39:2395–2402. doi: 10.1016/s0959-8049(03)00575-6. [DOI] [PubMed] [Google Scholar]
- 5.Grossmann J. Molecular mechanisms of “detachment-induced apoptosis—anoikis”. Apoptosis. 2002;7:247–260. doi: 10.1023/a:1015312119693. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Z, Sanchez-Sweatman O, Huang X, Wiltrout R, Khokha R, Zhao Q, Gorelik E. Anoikis and metastatic potential of Cloudman S91 melanoma cells. Cancer Res. 2001;61:1707–1716. [PubMed] [Google Scholar]
- 7.Chiarugi P, Giannoni E. Anoikis: a necessary death program for anchorage-dependent cells. Biochem Pharmacol. 2008;76:1352–1364. doi: 10.1016/j.bcp.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 8.Nagase H, Woessner JF., Jr Matrix metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 9.Lambert E, Dassé E, Haye B, Petitfrère E. TIMPs as multifacial proteins. Crit Rev Oncol Hematol. 2004;49:187–198. doi: 10.1016/j.critrevonc.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy K, Maguire T, McGreal G, McDermott E, O'Higgins N, Duffy MJ. High levels of tissue inhibitor of metalloproteinase-1 predict poor outcome in patients with breast cancer. Int J Cancer. 1999;84:44–48. doi: 10.1002/(sici)1097-0215(19990219)84:1<44::aid-ijc9>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 11.Murashige M, Miyahara M, Shiraishi N, Saito T, Kohno K, Kobayashi M. Enhanced expression of tissue inhibitors of metalloproteinases in human colorectal tumors. Jpn J Clin Oncol. 1996;26:303–309. doi: 10.1093/oxfordjournals.jjco.a023237. [DOI] [PubMed] [Google Scholar]
- 12.Chirco R, Liu XW, Jung KK, Kim HR. Novel functions of Timps in cell signaling. Cancer Metastasis Rev. 2006;25:99–113. doi: 10.1007/s10555-006-7893-x. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Fridman R, Kim HR. Tissue inhibitor of metalloproteinase-1 inhibits apoptosis of human breast epithelial cells. Cancer Res. 1999;59:6267–6275. [PubMed] [Google Scholar]
- 14.Jung KK, Liu XW, Chirco R, Fridman R, Kim HR. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006;25:3934–3942. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Orucevic A, Li M, Gorelik E. Nitric oxide NO, methylation and TIMP-1 expression in BL6 melanoma cells transfected with MHC class I genes. Clin Exp Metastasis. 2000;18:329–335. doi: 10.1023/a:1010867618014. [DOI] [PubMed] [Google Scholar]
- 17.Oba-Shinjo SM, Correa M, Ricca TI, Molognoni F, Pinhal MA, Neves IA, Marie SK, Sampaio LO, Nader HB, Chammas R, et al. Melanocyte transformation associated with substrate adhesion impediment. Neoplasia. 2006;8:231–241. doi: 10.1593/neo.05781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett DC, Cooper PJ, Hart IR. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer. 1987;39:414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 19.Mosmann T. Rapid colorimetric assay for growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalgo ML, Jones PA. Quantitative methylation analysis using methylation-sensitive single-nucleotide primer extension Ms-SNuPE. Methods. 2002;27:128–133. doi: 10.1016/s1046-2023(02)00064-6. [DOI] [PubMed] [Google Scholar]
- 21.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supiot S, Gouard S, Charrier J, Apostolidis C, Chatal JF, Barbet J, Davodeau F, Cherel M. Mechanisms of cell sensitization to a radioimmunotherapy by doxorubicin or paclitaxel in multiple myeloma cell lines. Clin Cancer Res. 2005;11:7047s–7052s. doi: 10.1158/1078-0432.CCR-1004-0021. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Gosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhee JS, Diaz R, Korets L, Hodgson JG, Coussens LM. TIMP-1 alters susceptibility to carcinogenesis. Cancer Res. 2004;64:952–961. doi: 10.1158/0008-5472.can-03-2445. [DOI] [PubMed] [Google Scholar]
- 25.Hawthorn L, Stein L, Varma R, Wiseman S, Lorre T, Tan D. TIMP1 and SERPIN-A overexpression and TFF3 and CRABP1 underexpression as biomarkers for papillary thyroid carcinoma. Head Neck. 2004;26:1069–1083. doi: 10.1002/hed.20099. [DOI] [PubMed] [Google Scholar]
- 26.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 27.Campos ACE, Molognoni F, Melo FH, Galdieri LC, Carneiro CR, D'Almeida V, Correa M, Jasiulionis MG. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia. 2007;9:1111–1121. doi: 10.1593/neo.07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frisch SM. E1a induces the expression of epithelial characteristics. J Cell Biol. 1994;127:1085–1096. doi: 10.1083/jcb.127.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP, Wagner SN, Ramaswamy S, Mesirov JP, Hynes RO. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6:760–769. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, Costa JL, Pinder SE, van de Wiel MA, Green AR, et al. High-resolution a CGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 32.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 33.Storz MN, van de Rijn M, Kim YH, Mraz-Gernhard S, Hoppe RT, Kohler S. Gene expression profiles of cutaneous B cell lymphoma. J Invest Dermatol. 2003;120:865–870. doi: 10.1046/j.1523-1747.2003.12142.x. [DOI] [PubMed] [Google Scholar]
- 34.Ki DH, Jeung HC, Park CH, Kang SH, Lee GY, Lee WS, Kim NK, Chung HC, Rha SY. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121:2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 35.Graudens E, Boulanger V, Mollard C, Mariage-Samson R, Barlet X, Grémy G, Couillault C, Lajémi M, Piatier-Tonneau D, Zaborski P, et al. Deciphering cellular states of innate tumor drug responses. Genome Biol. 2006;7:R19. doi: 10.1186/gb-2006-7-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrohl AS, Christensen IJ, Pedersen AN, Jensen V, Mouridsen H, Murphy G, Foekens JA, Brunner N, Holten-Andersen MN. Tumor tissue concentrations of the proteinase inhibitors tissue inhibitor of metalloproteinases-1 TIMP-1 and plasminogen activator inhibitor type 1 PAI-1 are complementary in determining prognosis in primary breast cancer. Mol Cell Proteomics. 2003;2:164–172. doi: 10.1074/mcp.M300019-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Kossakowska AE, Urbanski SJ, Edwards DR. Tissue inhibitor of metalloproteinases-1 (TIMP-1) RNA is expressed at elevated levels in malignant non-Hodgkin's lymphomas. Blood. 1991;77:2475–2481. [PubMed] [Google Scholar]
- 38.Yoshino Y, Kageshita T, Nakajima M, Funakubo M, Ihn H. Clinical relevance of serum levels of matrix metallopeptidase-2, and tissue inhibitor of metalloproteinase-1 and -2 in patients with malignant melanoma. J Dermatol. 2008;35:206–214. doi: 10.1111/j.1346-8138.2008.00446.x. [DOI] [PubMed] [Google Scholar]
- 39.Grossmann J, Walther K, Artinger M, Kiessling S, Schölmerich J. Apoptotic signaling during initiation of detachment-induced apoptosis (“anoikis”) of primary human intestinal epithelial cells. Cell Growth Differ. 2001;12:147–155. [PubMed] [Google Scholar]
- 40.Freedman VH, Shin SI. Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell. 1974;3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 41.Rak J, Mitsuhashi Y, Sheehan C, Krestow JK, Florenes VA, Filmus J, Kerbel RS. Collateral expression of proangiogenic and tumorigenic properties in intestinal epithelial cell variants selected for resistance to anoikis. Neoplasia. 1999;1:23–30. doi: 10.1038/sj.neo.7900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meredith JE, Jr, Schwartz MA. Integrins, adhesion and apoptosis. Trends Cell Biol. 1997;7:146–150. doi: 10.1016/S0962-8924(97)01002-7. [DOI] [PubMed] [Google Scholar]
- 43.Elezkurtaj S, Kopitz C, Baker AH, Perez-Cantó A, Arlt MJ, Khokha R, Gansbacher B, Anton M, Brand K, Krüger A. Adenovirus-mediated overexpression of tissue inhibitor of metalloproteinases-1 in the liver: efficient protection against T-cell lymphoma and colon carcinoma metastasis. J Gene Med. 2004;6:1228–1237. doi: 10.1002/jgm.637. [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi K, Ogata Y, Nagase H, Shirouzu K. Inhibition of liver metastasis from orthotopically implanted colon cancer in nude mice by transfection of the TIMP-1 gene into KM12SM cells. Surg Today. 2001;31:791–798. doi: 10.1007/s005950170049. [DOI] [PubMed] [Google Scholar]
- 45.Ikenaka Y, Yoshiji H, Kuriyama S, Yoshii J, Noguchi R, Tsujinoue H, Yanase K, Namisaki T, Imazu H, Masaki T, et al. Tissue inhibitor of metalloproteinases-1 (TIMP-1) inhibits tumor growth and angiogenesis in the TIMP-1 transgenic mouse model. Int J Cancer. 2003;105:340–346. doi: 10.1002/ijc.11094. [DOI] [PubMed] [Google Scholar]
- 46.Yamazaki M, Akahane T, Buck T, Yoshiji H, Gomez DE, Schoeffner DJ, Okajima E, Harris SR, Bunce OR, Thorgeirsson SS, et al. Long-term exposure to elevated levels of circulating TIMP-1 but not mammary TIMP-1 suppresses growth of mammary carcinomas in transgenic mice. Carcinogenesis. 2004;25:1735–1746. doi: 10.1093/carcin/bgh181. [DOI] [PubMed] [Google Scholar]
- 47.De Lorenzo MS, Ripoll GV, Yoshiji H, Yamazaki M, Thorgeirsson UP, Alonso DF, Gomez DE. Altered tumor angiogenesis and metastasis of B16 melanoma in transgenic mice overexpressing tissue inhibitor of metalloproteinases-1. In Vivo. 2003;17:45–50. [PubMed] [Google Scholar]
- 48.Shi Y, Parhar RS, Zou M, Al-Mohanna FA, Paterson MC. Gene therapy of melanoma pulmonary metastasis by intramuscular injection of plasmid DNA encoding tissue inhibitor of metalloproteinases-1. Cancer Gene Ther. 2002;9:126–132. doi: 10.1038/sj.cgt.7700426. [DOI] [PubMed] [Google Scholar]
- 49.Khokha R. Suppression of the tumorigenic and metastatic abilities of murine B16-F10 melanoma cells in vivo by the overexpression of the tissue inhibitor of the metalloproteinases-1. J Natl Cancer Inst. 1994;86:299–304. doi: 10.1093/jnci/86.4.299. [DOI] [PubMed] [Google Scholar]
- 50.Xu F, Carlos T, Li M, Sanchez-Sweatman O, Khokha R, Gorelik E. Inhibition of VLA-4 and up-regulation of TIMP-1 expression in B16BL6 melanoma cells transfected with MHC class I genes. Clin Exp Metastasis. 1998;16:358–370. doi: 10.1023/a:1006569631330. [DOI] [PubMed] [Google Scholar]
- 51.Airola K, Karonen T, Vaalamo M, Lehti K, Lohi J, Kariniemi AL, Keski-Oja J, Saarialho-Kere UK. Expression of collagenases-1 and -3 and their inhibitors TIMP-1 and -3 correlates with the level of invasion in malignant melanomas. Br J Cancer. 1999;80:733–743. doi: 10.1038/sj.bjc.6690417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacDougall JR, Bani MR, Lin Y, Muschel RJ, Kerbel RS. “Proteolytic switching”: opposite patterns of regulation of gelatinase B and its inhibitor TIMP-1 during human melanoma progression and consequences of gelatinase B overexpression. Br J Cancer. 1999;80:504–512. doi: 10.1038/sj.bjc.6690385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kopitz C, Gerg M, Bandapalli OR, Ister D, Pennington CJ, Hauser S, Flechsig C, Krell HW, Antolovic D, Brew K, et al. Tissue inhibitor of metalloproteinases-1 promotes liver metastasis by induction of hepatocyte growth factor signaling. Cancer Res. 2007;67:8615–8623. doi: 10.1158/0008-5472.CAN-07-0232. [DOI] [PubMed] [Google Scholar]
- 54.Goss KJ, Brown PD, Matrisian LM. Differing effects of endogenous and synthetic inhibitors of metalloproteinases on intestinal tumorigenesis. Int J Cancer. 1998;78:629–635. doi: 10.1002/(sici)1097-0215(19981123)78:5<629::aid-ijc17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 55.Guedez L, McMarlin AJ, Kingma DW, Bennett TA, Stetler-Stevenson M, Stetler-Stevenson WG. Tissue inhibitor of metalloproteinase-1 alters the tumorigenicity of Burkitt's lymphoma via divergent effects on tumor growth and angiogenesis. Am J Pathol. 2001;158:1207–1215. doi: 10.1016/S0002-9440(10)64070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kopitz C, Gerg M, Gansbacher B, Krüger A. Plasminogen activator inhibitor-2 but not cystatin C inhibits the pro-metastatic activity of tissue inhibitor of metalloproteinases-1 in the liver. Hum Gene Ther. 2008;19:1039–1049. doi: 10.1089/hum.2008.078. [DOI] [PubMed] [Google Scholar]
- 57.Aljada IS, Ramnath N, Donohue K, Harvey S, Brooks JJ, Wiseman SM, Khoury T, Loewen G, Slocum HK, Anderson TM, et al. Upregulation of the tissue inhibitor of metalloproteinase-1 protein is associated with progression of human non-small-cell lung cancer. J Clin Oncol. 2004;22:3218–3229. doi: 10.1200/JCO.2004.02.110. [DOI] [PubMed] [Google Scholar]
- 58.Hoashi T, Kadono T, Kikuchi K, Etoh T, Tamaki K. Differential growth regulation in human melanoma cell lines by TIMP-1 and TIMP-2. Biochem Biophys Res Commun. 2001;288:371–379. doi: 10.1006/bbrc.2001.5789. [DOI] [PubMed] [Google Scholar]
- 59.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 60.Heppner KJ, Matrisian LM, Jensen RA, Rodgers WH. Expression of most matrix metalloproteinase family members in breast cancer represents a tumor-induced host response. Am J Pathol. 1996;149:273–282. [PMC free article] [PubMed] [Google Scholar]
- 61.Baker EA, Bergin FG, Leaper DJ. Matrix metalloproteinases, their tissue inhibitors and colorectal cancer staging. Br J Surg. 2000;87:1215–1221. doi: 10.1046/j.1365-2168.2000.01531.x. [DOI] [PubMed] [Google Scholar]
- 62.Yoshikawa T, Tsuburaya A, Kobayashi O, Sairenji M, Motohashi H, Yanoma S, Noguchi Y. Intratumoral concentrations of tissue inhibitor of matrix metalloproteinase 1 in patients with gastric carcinoma a new biomarker for invasion and its impact on survival. Cancer. 2001;91:1739–1744. doi: 10.1002/1097-0142(20010501)91:9<1739::aid-cncr1192>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 63.Würtz SØ, Schrohl AS, Mouridsen H, Brünner N. TIMP-1 as a tumor marker in breast cancer—an update. Acta Oncol. 2008;47:580–590. doi: 10.1080/02841860802022976. [DOI] [PubMed] [Google Scholar]
- 64.Lipton A, Leitzel K, Chaudri-Ross HA, Evans DB, Ali SM, Demers L, Hamer P, Brown-Shimer S, Pierce K, Gaur V, et al. Serum TIMP-1 and response to the aromatase inhibitor letrozole versus tamoxifen in metastatic breast cancer. J Clin Oncol. 2008;26:2653–2658. doi: 10.1200/JCO.2007.15.4336. [DOI] [PubMed] [Google Scholar]
- 65.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 66.Gopisetty G, Ramachandran K, Singal R. DNA methylation and apoptosis. Mol Immunol. 2006;43:1729–1740. doi: 10.1016/j.molimm.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 67.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eckhardt F, Lewin Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;3812:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 70.Wada K, Maesawa C, Akasaka T, Masuda T. Aberrant expression of the maspin gene associated with epigenetic modification in melanoma cells. J Invest Dermatol. 2004;122:805–811. doi: 10.1111/j.0022-202X.2004.22308.x. [DOI] [PubMed] [Google Scholar]
- 71.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 72.Zucker S, Cao J, Chen WT. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene. 2000;19:6642–6650. doi: 10.1038/sj.onc.1204097. [DOI] [PubMed] [Google Scholar]
- 73.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 74.Stetler-Stevenson WG. Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities. Sci Signal. 2008;1:re6. doi: 10.1126/scisignal.127re6. [DOI] [PMC free article] [PubMed] [Google Scholar]