Abstract
RATIONALE: Early detection of tumor response to therapy is a key goal. Finding measurement algorithms capable of early detection of tumor response could individualize therapy treatment as well as reduce the cost of bringing new drugs to market. On an individual basis, the urgency arises from the desire to prevent continued treatment of the patient with a high-cost and/or high-risk regimen with no demonstrated individual benefit and rapidly switch the patient to an alternative efficacious therapy for that patient. In the context of bringing new drugs to market, such algorithms could demonstrate efficacy in much smaller populations, which would allow phase 3 trials to achieve statistically significant decisions with fewer subjects in shorter trials. MATERIALS AND METHODS: This consensus-based article describes multiple, image modality-independent means to assess the relative performance of algorithms for measuring tumor change in response to therapy. In this setting, we describe specifically the example of measurement of tumor volume change from anatomic imaging as well as provide an overview of other promising generic analytic methods that can be used to assess change in heterogeneous tumors. To support assessment of the relative performance of algorithms for measuring small tumor change, data sources of truth are required. RESULTS: Very short interval clinical imaging examinations and phantom scans provide known truth for comparative evaluation of algorithms. CONCLUSIONS: For a given category of measurement methods, the algorithm that has the smallest measurement noise and least bias on average will perform best in early detection of true tumor change.
Introduction
General Problem Description and History Review
Strategies for disease response assessment must be useful in a wide range of cancers, encompassing a large variety of image-based measurements and many different treatment options. Chemotherapy and neoadjuvant chemotherapy treatment protocols vary across the world and may include group protocol studies for novel agents or combinations, the application of best therapy in multicenter clinical trials, and many instances of therapy given off-study to individual patients. Many therapy plans now include surgery or radiation as additional therapy options. New biologic response modifiers (so-called targeted therapies) for diseases such as lung cancer have received increased interest recently. These generally less-toxic agents are targeted to affect the tumor blood supply or other critical pathways in cancer cell growth, differentiation, or metastatic processes. The end point of such therapies may not be cancer regression but stasis, that is, tumor growth cessation. Therefore, measures of tumor size may be an inappropriate early measure to evaluate useful change. For example, subtle changes in image-based measures in the cancer such as density, tumor margin alterations, or other pixel-based features may signal a useful response at an early stage of therapy; tumor blood flow may be an important measure for tumor vasculature-based changes; and metabolic changes may be measured by PET—all these changes preceding any change in tumor volume.
Critical to the image-based evaluation of either tumor growth or shrinkage (or some of the more subtle features mentioned already) in response to therapy is a much-improved understanding of the three-dimensional anatomic/pathologic structure of cancers. Current assessments based on two-dimensional pathology slides indicate that malignant cells occupy only a fraction of a tumor nodule's volume, whereas the remainder consists of inflammatory cells, edema, fibrosis, or necrosis. Understanding the three-dimensional structure of cancer pathologically is critical to the evaluation of three-dimensional imaging modalities. Future response assessment protocols could then target specifically the cancer component of a tumor.
The current standard method to measure tumor response using imaging is referred to as Response Evaluation Criteria in Solid Tumors (RECIST), which is based on unidimensional, linear measurements of tumor diameter [1–5]. In promoting the summed linear measurement of a limited number of target tumors, RECIST offers a simple approach that requires minimal effort. The RECIST guidelines, however, presume that tumors are spherical and change in a uniform symmetric manner. In actuality, tumors do not necessarily grow symmetrically; different portions may grow at different rates [6]. Significant variability in the RECIST measures exists among different observers [7–10], and published work generally focuses on the surrogate of “best overall response” with only a few methods addressing other end points such as “time to progression” and “disease-free survival.” As a therapy response measurement procedure, RECIST maps linear data into an established set of four discrete categories: complete response, partial response, stable disease, and progressive disease. These categorical bins, however, are quite coarse with most trial analyses critically pivoting on partial response (defined by a 30% linear sum reduction) and progressive disease (defined by a 20% increase in tumor dimension). Furthermore, if the cancer volume is mostly inflammation, then linear size change alone may give a false impression of therapy response (the inflammation was reduced, but the cancerous component was not); in fact, a tumor may slightly increase in size after initiation of therapy because of inflammatory reactions—although a beneficial response is occurring. As a consequence of observer measurement variability and the expectation that newer therapies will not cause initial size reduction, change in tumor volume is likely inadequate to assess early response to any therapy. Therefore, to improve the accurate assessment of response and to reduce observer variability, other lesion characteristics that may be tracked across temporally sequential scans are required. New imaging techniques and associated new image-processing algorithms allow for early assessment of response to therapy and are being introduced into human clinical trials as outcome measures.
In 2001, a National Institutes of Health working group's consensus was published defining a biomarker as a “characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to therapeutic intervention” [11]. Further, “a biomarker that is intended to substitute for a clinical endpoint” was defined by the same working group as a “surrogate endpoint”—in cancer clinical trials, the accepted criterion standard for clinical end point is overall survival. In a further paper [12], various subgroups of biomarkers are described including prognostic, predictive, and surrogate end point biomarkers. It is possible that imaging can provide biomarkers for all three of these functions, but only if the image-based measure is very well characterized, as indicated in these series of papers. As imaging matures as a measurable modality, we would propose an expanded definition of a biomarker as follows: A biomarker is a validated disease characteristic which can be reliably measured in a cost-effective, repeatable and generalizable manner, and which acts as a meaningful surrogate for disease presence, absence, activity, or outcome in individuals or groups with the disease process. Examples include questionnaires, biochemical measures in various biologic fluids, or image based metrics. Many disease processes have an established phenotype, but a phenotype is not necessarily a good biomarker, and these two terms are therefore not interchangeable. This expanded definition includes the notion that, to be useful in healthcare, validated biomarkers should have the additional properties of being cost effective and generalizable, that is, capable of being implemented at multiple sites with uniform results.
In this paper, we summarize a recent initiative to develop a consensus approach to the benchmarking of software algorithms for the assessment of tumor response to therapy and to provide a publicly available database of images and associated meta-data. The Reference Image Database to Evaluate Response to therapy in cancer (RIDER) project is generating a database of temporally sequential computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) scans of subjects with cancer collected longitudinally during the course of nonsurgical cancer therapy [13]. The database will also include phantom images of synthetic tumors and short-interval patient scans for the evaluation of the variance and bias of change analysis software algorithms. This project evolved from the Lung Image Database Consortium, which is finishing the creation of a publicly available database of annotated thoracic CT scans as a reference standard for the development, training, and evaluation of computer-aided diagnostic methods for lung cancer detection and diagnosis [14,15].
The RIDER project was initiated in 2005 as a collaboration among the National Cancer Institute's (NCI) Cancer Imaging Program, the NCI's Center for Bioinformatics, the National Institute of Biomedical Imaging and Bioengineering, and the Cancer Research and Prevention Foundation, with information technology support from the Radiological Society of North America. The RIDER project was designed and continues to evolve through a consensus process among members of the RIDER steering committee composed of academic researchers, program staff at NCI, and members of the Cancer Biomedical Informatics Grid, National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration (FDA), and the National Institute of Standards and Technology. The broad purpose of the RIDER project is to 1) develop a public resource of serial (i.e., temporally sequential) images acquired during the course of various drug and radiation therapy trials across multiple centers so that change analysis software algorithms may be optimized and benchmarked before use in future trials and 2) enable the development of appropriate evaluation strategies for these new algorithms. The data that will be available to academic researchers and to the device and pharmaceutical industries will include images from CT, MRI, and PET/CT along with relevant metadata. Images of physical phantoms and patient images acquired under situations in which tumor size or biology is known to be unchanged (in which the “true” change is known to be zero) also will be provided and will play a key role in the assessment of software algorithm performance. The RIDER project will highlight the importance of creating standardized methods for benchmarking software algorithms to reduce sources of uncertainty in vital clinical assessments such as whether a specific tumor is responding to therapy.
The longer-term goal of RIDER is to help identify image-based biomarkers to measure cancer therapy response. Such biomarkers could potentially be adopted in clinical trials submitted to the FDA for regulatory approval. Further, such image-based biomarkers could be used to more easily validate other biomarker algorithms in development, such as those from genomics, proteomics, or metabolomics projects. In addition, the Centers for Medicare and Medicaid Services seek evidence to support informed reimbursement decisions for image-based biomarkers that may eventually be used clinically. Consequently, the RIDER project is expected to accelerate 1) FDA approval of both software-based response assessment algorithms and therapeutic agents evaluated through clinical trials that use such algorithms and 2) reimbursement of Centers for Medicare and Medicaid Services for subsequent therapeutic decisions made using such software algorithms. (Text for General Problem Description and History Review is an edited version of Armato et al. [13].)
Specific Focus on Early Detection of Change
Early detection of tumor response to therapy is a key goal. Finding a measurement algorithm capable of early detection of tumor response could individualize therapy treatment as well as reduce the cost of bringing new drugs to market. On an individual basis, the urgency arises from the desire to prevent continued treatment of the patient with a high-cost and/or high-risk regimen with no demonstrated individual benefit and rapidly switch the patient to another therapy that may increase treatment efficacy for that patient. In the context of bringing new drugs to market, such algorithms could demonstrate efficacy in much smaller subject populations, which would allow phase 3 trials to achieve statistically significant decisions in shorter durations with fewer subjects.
The emphasis placed on the word “early” implies that most interest exists near the measurement regime of zero change, that is, the detection of truly small changes from whatever algorithm and parameter set is used and measured. Given that a tumor has a change trajectory over time, for a first-order approximation valid for a short interval, we need only the first two terms of a Taylor series [16] to model the change trajectory, that is, the nodule's current state and its initial time rate of change. Clearly, the patients' oncologists in cases of an individual's health care, or clinical trial designers in the case of drug efficacy studies, are motivated to choose the smallest imaging interval that accurately (ratio of true calls over all calls) assesses the presence or absence of real change. From detection theory, we understand that our ability to detect small changes rests on the signal-to-noise ratio (SNR), alternatively described as effect size to variance. As this ratio increases, we migrate from the condition of being able to detect changes in large populations by averaging to the condition of using fewer subjects until we are able to detect such changes in an individual with clinically useful statistical accuracy. Whereas in most cases we can increase the effect size and thus improve the SNR by increasing the interval between imaging examinations, we would much prefer to use as short an interval as possible. In the limit as the interval between imaging examinations approaches zero, we see that we are indeed operating near the regime of zero tumor change, and it is the noise (variance) in this regime that limits our ability to see small real changes (effect size) in short-interval examinations.
Given the large task required to implement these measurements across a broad spectrum of algorithms and measurement parameters in search of optimal combinations, this consensus group of authors addressing issues facing construction of the RIDER database has focused on ways of estimating a measurement algorithm's noise, that is, variance, under the condition of no change across several modalities and measurement techniques. Arguably, the most realistic and useful data sets representing zero change come from subjects with known tumors who are imaged, removed from scanner, and then are rescanned within a very short time frame. We refer to these interval examinations as “coffee break” examinations. These data sets then contain all of the realities of imaging within a short time window with whatever modality was used, that is, imaged tissue contrast-to-noise, patient motion artifacts, repositioning errors, and so on, that will be encountered in the real world. In addition, because the time interval between these scans is on the order of hours or less, we can safely assume that we know the truth, that is, there are no macroscopic changes to the tumor in the interval between these two examinations. Therefore, measurement of nonzero change by any algorithm using these coffee break data sets is an error. Note that data sets with expert annotations are not used as truth due to their demonstrated variability in segmentation and thus lack of certainty in associated change assumptions [9,17]. An alternative to collecting these coffee break examinations that contain all sources of short-term noise for an estimate of the null hypothesis against which treatment effects must be compared is the collection of a large database of treatment trials along with clinical end points that can be modeled to determine the sources of potentially multiparametric covariance; we suggest that the collection of coffee break data may be far more efficacious at much lower collection cost.
Materials and Methods
Assessing the Measurement of Tumor Volume Change from Anatomic Imaging
Many parameters could be exploited to measure tumor change. There are physical parameters that already have either established or suggested relationships to cancer including density, diffusivity, and elastic moduli. In addition, there are shape and composition parameters including volume, spicularity (typically quantified as the ratio of surface area to volume), heterogeneity, and vascularity (typically quantified as number of vessels intersected per unit area in a histology section). The following section describe methodologies only for assessing the accuracy of measuring tumor volume change as rendered in anatomic imaging. In the simplest case, the same techniques can be used for assessing accuracy of measuring other parameters, but should these parameters have interactions, the measurement methods will require the use of multiparametric estimators such as generalized linear models (GLMs) potentially including mixed effects models that are not addressed herein.
By way of introduction to the problem of measuring volume change of tumors, we describe three of possibly many methods of implementing such measurements. The purpose here is to show the generality of the possible solutions as well as to view the following discussion from a common viewpoint, that is, primarily that of the quantification of tumor volume change. Consider the following two of many possible methods for estimating tumor volume change:
Currently, the standard method of measuring change is the sequential segmentation of the tumor in interval examinations followed by subtraction of the value of the tumor volume of the previous examination from that derived from the current examination. This double segmentation is an indirect method in that volume change is not measured directly and will depend on the accuracy or consistency of the segmentation and the change assessment paradigm [18,19].
- Registrations that map the same, possibly complex, tumor geometry between two different interval examinations can be performed with the volume change estimated directly
- (a) by the product of the resulting anisotropic scaling factors for affine geometry or
-
(b) by integrating the Jacobian [20], that is, the spatially varying, local determinant of the first partial derivatives of the deformation otherwise called local scaling, over the region of support of the deformation created by nonlinearly warping the early tumor to look like the later.These are direct approaches that incorporate partial volume effects and inhomogeneities within lesions that are ignored by binary segmentation approaches.
In the following discussion of volume change, we are fundamentally addressing directly the problem of quantifying volume change. When we discuss random error variance or bias, we are not referring to just the segmentation problem that may or may not precede more sophisticated estimates of volume change but rather to the entire change analysis methodology.
Components of Error
In every problem, we face two basic components of error:
-
Variance, σ2, is a quantitative estimate of the random variability of the data about its mean in repeated measurements associated with noise from various sources, for example, data sensors and subsequent measurement methods, and is estimated as shown here
Here xi represents one of the N discrete measurements wemake to compute the quantity within the brackets as an unbiased estimate of the variable's variance, is the estimate of the discrete data's mean, and E is the estimate of the quantity inside the brackets as N approaches infinity. The quantity ( is the noise or error term from the expected mean estimate. Thus, we estimate variance by computing the terms inside the brackets, that is, the sample variance, for an N large enough to give us a sufficiently low noise estimate of the true variance for our purposes. Precision, a qualitative term frequently used in radiologic literature, is quantified by the measurement of SD, σ, also called standard uncertainty, which is the square root of variance. Precision improves as the SD and variance of the repeated measurements decrease.For example, if we need to measure the length of an object with a cloth measuring tape, we can measure the object multiple times and calculate the variance of the measurement. Here, the variability could be due to several components of error, for example, the measurement tool can be randomly stretched by differing amounts, each time we place the beginning of the tape at slightly different positions, and so on. These differences between repeated measurements can be characterized by the variability about the mean, that is, the variance; the smaller that variance is, the more precise these measurements are said to be.
-
Bias is a quantitative estimate of systematic measurement error, that is, even if the random error were zero, the measured number would be systematically different from the truth if bias were nonzero. Examples of this include systematic over/under estimation of some measured property (again, such as volume). Accuracy, a qualitative term also frequently used in radiologic literature, is quantified by the measurement of bias. Accuracy improves as the measured bias decreases. The measurement of bias is discussed in more detail in the section on estimating variance and bias for the case of no volume change.
Continuing the previous cloth tape analogy, all measurements would be positively biased, that is, longer, if the cloth tape we used had been unknowingly cut off at the beginning of the tape by 1 inch. Thus, we could average many measurements to reduce the variance and improve the precision, but still be wrong (biased or inaccurate) by 1 inch.
For tumor change measurements, we will begin with the assumption that for similar physical imaging characteristics and subject setup, the variance and bias estimates are likely dependent on the size of the tumor as well as its complexity which includes factors such as heterogeneity, shape, and location; specifically, the derived parameters that describe each of the errors may in general be a function of these enumerated independent parameters.
In most experiments, we observe both effects simultaneously as they are not easily separated and only through the collection of sufficient data and the use of statistical analysis techniques such as GLMs with selected mixed effects are we able to separate estimates of error components. Such models are especially important when the measured quantities are truly changing with time. The modeling is complicated by having to choose the specific mixed effects and degrees of freedom (DOF). Owing to the model's large DOF, the amount of test data needed and collected under known conditions also increases. When a single measured quantity is stationary, as in the section on Estimating Variance and Bias for the Case of No Volume Change, we can also approach the problem as a simpler, ordered discovery of the two separate components.
Estimating Variance and Bias for the Case of No Volume Change
In the following discussion, we will describe an ordered quantification of both random error and bias around the operating point of no change. This is a crucial operating point because in many practical clinical applications, we wish to discover real change in as short a time interval as possible to affirm or refute the assumption that the applied therapy is effective. The urgency arises from the desire to prevent continued treatment of the patient with a high-cost and high-risk regimen with no demonstrated benefit as well as from the need to rapidly switch an individual patient to another therapy that may increase individual efficacy. Thus, measurement noise observed in the case of no change for a specific patient is a sample of the null hypothesis that must be quantified before we can determine with some stated probability that any measured change represents true change.
Estimating variance for the case of no volume change
Under the simplifying assumption that the random error is additive, we can estimate its variance by using input data sets where we know the underlying truth is no change. There are two main types of experiments to be considered here: “coffee break” studies, that is, very short interval examinations, and longitudinal studies, both used for gathering input data sets from which we can estimate variance.
-
(a) Coffee break studies
In a coffee break study, the time interval between scans is small compared with the time required for the tumor to change macroscopically, that is, the subject is scanned more than once in a given session or day. Requirements for these studies typically include a special prospective image acquisition protocol with local institutional review board approval, the acquisition of no more than two scans per patient owing to radiation or contrast dose considerations and use of the same scanner for both scans so that confounding effects such as scanner calibration drift, change of physical scanner, change in image acquisition, or reconstruction protocols are minimized. Although these efforts may seem overly constraining to be required in clinical practice, to achieve clinical measurements of early tumor change during as short an interval as 1 to 2 weeks, patients should be reassigned to the same scanner with the same specified image acquisition protocol despite any implementation difficulties. We would expect that maximum sensitivity to the measurement of change would be obtained under these conditions because of the reduced number of possible noise sources. Although performance of repeated imaging on manufactured phantoms may provide useful information about imaging equipment variations such as calibration drift, obtaining this imaging on patients is advantageous for predicting variance in clinical applications such as measuring tumor change. Because of a variety of factors including homogeneity of the background, simplistic shapes of simulated tumors and lack of interfering adjacent structures such as penetrating vessels, using physical phantoms to estimate the variance of image-derived parameters will, as a rule, underestimate the variance of image-derived parameters obtained in clinical practice, but such phantoms will have use in estimating bias as described later.
-
(b) Long-term clinical surveillance studies
A second possible source of input data sets for estimating variance is imaging examinations taken during multiple quarterly, semiannual, or annual intervals in which no statistically significant trend is observed. We would expect to find more random variability in the measurement of tumor change in this setting due to effects unrelated to tumor change such as long-term physiological change in the subjects and scanner changes, for example, different 1) scanners, 2) acquisition protocols, and 3) hardware and software owing to upgrades including image reconstruction algorithm changes. The main advantage of this approach is that cases may be retrospectively selected from clinical archives and special prospective institutional review board protocols would not likely be required. The difficulty here is that 1) the analysis of these longitudinal data to verify that the tumor volumes are statistically stationary over time is slightly more complicated than the simplistic analysis we describe for the coffee break data, and 2) we are only studying tumors that are stable and these tumors are not necessarily representative of cancers as a whole. Because stable tumors are generally more homogeneous than tumors that are rapidly growing, the measurement task may be simpler and the variance reduced compared with that obtained in malignant tumors. In addition, because these tumors may actually be slowly changing, these data may be useful for testing the relative comparison of algorithm variance using the estimator described later.
Although we are limiting our consideration to the measurement of change near the operating point of no change, we need to make the measurement of variance for tumors of differing sizes. There is significant evidence from manual and semiautomatic segmentation that SD and therefore variance is a function of tumor size; see [17]. Thus, we need a source of truth data, for example, coffee break examinations, which contain a spectrum of scanned tumor sizes to characterize the performance of the change measurement analysis for different size tumors.
Because sample variance is a noisy measurement of the underlying distribution's variance, we will need many measurements of tumors with no size change. There are two possible approaches to increase the number of observations of variance to approximate the variance of the underlying distribution.
(c) For every patient with N interval examinations of a tumor that is conservatively judged to show no change, we can compute N! / ((N - 2)! x 2!) different but partially correlated, interval pairs of examinations from which we can estimate the variance of change measurements; DOF will need to be adjusted to account for the correlation in the data pairs.
(d) Because estimates of random error are potentially dependent on tumor characteristics (e.g., shape, content, surroundings, volume, acquired voxel size), we should only use interval examinations containing tumors of similar characteristics that are conservatively judged to show no change. These variance estimates can then be aggregated to decrease our confidence limits for estimating the underlying population variance for tumors characterized by that specific volume and constitutive complexity.
The estimate of the random error's variance may be sensitive to the estimator used, particularly in case of an error in classifying a tumor as having no size change. For example, for measurements Xi with mean , the obvious estimator is the sample variance determined by:
which is exactly the same estimator as
a U statistics-based estimator [21] as suggested in (c) above. However, estimators and are valid only under the assumption that there is no change and will be biased if the tumor varies with time. Other model-based methods are more appropriate should the tumor's volume vary with time. For long-term clinical surveillance studies of slowly varying nodules, the estimator should be less heavily biased and can be justified based on simple assumptions on the nodule growth and the homogeneity of the variance:
Robust estimation is especially useful for small data sets when outliers may make a big difference. Huber [22] and Hoaglin et al. [23] give extensive discussion of the pros of robust estimation in practice.
Estimating bias for the case of no-volume change
Once the random error's variance has been approximated, we can explicitly compute the number of observations we must obtain to test for the presence of a bias effect at some stated level of significance. The number of observations (experiments performed to measure the bias) depends on the variance of the previously determined random error distribution, the size of the bias effect we wish to measure and the probability that we will measure such an effect, that is, reject the null hypothesis, at a stated level of confidence. The required number of observations, that is, measurements of volume change, increases as
(a) the size of the bias we wish to measure decreases for a fixed variance,
(b) the variance as measured above for the random error component increases for a fixed bias, or
(c) the power, that is, likelihood of detecting the change at the given level of significance, increases (an interactive demonstration of powering a test is available at: http://wise.cgu.edu/power/power_applet.html).
The measurement of bias is important because if present it will lead to a propensity for false-positives/negatives, depending on whether the change measurement bias is positive/negative, respectively. As the name implies, bias is a systematic error whose cause can be discovered and removed or at least modeled and ameliorated.
Estimating Variance and Bias for the Case of True Volume Change
The determination of bias and variance in the presence of true volume change is needed if we want a completely generalized statistical characterization of a measurement method. Note that if we want to quantify volume change, not simply determine whether there was or was no change, the truth data required for this task are more difficult to obtain. Because estimates of bias and variance in volume change may be dependent on tumor volume as well as tumor volume change (along with other characteristics such as shape, type, acquisition/reconstruction protocols, and possible motion), we will want to regress both bias and variance as a function of both tumor volume and tumor volume change through GLMs. Truth data for this task can only be known from manufactured phantoms; a method for obtaining volume change truth for real tumors is difficult and has yet to be defined for RIDER. Here, the “coffee break” null change paradigm for real patient scan data is of little use because the “truth” of tumor size is not known (only the null change in tumor size is known); instead, we need estimates of true change from other accurate sources.
The key issue is that we currently have no measurement method that will provide the true change in size of an actual tumor. For real tumors that do change in size between interval scans, we are restricted to using image measurements made by expert radiologists, and this measurement method is itself subject to bias and random error [9,17]. The only way we can obtain scans with known truth for size change is to scan manufactured phantoms with known tumor characteristics and different sizes or to embed simulated, mathematically defined tumors in actual patient scan data; the critical concern here is how well such phantoms represent real tumors and their growth. To summarize:
-
Phantom studies could be used to obtain measurement bias for phantom tumors both for expert radiologists, assuming the phantoms are representative of real tumor characteristics, and computer-based measurement systems. However, the phantoms must represent as closely as possible the properties of clinical cases in terms of tumor signal intensities, sizes, shapes, etc. At least initially, we will assume that the measurement variance for tumors that do change size will be a linear combination of the no-change variances measured at the initial and final sizes of the tumor; as mentioned earlier, owing to reduced complexity typically seen in phantom data, we would expect the variance in measurement error of tumors that do change size to be affected. In addition, although not perfect, the phantom data may still serve as a means for comparing the relative performance of most algorithms.
Monte Carlo simulation studies provide stochastic models of imaging systems and could also be helpful. For the case of x-ray-based imaging, recent advances in simulation tools [24] allow the generation of images with realistic statistical properties by tracking the transport of particles from the x-ray source through the object of interest to the detector. Monte Carlo simulation can be used to generate thoracic CT images of realistic anthropomorphic phantoms while controlling for variables such as image acquisition parameters, nodule characteristics, and the complexity of surrounding structures. Such simulation packages potentially could be developed for other modalities.
We may conduct studies with real tumors to compare the results between expert radiologists and computer methods; however, the truth will not be known.
-
We can compare the results for random error between the phantom and real tumor experiments to examine if (at least for a selected subset of real tumors) the phantom results are comparable. Given some level of agreement, we may be able to conjecture (and potentially establish by later statistical analysis of large studies) that the bias results from the phantom experiments are predictive of the bias for real tumors.
These weak assumptions lead us to believe that the data collection and analysis required for accurate clinical estimates of variance in the presence of real change will be very expensive.
General Overview of Methods Useful for Assessing Tumor Change
In the preceding section, methods for assessing the relative performance of algorithms specifically for measuring tumor volume change for the purpose of early assessment of tumor response to therapy were discussed. Whereas the use of volume was explicitly examined, we could use exactly the same techniques to examine any other single parameter, for example, average mass, elasticity, etc., and the same techniques for assessment of performance would apply, that is, the measurement of variance and bias. There is, however, an explicit difference between volume and most other single parameters: volume is necessarily a singular, summary parameter whereas other parameters have tumor-dependent, heterogeneous spatial distributions of values within that volume which can be characterized in several ways including a one-dimensional histogram of its values and the histogram's summary statistics, that is, mean, variance, skew, kurtosis, and other higher moments.
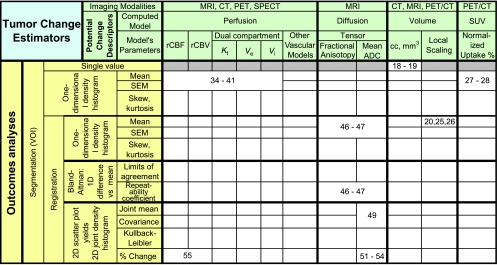
The function of the sparsely filled Table 1 is to demonstrate the relative relationships of some different outcomes analysis methods and computed parametric models previously contributed to NCI's public archive https://imaging.nci.nih.gov/ncia/, now called the National Biomedical Imaging Archive, through the efforts of previous RIDER groups as well as a few related methods previously published.
Table 1.
Shows Possible Ontological Relationships between Outcomes Analysis Methods (Leftmost Columns) and Parametric Models (Topmost Rows) Computed from Associated Modalities.
As seen in the rows of the Outcomes Analyses, Table 1, most processing is first subjected to segmentation, that is, defining the volume of interest (VOI) for further processing as the volume of clinical interest. Registration commonly follows segmentation in that registration of the whole, complex set of organ systems is very computationally intensive and challenging given that some organs deform and slide along slip walls, for example, lung compression and slippage along the pleural surface of the rib cage. Thus, registration of the lung alone is far simpler than attempting to register the lung and chest wall simultaneously owing to the discontinuity of velocity vectors at the pleural surface. Hence, registration of a segmented lesion with itself across interval examinations is typically preferred.
After segmentation and registration, differing outcomes analyses are applied to the following potential change descriptors for detecting/measuring response to therapy:
-
Volume: In the case of estimating a tumor's volume change, two of the methods we discussed in the previous section are shown in Table 1 in the second set of major columns from the right. Whereas the imaging modalities associated with volume estimation are typically those of CT and MRI owing to better spatial resolution, segmentation-based implementations can be applied to PET and single photon emission computed tomography (SPECT) as well.
The method of tumor segmentation followed by summing the volume of voxels inside a VOI yields a single numeric characterization of the volume of the tumor in the examination; subtraction of the results for any two interval examinations yields a single numeric characterization of the tumor's volumetric change. Although this method that can include manual as well as many sophisticated semiautomatic methods is the current method of choice of most groups for computing volume change, we have referenced only two papers here [18,19]. Note that this method yields only a single number, not a spatial distribution that can be summarized by other single metrics, such as the mean; because none of the other parametric methods we will discuss yields only a single number, the remainder of this row has been grayed out.
Another method we described previously that directly measures volume change is based on registration of the earlier interval examination onto the later. Assuming that the information content of the imaging modality is sufficient to support accurate registration, such methods provide a spatial distribution of local scale changes over the volume of the reference tumor as represented by the Jacobian matrix, that is, the determinant of the first partial derivatives of local change in all cardinal directions. The resulting scale distribution yields local measures of heterogeneous volume changes if they exist. Again we cite only a few reference examples using such methods [20,25,26].
Uptake: In PET, biologic chemists have had significant success in tagging specific physiological metabolites with radiotracers. Normalized standard uptake values (SUVs), calculated typically as the ratio of measured radioactivity concentration to injected dose divided by patient body weight, are proposed for quantifying tumor response to therapy [27]. In the accompanying article as well as in a predecessor article [28], Kinahan et al. describe methods of PET quality control and measurement for assuring that measured SUV changes are related to the tumor's physiological changes in response to therapy. The accompanying article additionally describes useful test data contributed to the RIDER data collection that help define tumor change effect sizes that are required to identify a meaningful change. Because SUV is a spatial distribution of values over the segmented tumor, its measurement is typically reported as the maximum and/or the mean and SD of the underlying one-dimensional histogram of values within the delineated VOI.
- Perfusion:
- (a) Many perfusion models exist, but in dynamic contrast enhancement (DCE) MRI, a simple, often used model is the two-compartment model: one compartment for the intravascular input contrast concentration, and the other for the extravascular-extracellular compartment. Common assumptions here are that the current gadolinium-based contrast agents do not penetrate cells and that the intravascular concentration of contrast only contributes to contrast enhancement in the extravascular-extracellular compartment by passive diffusion. The relationship between the change in signal amplitude due to T1 relaxivity and contrast concentration must first be established to convert voxel amplitudes into contrast concentrations. Then from Fick Law, the time rate of change for contrast material in the extracellular tissue is driven by the difference between the two concentrations, that is, the input from intravascular plasma and the loss from the tissue surrounding the capillaries back into the plasma. For a two-compartment model, this statement is typically written in equation form as
where Ct(t) is the time-dependent extracellular tissue concentration, Cp(t) is the time-dependent plasma concentration, Ktrans is the rate coefficient for contrast flow from the plasma into the tissue, and kep is the rate coefficient in the opposite direction. Because these rates are due to passive diffusion mechanisms through the capillary endothelial cells, that is, no active “pumps,” and volume normalization is applied, we can derive that kep = Ktrans/ve, where ve is the fractional extracellular-extravascular volume and vp is the fractional blood plasma volume; see Tofts et al. [29]. Computing coefficients from differential equations is very sensitive to noise so a more robust approach is to model the time integral of the equation given above, which converts the solution to the convolution of KtransCp(t) with the kernel , that is, , where noise is now attenuated owing to the averaging of the integral. Computing the coefficients for this simplified model is still fraught with some difficulties, for example, picking a good model of the plasma input function to determine the convolution kernel as well as attempting to avoid numerical instabilities in the discrete implementation of the convolution. Here, additional noise reduction can be achieved using the singular value decomposition in the discrete modeling of the convolution integral and elimination of the smaller eigenvalues in formulating the inverse [30]. -
(b) A simpler modeling approach is that the temporal integration of the T2* relaxivity change in the first pass of an intravenous bolus contrast injection at each voxel in brain yields the relative cerebral blood volume (rCBV) change [31] under the assumption that the blood-brain barrier is intact, or in cases of fenestration, that appropriate deconvolution modeling is used [32,33]. The relative mean transit time (rMTT) is obtained from the integral of the time-weighted concentration normalized by rCBV, and thus relative cerebral blood flow is defined by the ratio rCBV/rMTT. Owing to the rapid time rate of change of blood flow, dynamic susceptibility contrast MRI sampling is accomplished using rapid acquisition sequences such as echo-planar imaging. A detailed pictorial review accompanied by equations of these concepts and others that follow is provided in Jackson [34].Whereas MRI acquisition methods to obtain data in support of computing perfusion models described in sections (a) and (b) above were described, CT, PET, and SPECT are all capable of capturing images sufficiently rapidly to derive meaningful coefficients for modeling perfusion in section (a) as described by Tofts. But only CT and MRI are readily capable of the increased acquisition rates necessary to derive coefficients for the model in section (b) above. Coefficients for models described in both sections (a) and (b) can be computed on a voxel-by-voxel basis; thus, outcomes analyses can be computed in a number of ways. A good review of methods for perfusion (as well as diffusion) MRI is presented in Provenzale et al. [35]. By far, most outcomes analyses for perfusion coefficient models use summary statistics from VOIs to report a mean and standard error of the VOI mean (SEM); necessarily limited references to these approaches are included herein [36–41].More to the point of this article's emphasis, tumor change analysis in perfusion is often computed as the change in these summary statistics with t-tests performed to assess whether the treatment effect measured was different than the null hypothesis. In the case that there are multiple VOI pairs for an interval examination from which change is assessed, or where each voxel pair in registered interval data sets is treated as a separate “VOI,” the statistical test must be corrected for false-positives arising from multiple comparisons. If the one-sided level of significance were picked at α = 0.05 and the null hypothesis were true, that is, there was no tumor change, approximately 5 of 100 voxels would test as positive, that is, falsely changed, simply because we applied the test 100 times to the null Gaussian distribution. Good descriptions of possible correction methods (Bonferonni, family-wise error rate, false discovery rate, etc.) for multiple comparison tests are presented in Wiens [42], Perneger [43], and Genovese et al. [44]. A correction must be applied wherever multiple comparisons occur for the stated P values to be meaningful, whether related to perfusion diffusion or other metrics.
-
Diffusion:
In vivo assessment of organ system and tumor apparent diffusion coefficient (ADC) measures is available using MR diffusion-weighted imaging (DWI or DW-MRI). The formula for computing ADC in the direction of the diffusion gradient is
where Sh and Sl are the high and low signal amplitudes of the isotropic DW images corresponding to the use of bh and bl, respectively, where the high and low bx values are a function of the applied amplitude of the diffusion sensitizing gradient pulses, as well as the temporal duration and temporal separation of the pulses. Owing to the dipole nature of the coils used to apply the diffusion gradients, the results are anisotropic for all but the case where bl = 0, which is isotropic because there is no diffusion gradient applied. Many gradient directions can be acquired [45] subject to scan time constraints to improve the resulting SNR. Singular value decomposition of the amplitude response of all these components at each voxel yields the amplitude response for each of three principal axes, that is, the eigenvalues (λ1, λ2, λ3). The singular value decomposition result is the complete summary of all excitations at each voxel, but the fractional anisotropy (FA) is a normalized scalar that is commonly used to characterize the variation in the eigenvalues for each voxel. FA is expressed as where M is the vector magnitude, that is, and λ is the mean of the eigenvalues, that is, λ = (λ1 + λ2 + λ3) / 3. Note that FA varies between the limits of 0 and 1; 0 is obtained for the isotropic diffusion case (as in a pure cyst where λ1 = λ2 = λ3 = λ), whereas FA = 1 is obtained in the most anisotropic condition, that is, only one of the three principal axis magnitudes is nonzero (approximated by a straight segment of white matter tract in brain).
Alternative outcomes analyses
Instead of using manually drawn multiple VOIs as the only means to approximate following spatial changes in the same tumor across interval examinations, registration of the interval data sets can be implemented to reduce the increased variance associated with manual misplacement of VOIs drawn on interval examinations. More importantly depending on the accuracy of its implementation, registration is capable of supporting voxel-by-voxel change analysis. After registration, a single VOI may be used on registered interval examinations to limit voxel-by-voxel analysis to, or generate summary statistics from, registered differences considered important by the investigator. Such registered differences may come from the same enhancing region as used to define a VOI on a registered T1-postGad series that has been mapped, that is, warped to, the series (one or more) of analytical interest. Summary statistics, typically mean and variance, can be compiled
from the one-dimensional histograms of values from the VOI registered onto the pretherapy and posttherapy examinations and compared for statistically significant changes, or
from two one-dimensional histograms of values from the VOI registered onto two pretherapy baseline examinations to sample baseline noise; see [46,47] in Table 1 as examples. The experiment described in these references assesses the repeatability of measurements across patients for which we expect no difference, for example, between two baseline examinations acquired during a short interval. Bland and Altman [48] first described an appropriate method of making this assessment in their study based on plotting the difference between a pair of measurements on the same subject versus the mean of the two measurements. The 95% confidence interval for these differences is the definition of repeatability, that is, ±2σ or two times the SD of the differences, that is, repeatability improves as the SD and thus variance decrease. Importantly, the same study of Bland and Altman also describes in a similar fashion how to measure and characterize agreement between two methods, an assessment often mistakenly attempted through correlation or regression.
-
Further, the voxel-by-voxel analysis can consist of a two-dimensional, co-occurrence plot of the registration-paired voxel values, or its joint density histogram constructed by summing the number of co-occurrences in bins. This is just the usual t-test with the exception that the distribution is now two-dimensional instead of the usual one-dimensional distribution that we commonly use. The somewhat hidden issue here is what are the DOF of the estimate of the mean, that is, how many independent samples contributed to its computation? Typically, in acquisition of functional MRIs, the number of acquired data points (whose independence is also based on slice profiles) in k-space is zero padded and interpolated up to some desired array display matrix size several times larger than the actual data acquisition matrix. These data acquisition parameters can be gleaned only by careful reading of the data's DICOM header; but even then, additional vendor signal-processing specifics may remain hidden in vendor-specific encoded DICOM header regions. Clearly, when the data have been interpolated, either by the MR vendor or in the process of registration, the DOFs of the estimate of the mean are only indirectly related to the number of data samples. The multipliers associated with vendors' signal processing and user's interpolation associated with registration for voxel-by-voxel analysis must be used to correct DOF [49].
Summary statistics for the voxel-by-voxel analysis may be expressed by one or more of the following metrics:- (a) displacement of the joint mean relative to the covariance of the mean's null distribution, which, for this case, is tested for significance by the multivariate version of Student's t-test also known as Hotelling's T test,
- (b) Kullback-Leibler (KL)-directed “distance” [50] between the treatment effect and null distributions (this metric is sensitive to any differences between the two distributions), or
-
(c) percent change (%change) of tumor voxels that have a significant change in perfusion above a threshold, for example, the two-tailed 95th percentile determined from the null distribution.In (b) above, the KL-directed distance metric is defined as the log-weighted, average distance from distribution p1(a) to distribution p2(a), that is,
Note that this definition is sensitive to small differences in the two distributions wherever they occur but is weighted to be more sensitive near the mode of p1(a). In clinical applications, p1(a) could be that of the treatment effect and then p2(a) could be the null distribution. Note that this measure is not intrinsically symmetric, that is, the “distance” from p1(a) to p2(a) is typically not the same as from p2(a) to p1(a) when we exchange 1's and 2's in the definition but can be made symmetric by taking the average of both directed distances. The KL-directed distance metric can obviously be applied to any number of variables, for example, the univariate version as shown in the definition above assumes the variable a is a scalar, but a could be a vector as well.In (c) above, under the assumption that the VOI encompasses primarily the tumor, estimates of percent change are accomplished by applying a threshold to the treatment effect distribution where the threshold's parametric value is defined by selecting a percentile on the null distribution, for example, the 97.5th percentile to minimize false-positives. By measuring the percentage of the treatment effect above that threshold and subtracting the percentage expected for the null distribution, for example, 2.5% for the 97.5th percentile suggested above, the percent of voxels that have significantly changed in the tumor can be reported and their spatially coherent loci in the tumor demonstrated [51–54]. Further, given the current chaos in attempted change analysis generated from perfusion data, there is some hope from recent results [55] that suggest that the voxel-by-voxel analysis may more accurately support detection of change in heterogeneous tumors (such as glioblastoma multiforme) than simple, mean histogram VOI analysis applied in current practice.
Recommendations
Coffee Break Examinations as Sources of Truth for Assessing Relative Algorithm Performance in Measuring Early Therapeutic Tumor Response
Defining truth in realistic, complex data sets is difficult except in the case of multiple, short-interval examinations, that is, coffee beak examinations. Previously, expert physician annotations have been the accepted standard, but recent studies suggest that even among recognized experts, the variance of annotations is significantly large such that the expense for obtaining sufficient data to observe small standard error of the mean expert trends is prohibitive. Thus, sources of truly known imaging “truth” in realistic, complex settings are invaluable. Despite the costs of scanning including increased radiation burden for CT, PET, and SPECT, such known truth can be obtained from multiple short-interval examinations on consented patients where the known truth is that no macroscopic change in the tumor can have occurred in the sufficiently short interval between scans. The use of interval examinations having uncorrelated or even partially correlated noise contributes to the knowledge of the covariance of the null hypothesis distribution and thus allows probabilistic limits to be set on the chance that the observed outcome represents real change versus noise.
Without such data, the only apparent other option for investigators is to gather large population databases with low-noise outcome measures of truth such as length of disease-free survival or complete pathologic response. These databases typically need to be accumulated from a minimum of 50 subjects so that part of the database may be used for training the change detection algorithm and the remainder of the database may be used for testing through application of the tuned algorithm; bootstrapping may be used [56,57]. For univariate data, the test typically consists of finding an optimal cut point using receiver operating characteristic (ROC) analysis on the training data set component of the database, which can then be applied to a separate test set for unbiased assessment of performance [58,59]. Much larger databases (e.g., 200 or more subjects) are typically required to achieve sufficiently small confidence limits for the area under the curve (or Az) of the ROC to see statistically significant changes between competing algorithms. The number of subjects required is large because experimental truth gathered outside carefully designed clinical trials is itself noisy because clinical treatments across multiple subjects are typically not uniform owing to differing surgical and other unplanned life-saving interventions that can significantly alter individual patient outcomes but obfuscate the effects of the initial therapy and associated image-based change analysis used to define early therapeutic response. For multivariate analyses, ROC techniques can be supplanted by use of GLM regression.
The concluding assumption is that it is likely more cost-effective to collect multisubject, multiple short-interval coffee break examinations on which we can measure the variance for the null condition across competing algorithms for detecting early change, than it is to essentially perform a small (∼200 subjects) phase 2 study to obtain the necessary database to test the algorithms for efficacy. The noise in these larger studies may also be increased owing to a multitude of other clinical and multi-institutional factors not rigorously controlled such as variation in scanners across institutions, slight variations in scanning protocols, medications, and so on.
Experimental Design Evolution for Measurement of Tumor Volume Change from Anatomic Imaging: Start with Well-Controlled Experiments
In estimating variance and bias in the measurement of tumor volume change around the operating point of no volume change observed with coffee break data sets, the number of measured parameters is likely dependent on a number of factors including tumor size, structural complexity (e.g., inhomogeneity, shape, surroundings), the extent of change (in terms of size and morphology), and possible changes in scanner parameters between scans; that is, the problem space is of high dimensionality with respect to lesion size and complexity and scanner settings. Given realistic limitations on the number of available, finite input data sets, it will be necessary to assess complexity and control the number of variables to obtain statistically meaningful results. Investigators should initially consider pilot experiments that focus on a small number of selected points in this problem space (e.g., well-defined lesions of clinically meaningful size and size change with well-defined margins and very similar scanner parameters). Such experiments should provide insight on how to conduct experiments to characterize error for real lesions in the larger problem space.
In quantifying the accuracy of measuring true lesion change we will also need to know bias at “operating points” other than the no-volume change point described immediately above. As a first-order approximation, we can scan simple spherical phantoms of known volumes, and by using different combinations of phantom tumors for “early” and “late” interval pairs, we can evaluate multiple combinations of volume and volume change operating points. Both low and high contrast-to-noise acquisitions should be examined. Because the variance of the random error component for these measurements should be relatively small owing to the structural simplicity compared with the coffee break examinations, the ability to see small bias should be relatively easier to observe. Irregularly shaped tumor phantoms, for example, those with spiculations and random orientations in the field of view could be used to increase the complexity of the phantom measurement setting to more closely approximate outcomes in real data sets.
Measurement of Tumor Change from Voxel-by-Voxel Analysis May Be Necessary for Heterogeneous Tumors
All of the points discussed in the preceding paragraphs are valid for this topic as well. Additionally for heterogeneous tumors, change analysis based on one-dimensional histogram summary statistics accumulated over the volume of interest of the tumor may be misleading. Consider trying to measure response changes in a heterogeneous tumor where the changes over different regions of the tumor both increase and decrease with respect to the parameter's mean such as might be observed where therapeutic intervention is successful in some compartments while tumor growth temporarily continues in other, more isolated compartments. Under such conditions, changes in the summary statistic (mean or other moments such as variance, skew, and kurtosis) would be attenuated, and the detection of such changes, if present, would be less likely. Under these conditions, tracking of changes in individual tumor compartments supported by voxel-by-voxel change analysis has the possibility of demonstrating such confounding effects.
Discussion
The initial focus of the search for algorithms that provide early image-based markers of tumor change in response to therapy will likely use
the “coffee break” study paradigm, that is, multiple short-interval baseline examinations, to find algorithms yielding minimal measurement variance while observing data from the null change condition, and
physical or simulated known phantom scans to demonstrate adequate measurement accuracy, that is, small bias, for small effect sizes.
In addition to discovering which algorithms are low-noise estimators of tumor change and thus optimally suited to detect early change, in practice
we must also control other sources of noise that have the potential to be much larger sources of variance. Such sources include differences in image acquisition protocols, patient positioning, and physiological condition, which can create differences in the apparent response of the tumor not related to its biology/physiology. Further,
systematic quality control programs appropriate for the image-based biomarkers must be implemented to allow assessment of, and corrections for, scanner variations over time and across upgrades.
The motivation for these issues and modality-specific issues are discussed in more detail in the editorial and three companion articles of this issue [60–63]; modality-independent issues are enumerated in the Appendix.
Acknowledgments
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Appendix: Modality-Independent Sources of Bias and Variance
Introduction
There are many factors common to most medical imaging modalities that affect our ability to measure tumor change with little bias or variance error. In this appendix, we enumerate some of those factors in the context of measuring tumor volume change and discuss possible methods of mitigation. We list them here as separate factors with the understanding that there are likely significant interactions between different sources of random error, that is, their factors will have nonzero covariance. Generalization of the principles discussed here to the measurement of parameter changes other than volume is relatively straightforward. Modality-specific examples are presented in the companion articles [61,62,63].
Sources of Bias and Variance
Patient-related
(a) Motion: sources of voluntary and involuntary patient related motion include respiration, cardiac pulsatility, peristalsis, pain, stiffness, seizures, muscular twitching, prolonged discomfort, and so on. Clearly, all motion during imaging contributes to the acquisition of nonanatomic data sets such as those with extended or shortened organs and tumors [64]. Whereas breath holding is often practiced, breath holding at the same level of inspiration across interval scans is less frequently imposed. Interval imaging at the same respiratory phase is important because many abdominal organs move surprisingly large distances driven by respiration, for example, the prostate can move as much as 1 cm cranial-caudally [65]!
(b) Abnormalities: body habitus changes between examinations can affect signal attenuation of all modalities. Competing, concomitant disease such as inflammation may mimic tumor progression, whereas varied use of medications, for example, corticosteroids between interval examinations can change inflammation and result in cellular swelling.
Image analysis-related
Each measurement method for quantifying tumor volume change, whether based broadly on subtracted segmentations or registration, will likely have its own characteristic variance and bias. The following describes many of the variables that affect each of the two different volume change estimation methods:
- (a) for subtracted segmentations
- changing choices of manual, semiautomatic, or fully automatic segmentation,
- changing tuning parameters of semiautomatic or fully automatic segmentation algorithms: for example, thresholding, region growing, level sets, and associated penalty functions, and
- measurement software version changes
- (b) for registration-based estimates, the DOFs for the geometry model are limited by the mutual information content between the two interval examinations where the mutual information varies locally and is affected by
- local SNR, and
- tumor structural decorrelation over time, that is, different tumor compartments arise or decay during temporally under-sampled imaging intervals; shorter imaging intervals would observe these changes where longer intervals will miss the stages of demise of older and creation of newer subcompartments.
Mitigation Efforts
Level of breath hold can be partially or fully achieved several ways that vary from asking the patient to hold their breath, for example, at full inspiration (this is an example of partial control), to measurement of tidal phase through a flow meter, which actuates a valve to enforce breath holding [66]. Other possibilities include cardiac and respiratory gating of the image acquisition system or list mode acquisition where provided by the vendor followed by gated reconstruction and registration of the differently gated cycles.
The basic principle in change analysis is that whenever possible, keep all potential sources of bias and variance unchanged between interval examinations, that is, use the same segmentation method for both interval examinations, and if the segmentation method is semiautomatic or fully automatic, continue to use the same tuning parameters for both examinations. Use the same scanner with the same technical protocol and consistent patient factors (contrast dose, rate of delivery, flush, injection site, breath hold, table position, etc.). The scanner should have a rigorous quality assurance program in place to ensure consistent performance, and technical protocols should be user-locked.
Footnotes
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400.
References
- 1.James K, Eisenhauer E, Christian M, Terenziani M, Vena D, Muldal A, Therasse P. Measuring response in solid tumors: unidimensional versus bidimensional measurement. J Natl Cancer Inst. 1999;91:523–528. doi: 10.1093/jnci/91.6.523. [DOI] [PubMed] [Google Scholar]
- 2.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 3.Bogaerts J, Ford R, Sargent D, Schwartz LH, Rubinstein L, Lacombe D, Eisenhauer E, Verweij J, Therasse P. Individual patient data analysis to assess modifications to the RECIST criteria. Eur J Cancer. 2009;45:248–260. doi: 10.1016/j.ejca.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 4.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz LH, Bogaerts J, Ford R, Shankar L, Therasse P, Gwyther S, Eisenhauer EA. Evaluation of lymph nodes with RECIST 1.1. Eur J Cancer. 2009;45:261–267. doi: 10.1016/j.ejca.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Yankelevitz DF, Reeves AP, Kostis WJ, Zhao B, Henschke CI. Small pulmonary nodules: volumetrically determined growth rates based on CT evaluation. Radiology. 2000;217:251–256. doi: 10.1148/radiology.217.1.r00oc33251. [DOI] [PubMed] [Google Scholar]
- 7.Marten K, Auer F, Schmidt S, Kohl G, Rummeny EJ, Engelke C. Inadequacy of manual measurements compared to automated CT volumetry in assessment of treatment response of pulmonary metastases using RECIST criteria. Eur Radiol. 2006;16:781–790. doi: 10.1007/s00330-005-0036-x. [DOI] [PubMed] [Google Scholar]
- 8.Bobot N, Kazerooni E, Kelly A, Quint L, Desjardins B, Nan B. Inter-observer and intra-observer variability in the assessment of pulmonary nodule size on CT using film and computer display methods. Acad Radiol. 2005;12:948–956. doi: 10.1016/j.acra.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Erasmus JJ, Gladish GW, Broemeling L, Sabloff BS, Truong MT, Herbst RS, Munden RF. Interobserver and intraobserver variability in measurement of non-small-cell carcinoma lung lesions: implications for assessment of tumor response. J Clin Oncol. 2003;21:2574–2582. doi: 10.1200/JCO.2003.01.144. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz LH, Mazumdar M, Brown W, Smith A, Panicek DM. Variability in response assessment in solid tumors: effect of number of lesions chosen for measurement. Clin Cancer Res. 2003;9:4318–4323. [PubMed] [Google Scholar]
- 11.Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA, et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 12.Kelloff GJ, Sigman CC. New science based endpoints to accelerate oncology drug development. Eur J Cancer. 2005;41:491–501. doi: 10.1016/j.ejca.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Armato SG, III, Meyer CR, McNitt-Gray MF, McLennan G, Reeves AP, Croft BY, Clarke LP. The Reference Image Database to Evaluate Response to Therapy in Lung Cancer (RIDER) Project: a resource for the development of change-analysis software. Clin Pharmacol Ther. 2008;84:448–456. doi: 10.1038/clpt.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armato SG, III, McNitt-Gray MF, Reeves AP, Meyer CR, McLennan G, Clarke LP, Aberle DR, Kazerooni EA, MacMahon H, van Beek EJR, et al. The Lung Image Database Consortium (LIDC): an evaluation of radiologist variability in the identification of lung nodules on CT scans. Acad Radiol. 2007;14:1409–1421. doi: 10.1016/j.acra.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNitt-Gray MF, Armato SG, III, Meyer CR, Reeves AP, McLennan G, Pais RC, Freymann J, Brown MS, Engelmann RM, Bland PH, et al. The Lung Image Database Consortium (LIDC) data collection process for nodule detection and annotation. Acad Radiol. 2007;14:1464–1474. doi: 10.1016/j.acra.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Athans M, Dertouzos M, Spann R, Mason S, editors. Systems, Networks and Computation: Multivariable Methods. New York, NY: McGraw Hill; 1974. [Google Scholar]
- 17.Meyer CR, Johnson TD, McLennan G, Aberle DR, Kazerooni EA, MacMahon H, Mullan BF, Yankelevitz DF, van Beek EJR, Armato SG, III, et al. Evaluation of lung MDCT nodule annotations across radiologists and methods. Acad Radiol. 2006;13:1254–1265. doi: 10.1016/j.acra.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves AP, Chan AB, Yankelevitz DF, Henschke CI, Kressler B, Kostis WJ. On measuring the change in size of pulmonary nodules. IEEE Trans Med Imaging. 2006;25:435–450. doi: 10.1109/TMI.2006.871548. [DOI] [PubMed] [Google Scholar]
- 19.Zhao B, Schwartz LH, Moskowitz CS, Ginsberg MS, Rizvi NA, Kris MG. Lung cancer: computerized quantification of tumor response—initial results. Radiology. 2006;241:892–898. doi: 10.1148/radiol.2413051887. [DOI] [PubMed] [Google Scholar]
- 20.Thirion J-P, Calmon G. Deformation analysis to detect and quantify active lesions in 3D medical image sequences. IEEE Trans Med Imaging. 1999;18:429–441. doi: 10.1109/42.774170. [DOI] [PubMed] [Google Scholar]
- 21.Lee A, editor. U-statistics. New York, NY: Marcel Dekker Inc.; 1990. [Google Scholar]
- 22.Huber PJ, editor. Robust Statistics. New York, NY: Wiley; 1981. [Google Scholar]
- 23.Hoaglin DC, Mosteller F, Tukey JW, editors. Understanding Robust and Exploratory Data Analysis. New York, NY: Wiley; 1983. [Google Scholar]
- 24.Badano A, Sempau J. MANTIS: combined x-ray, electron and optical Monte Carlo simulations of indirect radiation imaging systems. Phys Med Biol. 2006;51:1545–1561. doi: 10.1088/0031-9155/51/6/013. [DOI] [PubMed] [Google Scholar]
- 25.Reinhardt JM, Ding K, Cao K, Christensen GE, Hoffman EA, Bodas SV. Registration-based estimates of local lung tissue expansion compared to xenon CT measures of specific ventilation. Med Image Anal. 2008;12:752–763. doi: 10.1016/j.media.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar S, Narayanan R, Park H, Ma B, Bland PH, Meyer CR. Quantitative growth measurement of lesions in hepatic interval CT exams. SPIE Med Imaging. 2008;6914 1G:1–10. [Google Scholar]
- 27.Shankar LK, Hoffman JM, Bacharach S, Graham MM, Karp J, Lammertsma AA, Larson S, Mankoff DA, Siegel BA, Van den Abbeele A, et al. Consensus recommendations for the use of 18FDG PET as an indicator of therapeutic response in patients in National Cancer Institute trials. J Nucl Med. 2006;47:1059–1066. [PubMed] [Google Scholar]
- 28.Doot RK, Christian PE, Mankoff DA, Kinahan PE. Reproducibility of quantifying tracer uptake with PET/CT for evaluation of response to therapy. IEEE Nuc Sci Symp Conf Rec. 2007;4:2833–2837. [Google Scholar]
- 29.Tofts P, Brix G, Buckley D, Evelhoch J, Henderson E, Knopp M, Larsson H, Lee T-Y, Mayr N, Parker G, et al. Estimating kinetic parameters from dynamic contrast-enhanced T1-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 30.Sourbron S, Luypaert R, Schuerbeek PV, Dujardin M, Stadnik T. Choice of the regularization parameter for perfusion quantification with MRI. Phys Med Biol. 2004;49:3307–3324. doi: 10.1088/0031-9155/49/14/020. [DOI] [PubMed] [Google Scholar]
- 31.Belliveau JW, Rosen BR, Kantor HL, Rzedzian RR, Kennedy DN, McKinstry RC, Vevea JM, Cohen MS, Pykett IL, Brady TJ. Functional cerebral imaging by susceptibility-contrast NMR. Magn Reson Med. 1990;14:538–546. doi: 10.1002/mrm.1910140311. [DOI] [PubMed] [Google Scholar]
- 32.Ostergaard L, Weisskoff RM, Chesler DA, Gyldensted C, Rosen BR. High resolution measurement of cerebral blood flow using intravascular tracer bolus passages. Part I: Mathematical approach and statistical analysis. Magn Reson Med. 1996;36:715–725. doi: 10.1002/mrm.1910360510. [DOI] [PubMed] [Google Scholar]
- 33.Ostergaard L, Sorensen AG, Kwong KK, Weisskoff RM, Gyldensted C, Rosen BR. High resolution measurement of cerebral blood flow using intravascular tracer bolus passages. Part II: Experimental comparison and preliminary results. Magn Reson Med. 1996;36:726–736. doi: 10.1002/mrm.1910360511. [DOI] [PubMed] [Google Scholar]
- 34.Jackson EF. MR biomarkers: current applications and unmet needs; Invited Presentation, Annual Meeting of the AAPM; July 2007; Minneapolis, MN. [October 13, 2009]. Available at: http://www.aapm.org/meetings/amos2/pdf/29-7934-87026-358.pdf. [Google Scholar]
- 35.Provenzale JM, Mukundan S, Barboriak DP. Diffusion-weighted and perfusion MR imaging for brain tumor characterization and assessment of treatment response. Radiology. 2006;239:632–649. doi: 10.1148/radiol.2393042031. [DOI] [PubMed] [Google Scholar]
- 36.Groves AM, Wishart GC, Shastry M, Moyle P, Iddles S, Britton P, Gaskarth M, Warren RM, Ell PJ, Miles KA. Metabolic-flow relationships in primary breast cancer: feasibility of combined PET/dynamic contrast-enhanced CT. Eur J Nucl Med Mol Imaging. 2009;36:416–421. doi: 10.1007/s00259-008-0948-1. [DOI] [PubMed] [Google Scholar]
- 37.Kamath A, Smith WS, Powers WJ, Cianfoni A, Chien JD, Videen T, Lawton MT, Finley B, Dillon WP, Wintermark M. Perfusion CT compared to H2 15O PET in patients with chronic cervical carotid artery occlusion. Neuroradiology. 2008;50:745–751. doi: 10.1007/s00234-008-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Rodiger LA, Shen T, Miao J, Oudkerk M. Perfusion MR imaging for differentiation of benign and malignant meningiomas. Neuroradiology. 2008;50:525–530. doi: 10.1007/s00234-008-0373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin W, Guo J, Rosen MA, Song HK. Respiratory motion-compensated radial dynamic contrast-enhanced (DCE)-MRI of chest and abdominal lesions. Magn Reson Med. 2008;60:1135–1146. doi: 10.1002/mrm.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Langen AJ, van den Boogaart VEM, Marcus JT, Lubberink M. Use of H2 15O PET and DCE-MRI to measure tumor blood flow. Oncologist. 2008;13:631–644. doi: 10.1634/theoncologist.2007-0235. [DOI] [PubMed] [Google Scholar]
- 41.Kennan RP, Jägar HR. T2- and T2*-W DCE-MRI: blood perfusion and volume estimation using bolus tracking. In: Tofts P, editor. Quantitative MRI of the Brain. Weinheim, Germany: Wiley-VCH; 2004. pp. 365–412. [Google Scholar]
- 42.Wiens BL. A fixed sequence Bonferroni procedure for testing multiple endpoints. Pharm Stat. 2003;2:211–215. [Google Scholar]
- 43.Perneger TV. What's wrong with Bonferroni adjustments. Br Med J. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 45.Papadakis NG, Xing D, Huang CLH, Hall LD, Carpenter TA. A comparative study of acquisition schemes for diffusion tensor imaging using MRI. J Magn Reson. 1999;137:67–82. doi: 10.1006/jmre.1998.1673. [DOI] [PubMed] [Google Scholar]
- 46.Paldino MJ, Phadke D, DesJardins A, Vredenburgh J, Friedman H, Barboriak DP. Repeatability of apparent diffusion coefficient and fractional anisotropy in patients with recurrent glioblastoma multiforme. ISMRM'08 Electronic Multimedia Posters 4 (ISMRM, Berkeley, CA) 2008:3492. [Google Scholar]
- 47.Paldino MJ, Barboriak D, Desjardins A, Friedman HS, Vredenburgh JJ. Repeatability of quantitative parameters derived from diffusion tensor imaging in patients with glioblastoma multiforme. J Magn Reson Imaging. 2009;29:1199–1205. doi: 10.1002/jmri.21732. [DOI] [PubMed] [Google Scholar]
- 48.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 49.Meyer CR, Schott AF, Chenevert TL, Galban C, Johnson TD, Rehemtulla A, Hamstra DA, Ross BD. Parametric Response Mapping (PRM): voxel-based analysis of quantitative diffusion MRI changes for individualized assessment of primary breast cancer response to therapy; World Molecular Imaging Congress (WMIC'08); 2008. [October 13, 2009]. Poster 500. Available at: http://www.abstractsonline.com/viewer/viewAbstractPrintFriendly.asp?CKey={A10D3708-C38A-4A86-9146- A522E431623E}&SKey={CADAF2CB-0224-482A-BEAE-3A276689C1F2}& MKey={B47BAE74-CCA9-4C27-80FB-0005AFC9E5C0}&AKey={A4C6DD8F- 4BF2-400D-97ED-20C14381CDBB} [Google Scholar]
- 50.Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22:79–86. [Google Scholar]
- 51.Moffat B, Chenevert T, Lawrence T, Meyer C, Johnson T, Dong Q, Tsien C, Mukherji S, Quint D, Gebarski S, et al. Functional diffusion map: a non-invasive MRI biomarker for early stratification of clinical brain tumor response. Proc Natl Acad Sci USA. 2005;102:5524–5529. doi: 10.1073/pnas.0501532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamstra DJ, Chenevert TL, Moffat BA, Johnson TD, Meyer CR, Mukherji S, Quint DJ, Gebarski SS, Xiaoying F, Tsien C, et al. Evaluation of the functional diffusion map as an early biomarker of time-to-progression and overall survival in high grade glioma. Proc Natl Acad Sci USA. 2005;102:16759–16764. doi: 10.1073/pnas.0508347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamstra DA, Galban CJ, Meyer CR, Johnson TD, Sundgren PC, Tsien C, Lawrence TS, Junck L, Ross DJ, Rehemtulla A, et al. The functional diffusion map (fDM): an early imaging biomarker for overall survival in high-grade glioma. J Clin Oncol. 2008;26:1–9. doi: 10.1200/JCO.2007.15.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer C, Chenevert T, Galban C, Johnson T, Hamstra D, Rehemtulla A, Ross B. Parametric response mapping: a voxel-based analysis of quantitative diffusion MRI changes for individualized assessment of primary breast cancer response to therapy; Proceedings 17th Scientific Meeting, International Society for Magnetic Resonance in Medicine (ISMRM, Berkeley, CA); 2009. Poster 2223 http://troll.rad.med.umich.edu/dipl/publications/ISMRM2009Poster.pdf. [Google Scholar]
- 55.Galban CJ, Chenevert TL, Meyer CR, Tsien C, Lawrence TS, Hamstra DA, Junck L, Sundgren PC, Johnson TD, Ross DJ, et al. The parametric response map is an imaging biomarker for early cancer treatment outcome. Nat Med. 2009;15:572–576. doi: 10.1038/nm.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinkley DV. Bootstrap methods. J R Stat Soc Series B Stat Methodol. 1988;50:321–337. [Google Scholar]
- 57.Wu CFJ. Jackknife, bootstrap and other resampling methods in regression-analysis — discussion. Ann Stat. 1986;14:1261–1295. [Google Scholar]
- 58.Swets JA, Pickett RM. Evaluation of Diagnostic Systems. New York, NY: Academic Press; 1982. [Google Scholar]
- 59.Metz CE, Starr SJ, Lusted LB. Observer performance in detecting multiple radiographic signals: prediction and analysis using a generalized ROC approach. Radiology. 1975;121:337–347. doi: 10.1148/121.2.337. [DOI] [PubMed] [Google Scholar]
- 60.Clarke LP, Croft BS, Nordstrom R, Zhang H, Kelloff G, Tatum J. Quantitative imaging for evaluation of response to cancer therapy. Transl Oncol. 2009;2(4):195–197. doi: 10.1593/tlo.09217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McNitt-Gray MF, Bidaut LM, Armato SG, III, Meyer CR, Gavrielides MA, McLennan G, Petrick N, Zhao B, Reeves AP, Beichel R, et al. Computed tomography assessment of response to therapy: tumor volume change measurement truth data, and error. Transl Oncol. 2009;2(4):216–222. doi: 10.1593/tlo.09226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinahan PE, Doot RK, Wanner-Roybal M, Bidaut LM, Armato SG, III, Meyer CR, McLennan G. PET/CT assessment of response to therapy: tumor change measurement, truth data, and error. Transl Oncol. 2009;2(4):223–230. doi: 10.1593/tlo.09223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson EF, Barboriak DP, Bidaut LM, Meyer CR. Magnetic resonance assessment of response to therapy: tumor change measurement, truth data and error sources. Transl Oncol. 2009;2(4):211–215. doi: 10.1593/tlo.09241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen GT, Kung JH, Beaudette KP. Artifacts in computed tomography scanning of moving objects. Semin Radiat Oncol. 2004;14:19–26. doi: 10.1053/j.semradonc.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Dawson LA, Litzenberg DW, Brock KK, Sanda M, Sullivan M, Sandler HM, Balter JM. A comparison of ventilatory prostate movement in four treatment positions. Int J Radiat Oncol Biol Phys. 2000;48:319–323. doi: 10.1016/s0360-3016(00)00751-3. [DOI] [PubMed] [Google Scholar]
- 66.Keall PJ, Mageras GS, Balter JM, Emery RS, Forster KM, Jiang SB, Kapatoes JM, Low DA, Murphy MJ, Murray BR, et al. The management of respiratory motion in radiation oncology report of AAPM Task Group 76. Med Phys. 2006;33:3874–3900. doi: 10.1118/1.2349696. [DOI] [PubMed] [Google Scholar]