Abstract
The malignant glioma is the most common primary human brain tumor. Its tendency to invade away from the primary tumor mass is considered a leading cause of tumor recurrence and treatment failure. Accordingly, the molecular pathogenesis of glioma invasion is currently under investigation. Previously, we examined a gene expression array database comparing human gliomas to nonneoplastic controls and identified several Rac guanine nucleotide exchange factors with differential expression. Here, we report that the guanine nucleotide exchange factor SWAP-70 has increased expression in malignant gliomas and strongly correlates with lowered patient survival. SWAP-70 is a multifunctional signaling protein involved in membrane ruffling that works cooperatively with activated Rac. Using a glioma tissue microarray, we validated that SWAP-70 demonstrates higher expression in malignant gliomas compared with low-grade gliomas or nonneoplastic brain tissue. Through immunofluorescence, SWAP-70 localizes to membrane ruffles in response to the growth factor, epidermal growth factor. To assess the role of SWAP-70 in glioma migration and invasion, we inhibited its expression withsmall interfering RNAs and observed decreased glioma cell migration and invasion. SWAP-70 overexpression led to increased levels of active Rac even in low-serum conditions. In addition, when SWAP-70 was overexpressed in glioma cells, we observed enhanced membrane ruffle formation followed by increased cellmigration and invasiveness. Taken together, our findings suggest that the guanine nucleotide exchange factor SWAP-70 plays an important role in the migration and invasion of human gliomas into the surrounding tissue.
Introduction
Malignant gliomas are the most common primary brain tumors in adults [1]. Despite some recent therapeutic advances in the treatment of this type of tumor, the overall patient survival rate remains disappointing. Treatment failures frequently arise from the inability to eliminate the invading front of tumor cells that extends beyond the margin of the primary tumor [2]. Therefore, to improve our ability to treat patients with malignant gliomas, we must increase our understanding of the migratory and invasive properties of these tumors [3].
Invasion is a complex cellular phenomenon that involves cell-to-cell and cell-to-extracellular matrix (ECM) interactions, enzymatic degradation of the ECM, and cell migration [4]. Cell movement involves the coordination of multiple signaling pathways that regulate cell-substratum adhesion and actin cytoskeletal dynamics [5]. To migrate, glioma cells must acquire a special polarized asymmetry. This enables them to transform intracellular forces into net cell body translocation [6]. Typically, a migrating glioma cell remodels its cytoskeleton and forms protrusions and focal adhesions at the leading edge of the migratory front. This generates contractile forces and traction, allowing the cell body to move forward [7,8].
The small GTPase Rac1 is known to play a crucial role in the cytoskeletal and structural changes during cell migration and has been reported by our group and others to be a major signaling node important for the migration and invasion of glioma cells [9–14]. In a previous study, we used gene expression profiling to identify guanine nucleotide exchange factors (GEFs), which act on Rac proteins and which may be important for glioma invasion. Three GEFs, namely, Trio, Ect2, and Vav3, were selected for further study because their increased expression was associated with poor patient outcome and higher tumor grade and were shown to be important for glioma migration and invasion. To further study the role of Rac-activating GEFs in human malignant gliomas, we selected SWAP-70 as another candidate promigratory and proinvasive gene from a data set of more than 26 Rac-activating GEFs [15] because it is more highly expressed in gliomas than in nonneoplastic brain tissue. SWAP-70 is a 70-kDa protein originally isolated from activated B lymphocytes [16] and is known to mediate lipid second messenger signals to the cytoskeleton-organizing role of Rac.
SWAP-70 is widely expressed in various cell types and tissues [16,17], and a high expression of SWAP-70 has been observed in activated B cells, immature mast cells, and B-cell neoplasms. SWAP-70 has been shown to regulate c-kit-induced mast cell activation, cell-cell adhesion, and migration [18–21], but to date, no studies have reported on the functional significance of SWAP-70 in human gliomas. Accordingly, in the present study, we have examined the role played by SWAP-70 in the invasive phenotype of human glioma cells.
Materials and Methods
Expression Profile Data Set of Rho GEFS in Human Gliomas and Nonneoplastic Brain
To identify candidate GEFs in human gliomas, we mined the publicly available Repository for Molecular Brain Neoplasia Data (REMBRANDT) gene expression microarray database containing 295 clinically annotated brain tumor specimens (National Cancer Institute, 2005; REMBRANDT home page: <http://rembrandt.nci.nih.gov>; accessed April 15, 2009). We specifically examined the gene expression data from nonneoplastic brain (NB, n = 27), low-grade astrocytomas (LGGs, n = 61), and glioblastoma multiformes (GBMs, n = 147). Gene expression profiling and normalization was conducted on all samples as previously described [15] (National Cancer Institute, 2005; REMBRANDT home page: <http://rembrandt.nci.nih.gov>; accessed April 15, 2009).
Cell Lines and Cell Culture
The permanent, well-characterized U251, U343, U118, SNB19, and T98 human glioma cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS; HyClone Laboratories, Inc, Logan, UT). The U87 cell line was maintained in minimum essential medium supplemented with 10% FBS. Cells were cultured in a 37°C, 5% carbon dioxide-humidified chamber.
Tissue Microarray and Immunohistochemistry
Immunohistochemical staining was done using the commercially available brain tumor tissue microarray GL803 (US Biomax, Inc, Rockville, MD), which consists of 55 astrocytic tumor specimens and 5 normal brain specimens. The patient population included 22 male and 33 female tumor specimens, with a mean patient age of 41.7 years (range, 4–68 years), and 3 male and 2 female normal control specimens, with a mean patient age of 45.8 years (range, 30–52 years). Of the astrocytic tumor specimens, 6 were grade 1, 9 were grade 2, 25 were grade 3, and 15 were grade 4. Tumor grade data were provided by US Biomax, Inc, and determined according to the 2007 WHO grading system.
The tissues were deparaffinized, endogenous peroxidase activity was blocked using 3% hydrogen peroxide in methanol, and microwave antigen retrieval was achieved using Antigen Unmasking Solution (Vector Laboratories, Inc, Burlingame, CA). Blocking was done using 10% normal horse serum, and endogenous biotin was blocked using an avidin/biotin kit (Vector Laboratories). The tissue and tumor sections were incubated with a mouse monoclonal anti-SWAP-70 antibody (1:500; Abcam, Cambridge, MA) followed by a biotinylated secondary antibody (Vector Laboratories) for 30 minutes, then a horseradish peroxidase-conjugated ultra-streptavidin labeling reagent (ID Labs Biotechnology, Inc, London, Ontario, Canada) for 30 minutes. After rinsing thoroughly in PBS, color development was performed using freshly prepared NovaRED solution (Vector Laboratories). Finally, sections were counterstained lightly with Mayer hematoxylin, dehydrated in alcohol, cleared in xylene, and mounted in Permount (Fisher Scientific, Ottawa, Ontario, Canada).
SWAP-70 immunostaining was graded semiquantitatively using a score ranging from 0 to 3 based on the extent of positive staining: 0 = 0%, 1 = 1% to 30%; 2 = 31% to 70%; and 3 ≥ 71% positive tumor cells. Sections of normal human spleen were used as a positive control for SWAP-70 immunoreactivity [22].
Western Blot Analysis
Whole cell lysates were prepared by harvesting cells in 50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, 10% glycerol, and a complete cocktail of protease inhibitors (Roche Molecular Biochemicals, Laval, Quebec, Canada). Protein concentrations were determined using the Micro BCA kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's directions. Proteins were separated by SDS-PAGE, transferred onto polyvinylidene fluoride membrane (Immobilon-P; Millipore, Billerica, MA), and immuno-blotted with a goat polyclonal anti-SWAP-70 antibody (1:1000; Abcam) or anti-FLAG M2 mouse monoclonal antibody (1:1000; F3165; Sigma-Aldrich, St Louis, MO) overnight at 4°C. A mouse monoclonal antibody against β-actin (1:1000; Sigma) or the human transferrin receptor (1:1000; Zymed Laboratories, South San Francisco, CA) was used to control for loading. Bound primary antibodies were visualized using horseradish peroxidase-conjugated donkey antigoat or goat antimouse immunoglobulin G antibodies (Bio-Rad Laboratories, Hercules, CA).
SWAP-70 Expression Construct and Transient Overexpressing Cell Line
A plasmid containing full-length human SWAP-70 complementary DNA was obtained from the Mammalian Gene Collection (SWAP-70-IRAU9-H2 in pOTB7). This complementary DNA was subcloned into the pFLAG-CMV2 (Sigma) expression vector using polymerase chain reaction cloning techniques to generate KpnI and BamHI restriction sites. The sequence was then verified. Empty pFLAG-CMV2 was used as a negative control. These constructs were transfected into the U87 cell line using FuGene6 (Roche Applied Science) reagent. Western blot was used to verify the protein expression level of SWAP-70 level in the transfected cells.
Small Interfering RNA Preparation and Transfections
For gene silencing, a small interfering RNA (siRNA) quadriplex specific to SWAP-70 (NM_015055) ON-TARGETplus SMARTpool (Dharmacon, Thermo Scientific, Waltham, MA) was purchased. The four target sequences were as follows: First sequence corresponds to nucleotides 1902 to 1920 (5′ CAGGAGAGCTTTACGCTAA); the second sequence corresponds to nucleotides 1725 to 1743 (5′ AGGATTAATTCGACTGATA); the third corresponds to nucleotides 2591 to 2609 (5′ CCGAGTATGTGCTGTTAAA); and the fourth to nucleotides 4317 to 4335 (5′ GCATAGTGTTACCGATTTA). A commercial ON-TARGETplus nontargeting pool (Thermo Scientific) was used to monitor off-target effects. Transient transfections were performed with DharmaFect reagent (Thermo Scientific) according to the manufacturer's protocol.
Immunofluorescence and Confocal Microscopy
Cells were plated onto coverslips 24 hours after transfection. At 72 hours after transfection, cells were washed three times with PBS and fixed with 4% paraformaldehyde and 750 µl of 1 M sucrose for 30 minutes, then permeabilized for 10 minutes using 0.2% Triton X-100 in PBS at room temperature. Nonspecific binding was blocked by 5% BSA in PBS for 30 minutes at room temperature. Subsequently, cells were incubated with goat polyclonal anti-SWAP-70 antibody (1:100; Abcam) for 30 minutes in a 37°C incubator. After washing, cells were then incubated with Alexa Fluor 488 (1:200; Molecular Probes, Eugene, OR)-conjugated donkey antigoat immunoglobulin G secondary antibody. For double labeling with F-actin, cells were coincubated with Texas Red-X phalloidin (1:50; Molecular Probes). To visualize fluorescence, a Zeiss Axiovert 200M Spinning Disk confocal microscope (Carl Zeiss Canada, Toronto, ON, Canada) equipped with a Hamamatsu Back-Thinned EM-CCD camera (Hamamatsu Corporation, Bridgewater, NJ) was used.
Treatment with 50 ng/ml epidermal growth factor (EGF) for 10 minutes induced membrane ruffling. After this, the morphologic changes associated with the expression and/or localization of SWAP-70 were examined.
Cell Attachment Assay
The cell attachment assay was performed as described previously [23,24] with some modifications. Briefly, 96-well plates were incubated at 4°C overnight with different ECM proteins (laminin [LN] and fibronectin [FN] at 10 µg/ml, and vitronectin [VN] at 1 µg/ml). The unbound sites were blocked with 0.1% BSA in PBS for 1 hour at 37°C. Control dishes were prepared by blocking with BSA alone. Glioma cells were detached 72 hours after transfection, resuspended in serum-free medium then plated at 5 x 104 cells per well on ECM protein- or BSA only coated plates. The cells were allowed to adhere for 3 hours at 37°C and 5% carbon dioxide in a humidified chamber. Unattached cells were removed by washing with PBS three times, and the remaining cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Cells were destained with water then allowed to air-dry overnight. The crystal violet bound to the attached cells was solubilized with 1% SDS and measured the absorbance of each well was measured at 595 nm using a microtiter plate reader (Molecular Devices, Sunnyvale, CA). All experiments were repeated three times with five replicates.
Radial Cell Migration Assay
Cell migration assays were performed using the micrometer-scale radial monolayer assay as described previously [9,25,26]. Briefly, 10-well Teflon-coated slides (CSM Inc, Phoenix, AZ) were coated with different ECM proteins and 0.1% BSA as described above. To establish a circular confluent monolayer at the center of the substrate-coated well, transfected cells were collected 24 hours after transfection and seeded through a cell sedimentation manifold (CSM Inc) at 2000 cells per well. Photographs were acquired 22 hours after plating, and a circle of best-fit circumscribing the cells was drawn representing the first time point. The cells were allowed to migrate for 24 hours, then another circle circumscribing the newly migrated cells was made. The average migration rate was calculated as the change in the diameter of the circle circumscribing the cell population during a 24-hour period (micrometers per 24 hours). Photomicrographs were taken with an inverted microscope (Leica DM IRE2; Leica Microsystems, Inc, Bannockburn, IL) and studied using an image analysis software (Scion Image, Frederick, MD). All experiments were done in triplicate and repeated three times.
Transwell Cell Invasion Assay
A cell invasion assay was carried out using modified Boyden chambers that consisted of Transwell-coatedMatrigel membrane filter inserts with 8-µm pores in 24-well tissue culture plates (BD Biosciences, Bedford, MA) as described previously [26]. Two days after transfection, 4 x 104 cells were plated onto the top of the chamber in DMEM with 5% FBS. The bottom chamber was filled with DMEM containing 20% FBS as a chemoattractant. Plates were incubated for 24 hours in a 5% carbon dioxide-humidified chamber at 37°C. Noninvading cells were wiped off the upper surface of the membrane with a cotton swab, and the filter membrane was fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. The degree of invasion was determined by counting the number of cells that had migrated through the membrane in at least 6 random fields (total magnification, x200) per filter. Experiments were repeated three times in triplicate.
GTPase ELISA-Based Rac Activation Assay
For GTPase ELISA-based Rac activation, 5 x 104 U87 and T98 cells were plated in six-well plates. Two days after transfection, cells were serum-starved for 24 hours and then stimulated with EGF (10 ng/ml) for 10 minutes. Cells were then washed twice with ice-cold PBS and lysed in lysis buffer provided by the manufacturer. Equal aliquots of lysate were used to interrogate Rac-GTP levels according to the manufacturer's instructions (Cytoskeleton, Denver, CO).
Statistical Analyses
Statistical analyses were performed using Student's t-test and the Mann-Whitney U test. P < .05 was considered statistically significant.
Results
Increased Expression of SWAP-70 in High-grade Gliomas Correlates with Poor Patient Outcome
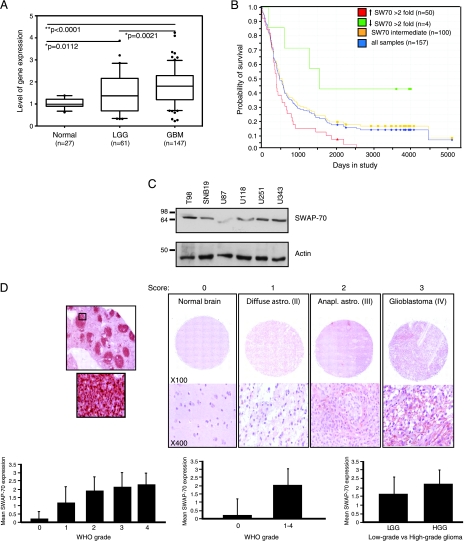
To determine the expression of SWAP-70 in human gliomas, we reviewed a more recent version of the previous data set used to mine GEFs in a recent report from our laboratory [15]. In this study, we used data collected for REMBRANDT (http://rembrandt.nci.nih.gov). Using this data set, we show that SWAP-70 (probe ID 209307_at) has a significantly higher expression in GBMs compared with nonneoplastic brain (P < .0001) and LGG (P = .0021). Statistical significance was also achieved between SWAP-70 expression in LGG and NB (P = .0112). Data are plotted as box plots of the median gene expression ratios between the tumor sample and the geometric mean of nontumor samples for the probe set selected in the query (Figure 1A). Given the strong correlation of SWAP-70 expression in high-grade gliomas, we evaluated patient survival outcome based on SWAP-70 expression from samples derived from the patient tumors. On plotting a Kaplan-Meier survival curve, we observed significantly decreased survival times for samples with more than two-fold increased SWAP-70 expression (n = 50) when compared with all other samples (log rank P value = 10-4). Conversely, extended patient survival times for those samples withmore than two-fold decreased SWAP-70 expression was observed; however, statistical significance was not achieved (P = .0506). This could be due to a very small number of samples in this group (n = 4; Figure 1B).
Figure 1.
Increasing SWAP-70 expression in glioma correlates with advanced disease and reduced survival. (A) Gene expression profiling analysis for human SWAP-70. Messenger RNA expression levels of SWAP-70 fromthe data set are significantly increased in GBMs (n = 147) or LGG (n = 61) when compared with nonneoplastic brain (n = 27). Data were obtained from the REMBRANDT gene expression microarray database containing 295 clinically annotated brain tumor specimens (National Cancer Institute, 2005; REMBRANDT home page: <http://rembrandt.nci.nih.gov>; accessed April 15, 2009). (B) Kaplan-Meier survival curve of patient samples (n = 157) based on SWAP-70 expression. (C) Western blot analysis of SWAP-70 expression in glioma cell lines. U87 has the lowest SWAP-70 expression and was selected for overexpression studies, and conversely, T98 and SNB19 express SWAP-70 at higher levels and were selected for subsequent RNA interference experiments. (D) Examination of SWAP-70 expression on tissue microarray. The tumors were immunostained for SWAP-70 monoclonal antibody and tested on positive control specimen (spleen, left panel). Tissuemicroarray sample of diffuse astrocytoma (WHO grade 2) shows weak cytoplasmic staining, score 1. Anaplastic astrocytoma (grade 3) with moderate SWAP-70 reactivity, score 2. Glioblastoma with strong staining in neoplastic cells, score 3 (right panel). Comparison of SWAP-70 expression in human gliomas showed statistically significant differences in SWAP-70 gene expression when different tumor grades were examined (P = .001). Comparison between normal brain and gliomas showed very significant high expression in gliomas (P = .0005). HGG showed higher expression level than LGG (P = .043; lower panels). Anaplas. indicates anaplastic; astro, astrocytoma; HGG, high-grade glioma; LGG, low-grade glioma.
Characterization of SWAP-70 Expression in Glioma Cell Lines
Western blot analysis to detect SWAP-70 protein expression was performed on different glioma cell lines, including U343, U251, U118, U87, SNB19, and T98. Cell lines demonstrated varying degrees of SWAP-70 expression as observed in the Western blot (Figure 1C). From these results, we selected U87 for SWAP-70 overexpression experiments given its low basal levels of SWAP-70. The T98 and SNB19 glioma cells lines were selected for gene silencing because these two cell lines demonstrated abundant levels of SWAP-70.
Validation of SWAP-70 Expression in Primary Patient Samples Using Tissue Microarray Analysis
Given the strong correlation of increased SWAP-70 transcript expression with increasing grade gliomas, we were interested in validating SWAP-70 gene expression at the protein level. We obtained a commercially prepared tissue microarray consisting of 56 glioma samples (grades 1 to 4) and 5 nonneoplastic controls. SWAP-70 immunoreactivity was significantly higher in high-grade gliomas (WHO grades 3 and 4) than in low-grade gliomas (WHO grades 1 and 2; P = .043). In addition, there is a significant difference in SWAP-70 expression when nonneoplastic brain and gliomas were compared (P = .0001; Figure 1D, lower panels). The comparison of SWAP-70 expression in different grades of glioma revealed a direct correlation between protein levels and increasing glioma grade (P = .001; Figure 1D, lower panels).
Cellular Redistribution of SWAP-70 after EGF Stimulation and Alteration of Cell Morphology on SWAP-70 Overexpression
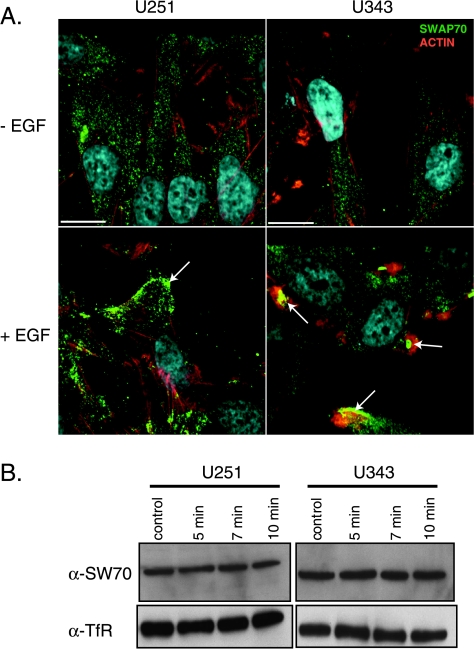
Previously SWAP-70 was described to interact directly with F-actin in COS-7 cells after EGF treatment [27]. Therefore, we examined the localization of SWAP-70 in the context of glioma cells using immunofluorescence before and after stimulation with EGF (Figure 2A). Before EGF stimulation, SWAP-70 was distributed throughout the cytosol, and after EGF treatment, SWAP-70 relocalized to the cell periphery in subcortical regions of the cell. The cell lines used for SWAP-70 knockdown studies, T98 and SNB19, shared similar cytosolic SWAP-70 distribution in unstimulated cells and relocalization to cell cortex after EGF treatment (Figure W1). Western blot confirmed that this change in SWAP-70 localization occurred without a change in the expression levels of SWAP-70 (Figure 2B).
Figure 2.
Characterization of SWAP-70 localization in glioma cell lines. (A) Redistribution of SWAP-70 after treatment of EGF. Glioma cells expressing SWAP-70 were treated with or without EGF (50 ng/ml) for 10 minutes and fixed with Alexa Fluor 488-conjugated donkey antigoat antibody and Texas Red-X phalloidin to visualize F-actin (red), followed by analysis with confocalmicroscopy. SWAP-70 is enriched at the cortical membrane and membrane ruffles after EGF stimulation (arrows). Arrows show staining for SWAP-70 at the leading edge in lamellipodia. (B) There was no change in total SWAP-70 protein expression levels after EGF treatment by Western blot (lower panels). SW70 indicates SWAP-70; TfR, transferrin receptor.
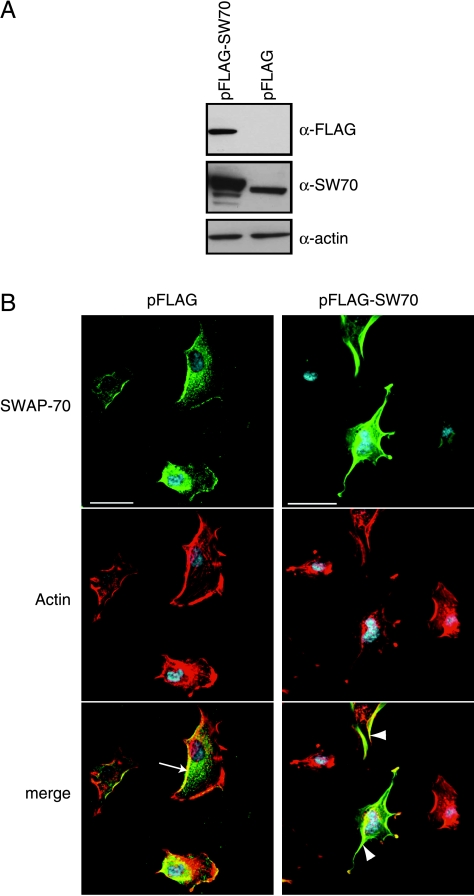
As described above, U87 glioma cells were transfected with FLAG-SWAP-70 and assessed by Western blot analysis (Figure 3A). Exogenous SWAP-70 was elevated in SWAP-70-transfected U87 glioma cells compared with pFLAG empty vector controls. Overexpression of SWAP-70 in U87 glioma cells did not cause a decrease in cell viability when measured at 24, 48, 72, and 96 hours after transfection in anMTS cell growth assay (data not shown). In serum-starved U87 cells transfected with the pFLAG empty vector control construct, SWAP-70 was mainly localized in the perinuclear cytoplasm and actin-rich membrane ruffles at the leading edge of the cells. SWAP-70 overexpression lead to cells having a more stellate morphology (Figure 3B).
Figure 3.
Overexpression of SWAP-70 induces changes in cell morphology. (A) U87 cells were selected for SWAP-70 overexpression given the relatively low endogenous levels of SWAP-70 and transient transfection with pFLAG-SWAP-70. (Single band in pFLAG lane is nonspecific cross-reacting protein that runs at a slightly lower molecular weight than SWAP-70.) (B) SWAP-70 was mainly localized to the perinuclear cytoplasm and actin-rich membrane ruffles at the leading edge (arrow) in serum-starved pFLAG-U87 cells. pFLAG-SWAP-70 cells showed slightly increased membrane ruffles and redistribution of SWAP-70, even in serum-starved condition, and cells appeared to have increased projections giving the cells a stellate appearance. Scale bar, 10 µm. SW70 indicates SWAP-70.
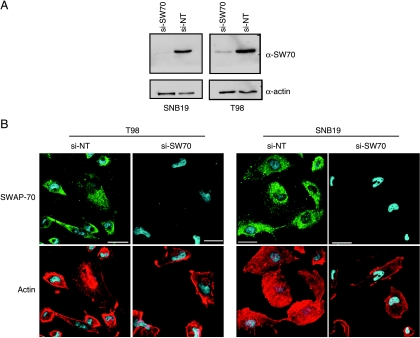
SWAP-70 knockdown with a cocktail of four independent SWAP-70 siRNA sequences resulted in a significant decrease in SWAP-70 protein levels within 48 hours after siRNA transfection (Figure 4A) and reached maximum depletion at 72 hours (data not shown). No reduction in SWAP-70 levels was observed with nontargeting siRNA controls. Depletion of SWAP-70 had no effect on cell viability in T98 or SNB19 cells when measured by an MTS cell growth assay (data not shown). In siRNA-treated T98 (Figure 4B, left panels) and SNB19 (Figure 4B, right panels) glioma cells, SWAP-70 depletion had modest effects on cell morphology but cells tended to be rounder and smaller.
Figure 4.
SWAP-70 knockdown alters glioma cell morphology. (A) T98 and SNB19 glioma cells were treated with either nontargeting or SWAP-70 siRNA and SWAP-70 expression was assessed 72 hours after transfection. Greater than 80% knockdown was observed in both cell lines. (B) The morphology of T98 (left panels) and SNB19 (right panels) glioma cells after SWAP-70 siRNA transfection tended to be rounder and smaller and had a reduced number of membrane ruffles compared with nontargeting siRNA controls (left panels). si-NT indicates non-targeting siRNA; si-SW70, siRNA-SWAP-70.
Modulation of SWAP-70 Expression Alters Glioma Cell Adhesion to ECM
To further examine the importance of Rac-GEFs during the invasive process of glioma cells, we selected SWAP-70 as another candidate proinvasive gene in gliomas. We based our selection by virtue of its grade-dependent increase in gene expression and its correlation to poor survival. Accordingly, we conducted experiments to measure cell adhesion, migration, and invasion when SWAP-70 is overexpressed or depleted in glioma cells. Furthermore, we examined the varying effects of different ECM proteins as a way to determine context-specific function, which may further lead us to clues to understanding the underlying mechanisms mediating SWAP-70 function.
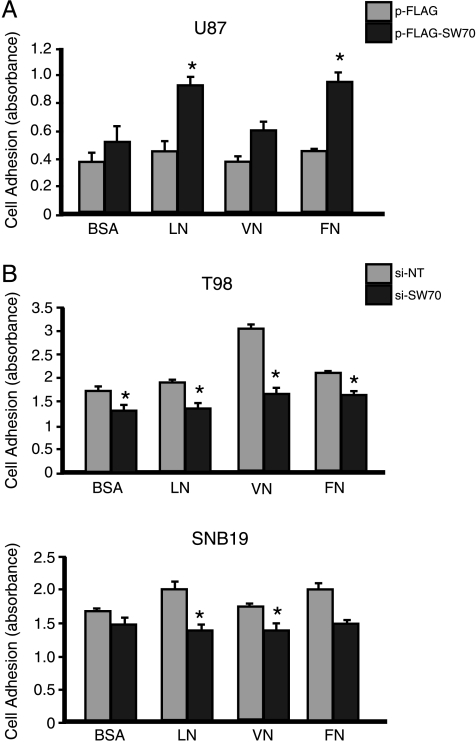
First, we performed a crystal violet cell attachment assay. The adherence of U87 glioma cells to LN, VN, and FN was tested by plating U87 cells transfected with empty vector or FLAG-SWAP-70 onto the appropriate ECM-coated substrates. Adhesion of empty vector-transfected U87 cells was not enhanced on LN, VN, or FN above that on BSA only. However, overexpression of SWAP-70 significantly increased the adhesion of U87 glioma cells on both LN and FN (Figure 5A, P = .001 and P = .006, respectively). In contrast, SWAP-70 knockdown in T98 cells inhibited glioma cell adhesion; however, only cells on VN and not those on LN or FN further inhibited the ability of cells to adhere (Figure 5B). SWAP-70 depletion in SNB19 cells had virtually no difference on the adhesion of cells on BSA when compared with NT control (P = .06). Although, in contrast to T98 cells, adhesion on LN and FN was slightly more attenuated in the absence of SWAP-70 cells when compared with cells on BSA. Overall, these data show that SWAP-70 has modest effects on cell adhesion, and the effects of ECM, if any, vary by cell line.
Figure 5.
Effects of SWAP-70 expression on glioma cell adhesion. (A) U87 cells overexpressing SWAP-70 or (B) T98 and SNB19 cells transfected with SWAP-70 siRNA were plated and allowed to adhere for 3 hours at 72 hours after transfection. *P < .05, statistically significant change in adhesion after modification of gene expression levels. Columns represent the average optical absorbance of three independent experiments. Bars, SE.
SWAP-70 Expression Alters Glioma Migration and Invasion
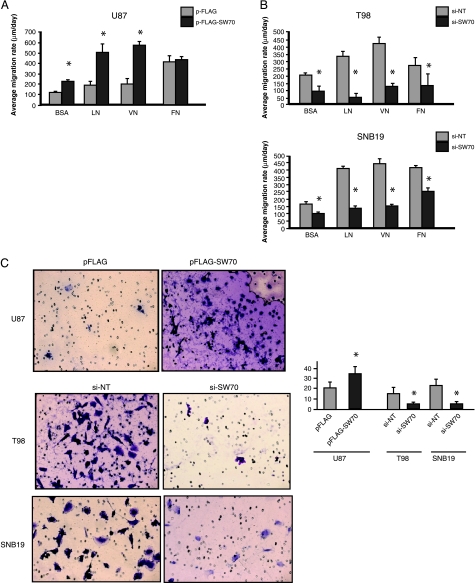
To investigate whether SWAP-70 plays a role in the migratory process of human glioma cells, we used radial migration assays with the same ECM macromolecules as in the attachment assays described above. As expected, the migration rate of all three glioma cell lines was slightly to greatly enhanced when placed on ECM macromolecules. In SWAP-70-transfected U87 glioma cells, the rate of migration was increased on BSA by almost two-fold when compared with empty vector control. However, SWAP-70-transfected cells plated on LN or VN resulted in a nearly three-fold increase in migration, whereas FN attenuated the migration rate of these cells (Figure 6A). Although migration in SWAP-70-depleted T98 cells was reduced nearly two-fold, migration rate of these cells was decreased nearly six-fold on LN and three-fold on VN (Figure 6B, upper panel), but no added effect was seen on FN when compared with cells on BSA. Similarly, in SNB19 glioma cells, SWAP-70 knockdown decreased the migration rate on BSA by slightly less than two-fold; however, migration was decreased by approximately three-fold on LN and VN, but no added effect was seen on FN (Figure 6B, lower panel). Together, these data suggest that SWAP-70 is required for enhanced cellmigration on LN and VN but is less important for enhanced migration seen on FN. Therefore, signaling through LN and VN proteins may be important components of the SWAP-70-Rac signaling node.
Figure 6.
Effects of SWAP-70 expression on glioma cell migration and invasion. Radial cell migration assays were performed on (A) U87 glioma cells overexpressing SWAP-70 or (B) T98 and SNB19 glioma cells transfected with SWAP-70 siRNA. Cells were transfected, then 48 hours after transfection, the cells were seeded and allowed to migrate for 24 hours. Images were acquired before and after migration, and the average migration rate was calculated during 24 hours. *P < .05, significant changes in migration after transfection. (C) Matrigel cell invasion assay in transient overexpressing and gene-silenced glioma cell lines. A modified Boyden chamber with Matrigel-precoated membrane filter insert was used to measure in vitro invasiveness. At 24 hours after seeding the cells, the invading cells were fixed and stained. Representative images are shown (left panels). Original magnification, x200. Invasive cells were counted in six random fields. Invasiveness was increased in U87 cells overexpressing SWAP-70. In contrast, the invasiveness was decreased in both T98 and SNB19 glioma cells in which SWAP-70 protein levels had been knocked down by RNA interference. *P < .05, significant changes in invasion after modulation of SWAP-70 expression levels.
Given that SWAP-70 plays a role in glioma cell adhesion and migration, we tested the effect of the modulation of SWAP-70 expression on glioma invasion. In U87 that overexpressed SWAP-70, the cells were more invasive through transwell filters than their parental counterpart (P = .0005; Figure 6C). Invasive potential decreased in SWAP-70-depleted T98 cells (P = .009) and, similarly, in SNB19 in which SWAP-70 was knocked down (P = .0003; Figure 6C).
Role of SWAP-70 Expression on Rac Activation
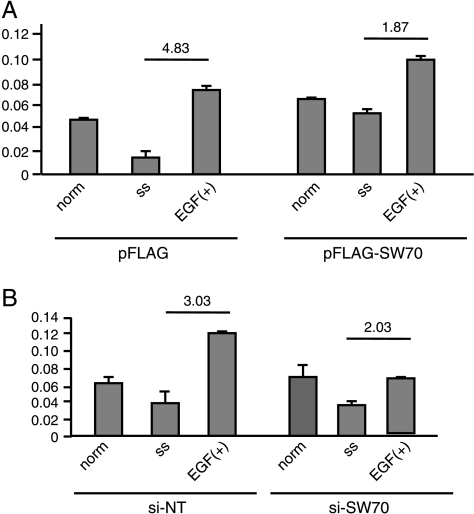
Given that SWAP-70 is a Rac-GEF, we were interested in determining the effect of SWAP-70 on Rac activation levels in glioma cells. To determine and compare the relative amounts of GTP-bound Rac in the different glioma cell lines before and after EGF stimulation, we measured active Rac with an ELISA-based method in SWAP-70 transfected U87 glioma cells. In addition, SWAP-70 siRNA-transfected T98 glioma cells and controls were measured in normal culture conditions. Initially, the maximal activation of Rac was optimized using EGF after 24 hours of serum starvation. In both U87 and T98 cells, the maximum activation peak of active Rac was observed at 12 minutes after treatment with 10 ng/ml EGF (Figure W2). Therefore, at this time point, we measured the amount of GTP-bound Rac in normal culture conditions, in serum-starved cells, and in EGF-treated cells (Figure 7). pFLAG-SWAP-70-transfected U87 cells showed a 3.5-times higher level of active Rac in serum-starved conditions than pFLAG U87 cells did. In addition, the extent of increase after EGF treatment was lower in pFLAG-SWAP-70-transfected cells: 4.83 in pFLAG and 1.87 in pFLAG-SWAP-70, respectively (Figure 7A). These findings suggest that Rac can be activated even in serum-starved conditions if SWAP-70 is overexpressed. In SWAP-70-depleted T98 cells, Rac-GTP levels increased more than two-fold after EGF treatment compared with nearly three-fold in nontargeting siRNA cells (Figure 7B), indicating that depletion of SWAP-70 protein reduces Rac activation in glioma cells.
Figure 7.
Rac1 activation assay in SWAP-70 transient-overexpressing and gene-silencing cell lines. Glioma cells were harvested at the optimized 12-minute time point after EGF treatment. The amount of GTP-bound Rac of normal culturing cells, serum-starved cells, and EGF-treated cells wasmeasured byRac1 ELISA.Upper number shows the extent of increase after EGF treatment. (A) Rac1 could be activated even in serum-starved condition in SWAP-70-overexpressing cell line. (B) In contrast, activation of Rac1 was inhibited in siRNA-SWAP-70 cells compared with nonsilencing controls. EGF indicates EGF treated after serum starvation; norm, normal culturing condition; SS, serum starvation.
Discussion
In this study, we show for the first time that the Rac-GEF SWAP-70 is an important mediator of glioma cell migration and invasion. The expression of SWAP-70 was increased in gliomas when compared with nonneoplastic brain, and this was correlated directly with increasing grade of astrocytic malignancy and poor patient survival. Overexpression of SWAP-70 in glioma cells increased adhesion to ECM, migration, and invasiveness, whereas elimination of SWAP-70 expression by siRNA had the opposite effects. The results of our Rac activation confirmed that SWAP-70 does indeed activate Rac and is associated with growth factor-mediated membrane ruffle formation.
As a Rac-GEF, SWAP-70 has been shown to activate Rac and is involved in signaling downstream of PI3-kinase-linked tyrosine kinase receptors [28]. Among the signal pathways activated by growth factors, PI3-kinase and Rac play important roles in regulating membrane ruffling. By stimulating tyrosine kinase receptors with growth factors, active PI3-kinase catalyzes to produce PtdIns(3,4,5)P3. It has been suggested that SWAP-70 acts as an effector or adaptor in response to PI3-kinase activity and is therefore important in regulating the actin cytoskeleton and cell motility by promoting PtdIns(3,4,5)P3-dependent activation of Rac [17,21,27,28]. The colocalization and direct binding of SWAP-70 with F-actin have been reported, especially when membrane ruffling or Rac activation is induced by growth factors such as EGF or platelet derived growth factor [27].
SWAP-70 contains an EF-hand motif at its N-terminal, a pleckstrin homology domain at the center, and an extended coiled-coil region at the C-terminal region [16,19]. Full-length SWAP-70 does not colocalize with F-actin unless cells are stimulated with growth factors, which suggests the presence of a stimuli-dependent regulatory mechanism for actin-binding activity in vivo. Gene mutation studies by Ihara et al. [27] showed that the binding and colocalization of SWAP-70 to F-actin depends on the C-terminal region of SWAP-70. In addition, a study by Wakamatsu et al. [29] revealed the crucial role that the pleckstrin homology domain of SWAP-70 plays in facilitating the binding of SWAP-70 to activated Rac through PtdIns(3,4,5)P3.
Previous studies have shown SWAP-70 to be expressed in various tissues, such as heart, kidney, spleen, and liver [17]. Interestingly, SWAP-70 is not expressed to any significant degree in the brain. Previous studies have demonstrated the translocation of SWAP-70 to membrane ruffles by EGF [27,29]. The morphology and Western blot studies performed in our study support the results of Ihara et al. Shinohara et al. [28] showed that overexpression of SWAP-70 resulted in an increase in activated Rac concentration. Previous studies have suggested that SWAP-70 cooperates with activated Rac [17,21,27,28]. Furthermore, SWAP-70 itself might increase the activation of Rac and facilitate the recruitment of active Rac to the regions of membrane ruffles.
In summary, in our study, a higher level of SWAP-70 was expressed in malignant gliomas, including glioblastomas, than in normal brain. In addition, through the use of gene manipulation studies, we have shown that SWAP-70 alters glioma cell motility and invasion. These results likely correlate with the level of GTP-bound activated Rac. Recently, SWAP-70 was found to enhance the invasive phenotype of Src-transformed mouse embryonic fibroblasts [30,31]. Without SWAP-70, the Src-transformed mouse embryonic fibroblasts were found to be significantly less invasive than their parental counterparts, and expression of both SWAP-70 and Src induced constant membrane ruffling not found with Src alone [31]. A study by Sharma and Mayer [32] found that Src phosphorylates p130Cas resulting in specific activation of Rac1 and induction of membrane ruffling and migration events. Future studies could include examination of this signaling complex to determine how the interaction of SWAP-70 and Rac relates to Src signaling through p130Cas.
In addition, our future studies will examine how SWAP-70 functions in vivo. To this end, it would be interesting to study mice harboring intracranial gliomas derived from cell lines that have SWAP-70 expression knocked down or overexpressed and examine in vivo tumor growth and invasiveness. SWAP-70 now joins the ranks of other GEFs, such as Trio, ECT2, and Vav3, as important elements in the Rho-GTPase pathway, which plays a role in glioma invasion and motility.
Supplementary Material
Footnotes
This work was supported by a grant from the Canadian Institutes of Health Research (MOP-74610) and by the Wiley Fund and b.r.a.i.n.child. Ho Jun Seol was supported by Kangwon National University.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.transonc.com.
References
- 1.Burger PC, Vogel FS, Green SB, Strike TA. Glioblastoma multiforme and anaplastic astrocytoma. Pathologic criteria and prognostic implications. Cancer. 1985;56(5):1106–1111. doi: 10.1002/1097-0142(19850901)56:5<1106::aid-cncr2820560525>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 2.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21(8):1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 3.Salhia B, Tran NL, Symons M, Winkles JA, Rutka JT, Berens ME. Molecular pathways triggering glioma cell invasion. Expert Rev Mol Diagn. 2006;6(4):613–626. doi: 10.1586/14737159.6.4.613. [DOI] [PubMed] [Google Scholar]
- 4.Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol. 2004;70(2):217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 5.Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84(3):371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- 6.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84(3):359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 7.Bolteus AJ, Berens ME, Pilkington GJ. Migration and invasion in brain neoplasms. Curr Neurol Neurosci Rep. 2001;1(3):225–232. doi: 10.1007/s11910-001-0022-x. [DOI] [PubMed] [Google Scholar]
- 8.Salhia B, Hwang JH, Smith CA, Nakada M, Rutka F, Symons M, Rutka JT. Role of myosin II activity and the regulation of myosin light chain phosphorylation in astrocytomas. Cell Motil Cytoskeleton. 2008;65(1):12–24. doi: 10.1002/cm.20240. [DOI] [PubMed] [Google Scholar]
- 9.Salhia B, Rutten F, Nakada M, Beaudry C, Berens M, Kwan A, Rutka JT. Inhibition of Rho-kinase affects astrocytoma morphology, motility, and invasion through activation of Rac1. Cancer Res. 2005;65(19):8792–8800. doi: 10.1158/0008-5472.CAN-05-0160. [DOI] [PubMed] [Google Scholar]
- 10.Chan AY, Coniglio SJ, Chuang YY, Michaelson D, Knaus UG, Philips MR, Symons M. Roles of the Rac1 and Rac3 GTPases in human tumor cell invasion. Oncogene. 2005;24(53):7821–7829. doi: 10.1038/sj.onc.1208909. [DOI] [PubMed] [Google Scholar]
- 11.Chuang YY, Tran NL, Rusk N, Nakada M, Berens ME, Symons M. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 2004;64(22):8271–8275. doi: 10.1158/0008-5472.CAN-04-2097. [DOI] [PubMed] [Google Scholar]
- 12.Walmod PS, Hartmann-Petersen R, Prag S, Lepekhin EL, Ropke C, Berezin V, Bock E. Cell-cycle-dependent regulation of cell motility and determination of the role of Rac1. Exp Cell Res. 2004;295(2):407–420. doi: 10.1016/j.yexcr.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Nomura N, Nomura M, Mizuki N, Hamada J. Rac1 mediates phorbol 12-myristate 13-acetate-induced migration of glioblastoma cells via paxillin. Oncol Rep. 2008;20(4):705–711. [PubMed] [Google Scholar]
- 14.Hu B, Shi B, Jarzynka MJ, Yiin JJ, D'Souza-Schorey C, Cheng SY. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res. 2009;69(3):794–801. doi: 10.1158/0008-5472.CAN-08-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, Ennis M, McDonough WS, Berens ME, Symons M, et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173(6):1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borggrefe T, Wabl M, Akhmedov AT, Jessberger R. A B-cell-specific DNA recombination complex. J Biol Chem. 1998;273(27):17025–17035. doi: 10.1074/jbc.273.27.17025. [DOI] [PubMed] [Google Scholar]
- 17.Hilpela P, Oberbanscheidt P, Hahne P, Hund M, Kalhammer G, Small JV, Bahler M. SWAP-70 identifies a transitional subset of actin filaments in motile cells. Mol Biol Cell. 2003;14(8):3242–3253. doi: 10.1091/mbc.E03-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivalenka RR, Jessberger R. SWAP-70 regulates c-kit-induced mast cell activation, cell-cell adhesion, and migration. Mol Cell Biol. 2004;24(23):10277–10288. doi: 10.1128/MCB.24.23.10277-10288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masat L, Caldwell J, Armstrong R, Khoshnevisan H, Jessberger R, Herndier B, Wabl M, Ferrick D. Association of SWAP-70 with the B cell antigen receptor complex. Proc Natl Acad Sci USA. 2000;97(5):2180–2184. doi: 10.1073/pnas.040374497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross B, Borggrefe T, Wabl M, Sivalenka RR, Bennett M, Rossi AB, Jessberger R. SWAP-70-deficient mast cells are impaired in development and IgE-mediated degranulation. Eur J Immunol. 2002;32(4):1121–1128. doi: 10.1002/1521-4141(200204)32:4<1121::AID-IMMU1121>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Li D, Cao B, Li YX, Herva R, Piao YS, Wang YL. Expression and localization of SWAP-70 in human fetomaternal interface and placenta during tubal pregnancy and normal placentation. J Histochem Cytochem. 2007;55(7):701–708. doi: 10.1369/jhc.6A7151.2007. [DOI] [PubMed] [Google Scholar]
- 22.Borggrefe T, Masat L, Wabl M, Riwar B, Cattoretti G, Jessberger R. Cellular, intracellular, and developmental expression patterns of murine SWAP-70. Eur J Immunol. 1999;29(6):1812–1822. doi: 10.1002/(SICI)1521-4141(199906)29:06<1812::AID-IMMU1812>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 23.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 24.Sriramarao P, Mendler M, Bourdon MA. Endothelial cell attachment and spreading on human tenascin is mediated by α2β1 and αvβ3 integrins. J Cell Sci. 1993;105(Pt 4):1001–1012. doi: 10.1242/jcs.105.4.1001. [DOI] [PubMed] [Google Scholar]
- 25.Hwang JH, Smith CA, Salhia B, Rutka JT. The role of fascin in the migration and invasiveness of malignant glioma cells. Neoplasia. 2008;10(2):149–159. doi: 10.1593/neo.07909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37(2):208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Ihara S, Oka T, Fukui Y. Direct binding of SWAP-70 to non-muscle actin is required for membrane ruffling. J Cell Sci. 2006;119(Pt 3):500–507. doi: 10.1242/jcs.02767. [DOI] [PubMed] [Google Scholar]
- 28.Shinohara M, Terada Y, Iwamatsu A, Shinohara A, Mochizuki N, Higuchi M, Gotoh Y, Ihara S, Nagata S, Itoh H, et al. SWAP-70 is a guaninenucleotide-exchange factor that mediates signalling of membrane ruffling. Nature. 2002;416(6882):759–763. doi: 10.1038/416759a. [DOI] [PubMed] [Google Scholar]
- 29.Wakamatsu I, Ihara S, Fukui Y. Mutational analysis on the function of the SWAP-70 PH domain. Mol Cell Biochem. 2006;293(1–2):137–145. doi: 10.1007/s11010-006-9236-1. [DOI] [PubMed] [Google Scholar]
- 30.Fukui Y, Tanaka T, Tachikawa H, Ihara S. SWAP-70 is required for oncogenic transformation by v-Src in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2007;356(2):512–516. doi: 10.1016/j.bbrc.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Murugan AK, Ihara S, Tokuda E, Uematsu K, Tsuchida N, Fukui Y. SWAP-70 is important for invasive phenotypes of mouse embryo fibroblasts transformed by v-Src. IUBMB Life. 2008;60(4):236–240. doi: 10.1002/iub.33. [DOI] [PubMed] [Google Scholar]
- 32.Sharma A, Mayer BJ. Phosphorylation of p130Cas initiates Rac activation and membrane ruffling. BMC Cell Biol. 2008;9:50. doi: 10.1186/1471-2121-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.