Abstract
5-Fluorouracil (5-FU) or 5-FU plus oxaliplatin (FOLFOX) remains the backbone of colorectal cancer chemotherapeutics but with limited success. This could partly be due to the enrichment of cancer stem cells (CSCs) that are resistant to conventional chemotherapy. Therefore, validation of a nontoxic agent that can either cause reversal of chemoresistance or promote the killing of CSCs would be highly desirable. The current study examines whether curcumin, the major active ingredient of turmeric, either alone or together with FOLFOX, would be an effective strategy to eliminate colon CSCs. Exposure of colon cancer HCT-116 or HT-29 cells to FOLFOX that inhibited their growth led to the enrichment of CSC phenotype as evidenced by increased proportion of CD133-, CD44-, and/or CD166-positive cells and epidermal growth factor receptor (EGFR) levels. Treatment of FOLFOX-surviving colon cancer cells with either curcumin alone or together with FOLFOX resulted in a marked reduction in CSCs, as evidenced by the decreased expression of CD44 and CD166 as well as EGFR and by their ability to form anchorage-dependent colonies. They also caused disintegration of colonospheres. Increased expression of EGFR in FOLFOX-surviving cells could be attributed to hypomethylation of the EGFR promoter, whereas an opposite phenomenon was observed when the FOLFOX-surviving cells were treated with curcumin and/or FOLFOX. These changes were accompanied by parallel alterations in the levels of DNA methyltransferase 1. In conclusion, our data suggest that curcumin by itself or together with the conventional chemotherapeutic could be an effective treatment strategy for preventing the emergence of chemoresistant colon cancer cells by reducing/eliminating CSCs.
Introduction
Colorectal cancer is the third most common cancer in both men and women constituting 10% of new cancer cases in men and 11% in women [1]. Despite the use of aggressive surgical resection and chemotherapy, nearly 50% of patients with colorectal carcinoma develop recurrent disease, highlighting the need for improved therapies [1].
There is a growing body of evidence that lend support to the concept that epithelial cancers including colorectal cancer are diseases driven by a subset of self-renewing cells, termed cancer stem cells (CSC) or cancer-initiating cells, that are distinct from bulk of the cells in the tumor [2,3]. CSCs possess the capacity for self-renewal, show the potential to develop into any cell in the overall tumor population, have the ability to drive continued expansion of the population of malignant cells, and invade and metastasize [2,3]. Thus, failure to eliminate them is thought to be one of the underlying causes for recurrence of malignancy. Therefore, therapeutic strategies that specifically target colon CSCs could be effective in eradicating colorectal cancer and in reducing the risk of relapse and metastasis.
F-Fluorouracil (5-FU) or 5-FU plus oxaliplatin (FOLFOX), which remains the backbone of colorectal cancer chemotherapeutics, produces incomplete response, resulting in survival of cells that often leads to cancer recurrence. Continued use of conventional chemotherapeutics is well known to be associated with added toxicities, some of which are even fatal. Therefore, validation of a nontoxic agent that could improve on the current chemotherapeutic regimen would be highly desirable.
Curcumin (diferuloylmethane), the major active ingredient of turmeric (Curcuma longa) with no discernable toxicity, inhibits the growth of transformed cells [4] and has also been shown to suppress initiation, promotion, and progression of colon carcinogenesis in carcinogen-induced rodent models [5,6]. In a phase 1 clinical trial, curcumin was found to be effective in inhibiting the growth of a variety of tumors [7]. We have recently demonstrated that curcumin, in combination with either 5-FU or FOLFOX, causes greater growth inhibition of HCT-116 or HT-29 colon cancer cells than each agent/regimen alone [8]. However, whether curcumin alone or in combination with FOLFOX would affect colon CSCs is unknown. The present investigation was, therefore, undertaken to examine 1) whether a portion of the FOLFOX-surviving colon cancer cells contain CSCs and 2) whether they could be eliminated by curcumin alone or together with FOLFOX.
Materials and Methods
Cell Lines and Cell Cultures
Human colon cancer HCT-116 and HT-29 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). Cells were maintained in Dulbecco's modified Eagle medium (DMEM; 4.5 g/L d-glucose) supplemented with 10% FBS and 1% antibiotic/antimycotic in tissue culture flasks in a humidified incubator at 37°C in an atmosphere of 95% air and 5% carbon dioxide. The medium was changed two times a week, and cells were passaged using 0.05% trypsin/EDTA.
Unless otherwise stated, FOLFOX-surviving colon cancer cells were generated by incubating HCT-116 or HT-29 cells with a mixture of 50 µM 5-FU and 1.25 µM oxaliplatin (FOLFOX) for 48 hours. The adherent cells, which survived the FOLFOX insult, were subjected to trypsin/EDTA treatment.
In some experiments, FOLFOX-resistant colon cancer cells were used. They were generated by exposing HCT-116 or HT-26 cells to FOLFOX at clinically relevant doses and schedules. The exposing schedule was for 12 cycles; each cycle lasted for a week. Briefly, the cells were first exposed to FOLFOX (25 µM 5-FU and 0.625 µM oxaliplatin) for 72 hours. The surviving cells were then cultured in normal medium without the drugs for 4 to 5 days. The cycle was repeated 12 times. The surviving cells were then split and exposed to higher doses of FOLFOX (50 µM 5-FU + 1.25 µM oxaliplatin or 100 µM 5-FU + 2.5 µM oxaliplatin) for 2 to 3 d/wk for approximately 4 weeks. Finally, the resistant cells were maintained in normal culture medium containing a low dose of FOLFOX (5 µM 5-FU + 0.125 µM oxaliplatin).
Growth Inhibition Assay
Inhibition of cell growth in response to curcumin and/or FOLFOX was assessed by 3-(4,5-dimethylthiazol-2yl)-2, 5-diphenyltetrazolium bromide (MTT) assay as described previously [8]. Briefly, cells were dispersed by trypsin-EDTA treatment and 2.5 x 104 cells per milliliter, resuspended in DMEM containing 10% FBS, and seeded into 96-well culture plates with six replicates. After 24 hours of plating, incubation was continued for another 48 hours in the absence (control) or presence of different testing agents as described in the legends to the figures. At the end of the 48-h incubation period, the reaction was terminated by adding 20 µl of 5-mg/ml stock of MTT to each well. The reaction was allowed to proceed for 3 to 4 hours at 37°C. The culture medium was then removed. The formazan crystals were then dissolved by adding 0.1 ml of dimethyl sulfoxide. The intensity of the color that developed, which is the reflection of number of live cells, was measured at a wavelength of 570 nm. All values were compared with the corresponding controls. All assays were performed with six replicates.
Western Blot Analysis
Western blot analysis was performed essentially according to our standard protocol [8,9]. Briefly, the cells were solubilized in lysis buffer (50 mM Tris, 100 mM NaCl, 2.5 mM EDTA, 1% Triton X-100, 1% Nonidet P-40, 2.5 mM Na3VO4, 25 µg/ml aprotinin, 25 µg/ml leupeptin, 25 µg/ml pepstatin A, and 1 mM phenylmethylsulfonyl fluoride). After clarification at 10,000g for 15 minutes, the supernatant was used for Western blot analysis. In all analyses, protein concentration, determined by the Bio-Rad Protein Assay kit (Bio-Rad, Hercules, CA), was standardized among the samples. Aliquots of cell lysates containing 50 µg of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After electrophoresis, proteins were transferred electrophoretically onto supported nitrocellulose membranes (Osmonics, Gloucester, MA). Membranes were incubated for 1 hour at room temperature with blocking buffer, TBS-T (20 mM Tris, pH7.6, 100 µM NaCl, 0.1% Tween-20) and 5% nonfat dry milk with gentle agitation. After washing the membranes with TBS-T, they were incubated overnight at 4°C in TBS-T buffer containing antibody dilution buffer as suggested by the manufacturer and with antibodies (1:1000 dilution) to CD44, CD166 (Santa Cruz Biotechnology, Santa Cruz, CA), or epidermal growth factor receptor (EGFR; Cell Signaling, Beverly, MA). The membranes were washed three times with TBS-T and subsequently incubated with appropriate secondary antibodies (1:5000 dilutions) in TBS-T containing 5% milk for 1 to 2 hours at room temperature with gentle agitation. The membranes were washed again with TBS-T, and the protein bands were visualized by enhanced chemiluminescence (ECL) detection system (Amersham, Piscataway, NJ). The membranes containing the electrophoresed proteins were exposed to X-Omat film (Sigma-Aldrich, St Louis, MO). The membranes were stripped (2 x for 15 minutes at 55°C) in stripping buffer containing 100 mM 2-mercaptoethanol, 2% sodium dodecyl sulfate, and 62.5 mM Tris-HCl pH 6.7, and reprobed for β-actin using corresponding antibodies, which was used as a loading control. All Western blots were performed at least three times for each experiment.
Isolation of RNA and Quantitative Polymerase Chain Reaction Analysis
Total RNA was extracted from parental and FOLFOX-surviving HCT-116 cells using RNA-STAT solution (Tel Test, Friendswood, TX) according to the manufacturer's instruction. The total RNA was treated with DNase I to remove contaminating genomic DNA, subsequently purified using RNAeasy Mini Kit (Qiagen, Valencia, CA). RNA concentration was measured spectrophotometrically at an optical density of 260 nm.
Quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed using the GeneAmp RNA PCR Kit (Applied Biosystems, Foster City, CA). Briefly, 1 µg of purified RNA was reverse-transcribed in the presence of 2.5 mM MgCl2, 1x RT-PCR buffer, 1 mM dNTPs, 10 mM dithiothreitol, 10 U of RNase inhibitor, 1.25 µM random hexamers, and 15 U of MultiScribe Reverse Transcriptase in a final reaction volume of 20 µl. Mix the components, briefly spin down, and incubate at 25°C for 10 minutes for hybridization. The reactions were carried out at 42°C for 15 minutes in a GeneAmp PCR System 9600 (Perkin-Elmer) and then by cooling to 4°C. The RT reactions were subjected to quantitative PCR amplification. Five microliters of complementary DNA products was amplified with SYBR Green Quantitative PCR Master Mix (Applied Biosystems). PCR primers were used as follows: CD44 forward: 5′-aaggtggagcaaacacaacc-3′ and reverse: 5′-aactgcaatgcaaactgcaag-3′, CD166 forward: 5′-tagcaggaatgcaactgtgg-3′ and reverse: cgcagacatagtttccagca-3′, and β-actin forward: 5′-cccagcacaatgaagaatcaa-3′ and reverse 5′-acatctgctggaaggtggtggac-3′. Reactions were carried out in Applied Biosystems 7500 Real-Time PCR System; first, hold for 10 minutes at 95°C for activated DNA polymerase and were followed by 15 seconds at 95°C and 60 seconds at 60°C for 40 cycles.
Methylation-Specific PCR of Human EGFR Promoter
Genomic DNA was extracted from HCT-116 Cells, FOLFOX-Surviving HCT-116, and the treated survivor cells as described previously [10]. Bisulfite modification of DNA to convert all unmethylated cytosines to uracil and then to thymidine during the subsequent PCR step while leaving the methylated cytosines unaffected was done as described by Herman et al. [11]. Modified DNA was desulfonated and purified using the Zymo-Spin IC columns according to the manufacturer's instructions (ZYMO Research, Orange, CA) eluted into 10 µl of water. The bisulfite-treated DNA was used for PCR amplification of a human EGFR promoter fragment. Amplification of a methylated 103-bp product (from positions 573 to 675; accession no. gi3118) was carried out using a combination of forward (5′-TTTAGAGGGGTAGTGTTGGGAACGT-3′) and reverse primers (5′-GCGAAACCTAATCTCCGACGAA-3′). The primers for amplification of the unmethylated 107-bp DNA fragment were 5′-GTTTAGAGGGGTAGTGTTGGGAATGT-3′ (forward primer) and 5′-CCCACAAAACCTAATCTCCAACAAA-3′ (reverse primer). The forward primer for the methylated template had a single mismatch at the 3′-end, whereas the reverse primer for the methylated template has two mismatches at the 3′-end. These mismatches were expected to confer sufficient selectivity and specificity for amplification of the methylated template. For PCR amplification, the annealing temperature was 65°C. PCR amplification was done by using 10 ng of treated DNA as the template in a final volume of 25 µl. Reactions were hot-started at 95°C for 5 minutes before amplification in an Eppendorf thermal cycler for 40 cycles (30 seconds at 95°C, 30 seconds at 65°C, and 30 seconds at 70°C) followed by a final 4-minute extension at 72°C. Controls without DNA were done for each set of PCRs. Each PCR product (10 µl) was directly electrophoresed and visualized after ethidium bromide staining of 3% agarose gels. Densitometric measurements of the scanned bands were performed using the digitized scientific software program UN-SCAN-IT (Silk Scientific, Orem, UT). Data were normalized to β-actin.
Flow Cytometric Analyses
Parental (control) and FOLFOX-surviving colon cancer cells were subjected to direct immunofluorescence staining followed by flow cytometric analyses. Briefly, the cells were harvested and washed with PBS. Half a million cells were suspended in 90 µl of PBS containing 0.5% BSA for 10 minutes at room temperature followed by the addition of 10 µl of Alexa Fluor 488 fluorescent dye conjugated to CD44 monoclonal antibody (Cell Signaling) and incubated for 30 minutes in the dark at room temperature. The samples were then washed and analyzed using a FACS DiVa (BD, San Jose, CA).
Colonosphere Formation or Disintegration
To examine the effects of different agents on the formation of colonospheres, the cells were suspended in serum-free stem cell medium containing DMEM/F12 (1:1) supplemented with B27 (Life Technologies, Gaithersburg, MD), 20 ng/ml EGF (Sigma, St Louis, MO), 10 ng/ml fibroblast growth factor (Sigma), and antibiotic-antimycotic. The cells (200 per well) were then plated in an ultra low-attachment 96-well plate (Corning Inc, Lowell, MA). Once the colonospheres were formed after 10 days, they were incubated in the absence (controls) or presence of the testing agent(s) for 5 days and evaluated for the number of colonospheres by light microscopy.
Statistical Analysis
Unless otherwise stated, data are expressed as mean ± SEM. Where applicable, the results were analyzed using analysis of variance followed by Fisher protected least significant differences or Scheffé test. P < .05 was designated as the level of significance.
Results
Currently, CSCs are identified by specific surface epitopes. Colon CSCs have been shown to express surface markers CD44, CD166, CD133, and epithelial-specific antigen (also known as EpCAM) [12,13]. CSCs also show resistance to a number of conventional therapies [14], which may explain why it is difficult to completely eradicate cancer and why recurrence is an ever-present threat.
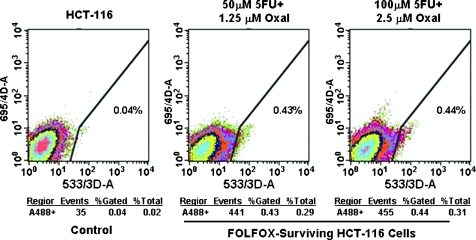
The first set of experiments was undertaken to determine whether colon CSCs are present in HCT-116 cells and whether their proportion changes by FOLFOX. The FOLFOX-resistant colon cancer HCT-116 cells, which were generated by prolonged exposure to two different doses of FOLFOX and the parental HCT-116 cells (untreated controls), were analyzed for colon CSCs by tagging them with CD44 antibodies and subsequently sorting them by flow cytometry. The results revealed that the proportion of CD44-positive cells, representing colon CSCs, in untreated parental control cells consisted only 0.04% of total cells, whereas in FOLFOX-resistant HCT-116 cells, their proportion was increased by at least 10-fold (Figure 1). The results show a marked rise in colon cancer-like cells in FOLFOX-resistant colon cancer cells.
Figure 1.
Sorting of anti-CD44 antibodies-tagged untreated (control) and FOLFOX-resistant HCT-116 cells by flow cytometry. FOLFOX-resistant HCT-116 cells were generated by exposing HCT-116 to 25 µM 5-FU and 0.625 µM oxaliplatin for 48 to 72 hours and subsequently cultured them in normal medium without the drugs for 4 to 5 days. The cycle was repeated 12 times. The surviving cells were then split and exposed to higher doses of FOLFOX (50 µM 5-FU + 1.25 µM oxaliplatin or 100 µM 5-FU + 2.5 µM oxaliplatin) for 2 to 3 days a week for approximately 4 weeks. The FOLFOX-surviving cells were maintained in normal culture medium containing a low dose of FOLFOX (5 µM 5-FU + 0.125 µM oxaliplatin).
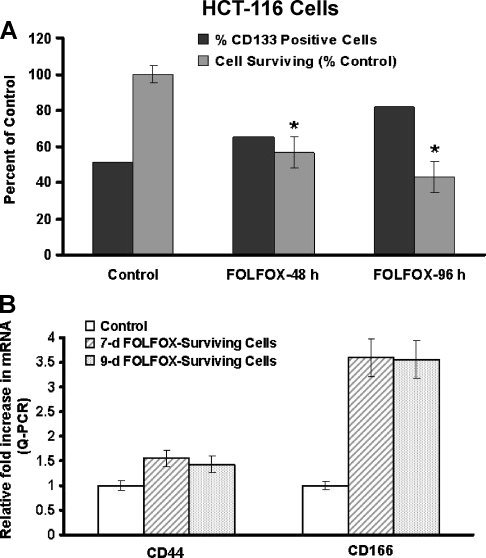
Previously, we have demonstrated that FOLFOX inhibits the growth of colon cancer cells in vitro [8]. However, whether and to what extent FOLFOX affects the growth of colon CSCs is unknown. To examine the effect of FOLFOX on the growth of colon CSCs, HCT-116 cells were incubated in the absence (controls) or presence of FOLFOX for 48 and 96 hours. They were then assayed for overall cell growth by MTT assay and for changes in the proportion of colon CSCs by examining the expression of CD133, CD44, and/or CD166. As expected, FOLFOX at a dose of 50 µM 5-FU + 1.25 µM oxaliplatin inhibited the growth of HCT-116 cells by 50% and 60% at 48 and 96 hours, respectively, and this inhibition was accompanied by a concomitant increase in the levels of CD133 when compared with the corresponding controls (Figure 2A). The relative concentrations of CD44 and CD166 messenger RNA (mRNA) were also found to be markedly higher (1.5- to 3.7-fold) in FOLFOX-surviving HCT-116 cells when compared with the corresponding controls (Figure 2B). Taken together, the results indicate that whereas FOLFOX inhibits the growth of HCT-116 cells, the proportion of colon CSCs increases after FOLFOX treatment.
Figure 2.
(A) Exposure of HCT-116 cells to FOLFOX (50 µM 5-FU + 1.12 µM oxaliplatin) for 48 and 96 hours inhibits growth but increases the proportion of CD133-positive cells as analyzed by flow cytometry after tagging with anti-CD133 antibodies. (B) Levels of CD44 and CD166 mRNA in HCT-116 cells that survived the continuous exposure of FOLFOX (50 µM 5-FU + 1.12 µM oxaliplatin) for 7 and 9 days. The controls represent the parental cells that were incubated with vehicle only. *P < .01, compared with the corresponding control.
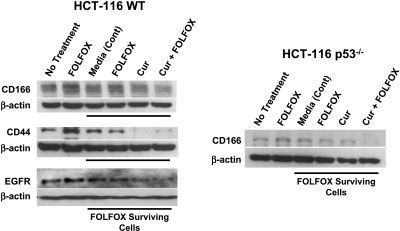
The primary objective of the current investigation was to test our hypothesis whether curcumin either alone or together with FOLFOX would eliminate colon CSCs. To accomplish this objective, HCT-116 (wt) and HCT-116 (p53-/-) cells, which survived the 72-hour exposure to FOLFOX, were incubated for another 72 hours with the media (controls), FOLFOX, curcumin, or a combination of curcumin and FOLFOX and subsequently analyzed for protein levels of CD44 and CD166. The FOLFOX-surviving p53-positive (wt) and p53-null (p53-/-) HCT-116 cells showed a marked increase in CD166 levels compared with untreated parental controls (Figure 3). The levels of CD44 in HCT-116 (wt) cells were also found to be higher in FOLFOX-surviving cells than in untreated controls (Figure 3). Likewise, EGFR levels were also found to be higher in FOLFOX-surviving HCT-116 (wt) cells compared with untreated parental controls (Figure 3). Exposure of FOLFOX-surviving HCT-116 (wt or p53-/-) cells to the same concentration of FOLFOX for another 72 hours produced no apparent change in CD166, CD44, or EGFR levels when compared with those incubated with the medium alone (Figure 3). In contrast, curcumin either alone or together with FOLFOX caused a marked reduction in CD166, CD44, and EGFR levels in FOLFOX-surviving HCT-116 cells when compared with the corresponding media-treated or untreated parental controls (Figure 3). These observations suggest that the combination of curcumin and FOLFOX is highly effective in eliminating most colon CSCs. Moreover, our observation that the combination of curcumin and FOLFOX was equally effective in inhibiting CD166 expression in HCT-116 (WT) and HCT-116 (p53-/-) cells suggests that the effect of the combination treatment on the elimination of colon CSCs is independent of p53 status.
Figure 3.
Western blot showing changes in the levels of CD166, CD44, and EGFR in parental (untreated control) and FOLFOX-surviving HCT-116 (wt) cells and CD166 in FOLFOX-surviving p53-/- HCT-116 cells after further incubation with the media, FOLFOX (50 µM 5-FU + 1.12 µM oxaliplatin), curcumin (20 µM), or curcumin + FOLFOX.
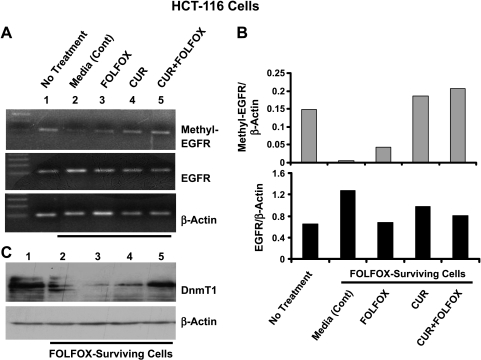
Figure 4.
(A) Representative photograph showing PCR-amplified methylated human EGFR promoter (103 bp; upper panel) and RT-PCR-generated unmethylated human EGFR (107 bp; middle panel) and β-actin (bottom panel). (B) Histogram of data from densitometric analyses of the bands in panel A, normalized for the corresponding β-actin. (C) Western blot showing changes in the levels of Dnmt1 (DNA 5-cytosine methyltransferase) in parental and FOLFOX-surviving HCT-116 cells that were incubated in the absence (control), or presence of curcumin (20 µM), FOLFOX (50 µM 5-FU + 1.12 µM oxaliplatin), or curcumin + FOLFOX.
Our current observation showing higher expression of CSC markers and EGFR in FOLFOX-surviving colon cancer HCT-116 cells over the parental cells suggests a relationship between colon CSCs and EGFR. The latter is known to play a crucial role in regulating several pathways that affect tumor cell survival, angiogenesis, motility, and invasiveness [15–17]. Although the mechanisms for increased expression of EGFR in FOLFOX-surviving HCT-116 cells and attenuation of the same with curcumin and/or FOLFOX are not known, we examined the possibility of whether they could, in part, be attributed to changes in methylation status of the EGFR promoter. We observed that the levels of EGFR mRNA, when normalized for the corresponding β-actin, was approximately 80% higher in media-treated FOLFOX-surviving HCT-116 cells than in untreated parental cells (Figure 4, A and B, lower panel). This increase was associated with a marked reduction (>90%) in methylation of the EGFR promoter when compared with the untreated parental cells (Figure 4, A and B, upper panel). Treatment of FOLFOX-surviving HCT-116 cells with either curcumin or FOLFOX, which inhibited EGFR expression, caused a partial or complete reversal of the EGFR methylation status when compared with the media-treated cells (Figure 4, A and B, upper panel). We have observed that whereas further treatment of FOLFOX-surviving cells with FOLFOX increased methylation of the EGFR promoter to only approximately 30% of the untreated parental cells, treatment with either curcumin alone or together with FOLFOX stimulated the same to 120% to 130% of the untreated parental cells (Figure 4, A and B, upper panel). To further determine the underlying mechanisms for the aforementioned changes in methylation status of the EGFR promoter in FOLFOX-surviving HCT-116 cells, we carried out Western blot analysis to examine the expression of DNA 5-cytosine methyltransferase 1 (Dnmt1), an enzyme that catalyzes methylation of DNA at cytosine residues in mammalian cells. Although three families of DNA methyltransferases have been identified, Dnmt1 is the most abundant, and is constitutively expressed in proliferating cells and functions as maintenance methyltransferase [18–20]. We have observed that the marked reduction (>90%) in methylation of the EGFR promoter in media-treated FOLFOX-surviving HCT-116 cells was associated with a concomitant decrease in Dnmt1 levels when compared with the untreated parental cells (Figure 4C). Further treatment of FOLFOX-surviving cells with FOLFOX, which produced only a small reversal of methylation of EGFR, did not produce any appreciable change in Dnmt1 levels over the media-treated FOLFOX-surviving cells (Figure 4C). In contrast, curcumin alone or together with FOLFOX, each of which completely reversed the methylation status of the EGFR promoter, increased Dnmt1 to the levels of untreated parental cells (Figure 4C).
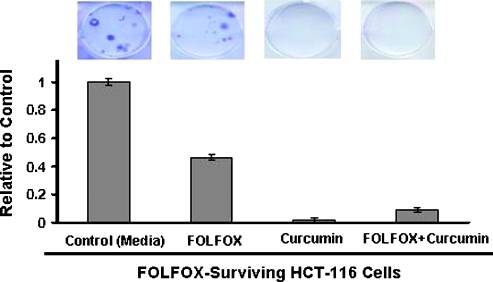
Use of surface epitopes as authentic markers of CSC remains controversial because of their presence in early progenitor cells [21]. In view of this, the next set of experiments was conducted where transformation properties of CSCs were examined. One of the transformation properties of CSCs is their ability to form colonies. Using anchorage-dependent colony formation as one of the properties of CSCs, we found FOLFOX-surviving HCT-116 cells to form colonies within approximately 3 weeks (Figure 5). Incubation of FOLFOX-surviving cells with either curcumin alone or together with FOLFOX resulted in a 95% to 99% reduction in colony formation (Figure 5). However, FOLFOX alone produced only approximately 50% reduction in colony formation when compared with the media-incubated controls (Figure 5).
Figure 5.
Representativephotograph showing anchorage-dependent colony formation by FOLFOX-surviving HCT-116 cells in the absence (control) or presence of FOLFOX (50 µM 5-FU + 1.12 µM oxaliplatin), curcumin (20 µM), or curcumin + FOLFOX.
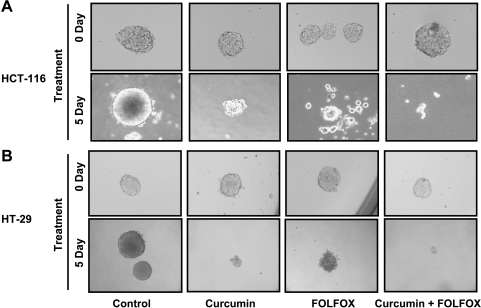
Another property of CSC is their ability to form spheroids [22]. Indeed, incubation of colon cancer HCT-116 and HT-29 cells in serum-free stem cell medium resulted in the formation of colonosphere during a period of 10 days. Incubation of colonosphere(s) with FOLFOX, curcumin, or curcumin together with FOLFOX for 5 days resulted in a marked disintegration of colonosphere(s) when compared with the corresponding controls (Figure 6A). Highest disintegration was observed in response to the combination of curcumin and FOLFOX (Figure 6).
Figure 6.
Representative photograph showing disintegration of colonospheres of HCT-116 (A) and HT-29 (B) cells after incubating with FOLFOX (50 µM 5-FU + 1.12 µM oxaliplatin), curcumin (20 µM), or curcumin + FOLFOX for 5 days. The controls were incubated with the medium only.
Discussion
The long-held view of tumor development, described in the stochastic model, is that every tumor cell is capable of initiating neoplastic growth. However, this theory has been challenged by the CSC theory, which suggests that only a small proportion of cells within a tumor, which are termed as CSCs, actually possess cancer-initiating potential and are thought to be responsible for tumor development and sustain growth [3]. CSCs are widely believed to arise from the normal stem cells or progenitor cells on mutation(s) [14]. They possess the ability to self-renew and differentiate, giving rise to heterogeneous tumors. Currently, CSCs are identified by specific surface epitopes. Colon CSCs have been shown to express surface markers CD44, CD166, CD133, and epithelial-specific antigen (also known as EpCAM) [12,13]. Cells expressing these surface epitopes have the ability to form tumors at a much diluted concentration in severe combined immunodeficient mice and histologically resemble the primary tumor from which they were derived [12,13].
Our current observation that only 0.04% of the total colon cancer HCT-116 cells represent CD44-positive cells indicates that only a very small proportion of HCT-116 cells are CSCs. Such an observation is in agreement with the CSC theory, which states that only a small proportion of cells within the tumor are responsible for initiating and maintaining the processes of carcinogenesis. It has been suggested that CSCs represent the source of malignant cells in a primary tumor and are also thought to constitute a reservoir of drug-resistant cells that are responsible for the recurrence of cancer. Our observation showing a 10-fold increase in the proportion of CD44-positive HCT-116 cells after prolonged treatment with FOLFOX could be interpreted as due to sparing of colon CSCs by this chemotherapy regimen. This inference is supported by the observation that the short-term (48–72 h) treatment of HCT-116 cells also leads to a further increase in colon CSCs as evidenced by increased expression of CD44 and CD166. Our observation that the levels of EGFR are also increased in FOLFOX-surviving HCT-116 cells suggests that induction of certain growth factor receptors particularly EGFR may contribute to CSC growth and/or survival, leading to a greater chance of malignant progression. Numerous studies have indicated a role for EGFR in the development of colonic neoplasia, an event that has been observed in almost all epithelial cancers [23,24]. In fact, 40% to 50% colon tumors show increased expression of EGFR [24].
That CSCs show resistance to a number of conventional therapies may explain why it is difficult to completely eradicate cancer and why recurrence is an ever-present threat [2,14]. Thus, therapeutic strategies that specifically target colon CSCs are likely to be effective in eradicating tumors and in reducing the risk of relapse and metastasis. 5-Fluorouracil or FOLFOX, which remains the backbone of colorectal cancer chemotherapeutics, shows limited success. In addition to the lack of response of advanced colorectal cancers to conventional chemotherapeutics, there are also costs of additional toxicities, some of which are even fatal. Therefore, validation of a nontoxic agent that could improve on the current chemotherapeutic regimen would be highly desirable. Recently, we reported that curcumin, the major active ingredient of turmeric with no discernable toxicity, inhibits the growth of transformed cells, suppresses initiation, promotion, and progression of colon carcinogenesis [5,6], and synergizes with FOLFOX to inhibit the growth of colon cancer cells [8]. Our current data further demonstrate that curcumin either alone or together with FOLFOX could be effective in eliminating colon CSCs. Support for this inference comes from the observation that curcumin alone or together with FOLFOX not only attenuates the expression of several colon CSC markers and EGFR in FOLFOX-surviving colon cancer cells but also inhibits colony formation and disintegrates colon cancer cell spheres formed by the FOLFOX-surviving cells. Furthermore, the effect of the combination therapy on elimination of colon CSCs seems to be independent of p53 status because the magnitude of reduction of CD166 levels is similar in both p53-positive (WT) and p53-null HCT-116 cells. Additional support for this inference comes from the observation that the extent of disintegration of colonosphere(s) formed by HCT-116 (WT) and p53-mutant HT-29 cells is also very similar in response to the combination therapy.
Although the mechanisms for increased expression of different markers of CSCs and EGFR in FOLFOX-surviving cells and their inhibition by curcumin and/or FOLFOX are poorly understood, our data suggest that, at least for EGFR, this could be due to changes in methylation of the gene. DNA methylation is the addition of a hydrophobic methyl group to cytosine in a CG sequence. Methylation of CpG sequences stabilizes chromatin, prevents binding of transcription factors, and is thought to be one of the primary mechanisms by which genes are turned on and off developmentally [25–27]. For example, hypermethylation of the EGFR promoter by folic acid and its metabolites has been shown to result in decreased expression of the receptor [28]. Hypermethylation has also been shown to cause functional inactivation of p16INK4A [29]. Our current observation showing increased EGFR expression in FOLFOX-surviving colon cancer HCT-116 cells and reduction of the same in response to curcumin and/or FOLFOX could be attributed to hypomethylation and hypermethylation of the EGFR promoter, respectively. Although the precise regulatory mechanisms for changes in methylation status of EGFR promoter in FOLFOX-surviving cells in response to different treatments are not fully understood, our observation of parallel alterations in DNA 5-cytosine methyltransferase levels with hypomethylation and hypermethylation of the promoter suggests a role for the enzyme in this process.
In conclusion, our data suggest that curcumin by itself or in combination with the conventional colon cancer chemotherapeutic regimen could be an effective therapeutic strategy to prevent the emergence of chemoresistant colon cancer cells by reducing/eliminating the CSCs.
Footnotes
This work was supported by grants (A.P.N.M.) from the National Institutes of Health/National Institute on Aging (AG014343) and the Department of Veterans Affairs.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15(9):494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Hanif R, Qiao L, Shiff SJ, Rigas B. Curcumin, a natural plant phenolic food additive, inhibits cell proliferation and induces cell cycle changes in colon adenocarcinoma cell lines by a prostaglandin-independent pathway. J Lab Clin Med. 1997;130(6):576–584. doi: 10.1016/s0022-2143(97)90107-4. [DOI] [PubMed] [Google Scholar]
- 5.Rao CV, Simi B, Reddy BS. Inhibition by dietary curcumin of azoxymethane-induced ornithine decarboxylase, tyrosine protein kinase, arachidonic acid metabolism and aberrant crypt foci formation in the rat colon. Carcinogenesis. 1993;14(11):2219–2225. doi: 10.1093/carcin/14.11.2219. [DOI] [PubMed] [Google Scholar]
- 6.Shishodia S, Chaturvedi MM, Aggarwal BB. Role of curcumin in cancer therapy. Curr Probl Cancer. 2007;31(4):243–305. doi: 10.1016/j.currproblcancer.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, Marczylo TH, Morgan B, Hemingway D, Plummer SM, et al. Phase I clinical trial of oral curcumin: biomarkers of systemic activity and compliance. Clin Cancer Res. 2004;10(20):6847–6854. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 8.Patel BB, Sengupta R, Qazi S, Vachhani H, Yu Y, Rishi AK, Majumdar AP. Curcumin enhances the effects of 5-fluorouracil and oxaliplatin in mediating growth inhibition of colon cancer cells by modulating EGFR and IGF-1R. Int J Cancer. 2008;122(2):267–273. doi: 10.1002/ijc.23097. [DOI] [PubMed] [Google Scholar]
- 9.Majumdar AP, Du J, Yu Y, Xu H, Levi E, Patel BB, Rishi AK. Cell cycle and apoptosis regulatory protein-1: a novel regulator of apoptosis in the colonic mucosa during aging. Am J Physiol Gastrointest Liver Physiol. 2007;293(6):G1215–G1222. doi: 10.1152/ajpgi.00324.2007. [DOI] [PubMed] [Google Scholar]
- 10.Nagothu KK, Jaszewski R, Moragoda L, Rishi AK, Finkenauer R, Tobi M, Naumoff JA, Dhar R, Ehrinpreis M, Kucuk O, et al. Folic acid mediated attenuation of loss of heterozygosity of DCC tumor suppressor gene in the colonic mucosa of patients with colorectal adenomas. Cancer Detect Prev. 2003;27(4):297–304. doi: 10.1016/s0361-090x(03)00100-4. [DOI] [PubMed] [Google Scholar]
- 11.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 14.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355(12):1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 15.Messa C, Russo F, Caruso MG, Di Leo A. EGF, TGF-alpha, and EGF-R in human colorectal adenocarcinoma. Acta Oncol. 1998;37(3):285–289. doi: 10.1080/028418698429595. [DOI] [PubMed] [Google Scholar]
- 16.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19(3):183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 17.Harari PM, Huang SM. Modulation of molecular targets to enhance radiation. Clin Cancer Res. 2000;6(2):323–325. [PubMed] [Google Scholar]
- 18.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279(46):48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 19.Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10(4):631–647. [PubMed] [Google Scholar]
- 20.Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270(3):385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- 21.Willis ND, Przyborski SA, Hutchison CJ, Wilson RG. Colonic and colorectal cancer stem cells: progress in the search for putative biomarkers. J Anat. 2008;213(1):59–65. doi: 10.1111/j.1469-7580.2008.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, II, Samuel S, Kim MP, Lim SJ, Ellis LM. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69(5):1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel BB, Majumdar APN. HER Family of Receptors as Treatment Targets. 1st ed. New York, NY: Springer; 2008. [Google Scholar]
- 24.Nautiyal J, Rishi AK, Majumdar AP. Emerging therapies in gastrointestinal cancers. World J Gastroenterol. 2006;12(46):7440–7450. doi: 10.3748/wjg.v12.i46.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 26.Lu S, Davies PJ. Regulation of the expression of the tissue transglutaminase gene by DNA methylation. Proc Natl Acad Sci USA. 1997;94(9):4692–4697. doi: 10.1073/pnas.94.9.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4(2):255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 28.Nagothu KK, Rishi AK, Jaszewski R, Kucuk O, Majumdar AP. Folic acid-mediated inhibition of serum-induced activation of EGFR promoter in colon cancer cells. Am J Physiol Gastrointest Liver Physiol. 2004;287(3):G541–G546. doi: 10.1152/ajpgi.00365.2003. [DOI] [PubMed] [Google Scholar]
- 29.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]