Abstract
The main circulating estrogen hormone 17β-estradiol (E2) contributes to the initiation and progression of breast cancer. Estrogen receptors (ERs), as transcription factors, mediate the effects of E2. Ablation of the circulating E2 and/or prevention of ER functions constitute approaches for ER-positive breast cancer treatments. These modalities are, however, ineffective in de novo endocrine-resistant breast neoplasms that do not express ERs. The interaction of E2-ERs with specific DNA sequences, estrogen responsive elements (EREs), of genes constitutes one genomic pathway necessary for cellular alterations. We herein tested the prediction that specific regulation of ERE-driven genes by an engineered monomeric and constitutively active transcription factor, monotransregulator, provides a basis for the treatment of ER-negative breast cancer. Using adenovirus infected ER-negative MDA-MB-231 cells derived from a breast adenocarcinoma, we found that the monotransregulator, but not the ERE-binding defective counterpart, repressed cellular proliferation and motility, and induced apoptosis through expression of genes that required ERE interactions. Similarly, the monotransregulator suppressed the growth of ER-negative BT-549 cells derived from a breast-ductal carcinoma. Moreover, the ERE-binding monotransregulator repressed xenograft tumor growth in a nude mice model. Thus, specific regulation of genes bearing EREs could offer a therapeutic approach for de novo endocrine-resistant breast cancers.
Keywords: Estrogen, Estrogen receptor, Estrogen responsive element, Designer Transcription factor, Gene expression, Phenotypic alterations
INTRODUCTION
The dysregulation of integrated systems that orchestrate the growth and death of breast-ductal epithelial cells leads to breast cancer. 17β-estradiol (E2), the main circulating estrogen hormone, is one agent involved in the initiation and progression of breast cancer (1, 2). The effects of E2 are primarily mediated by the estrogen receptors (ERs) α and β (1, 2). ERs, as with other transcription factors, are modular proteins in which distinct structural regions display unique functional features of the receptors (1, 2). The amino-termini of ERs contain a ligand-independent transactivation function. The central regions are the DNA binding domains followed by flexible hinge domains that contain a nuclear localization signal. The multifunctional carboxyl-termini mediate ligand binding, dimerization and ligand-dependent transactivation functions of the receptors.
Following synthesis, ERs dimerize and mainly translocate to the nucleus independently of E2 (3). The binding of E2 induces major conformational changes in ERs, which expose surfaces that support co-regulatory protein interactions critical for transcription activity (1, 2). One pathway for E2-ERs to regulate gene expression involves the interaction of E2-ERs with estrogen responsive elements (EREs), which are permutations of a palindromic, GGTCAnnnTGACC, DNA sequence (1, 2). This signaling is referred to as the ERE-dependent signaling pathway (1, 2). E2-ERs also regulate gene expression through interactions with transcription factors bound to their cognate regulatory elements on DNA, hence the ERE-independent signaling pathway (1, 2).
Current treatments for ER-positive breast cancer include inhibitors of enzymes involved in estrogen biosynthesis to reduce/ablate the circulating hormone and on anti-estrogenic compounds to alter/prevent ER functions (4–6). These approaches are initially successful in producing remission of established tumors (4, 5). However, tumors eventually develop resistance to such therapies resulting in the acquired endocrine-resistance phenotype (4–6).
Endocrine modalities are also ineffective in the treatment of de novo endocrine-resistance breast cancer due to the absence, or loss, of ER expression (4–6). An autonomous regulation of aberrant cell growth signifies de novo resistance malignancies (4, 5). Several growth factors and their receptors, which are trans-membrane tyrosine kinases, are over-expressed and act as autocrine growth stimulators (7). While treatment options for de novo endocrine-resistance neoplasms are limited, inhibition of growth factor signaling, including antibody therapy to block ligand binding to receptors and kinase inhibitors to inhibit receptor activity, are being explored as therapeutic approaches (7). In addition, re-expression of ERs by epigenetic approaches or introduction of ERs by gene delivery could provide therapeutic benefits by restoring ligand signaling for ER-negative breast neoplasms (8, 9).
Our recent studies aimed at dissecting nuclear estrogen signaling pathways suggest that genomic responses from the ERE-dependent signaling pathway to E2-ER are required to induce phenotypic changes (10, 11). These findings raise the possibility that targeted regulation of ERE-driven genes could be a basis for the development of a therapy for ER-negative breast cancers. We herein examined this prediction by using engineered transcription factors specifically designed to regulate the expression of ERE-driven genes independently of ligands, ER status and cell type (12). We previously showed that the intrinsic specificity of the DNA binding domain of ERα to interact with ERE sequences can be exploited to engineer a monomeric ERE-binding module by co-joining two DNA binding domains with the hinge domain (12). Integration of strong transcription activation domains from other transcription factors into the ERE binding module generated monomeric transcription factors, or monotransregulators, with constitutive activity at ERE-driven gene promoters (12). Our present findings from adenovirus infected cells emulating de novo endocrine-resistant tumors indicate that a monotransregulator, but not the ERE-binding defective counterpart, altered cellular proliferation, death and motility by mimicking the effects of E2-ERα on gene transcriptions that required ERE interactions. Furthermore, the monotransregulator effectively repressed xenograft tumor growth in nude mice. Emphasizing the critical importance of the ERE-dependent signaling pathway in the induction cellular responses, these findings provide a basis for the development of transcriptional approaches that target EREs of estrogen-responsive genes for the treatment of de novo endocrine-resistant breast cancer.
MATERIALS & METHODS
Engineering of ERE binding proteins
Expression vectors were previously described (12, 13). ERαEBD contains amino-acid substitutions (Glu203Ala, Gly204Ala and Arg211Glu) in the DNA recognition helix of the DNA binding domain (11). The construction of PV and PVEBD was also previously described (12). Due to an earlier engineering strategy (12), PVEBD contains a CDC with amino acid substitutions replacing Cys202 and Cys205 with His residues. These mutations completely prevent ERE interactions (12, 14).
Restriction and DNA modifying enzymes were obtained from New England Bio-Labs (Beverly, MA) or Invitrogen (Carlsbad, CA). 17β-estradiol (E2) was purchased from Sigma-Aldrich (St. Louis, MO).
Cell culture and biochemical assays
MDA-MB-231 cells were cultured as described (10, 11). BT-549 cells obtained from ATCC (Manassas, VA) were maintained in RPMI-164 supplemented with 10 μg/ml insulin (Sigma-Aldrich) and with 10% fetal bovine serum (FBS, Invitrogen). Recombinant adenoviruses were generated and tittered as described (10, 11).
To assess the functional protein synthesis, we used immunocytochemistry (ICC), Western Blot (WB) and Electrophoretic Mobility Shift Assay (EMSA) (13).
Endogenous Gene Expression
MDA-MB-231 cells (100,000 cells) were plated in 6-well culture plates in phenol-red free medium containing 10% CD-FBS without E2 for 24h. Cells were then infected with Ad-ERα or Ad5-ERαEBD and maintained with or without 10−9 M E2 for 48h, while Ad5-PV or Ad5-PVEBD infected cells were maintained for 48h in the presence of vehicle. The total RNA was subjected to quantitative PCR (qPCR) using custom TaqMan Low-Density Arrays (Foster City, CA) (10, 11). Relative Quantification Analysis was performed using the Comparative CT Method (15).
Cellular proliferation
MDA-MB-231 (5,000 cells) or BT-549 (10,000 cells) cells were plated in 24-well culture plates in phenol red-free medium containing 10% CD-FBS for 24h. Cells were infected with or without 10−9 M E2 for 6 days. At termination, cells were subjected to counting using a hemocytometer and an (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Invitrogen) (10, 11).
Cell cycle analysis
Cells (50,000 cells) in 6-well culture plates were infected with or without E2 for 48h. Cells were subjected to a fluorescence-activated cell sorting (FACS) using the EPICS Elite (Coulter Corp., Miami, FL) (10, 11).
Annexin V and TUNEL assays
Cells (100,000 cells) in 6-well culture plates infected with or without E2 for 96h were subjected to an Annexin V assay (Invitrogen) as described (10, 11). Infected cells (25,000 cells) in poly-D-lysine coated 48-well plates with or without E2 for 96h were also subjected to a TUNEL assay (Promega, Madison, Wisconsin) as described (10, 11).
Motility assays
Infected cells (200,000 cells) in 12-well plates were maintained for 48h with or without E2 to allow cells to reach near confluence. A wound was then generated and the gap closure was imaged at 24h intervals (10, 11).
For the invasion assay, equal numbers of infected cells (25,000) for 48h from each treatment group were seeded into BD Matrigel Invasion Chambers (San Diego, CA) for 24h. Cells on the bottom of the chamber membrane were stained with Diff-Quik (Dade Behring, Newark, DE). Images were captured, and stained cells were counted from images as described (10, 11).
Tumor Studies
All animal experiments were carried out under an Institutional Animal Care and Use Committee-approved protocol. MDA-MB-231 cells infected with an adenovirus for 48h were collected. Live cells (4×106 cells/mouse) were re-suspended in 100 μl base medium and Matrigel (BD Biosciences, Bedford, MA) at a 1:1 ratio to support initial tumor growth. Cell suspensions were subcutaneously inoculated using a 27-gauge needle into the left mammary fat pad of 6-week-old female athymic NCr-nu/nu nude mice (NCI, Frederick, Maryland) housed in micro-isolator cages and given food and water ad libitum. Mice were divided into five groups of 10 mice, with the exception of the Ad5-PV-100 group that had nine mice due to the death of a mouse at an early stage of the study. Groups of mice were inoculated with cells infected with 1) the parent Ad5 at 900 MOI, 2) Ad5-PVEBD at 900 MOI, 3) Ad5-PV at 100 MOI together with Ad5 at 800 MOI, 4) Ad5-PV at 200 MOI together with Ad5 at 700 MOI, or 5) Ad5-PV at 400 MOI together with Ad5 at 500 MOI. The initial tumor growth was monitored by palpation, and tumor size was subsequently measured weekly with calipers. The tumor volume was estimated by using an ellipsoid approximation and the formula of V = (π/6)abc, wherein a b and c denotes length, width and height, respectively. At termination, tumors were excised, weighed, fixed in 4% paraformaldehyde and embedded in paraffin. Sections of 4 μm thickness were stained with hematoxylin/eosin, and images were captured by light microscopy.
Statistical Analysis
Results were presented as the mean ± s.e.m. Significance was determined using a 2-tailed unpaired t test with a confidence interval of 95%.
RESULTS
Synthesis of monotransregulators
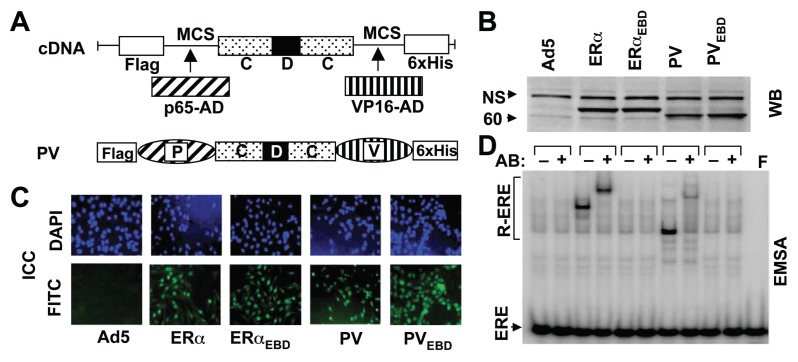
Our previous studies have demonstrated that genomic responses from the ERE-dependent pathway induced by E2-ERs are required to elicit cellular alterations (10, 11). We therefore anticipated that the regulation of ERE-bearing genes by monotransregulators would emulate the effects of E2-ERα on cellular phenotypes. To examine this prediction, we used the montransregulator p65-CDC-VP16, or PV (12). PV bears single copies of the activation domains of the p65 subunit of nuclear factor κB (16) and the herpes simplex virion protein-16 (17) genetically fused to the amino- and carboxyl-termini of the ERE binding module CDC (Fig 1A). CDC is composed of two DNA binding (C) domains co-joined with the hinge (D) domain to simulate the interaction of each ERα monomer in the ERα dimer with one ERE half-site (18, 19). We also used the ERE binding defective PV (PVEBD), which contains amino-acid replacements in the ERE binding module to prevent ERE interactions (12).
Fig. 1.
Engineering of monotransregulators. (A) The activation domains (ADs) of p65 and VP16 proteins were genetically joined through multiple cloning sites (MCS) to the 5′ and 3′ ends, respectively, of the CDC-cDNA to generate the monotransregulator PV. The construct also contains sequences encoding the Flag and 6xHis epitopes at the amino- and carboxyl-termini, respectively. (B) MDA-MB-231 cells were infected with the parent recombinant adenovirus (Ad5) at MOI 900, a recombinant adenovirus bearing cDNA for ERα at 50, ERαEBD at 150, PV at 200 and PVEBD at 900 MOI. For all infections, the total MOI was adjusted to 900 by supplementing with the parent Ad5. Extracts (10 μg) of infected MDA-MB-231 cells at 48h were subjected to western blotting (WB) using a horseradish peroxidase-conjugated monoclonal Flag antibody. Molecular mass in kDa is indicated. NS denotes non-specific protein detection. (C) Intracellular localization of receptor proteins was examined by immunocytochemistry (ICC). A fluorescein isothiocyanate-conjugated Flag antibody (FITC) localizes constructs to the nucleus stained with 4′,6-diamidino-2-phenylindole (DAPI) at 48h post-infection. (D) Cell extracts (10 μg) at 48h post-infection were also subjected to electrophoretic mobility shift assay (EMSA) with (+) or without (−) a Flag antibody (Ab). ERE specifies unbound and R-ERE denotes receptor-bound radiolabeled ERE. F indicates the radiolabeled ERE only. In all experiments a representative result from three independent experiments is shown.
We utilized ER-negative MDA-MB-231 cells derived from human breast adenocarcinoma as a model (8, 10, 11). The recombinant adenovirus bearing ERα-cDNA (Ad5-ERα) was used at 50 multiplicity of infection (MOI), with which infected cells synthesize ERα at a level that require E2 to function (11). The concentration of Ad5-PV was based on preliminary studies (Supplemental Fig. 1A–E). Ad5-PV at 200 MOI synthesized an amount of functional PV that altered the growth and cell cycle phases similar to those observed with E2-ERα (Supplemental Fig. 1A–E). The adenovirus expressing ERE binding defective ERα (ERαEBD) or PV (PVEBD) cDNA was used at 150 or 900 MOI, respectively. At these MOIs, infected cells produced mutants at levels comparable to those of parent proteins that reached to maximal amounts at 48h post-infection (Supplemental Fig. 1). For all experiments, the total MOI was adjusted to 900 by supplementing with the parent adenovirus (Ad5).
Western blot (WB) analysis using an antibody specific to the Flag epitope at the amino-terminus of the each construct indicated that cells synthesize monotransregulators with a molecular mass of ~63 kDa (Fig. 1B). As ERs, PV and PVEBD locate to the nucleus as assessed by immunocytochemistry (ICC) (Fig. 1C). An electrophoretic mobility shift assay (EMSA) further verified that PV, like ERα, but not PVEBD or ERαEBD, interacts with ERE (Fig. 1D).
Effects of monotransregulators on gene expression
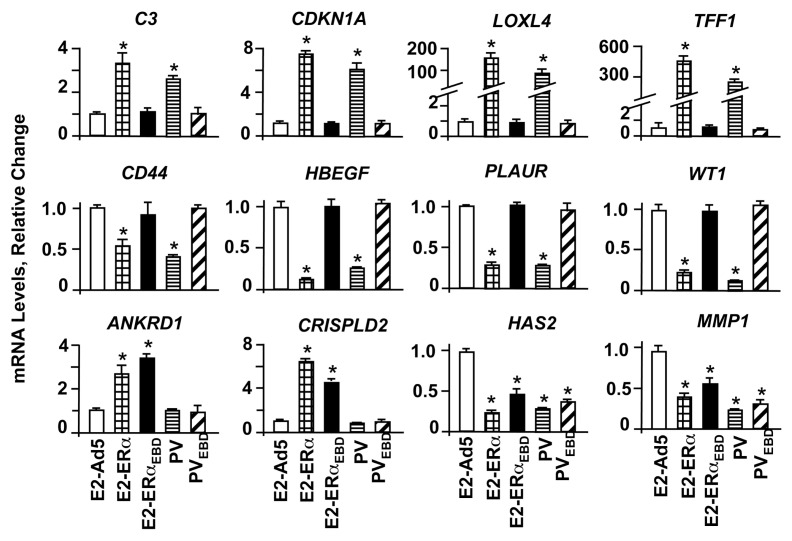
E2-ERs activate or repress the expression of responsive genes through the ERE-dependent and ERE-independent signaling pathways. To assess the effects of monotransregulators on gene expression, total RNA from MDA-MB-231 cells at 48h post-infection was subjected to quantitative PCR. E2-ERα induced the expression of the C3 (complement component 3) CDKNA1 (Cyclin dependent kinase inhibitor, p21), LOXL4 (Lysyl oxidase-like 4) and TFF1 (Trefoil factor 1, pS2) genes, as we previously reported (11). On the other hand, E2-ERα repressed the transcription of the CD44 (CD44 antigen), HBEGF (Heparin-binding EGF-like growth factor), PLAUR (Plasminogen activator, urokinase receptor) and WT1 (Wilms tumor) genes (Fig. 2). We found that PV mirrored the effects of E2-ERα on these gene expressions, while E2-ERαEBD or PVEBD did not affect the transcription of these genes. Thus, the regulation of this subset of genes by PV, like E2-ERα, requires ERE interactions.
Fig. 2.
Regulation of endogenous gene expression by monotransregulators. MDA-MB-231 cells infected with the adenovirus bearing none (Ad5), ERα or ERαEBD cDNA were treated without (data not shown) or with 10−9 M E2 for 48h, while cells infected with the adenovirus bearing Ad5-PV or Ad5-PVEBD were treated with vehicle. Total RNA was subjected to qPCR for the analysis of estrogen responsive gene expressions. Results, which are the mean ± s.e.m. of three independent determinations, depict relative change in mRNA levels compared to those observed in cells infected with Ad5 in the absence of E2, which is set to 1. Asterisk (*) indicates significant change.
We also observed that the expression of ANKRD1 (Ankyrin repeat domain 1), CRISPLD2 (Cystein-rich secretory protein LCCL domain containing 2), HAS2 (Hyaluronan synthase 2) and MMP1 (Matrix metallopeptidase 1) genes is independent of ER-ERE interactions as E2-ERαEBD and E2-ERα modulated the transcription of these genes, as we showed previously (11). In contrast, PV or PVEBD had no effect on the ANKRD1 or CRISPLD2 gene expression. This suggests that the expression of these genes is ERα-specific. All constructs, however, repressed the transcription of the HAS2 or MMP1 gene, implying the generality of action.
Mediation of cellular growth
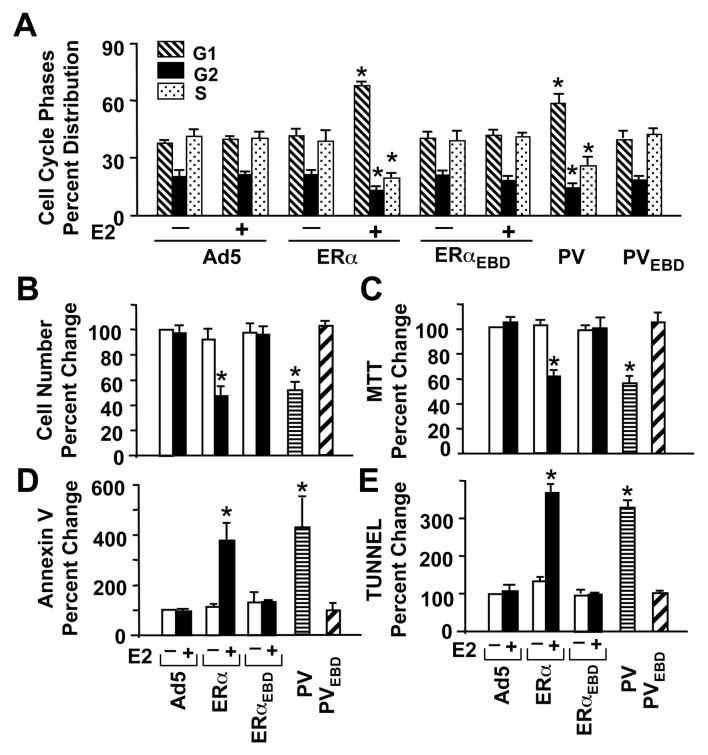
Our previous studies indicated that the ERE-dependent genomic responses to E2-ERα are necessary for the induction of phenotypic alterations (10, 11). Since PV mirrored the effects of E2-ERα on gene expressions that require ERE interactions, we asked whether PV alters cellular growth similar to E2-ERα. Analysis of histograms generated by cell sorting of MDA-MB-231 cells at 48h post-infection revealed that ERα only in the presence of 10−9 M E2, but not ERαEBD, altered cell cycle kinetics compared to the parent Ad5 (Fig. 3A & Supplemental Fig. 2A). PV also increased the number of cells accumulated in G1 phase and decreased cell numbers in S and G2 phases, whereas PVEBD had no effect on the distribution of cycle phases compared to Ad5.
Fig. 3.
Effects of monotransregulators on cellular growth. (A) MDA-MB-231 cells infected with recombinant adenoviruses in the presence (+) or absence (−) of 10−9 M E2 for 48h were subjected to fluorescence-activated cell sorting for the cell cycle analysis. Results depicted as the percent of cells in G1, G2 and S phases are the mean ± s.e.m. of three independent experiments. (B & C) Infected cells maintained in the presence (+) or absence (−) of 10−9 M E2 (E2) for six days were subjected to (B) cell counting or (C) MTT assay. The mean ± s.e.m., which depicts three independent experiments performed in duplicate, indicates percent (%) change in cell number compared to those observed in cells infected with Ad5 in the absence of E2, which is set to 100. (D & E) MDA-MB-231 cells were also subjected to (D) Annexin V and (E) TUNEL assays at 96h post-infection for the effects of proteins on cellular death. Results, which are the mean ± s.e.m. of three independent experiments, are summarized as percent (%) change in the number of cells bound to Annexin V or as the number of cells that incorporated fluorescein isothiocyanate-conjugated dUTP into the fragmented DNA (TUNEL) obtained in comparison with the cells infected with Ad5 in the absence of E2, which is set to 100. Asterisk (*) indicates significant change.
These results suggest that alterations in cell cycles require ERE interactions, which was also reflected in the cellular proliferation. ERs without E2 did not affect cell growth assessed by cell counting (Fig. 3B) or MTT assay (Fig. 3C), shown at day six of post-infection. However, in response to 10−9 M E2, ERα, but not ERαEBD, effectively suppressed proliferation. Similarly, PV, but not PVEBD, repressed cellular growth in comparison with Ad5.
Regulation of cell death
The effect of PV on proliferation was the inverse of its effect on apoptosis. PV, like E2-ERα, augmented apoptosis, as revealed by an increase in cell number stained with Annexin V. As a marker for the mid-stages of apoptosis (20), Annexin V assesses the integrity of the cellular membrane, with significant changes occurring at 96h post-infection (Fig. 3D & Supplemental Fig. 2B). A TUNEL assay, which catalytically incorporates fluorescein-conjugated nucleotides into the fragmented DNA as one late-stage characteristic of apoptosis (20), further corroborated the results. PV and E2-ERα, but not ERE binding defective counterparts, induced DNA fragmentation at 96h post-infection (Fig. 3D & Supplemental Fig. 2C) compared to Ad5.
Effects of monotransregulators on cell-type
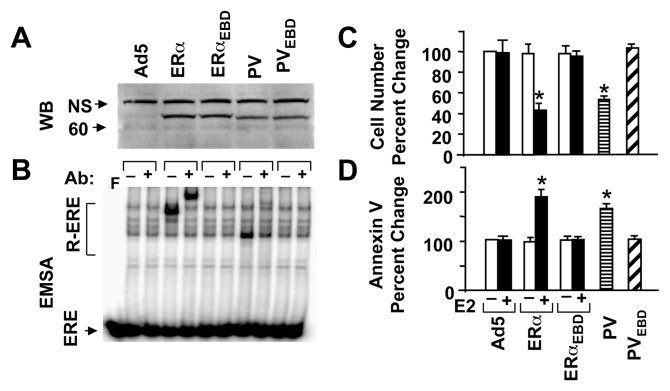
To ensure that the effect of PV on cellular growth is not cell-type specific, we also infected ER-negative BT-549 cells derived from a breast-ductal carcinoma. While mutants were synthesized at levels comparable to those of the parent proteins (Fig. 4A), ERα and PV, but not ERαEBD or PVEBD, interacted with ERE (Fig. 4B). As with MDA-MB-231 cells, ERα in the presence of 10−9 M E2 and PV repressed the growth of BT-549 cells (shown at day 6 of post-infection). On the other hand, ERαEBD with or without E2 or PVEBD had no effect on cell growth (Fig. 4C). Repression of cellular proliferation by E2-ERα or PV involves apoptosis since both increased the cell population stained with Annexin V compared to Ad5 (Fig. 4D).
Fig. 4.
. PV mimics the effects of E2-ERα on responses from BT-549 cells. (A & B) Synthesis of functional receptor proteins. BT-549 cells were infected with an adenovirus bearing none (Ad5) or a cDNA for ERα, ERαEBD, PV, ERαEBD at 80, 20, 20, 60 and 80 MOIs, respectively. The total MOI was adjusted to 80 MOI by supplementing with Ad5. (A) Extracts of infected BT-549 (25 μg) cells at 48h post-infection were subjected to WB using the horseradish peroxidase-conjugated monoclonal Flag antibody. Molecular mass in kDa is indicated. (B) Cell extracts were also subjected to EMSA in the presence (+) or absence (−) of a Flag antibody. ERE indicates the unbound and R-ERE depicts the receptor-bound radiolabeled ERE. F denotes the radiolabeled ERE only. (C & D) Infected cells in the presence (E2) or absence (−) of 10−9 M E2 for six days were subjected to (C) cell counting for cellular proliferation or (D) Annexin V assay at 96h post-infection for cellular death. The mean ± s.e.m., which represents three independent experiments, indicate percent (%) change in cell number compared to those observed in cells infected with Ad5 in the absence of E2, which is set to 100. Asterisk (*) indicates significant change.
Modulation of cellular motility
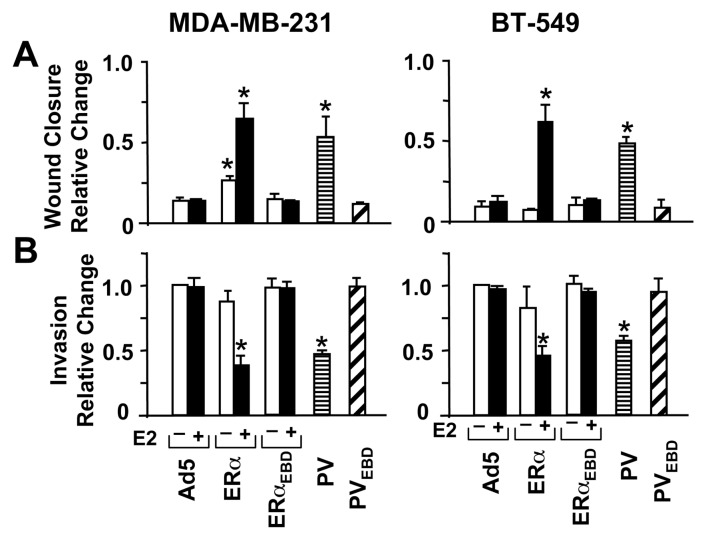
Breast cancer cells that synthesize ERs endogenously are less motile and invasive than ER-negative cells (8). Introduction of ERs into ER-negative cells hinders cellular motogenesis in an ERE-dependent manner as the ERE binding defective ERs induce no effect (8, 10, 11). We therefore anticipated that PV, like E2-ERα, would inhibit the motility and invasiveness of MDA-MB-231 or BT-549 cells. Infected cells with or without 10−9 M E2 were grown for 48h to near confluence. A wound was then created and the rate of wound closure was assessed as a function of time. The unliganded ERα delayed the wound closure of MDA-MB-231 cells, which was further delayed with E2, shown at 96h, in comparison with Ad5 (Fig. 5A & SI Fig. 2D). Similarly, PV hindered the wound healing, whereas PVEBD or ERαEBD had no effect. PV and E2-ERα also repressed the motility of BT-549 cells as compared to Ad5 at 48h post-infection (Fig. 5A).
Fig. 5.
Effects of receptor proteins on cellular motility. (a) Wound-healing assay. Infected MDA-MB-231 or BT-549 cells were incubated in the presence (+) or absence (−) of 10−9 M E2 as indicated for 48h to allow cells to reach near confluence. A wound was then created and images were captured at 0h and at 24h intervals. Results, representing the mean ± s.e.m. of three independent experiments performed in duplicate, summarize the wound closure at 96h for MDA-MB-231 cells and 48h for BT-549 cells relative to 0h, which is set to 1. (b) Invasion assay. Cells were infected with recombinant adenoviruses with (+) or without (−) 10−9 M E2 for 48h as depicted. Cells were collected and counted. The same number of cells from each group was then seeded on the upper section of the invasion chamber membrane with the corresponding treatment. After 24h incubation, cells on the bottom of chamber membrane as an indication of invasiveness were stained and counted. Results, which represent the mean ± s.e.m. of three independent experiments performed in duplicate, are relative changes compared to cells infected with Ad5 in the absence of E2, which is set to 1. Asterisk (*) indicates significant change.
We also examined the effects of receptor proteins on cellular invasiveness using an invasion assay that assesses the capacity of cells to migrate through a reconstituted basement membrane. Equal numbers of infected cells were seeded for 48h on the top of membranes of invasion chambers (Fig. 5B, Supplemental Fig. 2E). After 24h, cells on the bottom of the membrane were stained and imaged. PV, like E2-ERα, suppressed the invasiveness of both cell lines, which required ERE interactions, as ERαEBD or PVEBD did not affect migration of infected cells compared to Ad5.
Effects of monotransregulators on tumor formation
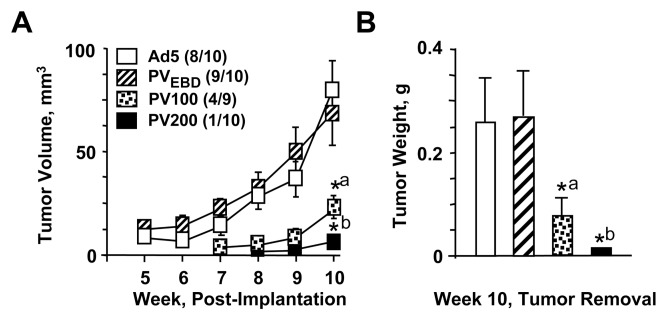
Our collective results in situ indicated that PV effectively represses the growth of ER-negative cells. We therefore anticipated that the introduction of PV to MDA-MB-231 cells would impair tumor formation in vivo. Since the extent by which PV affects cellular growth correlated with the amount of protein synthesized as a result of the Ad5-PV concentration used (Supplemental Fig. 1), we also wanted to address whether graded doses of Ad5-PV would influence the number and growth of tumors. To accomplish this, we infected cells with Ad5 or Ad5-PVEBD at 900 MOI. We also infected cells with Ad5-PV at 100, 200 or 400 MOI supplemented with Ad5 to bring the total MOI to 900. At 48h post-infection, 4×106 cells/mouse were implanted subcutaneously into the left mammary pads of athymic NCr-nu/nu mice (10 mice/group). The implantation site was monitored weekly for 10 weeks. Solid tumors in mice implanted with cells infected with Ad5 or PVEBD were palpable by week three, and the experiments were terminated at week 10. Tumor volumes (Fig. 6A) weights (Fig. 6B) were determined at this time, and tumors were removed for sectioning and histological preparations using hematoxylin/eosin staining (Supplemental Fig. 3). The incidences of primary tumors (indicated in parentheses, Fig. 6A) in the Ad5 or Ad5-PVEBD group were 80% and 90%, respectively. On the other hand, mice implanted with cells infected with Ad-PV at various MOIs resulted in slower onset and progression of tumor growth and showed a substantially reduced tumor incidence. The Ad5-PV-100 group had a 44% tumor incidence, versus 10% for the Ad5-PV at 200 MOI. No tumor growth was detected at any time in the Ad5-PV-400 group, nor were post-mortem tumors observed at the implantation site. These results indicate that PV also represses the development of tumors in xenograft models through a signaling pathway that requires ERE interactions.
Fig. 6.
Effects of monotransregulators on tumor growth in xenografts. MDA-MB-231 cells were infected with Ad5 or Ad5-PVEBD at 900 MOI for 48h. A group of cells was also infected with Ad5-PV at 100 or 200 MOI supplemented with 800 or 700 MOI of Ad5, respectively. Infected cell suspensions (4×106 cells/mouse) were subcutaneously inoculated into the left mammary fat pads of female athymic NCr-nu/nu mice (ten mice/group, with the exception of the Ad5-PV-100 group that contained nine mice). (A) The tumor size was measured with calipers weekly and the tumor volume was calculated. Tumor incidence for each group is shown in parenthesis. Points denote the mean ± s.e.m. of tumor volume of each group. *a denotes significant change (P<0.041) compared to Ad5. *b indicates significant change (P<0.0161) compared to Ad5. (B) At week ten mice were sacrificed. Tumors were removed and weighed. Bars are the mean ± s.e.m. of tumor weight from each group. *a denotes significant change (P<0.008) compared to Ad5. *b indicates significant change (P<0.001) compared to Ad5.
DISCUSSION
Current endocrine approaches for ER-positive breast cancer that target E2 synthesis and/or ER functions are successful in the management of the disease in an adjuvant setting or at advanced stages. However these modalities are ineffective in the treatment of ER-negative endocrine-resistant cancer that accounts for about one-third of all breast neoplasms. An insufficient understanding of the underlying mechanisms responsible for the disease limits treatment options.
Studies have shown that prevention of ER binding to ERE sequences of responsive genes by exogenously introduced ERE decoys (21) or electrophilic agents that disrupt zinc-fingers of ERα (22–24) represses E2-mediated growth of ER-positive breast cancer cells. Complementing this, we recently reported that genomic responses from the ERE-dependent pathway mediated by E2-ER are required to suppress the growth of ER-negative cells as well (10, 11). Collectively, these findings indicate the importance of ER-ERE interactions in the proliferation of cells synthesizing ERs. We therefore anticipated that targeted regulation of ERE-driven genes independent of ligand-ER signaling and cellular context could provide a basis for developing a therapeutic approach for endocrine-resistant breast cancers. Consistent with this prediction, we demonstrate here that monotransregulator PV suppressed in situ and in vivo growth of cells derived from ER-negative breast neoplasms by regulating gene expression that rely on ERE interactions.
Studies using global gene expression profiling have revealed that E2-ER regulates the expression of genes involved in various cellular functions, including signal transduction, cellular proliferation, apoptosis and motility (10, 11, 25). ER, as with other transcription factors, regulates gene transcription through events that involve covalent modifications of chromatin within which responsive gene promoter resides (26). The binding of E2-ER to an ERE sequence of a responsive gene generates a platform for an ordered and combinatorial recruitment of co-regulator complexes, including protein and chromatin modifiers (26). The integrated effects of these complexes induce local chromatin alterations that restructure the promoter for the recruitment of basal transcription machinery to activate gene expression (26). E2-ER, as with other transcription factors, also represses the expression of genes. Although the mechanisms are unclear, recruitment of co-repressor complexes or squelching of co-activators by the ERE-bound E2-ER is suggested to induce chromatin modifications that counteract the effects of activating signals, thereby repressing the gene expression (27, 28). Our results demonstrate that PV mirrored the ability of E2-ERα to regulate the transcription of responsive genes that rely on ERE-interactions. While PV and E2-ERα enhance the expression of the C3, CDKNA1, LOXL4 and TFF1 genes, both suppress the CD44, HBEGF, PLAUR or WT1 gene expression. These findings suggest that PV and ERα utilize common mechanisms to activate as well as repress the expression of ERE-driven genes. Studies showed that the activation domains of p65 or VP16 induce transcription by orchestrating the recruitment of co-activator proteins, the majority of which also partner with E2-ER (29, 30). Since PV and E2-ERα activate or repress the expression of the same genes by interacting with ERE sequences, it appears that the nature of the promoter of the estrogen responsive gene within which an ERE is located determines the direction of gene expression.
PV also regulated the expression of a set of estrogen responsive genes that do not require ERE interactions. In the ERE-independent signaling pathway, E2-ER interacts with transcription factors bound to their cognate regulatory elements on DNA to modulate gene expression in a ligand-, promoter- and cell-context dependent manner (31, 32). Although mechanisms are unclear, direct interactions between surfaces in the amino- and/or carboxyl-termini of ERα and the DNA-bound transcription factor or indirect interactions with co-regulators recruited with either transcription factor could be responsible for gene regulation (31, 32). We observed that PV or PVEBD had no effect on the transcription of the ANKRD1 or CRISPLD2 gene, whereas both ERα and ERαEBD induced the expression of these genes in response to E2. This suggests that the regulation of these genes is dependent upon ER-specific structural features. On the other hand, PV and PVEBD, as ERα and ERαEBD, modulated the expression of the HAS2 and MMP1 genes. This implies a generality of action, which could involve structural/mechanistic features shared by other transcription factors or hormone receptors. Indeed, HAS2 gene expression is also regulated by retinoic acid signaling (33, 34). Similarly, glucocorticoid, progesterone, retinoic acid and thyroid hormone signaling pathways modulate MMP1 gene expression (35–38).
Despite the ability of PVEBD to modulate the expression of a subset of estrogen responsive genes, the ERE binding defective monotransregulator had no effect on the proliferation, apoptosis or motility of model cells. Cell proliferation, for example, is dependent upon transcriptional as well as non-transcriptional events to activate the cascade of cyclin-dependent kinases critical for progression through cell cycle phases (39). The activities of cyclin-dependent kinases are modulated by kinase inhibitors that include p21 protein encoded by the CDKNA1 gene. p21 suppresses proliferation by promoting cell cycle arrest in the G1/S phase transition (39). Increased expression of CDKNA1 mediated by PV and E2-ERα but not the ERE-binding defective counterparts may be one key event that contributes to the arrest in cell cycle progression, and consequently, the repression of cellular proliferation. Hyaluronan (HA), a major glycosaminoglycan present in the extra cellular matrix, is critical for structural integrity of cells (40). HA is synthesized by the hyaluronan synthase family of transmembrane glycosyltransferases that include hyaluronan synthase 2, a product of the HAS2 gene (40). The interaction of HA with its surface receptor CD44 encoded by the CD44 gene initiates a signaling cascade that is critical for cellular proliferation, adhesion and migration (40). PV as E2-ERα regulated the expression of both the CD44 and HAS2 genes. On the other hand, the transcription of only the HAS2 gene was modulated by PVEBD and E2-ERαEBD, both of which had no effect on phenotypic features of cell models. These results further highlight the canonical importance of the ERE-driven gene network in the induction of cellular responses. Thus, the ability of the DNA binding domain of ERα to decode ERE sequences allowed PV to induce cellular responses by mimicking E2-ERα to modulate the transcription of genes whose regulations require ERE interactions independent of ligands. The monotransregulator approach therefore provides proof-of-principle for the development of a transcription therapy for de novo endocrine-resistant breast neoplasms.
Counteraction of the beneficial effects of endocrine approaches by the tumor cells that synthesize ERs leads to acquired endocrine-resistance phenotypes, in which anti-estrogenic compounds are no longer capable of inhibiting cellular growth (41, 42). Although mechanisms are unclear, changes in the relative levels of ER subtypes, ER-isoforms or co-regulatory proteins, alterations in the pharmacokinetics of anti-estrogenic compounds or aberrations in signaling pathways that cross-talk with E2-ER could contribute to the development of acquired endocrine-resistance (4–6). Since ER plays a pivotal role in both the initial response and subsequent resistance to anti-estrogenic compounds, additional therapeutic benefits could be achieved by preventing ER synthesis or function. Repression of ER synthesis by inhibitory RNA technologies (43) or interference with endogenous ER functions by future generations of anti-estrogenic compounds (44) or ER-specific electrophilic agents (22–24) could constitute strategies for the treatment of acquired endocrine-resistant breast cancers. Since, these approaches target ER, however, circumvention of ER-dependent events by growth signaling pathways could lead to tumor progression. Monotransregulators with repressor functions that effectively suppress the expression of ERE-driven genes, independent of ligand, receptor status or other signaling pathways could provide an approach for the treatment of acquired endocrine-resistant neoplasms as well. The monotransregulator design could also be extended to steroid/thyroid hormone receptors that show modular structure. Targeted regulation of endogenous genes by specific responsive element binders with activator or repressor functions may provide novel approaches to biology and medicine.
Supplemental Data
ACKNOWLEDGMENT
We thank Russell Hilf for his critical reading of the manuscript. We also thank Andrew Cardillo for providing guidance and execution of the qPCRs. We express our gratitude to Peter Keng and Michael Strong for FACS studies. We thank Linda Johnstone for histological preparations. This work was supported by an NCI grant CA113682 (M.M.) and a grant from the University of Rochester Clinical and Translational Science Institute (M.M.).
Footnotes
The authors have nothing to disclose.
REFERENCES
- 1.Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Li X, Hilf R, Bambara RA, Muyan M. Molecular basis of therapeutic strategies for breast cancer. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:379–396. doi: 10.2174/156800805774912944. [DOI] [PubMed] [Google Scholar]
- 3.Bai Y, Giguere V. Isoform-selective interactions between estrogen receptors and steroid receptor coactivators promoted by estradiol and ErbB-2 signaling in living cells. Mol Endocrinol. 2003;17:589–599. doi: 10.1210/me.2002-0351. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson RI, Gee JM, Knowlden J, et al. The biology of antihormone failure in breast cancer. Breast Cancer Res Treat. 2003;80(Suppl 1):S29–34. doi: 10.1023/a:1025467500433. discussion S35. [DOI] [PubMed] [Google Scholar]
- 5.Clarke R, Liu MC, Bouker KB, et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 6.Sengupta S, Jordan VC. Selective estrogen modulators as an anticancer tool: mechanisms of efficiency and resistance. Adv Exp Med Biol. 2008;630:206–219. doi: 10.1007/978-0-387-78818-0_13. [DOI] [PubMed] [Google Scholar]
- 7.Jones HE, Gee JM, Hutcheson IR, Knowlden JM, Barrow D, Nicholson RI. Growth factor receptor interplay and resistance in cancer. Endocr Relat Cancer. 2006;13(Suppl 1):S45–51. doi: 10.1677/erc.1.01275. [DOI] [PubMed] [Google Scholar]
- 8.Garcia M, Derocq D, Freiss G, Rochefort H. Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc Natl Acad Sci U S A. 1992;89:11538–11542. doi: 10.1073/pnas.89.23.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zajchowski DA, Sager R, Webster L. Estrogen inhibits the growth of estrogen receptor-negative, but not estrogen receptor-positive, human mammary epithelial cells expressing a recombinant estrogen receptor. Cancer Res. 1993;53:5004–5011. [PubMed] [Google Scholar]
- 10.Li X, Nott SL, Huang Y, et al. Gene expression profiling reveals that the regulation of estrogen-responsive element-independent genes by 17β-estradiol-estrogen receptor β is uncoupled from the induction of phenotypic changes in cell models. J Mol Endocrinol. 2008;40:211–229. doi: 10.1677/JME-07-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nott SL, Huang Y, Li X, et al. Genomic responses from the estrogen responsive element-dependent signaling pathway mediated by estrogen receptor α are required to elicit cellular alterations. J Biol Chem. 2009;284:15277–15288. doi: 10.1074/jbc.M900365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Li X, Yi P, Hilf R, Bambara RA, Muyan M. Targeting estrogen responsive elements (EREs): design of potent transactivators for ERE-containing genes. Mol Cell Endocrinol. 2004;218:65–78. doi: 10.1016/j.mce.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Muyan M, Yi P, Sathya G, et al. Fusion estrogen receptor proteins: toward the development of receptor-based agonists and antagonists. Mol Cell Endocrinol. 2001;182:249–263. doi: 10.1016/s0303-7207(01)00493-2. [DOI] [PubMed] [Google Scholar]
- 14.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2Δ ΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Moore PA, Ruben SM, Rosen CA. Conservation of transcriptional activation functions of the NF-κB p50 and p65 subunits in mammalian cells and Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1666–1674. doi: 10.1128/mcb.13.3.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 18.Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 19.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 20.Gorczyca W, Melamed MR, Darzynkiewicz Z. Analysis of apoptosis by flow cytometry. Methods Mol Biol. 1998;91:217–238. doi: 10.1385/0-89603-354-6:217. [DOI] [PubMed] [Google Scholar]
- 21.Wang LH, Yang XY, Zhang X, Mihalic K, Xiao W, Farrar WL. The cis decoy against the estrogen response element suppresses breast cancer cells via target disrupting c-fos not mitogen-activated protein kinase activity. Cancer Res. 2003;63:2046–2051. [PubMed] [Google Scholar]
- 22.Wang LH, Yang XY, Zhang X, et al. Suppression of breast cancer by chemical modulation of vulnerable zinc fingers in estrogen receptor. Nat Med. 2004;10:40–47. doi: 10.1038/nm969. [DOI] [PubMed] [Google Scholar]
- 23.Wang LH, Yang XY, Zhang X, et al. Disruption of estrogen receptor DNA-binding domain and related intramolecular communication restores tamoxifen sensitivity in resistant breast cancer. Cancer Cell. 2006;10:487–499. doi: 10.1016/j.ccr.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Mao C, Patterson NM, Cherian MT, et al. A new small molecule inhibitor of estrogen receptor α binding to estrogen response elements blocks estrogen-dependent growth of cancer cells. J Biol Chem. 2008;283:12819–12830. doi: 10.1074/jbc.M709936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 26.Reid G, Gallais R, Metivier R. Marking time: The dynamic role of chromatin and covalent modification in transcription. Int J Biochem Cell Biol. 2009;41:155–163. doi: 10.1016/j.biocel.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 27.Ellison-Zelski SJ, Solodin NM, Alarid ET. Repression of ESR1 through actions of estrogen receptor α and Sin3A at the Proximal Promoter. Mol Cell Biol. 2009;29:4949–4958. doi: 10.1128/MCB.00383-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stossi F, Madak-Erdogan Z, Katzenellenbogen BS. Estrogen receptor α Represses Transcription of Early Target Genes via p300 and CtBP1. Mol Cell Biol. 2009;29:1749–1759. doi: 10.1128/MCB.01476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 30.O'shea JM, Perkins ND. Regulation of the ReIA (p65) transactivation domain. Biochem Soc Trans. 2008;36:603–608. doi: 10.1042/BST0360603. [DOI] [PubMed] [Google Scholar]
- 31.Kushner PJ, Agard DA, Greene GL, et al. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 32.Safe S. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- 33.Saavalainen K, Pasonen-Seppanen S, Dunlop TW, Tammi R, Tammi MI, Carlberg C. The human hyaluronan synthase 2 gene is a primary retinoic acid and epidermal growth factor responding gene. J Biol Chem. 2005;280:14636–14644. doi: 10.1074/jbc.M500206200. [DOI] [PubMed] [Google Scholar]
- 34.Saavalainen K, Tammi MI, Bowen T, Schmitz ML, Carlberg C. Integration of the activation of the human hyaluronan synthase 2 gene promoter by common cofactors of the transcription factors retinoic acid receptor and nuclear factor κB. J Biol Chem. 2007;282:11530–11539. doi: 10.1074/jbc.M607871200. [DOI] [PubMed] [Google Scholar]
- 35.Saatcioglu F, Bartunek P, Deng T, Zenke M, Karin M. A conserved C-terminal sequence that is deleted in v-ErbA is essential for the biological activities of c-ErbA (the thyroid hormone receptor) Mol Cell Biol. 1993;13:3675–3685. doi: 10.1128/mcb.13.6.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson DW, Dodge GR. Dose-dependent effects of corticosteroids on the expression of matrix-related genes in normal and cytokine-treated articular chondrocytes. Inflamm Res. 2003;52:39–49. doi: 10.1007/s000110300012. [DOI] [PubMed] [Google Scholar]
- 37.Stojadinovic O, Lee B, Vouthounis C, et al. Novel genomic effects of glucocorticoids in epidermal keratinocytes: inhibition of apoptosis, interferon-gamma pathway, and wound healing along with promotion of terminal differentiation. J Biol Chem. 2007;282:4021–4034. doi: 10.1074/jbc.M606262200. [DOI] [PubMed] [Google Scholar]
- 38.Leo JC, Lin VC. The activities of progesterone receptor isoform A and B are differentially modulated by their ligands in a gene-selective manner. Int J Cancer. 2008;122:230–243. doi: 10.1002/ijc.23081. [DOI] [PubMed] [Google Scholar]
- 39.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nature Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson RI, Gee JM. Oestrogen and growth factor cross-talk and endocrine insensitivity and acquired resistance in breast cancer. Br J Cancer. 2000;82:501–513. doi: 10.1054/bjoc.1999.0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarke R, Skaar TC, Bouker KB, et al. Molecular and pharmacological aspects of antiestrogen resistance. J Steroid Biochem Mol Biol. 2001;76:71–84. doi: 10.1016/s0960-0760(00)00193-x. [DOI] [PubMed] [Google Scholar]
- 43.Madden TA, Barrow D, McClelland RA, Gee JM, Nicholson RI. Modulation of oestrogen action by receptor gene inhibition. Eur J Cancer. 2000;36(Suppl 4):S34–35. doi: 10.1016/s0959-8049(00)00216-1. [DOI] [PubMed] [Google Scholar]
- 44.McDonnell DP. Mechanism-based discovery as an approach to identify the next generation of estrogen receptor modulators. FASEB J. 2006;20:2432–2434. doi: 10.1096/fj.06-1202ufm. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.