Abstract
Excess dietary vitamin A is esterified with fatty acids and stored in the form of retinyl ester (RE) predominantly in the liver. According to the requirements of the body, liver RE stores are hydrolyzed and retinol is delivered to peripheral tissues. The controlled mobilization of retinol ensures a constant supply of the body with the vitamin. Currently, the enzymes catalyzing liver RE hydrolysis are unknown. In this study, we identified mouse esterase 22 (Es22) as potent RE hydrolase highly expressed in the liver, particularly in hepatocytes. The enzyme is located exclusively at the endoplasmic reticulum (ER), implying that it is not involved in the mobilization of RE present in cytosolic lipid droplets. Nevertheless, cell culture experiments revealed that overexpression of Es22 attenuated the formation of cellular RE stores, presumably by counteracting retinol esterification at the ER. Es22 was previously shown to form a complex with β-glucuronidase (Gus). Our studies revealed that Gus colocalizes with Es22 at the ER but does not affect its RE hydrolase activity. Interestingly, however, Gus was capable of hydrolyzing the naturally occurring vitamin A metabolite retinoyl β-glucuronide. In conclusion, our observations implicate that both Es22 and Gus play a role in liver retinoid metabolism.
Keywords: vitamin A, retinol, retinyl ester hydrolase, endoplasmic reticulum
Retinoids (vitamin A and metabolites) are essential micronutrients in mammals (1). Dietary retinoids are readily absorbed by the intestine. In the intestinal lumen, retinyl esters (REs) are hydrolyzed to retinol by the action of pancreatic triglyceride lipase (2, 3). Within the enterocytes, retinol is reesterified (3–5) for the incorporation into chylomicrons and secreted (6). In the circulation, chylomicrons are depleted from triglycerides by lipoprotein lipase and are thereby transformed to chylomicron remnants (7). These remnants acquire apolipoprotein E and are then cleared mostly by parenchymal cells of the liver (i.e., hepatocytes) (8–10). In hepatocytes, REs are hydrolyzed, and unesterified retinol is associated with retinol-binding protein 4 (RBP4) for secretion (11, 12) or transferred to hepatic stellate cells for storage (10, 13, 14). These stellate cells store most of the total body vitamin A reserves (∼80%) in the form of retinyl palmitate in cytosolic lipid droplets (14).
According to the body's demand, stored retinoids are released from the liver to facilitate a constant supply. In the circulation, the biologically inactive retinol is transported bound to RPB4 and delivered to target tissues. There, retinol is converted into its biologically active metabolites 11-cis retinaldehyde and retinoic acids, which act as hν acceptor in the visual cycle and as ligand of nuclear receptors, respectively.
The dynamic balance between synthesis and hydrolysis of RE determines the concentration of retinol in the circulation and also the availability of retinol for conversion in active metabolites in various cell types. Retinol is esterified by the action of acyl-CoA:retinol acyltransferase (ARAT) or lecithin:retinol acyltransferase (LRAT) for storage in lipid droplets. Much work has focused on the understanding of how REs are released from lipid droplets and which retinyl ester hydrolases (REHs) are involved in this process. A number of potential candidates for the hydrolysis of REs in the liver has been studied so far in more detail. For instance, bile salt-activated carboxyl ester lipase (CEL) (15, 16) has been demonstrated to hydrolyze RE. However, CEL-deficient mice generated by targeted disruption of the CEL gene failed to show any effect on RE metabolism in the liver, on serum levels of retinol and RBP4, or on levels of retinoids in various tissues (17). Two distinct bile salt-independent REHs, one active at neutral (18), the other at acid pH (19), have been characterized in rat liver; in addition, three rat liver carboxyl esterases [Es2 (20), Es4, and Es10 (21, 22)] exhibit REH activity in vitro. Beside their expression in liver, no convincing evidence has been reported that any of these enzymes may play a key role in the mobilization of REs.
In this study, we compared the potency of several members of the mouse carboxyl esterase superfamily to hydrolyze REs. Among other enzymes, we identified esterase 22 (Es22) as a potent REH. Notably, expression of Es22 attenuated cellular RE accumulation, indicating that this enzyme affects retinol metabolism in living cells. The enzyme is highly expressed in liver cells and localizes to the endoplasmic reticulum (ER), suggesting that it counteracts the formation of RE by LRAT and ARAT. Es22 has been shown to interact with β-glucuronidase (Gus) at the ER (23). We found that Gus colocalizes with Es22 at the ER and is capable of hydrolyzing retinoyl β-glucuronide, a naturally occurring retinoid. In conclusion, our data indicate that both Es22 and Gus play a role in liver retinoid metabolism.
MATERIALS AND METHODS
Materials
All-trans-retinol, all-trans-retinyl-palmitate, and palmitic acid were from Sigma-Aldrich Chemie GmbH (Taufkirchen, Germany). All-trans-retinoyl β-glucuronide (RAG) was kindly provided by Prof. Arun B. Barua, Iowa State University, Ames, IA. pCRCU, a self-made yeast expression-vector containing a monomeric RFP sequence, was a kind gift from Julia Petschnigg, Kohlwein Laboratory, University of Graz, Graz, Austria. Retinyl [9,10(n)-3H] palmitate was prepared according to Boechzelt et al. (24) using palmitic acid (2 mCi, 0.033 µmol, GE Healthcare, Piscataway, NJ) as radiolabel. All reagents were of per analysis grade.
Animals
Adult male C57BL/6 mice between 12 and 16 weeks of age were used in this study. Mice were maintained on a regular light-dark cycle (14 h light, 10 h dark) and fed a standard laboratory chow diet (4.5% wt/wt fat).
cDNA cloning of recombinant His-, GFP-, or RFP-tagged proteins
Total RNA was isolated from mouse tissue using the Trizol® Reagent procedure according to the manufacturer's instruction (Invitrogen™, Carlsbad, CA). Poly A+ RNA was isolated from liver total RNA using the Oligotex® mRNA Mini Kit from Qiagen GmbH (Hilden, Germany). Liver mRNA was transcribed into first-strand cDNA using SuperScript™ Reverse Transcriptase protocol from Invitrogen™. Second-strand cDNA was obtained from first-strand cDNA by addition of Escherichia coli DNA ligase buffer, E. coli DNA polymerase, E. coli DNA ligase (all chemicals from New England Biolabs, Inc., Beverly, MA), and deoxyribonucleotide triphosphates (Carl Roth GmbH and Co. KG, Karlsruhe, Germany) to the mixture and subsequent incubation at 16°C for 3 h. Thereafter, T4 DNA polymerase (New England Biolabs) was added and further incubated for 20 min to give blunt end cDNA. The coding sequences of various genes (see Table 1) were amplified by PCR from liver cDNA using Advantage® cDNA Polymerase Mix (BD Biosciences Clontech, Palo Alto, CA). Respective primers were designed to create 5′ and 3′ restriction endonuclease cleavage sites for in-frame ligation into expression vector pcDNA4/HisMax (Invitrogen™). A control pcDNA4/HisMax vector expressing β-galactosidase (LacZ) was provided by the manufacturer (Invitrogen™).
TABLE 1.
List of cloned genes
| GI Number | Accession Number | Identifier | Description | Primer Forward/Reverse |
|---|---|---|---|---|
| 117553603 | NM_053200 | Ces3 | Carboxylesterase 3 | 5′-GGAATTCCGCCTCTACCCTCTGATATGGC-3′ |
| 5′-CCTCGAGTCAGAGCTCAACATGTTCCCTG-3′ | ||||
| 141802881 | NM_172759 | Ces5 | Carboxylesterase 5 | 5′-GGAATTCCCACTATACAAACTTCTTGGATGG-3′ |
| 5′-CCTCGAGCTACAACTCTTTGTGCCTCTCCTG-3′ | ||||
| 10946841 | NM_021456 | Ces1 | Carboxylesterase 1 | 5′-GGAATTCTGGCTCTGTGCTTTGAGTCTGA-3′ |
| 5′-CCTCGAGTCACAGTTCAACATGTTCCCTATG-3′ | ||||
| 21362300 | NM_144511 | EG13 | predicted gene, EG13909 | 5′-GGGTACCAGGACAATGATACCAGCTGGGT-3′ |
| 5′-CCTCGAGTCAGAGCTCCTCTGGAACTTTCC-3′ | ||||
| 28279460 | BC046327 | ACE | Acetylcholinesterase | 5′-GGAATTCAGGCCTCCCTGGTATCCCCT-3′ |
| 5′-CTCTAGATCACAGGTCTGAGCAGCGCT-3′ | ||||
| 22122766 | NM_146213 | BC026 | cDNA seq. BC026374 | 5′-GGAATTCAAGTGGATTCTGGGCTTGAGC-3′ |
| 5′-CTCTAGACTAAGGTTTCTGAGATTGGCGA-3′ | ||||
| 20886282 | XM_146488 | sim Ces2 | similar to Ces 2 | 5′-GGAATTCAGACTGGAACAAATTCATGCTCG-3′ |
| 5′-CCTCGAGCTACAACTCTTTATGTCTGTCCTCAAT-3′ | ||||
| 124487349 | NM_009738 | Bche | Butyrylcholinesterase | 5′-GGAATTCCAGACTCAGCATACCAAGGTAACA-3′ |
| 5′-CCTCGAGTTAGAGAGCTGTACAGCTCTCTTTCTT-3′ | ||||
| 145301632 | NM_133660 | Es22 | Es22 (sim rat Es3) | 5′-GGAATTCTGCCTCTCTGCTCTGATCCTG-3′ |
| 5′-CCTCGAGTCACAGCTCAGTGTGTTCTGTCG-3′ | ||||
| 21450338 | NM_144930 | TGH2 | TGH2 (sim rat Es4) | 5′-GAATTCTGGCTCTTTGCTCTGGC-3′ |
| 5′-CTCGAGTCTCTGAGTGTCTCCCTTGGT-3′ | ||||
| 142348402 | NM_007954 | Es1 | Es1 (sim rat Es2) | 5′-GAATTCTGGCTCCATGCTCTGGTCT-3′ |
| 5′-CTCGAGTTTGTGTTCTCTGTGCTCAGTAGG-3′ | ||||
| 146134463 | NM_016903 | Es10 | Esterase 10 | 5′-GGAATTCGCGCTCAAACAGATTTCCAGCAC-3′ |
| 5′-CCTCGAGTCATGCATTCAGGTACTTAGCA-3′ | ||||
| 6754097 | NM_010368 | Gus | β-Glucuronidase | 5′-GGAATTCTCCCTAAAATGGAGTGCGTGT-3′ |
| 5′-CTCGAGTTAGAACGTGAACGGTCTGCTT-3′ |
For generation of Es22-green fluorescent protein (GFP) and Gus-red fluorescent protein (RFP) fusion constructs full-length Es22 and Gus coding sequences were amplified using liver cDNA as template and respective primers: Es22_forward, 5′- ATCTCGAGCCACCATGTGCCTCTCTGCTCTGATCC-3′; Es22_reverse, 5′- TCGAATTCGCAGC-TCAGTGTGTTCTGTCGG-3′; Gus_forward, 5′-TCTCGAGCC-ACCATGTCCCTAAAATGGAGTGCGT-3′; Gus_reverse, 5′-CGAATTCGAACGTGAACG-GTCTGCTTCC-3′. RFP coding sequence was amplified from the pCRCU vector with the following primers: RFP_forward, CGAATTCGCCTCCTCCGAGGATGTCAT; RFP_reverse, GGATCCTTAGGCGCCGGTGGAGTG. The PCR mixture contained 1 µl cDNA or pTR-CU vector (10 ng/µl), 10 pmol primers, 10 nmol dNTPs, 1 U PhusionTM High-Fidelity DNA Polymerase (Finnzymes Oy, Espoo, Finland), and 6 µl PhusionTM high fidelity buffer in a total volume of 30 µl. The amplified Es22 and Gus coding sequences were digested with XhoI/EcoRI and ligated into respective sites of the pEGFP-N1 vector (Takara Bio Inc., Otsu, Japan). The amplified RFP coding sequence was digested with EcoR1/BamH1 and ligated into respective sites of the pEGFP-N1 vector already containing the Gus coding sequence. The resulting fusion constructs encoded GFP and RFP at the C terminus of Es22 (Es22-GFP) and Gus (Gus-RFP), respectively.
Expression of recombinant proteins in cultured cells
Monkey embryonic kidney cells (COS-7, ATCC CRL-1651) were maintained in DMEM (Gibco® from Invitrogen™) containing 10% fetal calf serum (FCS) (Sigma-Aldrich Chemie GmbH) and antibiotics at 37°C in humidified air (89–91% saturation) and 5% CO2. The day before transfection, COS-7 cells were collected in logarithmic phase, seeded in 6-wells dishes at a density of 150,000 cells/well, and cultured overnight. Transient transfection of COS-7 cells with pcDNA4/HisMax encoding respective His-tagged recombinant proteins or as a control LacZ was performed with Metafectene™ (Biontex GmbH, Munich, Germany). One µg purified DNA (NucleoBond® AX, Macherey-Nagel GmbH and Co. KG, Düren, Germany) was mixed with 5 µl Metafectene™ in a total volume of 100 µl serum- and antibiotics-free DMEM and incubated for 20 min at room temperature to allow formation of the DNA/Metafectene™ complex. Then, 100 µl/well of the DNA/Metafectene™ mix was added to COS-7 cells and incubated for 4 h in serum and antibiotics-free DMEM. Thereafter, the medium was removed and cells were cultured in DMEM containing 10% FCS and antibiotics. Cells were analyzed two days after transfection.
Preparation of COS-7 cell lysates
Cells were collected by trypsination and washed three times with PBS. Then, cells were disrupted on ice in buffer A (0.25 M sucrose, 1 mM EDTA, 1 mM dithiothreitol, 20 µg/ml leupeptine, 2 µg/ml antipain, 1 µg/ml pepstatin, pH 7.0) by sonication (Virsonic 475, Virtis, Gardiner, NJ). Nuclei and unbroken cells were removed by centrifugation at 1,000 g at 4°C for 5 min.
Determination of lipid hydrolase activities
For determination of REH activity of recombinant proteins, 100 µl of COS-7 cell lysates (=100 µg cell protein) containing recombinant proteins and 100 µl substrate was incubated in a water bath at 37°C for 60 min. The reaction was terminated by addition of 3.25 ml of methanol-chloroform-heptane (10/9/7, v/v/v) and 1 ml of 0.1 M potassium carbonate, 0.1 M boric acid, pH 10.5. After vigorous vortexing and centrifugation (800 g, 10 min), 1 ml of the upper aqueous phase was aspirated and radioactivity was determined by liquid scintillation counting (Tri-Carb 2100TR, Packard Instrument Co., Downers Grove, IL). Blank incubation was performed with 100 µl buffer A under identical conditions as for cell lysates. Counts obtained for blank incubation were used for background corrections. Substrate for REH assay contained 10 nmol retinyl palmitate/assay and retinyl [9,10(n)-3H]palmitate, 50,000 cpm/nmol as tracer) and was either emulsified in 100 mM potassium phosphate buffer pH 7.4, containing 90 µM phosphatidyl-choline/-inositole (PC:PI; 3:1) or 100 mM Tris/Maleate buffer pH 8.0, containing 40 mM cholate. Other lipid substrates were prepared in 100 mM Tris/Maleate buffer pH 8.0, containing 40 mM cholate and either 33 nmol triglyceride/assay (TG; glycerol tri[9,10(n)-3H]oleate, 40,000 cpm/nmol) or 20 nmol/assay of cholesteryl oleate (CE; cholesteryl [9,10(n)-3H]oleate, 50,000 cpm/nmol). All lipid substrates were prepared by sonication on ice (Virsonic 475, Virtis). In some cases, substrate was prepared using buffer systems and detergents as indicated in respective figure.
Determination of RAG hydrolase activity
Cell lysates containing recombinant Gus, Es22, or LacZ were incubated in 50 mM Tris/Maleate buffer pH 8.0, containing 25 µM RAG for 30 min at 37°C. Then, reaction was stopped by addition of methanol and RAGs were extracted using three different organic solvents (n-hexane, ethyl acetate, and chloroform). Extracts were combined and RAG content was determined by HPLC as described below.
Intracellular retinyl palmitate accumulation
COS-7 cells were plated in 6-well plates and transfected with Es22 or LacZ as described above. After 48 h of incubation, the media was replaced with DMEM containing 10% FCS, antibiotics, 500 µM palmitic acid (10 mM, solubilized in sterile PBS, containing 50 mg defatted BSA/ml), and 100 µM all-trans-retinol (350 mM in ethanol). At various time points, cells were harvested by trypsination, washed three times with PBS, and subjected to determination of retinyl palmitate content by HPLC.
Extraction of retinoids
All operations were carried out with precooled solvents; whenever possible, samples were placed on ice and protected from light. For the extraction of retinyl palmitate, 400 µl of COS-7 cells (2 mg cell protein/ml) suspended in buffer A were treated with 400 µl 100% methanol containing 0.1% butylated hydroxytoluene (BHT), 1 mM EDTA, and 800 µl water-washed n-hexane. Thereafter, the mixture was vortexed (15 s) and centrifuged at 2,000 g for 3 min. An aliquot of the supernatant (600 µl) was removed, dried down using a speedvac (Heto-Holten, Allered, Denmark), and resuspended in 100 µl of methanol. An aliquot of 20 µl was analyzed by HPLC. Extraction of RAG was performed according to the procedure as outlined by Barua et al. (25). Briefly, 200 µl of lysates were treated with 200 µl ethanol containing 0.1% BHT and 400 µl ethyl acetate. After vortexing (15 s) and centrifugation (2,000 g for 2 min), 200 µl of water was added and extraction was performed by vortexing and centrifugation as described. Then, the upper organic phase was saved and the aqueous phase was acidified with 5 µl of 10% glacial acetic acid in water. The aqueous phase was subsequently extracted with 200 µl ethyl acetate and then extracted with 200 µl n-hexane by vortexing and centrifugation as described above. All organic solvents were pooled and brought to dryness using a speedvac (Heto-Holten). The residue was dissolved in 100 µl methanol and 40 µl were analyzed by HPLC.
HPLC analysis of retinoids
Retinoids were separated on a reverse-phase Lichrospher® 125-4 5 µm C18 column (125 × 4 mm) preceded by a Superspher® RP-18 guard column (Merck, Darmstadt, Germany). For the separation of retinyl palmitate 100% methanol and for RAG methanol-water (7.5:2.5, v/v) containing 10 mM ammonium acetate was used isocratically as the eluant at a flow rate of 1.5 and 1 ml/min, respectively. The HPLC system used was System Gold® from Beckmann Coulter Inc. (Fullerton, CA), consisting of a 125 solvent module, a 508 autosampler, and a 168 diode-array detector. Detections of retinyl palmitate and RAG were performed at 325 nm and 350 nm, respectively. The areas under peaks were standardized against known amounts of reference compounds. For the determination of molar concentrations of reference compounds, the following molar extinction coefficients were used: all-trans-retinyl palmitate (ϵ325nm = 52,275 M−1cm−1) (26); RAG (E1cm, 1% = 1065 at 360 nm) (27).
Isolation of various liver cell-types
Mice were anesthetized and the abdomen was surgically opened by a vertical incision. Then, the liver was perfused via the portal vein with Krebs-Henseleit buffer (without Ca2+ and SO42−) for 10 min followed by a perfusion with Krebs-Henseleit buffer containing 30 mg collagenase type II (0.2 mg/ml, Worthington Biochemical Corporation, Lakewood, NJ), 2% BSA, and 0.1 mM CaCl2 for 10–15 min. Thereafter, the liver was excised, disrupted, and the cell suspension passed through gauze, followed by filtration through a 70 µm cell strainer. Parenchymal cells were separated from nonparenchymal cells by centrifugation at 50 g for 3 min at 4°C. The remaining supernatant was used for the isolation of various nonparenchymal cell-types using OptiPrep™ self-forming density gradient solutions (Axis-Shield PoC AS, Rodeløkka, Norway) according to manufacturer's instructions. Briefly, nonparenchymal cell suspension was adjusted to a density of 24% iodixanol, overlaid with 17%, 11.5%, 8.4%, and 0% iodixanol in Krebs-Henseleit buffer containing 1.25 mM CaCl2 and 1.2 mM NaSO4. After centrifugation at 1,400 g for 20 min at 4°C, Kupffer and stellate cells were isolated at 11.5/8.4% and 8.4/0% iodixanol interphases, respectively. RNA of hepatocytes and Kupffer cells was isolated immediately or after cultivation of the cells overnight. RNA of stellate cells was obtained from freshly prepared cells or after cultivation for 7 days [selective detachment according to Trøen et al. (28)]. Cell lysates for Western blotting analysis of different liver cells were washed twice with PBS after respective cultivation, scraped off, and disrupted as described for COS-7 cells.
Northern blotting analysis
Total RNA was extracted from various mouse tissues or liver cell types using Trizol® Reagent (Invitrogen™), separated by formaldehyde/agarose gel electrophoresis and blotted onto a Hybond-N+ membrane (GE Healthcare) by vacuum blotting (Bio-Rad 785, BioRad Laboratories, Hercules, CA). The membrane was probed with [α-32P]dCTP-labeled cDNA specific for mouse Es22, Gus, or α-smooth muscle actin (α-SMA). The following probes were used: for Es22, the first 416 bp and for Gus, the first 606 bp of the coding region were obtained by restriction digestion using BamHI and HindIII, respectively; for α-SMA, the fragment between 66 and 638 bp of the coding sequence was amplified using following primers: forward, 5′-CTTCGCTGGTGATGATGCTC-3′; reverse, 5′-TCACGGACAATCTCACGCTC-3′), ligated into pCR2.1 vector (Invitrogen™) and isolated from the multiple cloning site of the vector. After hybridization and washing, signals were visualized by exposure to a PhosphorImager Screen (ApBiotech, Freiburg, Germany) and analyzed using ImageQuant Software (Molecular Dynamics). As a loading control, RNA separated by the formaldehyde/agarose gel was visualized with ethidium bromide staining.
Western blotting analysis
Cell lysates were solubilized in SDS-PAGE sample buffer and proteins separated on a 10% SDS-PAGE gel using the Laemmli discontinuous buffer system (29). Then, proteins were transferred onto a polyvinylidene fluoride transfer membrane (Pall Life Sciences, Pensacola, FL). The membrane was blocked with 10% blotting grade milk powder (Carl Roth GmbH and Co. KG) in Tris/NaCl/Tween-20 (TBST) and incubated with mouse anti-keratin-18 monoclonal antibody (Progen, Heidelberg, Germany) at a dilution of 1:1,000 or rat anti-mouse MOMA-2 antibody (Acris antibodies, Hiddenhausen, Germany) at a dilution of 1:5,000. After washing with TBST, the membrane was incubated with horseradish peroxidase-conjugated sheep anti-mouse (GE Healthcare) antibody at a dilution of 1:10,000 and with horseradish peroxidase-conjugated rabbit anti-rat (Dako, Glostrup, Denmark) antibody at a dilution of 1:1,000. After washing with TBST, the membrane was developed with enhanced chemiluminescence (ECL plus, GE Healthcare) and exposed to X-ray film (Hyperfilm™ ECL, GE Healthcare).
Live cell imaging
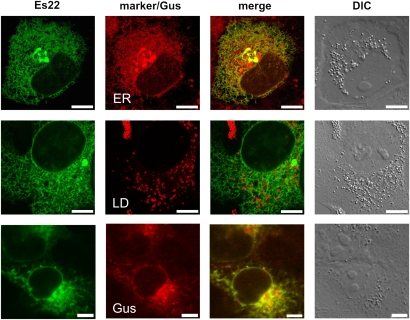
COS-7 cells were cultivated on 24 × 24 mm coverslips and transiently transfected with GFP-tagged Es22, RFP-tagged Gus, or both (for transfection see above). ER was stained with 4 µM ER Tracker RedTM for 15 min at 37°C. Lipid droplets were stained with 2 µM Bodipy® 558/568 C12 for 30 min at 37°C. After staining, cells were washed twice with PBS. For microscopy, coverslips with attached cells were mounted on standard microscope slides. Microscopy was performed using a Leica SP2 confocal microscope (Leica Microsystems, Mannheim, Germany) with spectral detection and a Leica 63× water immersion objective (HCX PL APO W Corr CS, 1.2 NA). GFP or RFP fluorescence was excited at 488 or 555 nm and detected in the range between 500 and 535 or 580 and 620, respectively. ER-Tracker RedTM fluorescence was excited at 543 nm and detected in the range from 600 to 650 nm. Bodipy® 558/568 C12 fluorescence was excited at 543 nm and detected in the range between 550 and 650 nm. Fluorescence emissions of GFP and RFP or ER Tracker RedTM or Bodipy® 558/568 C12 were detected simultaneously as indicated in the legend to Fig. 7.
Fig. 7.
Es22 and Gus colocalize to the endoplasmic reticulum. COS-7 cells were transfected with plasmids encoding either Es22-GFP or both Es22-GFP and Gus-RFP fusion proteins. The next day, cells expressing Es22-GFP alone were incubated with 400 µM oleate for 18 h to promote lipid droplet formation. In some cases, Es22-GFP expressing cells were incubated with ER-TrackerTM Red dye (upper panels) or Bodipy® 558/568 C12 (middle panels) for staining of the ER or lipid droplets, respectively. Localization of Es22-GFP (green), Gus-RFP (red), and the ER/lipid droplets staining (red) as well as colocalizations (yellow) were visualized by confocal laser scanning microscopy. The right column displays a three dimensional picture of respective cells as obtained by differential interference contrast microscopy. Bars = 10 µm.
Determination of protein concentration
Protein concentrations of cell lysates were determined with Bio-Rad protein assay according to manufacturer's instructions (Bio-Rad Laboratories) using BSA as standard.
Statistical analysis
All data are expressed as means ± SD. Statistical significance was determined by the Student's unpaired t-test (two-tailed). Group differences were considered significant for P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
RESULTS
REH activities of mouse carboxyl esterases
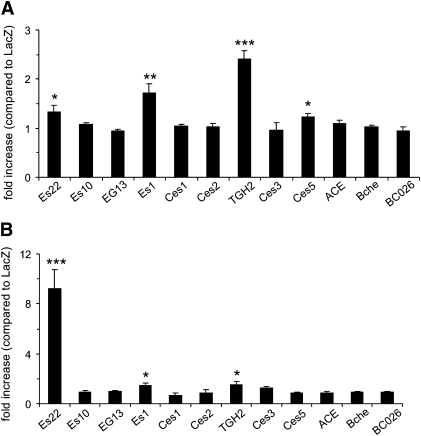
Some of the most promising enzymes thought to be involved in the hydrolysis of RE in the liver belong to the group of the carboxyl esterase super family (30). To compare REH activities of different carboxyl esterases, we cloned respective coding sequences of mouse carboxyl esterases (Table 1) into a mammalian expression vector and expressed the recombinant His-tagged proteins in COS-7 cells. A vector encoding LacZ was used for control transfections. REH activities were determined in cell lysates containing the respective recombinant proteins (1,000 g supernatant). Using retinyl palmitate emulsified with phospholipids as substrate, we found significantly increased REH activities in cells expressing Es22 (∼1.3-fold), Es1 (∼1.7-fold), TGH2 (∼2.4-fold), and Ces5 (∼1.2-fold), compared with LacZ expressing cells (Fig. 1A). To investigate whether the activity of carboxyl esterases is dependent on the presence of bile-salts, we also determined REH activities using retinyl palmitate emulsified in the presence of cholate. Under these conditions, expression of Es22 resulted in an ∼9-fold increase in REH activity compared with the LacZ control (Fig. 1B). A minor yet statistically significant increase was also observed for Es1 and TGH2.
Fig. 1.
Retinyl ester hydrolase activity of various carboxyl esterases using (A) phospholipids or (B) cholate as detergents for substrate preparation. Carboxyl esterases (see Table 1) were cloned into the mammalian expression vector pcDNA4/HisMax for expression in COS-7 cells. As a control, COS-7 cells were transfected with a plasmid encoding β-galactosidase (LacZ). Lysates of COS-7 cells (1,000 g supernatant) were subjected to REH assays using radiolabeled retinyl (3H) palmitate as tracer. Data are mean ± SD of triplicate determination in relation to the activity detected in control transfected cells (LacZ) and are representative for three independent determinations. * P < 0.05; ** P < 0.01; *** P < 0.001.
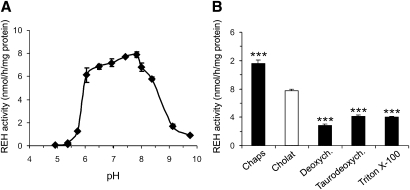
Es22 REH activity is highest in the neutral pH range and affected by the use of different detergents
For further characterization of Es22 REH activity, we determined the pH optimum of the enzyme using RE emulsified with cholate as substrate. As shown in Fig. 2A, Es22 REH activity exhibited a rather broad pH maximum, giving the highest rates in the neutral range. Next, we determined whether Es22 REH activity is dependent on the detergent cholate or whether the enzyme is also active in the presence of ionic or nonionic detergents. As shown in Fig. 2B, Es22 REH activity was highest with the detergent CHAPS (∼150%) and decreased to 36, 53, and 52% when the detergents deoxycholate, taurodeoxycholate, or Triton X-100 were used, respectively. These data indicate that Es22 REH activity is affected by the use of different detergents. REH activity was also detected in the presence of the nonionic detergent Triton X-100, which indicates that enzyme activity is not critically dependent on the presence of bile-salts.
Fig. 2.
Dependence of Es22 REH activity on pH (A) and on various detergents (B). A: For the determination of pH dependence, lysates of COS-7 cells expressing Es22 were incubated with retinyl (3H) palmitate substrate emulsified in cholate using the following buffer systems: 100 mM Na-acetate buffer (pH 4.5 and 5), 100 mM Tris/Maleate buffer (pH 5 to 8), and 100 mM Glycin/NaOH buffer (pH 8 to 9.5). B: REH activity of Es22 in 100 mM Tris-maleate, pH 8.0 using the following detergents (final concentration): CHAPS (30 mM), cholate (20 mM), deoxycholate (20 mM), taurodeoxycholate (2 mM), and Triton X-100 (0.4%).). In A and B, the activity detected in control transfected cells (LacZ) under identical conditions was set as blank to account for endogenous activity in COS-7 cells. Data are mean ± SD of triplicate determination (LacZ) and are representative for three independent determinations. *** P < 0.001.
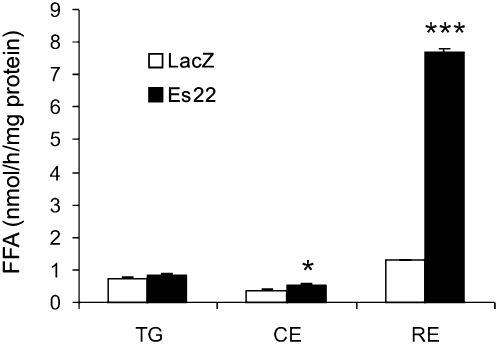
Es22 specifically hydrolyzes RE
To determine the substrate specificity of Es22, lysates of COS-7 cells expressing Es22 or LacZ were prepared and incubated with CE, TG, or retinyl palmitate as substrate. All substrates were emulsified in 20 mM cholate and contained the respective radiolabeled compounds as tracer. Compared with LacZ, Es22 exhibited substantial activity using RE as substrate (Fig. 3). Furthermore, we found a moderate but significant increase by ∼50% using CE as substrate whereas no difference was observed with TG as substrate. These results indicate that Es22 specifically hydrolyzes retinyl palmitate and, to a much lower extent, (∼30 times less efficient) cholesteryl oleate.
Fig. 3.
Es-22 specifically hydrolyzes retinyl palmitate. COS-7 cells were transfected with pcDNA4/HisMax encoding Es22 or LacZ. Then, lysates were prepared and incubated with the following substrates containing radiolabeled substrates as tracer: triolein (TG), cholesteryl oleate (CE), and retinyl palmitate. All substrates were prepared in 100 mM Tris-maleate, pH 8.0 and 40 mM cholate as detergent. Data are mean ± SD and representative for at least three independent experiments. * P < 0.05; *** P < 0.001.
Es22 inhibits accumulation of RE in COS-7 cells
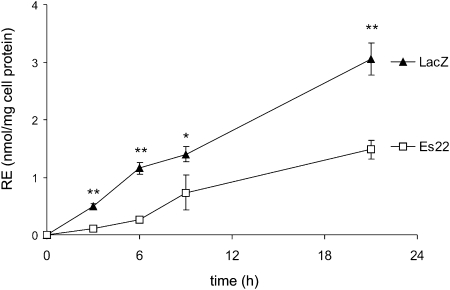
To demonstrate that Es22 affects retinol metabolism in living cells, we used COS-7 cells as a model system. HPLC analysis of endogenous RE content indicated that this cell line does not contain detectable amounts of this compound (detection limit ∼0.5 µM). Therefore, artificial loading of COS-7 cells by incubation with retinol and palmitic acid was necessary to promote retinyl palmitate synthesis. First, COS-7 cells were transfected with either Es22 or LacZ and subsequently incubated with retinol and palmitic acid. The formation of intracellular retinyl palmitate content was determined by HPLC. Incubation of LacZ transfected COS-7 cells with retinol and palmitic acid led to a linear increase in the cellular RE content over a period of 20 h (Fig. 4). In contrast, under identical conditions, Es22 transfected COS-7 cells exhibited decreased RE accumulation by ∼50% (Fig. 4). This effect was more pronounced during the initial 6 h of incubation.
Fig. 4.
Es22 inhibits the accumulation of retinyl esters in COS-7 cells. COS-7 cells were transfected with Es22 or LacZ. After 48 h of transfection (time point = 0 h), cells were incubated with 500 µM palmitic acid and 100 µM all-trans-retinol for up to 20 h. At various times, cells were harvested and cellular retinyl palmitate content was determined by RP-HPLC. Data are mean ± SD of three determinations and are representative for three independent experiments. * P < 0.05; ** P < 0.01.
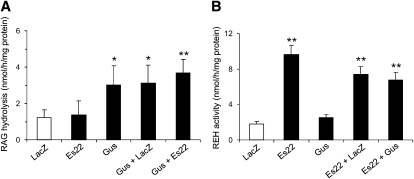
Gus hydrolyzes RAG
It has been shown that Es22 can form a complex with Gus, thereby sequestering Gus at the ER. The interaction of Es22 and Gus raised the question whether Gus might be involved in retinoid metabolism by hydrolyzing RAGs, which are naturally occurring metabolites of vitamin A. For the determination of glucuronide hydrolase activity, we incubated COS-7 cell lysates containing Gus with RAG as substrate. Before and after incubation, the RAG concentration was determined by HPLC. As shown in Fig. 5A, Gus containing lysates exhibited increased RAG hydrolyzing activity as compared with cells expressing LacZ or Es22. Coincubation of Gus and Es22 did not affect glucuronide hydrolase activity of Gus (Fig. 5A). Conversely, coincubation of Gus and Es22 had no effect on REH activity of Es22 (Fig. 5B). However, the complex forming nature of these enzymes suggests that Es22/Gus act in concert in mobilizing retinol from RE and RAG.
Fig. 5.
β-Glucuronidase hydrolyzes retinoyl β-glucuronide (RAG). COS-7 cells were transfected with plasmids encoding Gus, Es22, or LacZ. Lysates were subjected to activity assays using (A) retinoyl β-glucuronide or (B) retinyl (3H) palmitate as substrate. To investigate whether Gus affects the activity of Es22 and vice versa, mixtures of lysates containing Gus and Es22 or Gus and LacZ were incubated with the substrates. A: Activity assays were performed in 50 mM Tris/Maleate buffer, pH 8.0. After 30 min of incubation, the content of RAG was determined by RP-HPLC. Loss in RAG content was expressed as nmol/h/mg protein. Data are mean ± SD of three determinations and are representative for two independent experiments. * P < 0.05; ** P < 0.01. B: β-Glucuronidase does not affect Es22 REH activity. Activity assays were performed with radiolabeled retinyl palmitate substrate in 50 mM Tris-maleate, pH 8.0 and 20 mM cholate. Data are mean ± SD of triplicate determinations and are representative for two independent experiments. ** P < 0.01.
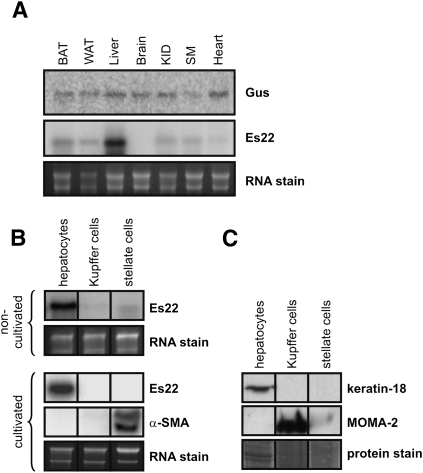
Es22 is predominantly expressed in hepatocytes
Tissue expression patterns of Es22 and Gus were analyzed by Northern blotting using respective DNA probes. As shown in Fig. 6A, Es22 was found to be highly expressed in liver compared with other tissues, including white/brown adipose tissue, skeletal/cardiac muscle, and kidney. No signal was detected in the brain. In contrast, Gus expression levels were comparable in all tissues examined. Next, we investigated Es22 mRNA expression levels in specific liver cell types, such as hepatocytes, Kupffer, or stellate cells. These liver cell types were isolated from collagenase perfused mouse livers using iodixanol self-forming density gradient. Subsequently, RNA was isolated immediately (noncultivated cells) or after cultivation of cells and used for Northern blotting experiments. As depicted in Fig. 6B, in noncultivated liver cells, Es22 mRNA expression level was highest in hepatocytes and was barely detectable in Kupffer and stellate cells. Interestingly, after cultivation of liver cells, Es22 mRNA expression diminished in Kupffer and stellate cells. As a control, we also used a probe for α-SMA, which is known to be highly selective for stellate cells. α-SMA positively hybridized only with RNA obtained from stellate cells, confirming purity of the stellate cell preparations (Fig. 6B). Similarly, purity of hepatocyte and Kupffer cell preparations were analyzed using α-keratin 18 and monocyte-macrophage antibody 2 (MOMA-2) as marker proteins, respectively. As depicted in Fig. 6C, Western blotting analyses of cell preparations showed that keratin-18 and MOMA-2 were exclusively expressed in hepatocytes and Kupffer cells, respectively, demonstrating purity of these cell preparations.
Fig. 6.
Es22 is highly expressed in liver and specifically in hepatocytes. Total RNA was isolated from (A) mouse tissues or (B) different liver cell types obtained from perfused mouse liver using self-forming iodixanol gradient separation. Various liver cell types were either used immediately for RNA isolation (noncultivated) or cultured overnight (cultivated; hepatocytes and Kupffer cells). Stellate cells were purified by selective detachment for 7 days of culture (cultivated). RNA of various cell types was subjected to Northern blotting analysis using 32P-labeled probes specific for Es22, Gus, or α-smooth muscle actin (α-SMA). RNA loaded on the gels was stained with ethidium bromide. C: Lysates of isolated liver cells (20 µg total protein) were subjected to Western blotting analysis using anti-keratin-18 as hepatocyte and anti-MOMA-2 as macrophage/Kupffer cell marker. Proteins on the membrane were stained with coomassie brilliant blue. Abbreviations: BAT, brown adipose tissue; KID, kidney; SM, skeletal muscle; WAT, white adipose tissue.
Es22 and Gus colocalize at the ER
To determine the subcellular localization of Es22, a GFP fusion protein (Es22-GFP) was expressed in COS-7 cells. Laser scanning microscopy revealed that the Es22-GFP fusion protein distributed throughout the cell, thereby forming a network-like structure (Fig. 7 upper panel, left picture). To investigate if this structure represents the ER, we stained cells with an ER specific dye (ER-tracker). As shown in Fig. 7 (upper panel), the signals obtained for Es22-GFP and ER-tracker resulted in a perfect overlay, demonstrating that Es22 localizes to the ER. We also studied any localization of Es22-GFP at lipid droplets using the neutral-lipids staining dye Bodipy®. However, we could not detect any association of Es22 with lipid droplets, as evident from the merge (Fig. 7, middle panel). Next, we coexpressed Es22-GFP and Gus-RFP in COS-7 cells. As shown in Fig. 7 (lower panel), Es22 and Gus were found to mainly colocalize at the ER whereas Gus also localized to circular structures, presumably lysosomes (31).
DISCUSSION
Vitamin A has been known for almost a century as a fat soluble factor that is essential for normal growth (32, 33) and affects multiple physiological processes, including vision, cell differentiation, reproduction, and immunity. The retinoid 11-cis retinaldehyde functions as the active chromophore in rhodopsin and is crucial for vision. The metabolites all-trans and 9-cis retinoic acid interact with a number of nuclear receptors of the retinoic acid receptor and the retinoid X receptor family. These receptors function as ligand-activated factors and are involved in the regulation of gene expression. Up-to-date chronic undersupply of vitamin A affects millions of people, especially in the “third world” countries. The deleterious effects of chronic vitamin A deficiency include, most importantly, increased susceptibility to infectious diseases, which represents an enormous threat for these populations. Yet, apart from its function in vision and as a ligand for nuclear receptors, little is known about mechanisms facilitating constant supply of the body, in particular the release of retinol from the liver.
In this study, we identified murine Es22 as a potent liver REH. Earlier studies suggested that the rat homolog of Es22, esterase 3 (Es3, 93% homology), might possess REH activity. Rat Es3 was first identified as “esterase with pI 5.6” and originally found to hydrolyze retinyl acetate and, to a much lower extent, retinyl palmitate (21). In another study, Es3 was shown to hydrolyze REs with low efficiency compared with rat Es4 and Es10 (34). In addition to its putative role in retinoid metabolism, two studies suggested that Es3 is involved in the detoxification of xenobiotics. Rat Es3 has been found to hydrolyze acetanilide (35, 36), a compound that exhibits analgesic and antipyretic properties (37). Our studies suggest that murine Es22 is a potent REH compared with the other members of the carboxyl esterase family investigated in this study. These observations are in contrast to the results obtained with rat homologs. Explanations for this discrepancy could be either species differences in enzyme specificity or the use of different assay conditions. For example, the influence of assay conditions on Es22 activity is apparent from assays using cholate or phospholipids as detergent. Es22 was highly active using cholate or CHAPS but exhibited only poor activity when retinyl palmitate was emulsified in the presence of phospholipids. Thus, different assay conditions substantially affect enzyme activity of Es22 and may explain the discrepancy in REH activities of the rat and mouse homologs.
Es22 is highly expressed in the liver, which is the primary storage site of RE. Interestingly, the highest expression was found in hepatocytes and, to a much lesser extent, in Kupffer and hepatic stellate cells, which contain most of the body's vitamin A reserves. Yet, in cell culture experiments, expression of Es22 attenuated the accumulation of REs, suggesting that the enzyme affects the deposition of cellular RE stores. In general, RE hydrolyzing enzymes can attenuate RE accumulation by two independent mechanisms. First, increased RE hydrolase activity at the ER can counteract the esterification of retinol by ARAT or LRAT. Second, enzymes capable of binding to lipid droplets can increase the hydrolysis of stored RE. Our studies demonstrate that Es22 does not attach to lipid droplets, implying that the enzyme does not play a role in the mobilization of RE stores contained in lipid droplets. Es22 is localized exclusively at the ER, indicating that the enzyme affects RE synthesis by counteracting retinol esterifying enzymes, resulting in reduced RE formation. The liberated retinol most likely is subsequently secreted, converted into bioactive vitamin A metabolites, or reesterified. Thus, Es22 presumably inhibits RE accumulation in hepatocytes and possibly other cell types. From this point of view, the low expression of Es22 in stellate cells is not surprising because, specifically, these cells deposit large amounts of RE in lipid droplets.
Es22 was first identified 30 years ago and named egasyn (from the Greek root meaning “to hold together”) (38). This name was chosen because it was found to sequester up to 25% of total Gus at the ER. The rest is delivered to lysosomes. Lysosomal Gus plays an important role in the catabolism of glycosaminoglycans as evident from the mucopolysaccharidose type VII (MPS VII) mouse (39) and from human patients with Gus deficiency (MPS VII or Sly syndrome) (40). The physiological role of microsomal Gus is less understood. Yet, some evidence suggests a possible role in the hydrolysis of endogenous and xenobiotic glucuronides at the ER (41, 42). Our data demonstrate that Gus colocalizes with Es22 at the ER. In addition, we found that RAGs represent an additional physiological substrate for Gus. β-Glucuronides of all-trans retinol and of all-trans retinoic acid have been shown to be formed by liver microsomes in vitro (43) and in vivo (25, 42, 45) by the action of microsomal UDP-glucuronosyl transferase (45). Retinoid β-glucuronides have been detected in various mouse tissues (44) and in the circulation (46), suggesting that these compounds could represent a water-soluble transport form of vitamin A channeling retinol through the ER or through the circulation. In analogy to the formation and hydrolysis of RE, Gus could counteract the formation of retinoyl β-glucuronides by UDP-glucuronosyl transferase at the ER.
In summary, our studies demonstrate that murine Es22 and Gus are capable of hydrolyzing retinoids and colocalize at the ER. Our data indicate that these enzymes affect retinoid storage and mobilization by counteracting the formation of RE and retinoid glucuronides at the ER.
Acknowledgments
The authors thank Prof. Arun B. Barua for providing all-trans-retinoyl β-glucuronide, Prof. Michael Trauner for keratin-18 antibody, and Dr. Dagmar Kratky for MOMA-2 antibody.
Footnotes
Abbreviations:
- ARAT
- acyl-CoA:retinol acyltransferase
- BHT
- butylated hydroxytoluene
- CE
- cholesteryl oleate
- CEL
- carboxyl ester lipase
- ER
- endoplasmic reticulum
- Es22
- esterase 22
- GFP
- green fluorescent protein
- Gus
- β-glucuronidase
- KID
- kidney
- LacZ
- β-galactosidase
- LRAT
- lecithin:retinol acyltransferase
- MOMA
- monocyte-macrophage antibody
- RAG
- retinoyl β-glucuronide
- RBP4
- retinol binding protein 4
- RE
- retinyl ester
- REH
- retinyl ester hydrolase
- RFP
- red fluorescent protein
- α-SMA
- alpha smooth muscle actin
- TBST
- Tris/NaCl/Tween-20
- TG
- triglyceride
This research was supported by the grants ‘GOLD - Genomics of Lipid-Associated Disorders’, which is part of the Austrian Genome Project ‘GEN-AU Genome research in Austria’ funded by the Austrian Ministry of Science and Research, W901-B05 DK (Molecular Enzymology), and P21296-B19, which are funded by the Austrian Science Foundation (FWF).
REFERENCES
- 1.Blomhoff R., Green M. H., Green J. B., Berg T., Norum K. R. 1991. Vitamin A metabolism: new perspectives on absorption, transport, and storage. Physiol. Rev. 71: 951–990 [DOI] [PubMed] [Google Scholar]
- 2.van Bennekum A. M., Fisher E. A., Blaner W. S., Harrison E. H. 2000. Hydrolysis of retinyl esters by pancreatic triglyceride lipase. Biochemistry. 39: 4900–4906 [DOI] [PubMed] [Google Scholar]
- 3.Ruiz A., Winston A., Lim Y. H., Gilbert B. A., Rando R. R., Bok D. 1999. Molecular and biochemical characterization of lecithin retinol acyltransferase. J. Biol. Chem. 274: 3834–3841 [DOI] [PubMed] [Google Scholar]
- 4.MacDonald P. N., Ong D. E. 1988. A lecithin:retinol acyltransferase activity in human and rat liver. Biochem. Biophys. Res. Commun. 156: 157–163 [DOI] [PubMed] [Google Scholar]
- 5.MacDonald P. N., Ong D. E. 1988. Evidence for a lecithin-retinol acyltransferase activity in the rat small intestine. J. Biol. Chem. 263: 12478–12482 [PubMed] [Google Scholar]
- 6.Vogel S., Gamble M. V., Blaner W. S. 1999Biosynthesis, absorption and transport of retinoids. Handbook of Experimental Pharmacology. Retinoids Nau H., Blaner W. S., Springer Verlag Publishing, Heidelberg: 31–95 [Google Scholar]
- 7.Goldberg I. J.1996. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J. Lipid Res. 37: 693–707 [PubMed] [Google Scholar]
- 8.Blaner W. S., Olson J. A. 1999Retinol and retinoic acid metabolism. The Retinoids, Biology, Chemistry and Medicine. Sporn M. B., Roberts A. B., Goodman D. S., Raven Press, New York, NY: 229–256 [Google Scholar]
- 9.Cooper A. D.1997. Hepatic uptake of chylomicron remnants. J. Lipid Res. 38: 2173–2192 [PubMed] [Google Scholar]
- 10.Blomhoff R., Helgerud P., Rasmussen M., Berg T., Norum K. R. 1982. In vivo uptake of chylomicron. Proc. Natl. Acad. Sci. USA. 79: 7326–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newcomer M. E., Ong D. E. 2000. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochim. Biophys. Acta. 1482: 57–64 [DOI] [PubMed] [Google Scholar]
- 12.Ronne H., Ocklind C., Wiman K., Rask L., Obrink B., Peterson P. A. 1983. Ligand-dependent regulation of intracellular protein transport: effect of vitamin a on the secretion of the retinol-binding protein. J. Cell Biol. 96: 907–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blomhoff R., Holte K., Naess L., Berg T. 1984. Newly administered [3H]retinol is transferred from hepatocytes to stellate cells in liver for storage. Exp. Cell Res. 150: 186–193 [DOI] [PubMed] [Google Scholar]
- 14.Senoo H.2004. Structure and function of hepatic stellate cells. Med. Electron Microsc. 37: 3–15 [DOI] [PubMed] [Google Scholar]
- 15.Fredrikzon B., Hernell O., Blackberg L., Olivecrona T. 1978. Bile salt-stimulated lipase in human milk: evidence of activity in vivo and of a role in the digestion of milk retinol esters. Pediatr. Res. 12: 1048–1052 [DOI] [PubMed] [Google Scholar]
- 16.Lombardo D., Guy O. 1980. Studies on the substrate specificity of a carboxyl ester hydrolase from human pancreatic juice. II. Action on cholesterol esters and lipid-soluble vitamin esters. Biochim. Biophys. Acta. 611: 147–155 [DOI] [PubMed] [Google Scholar]
- 17.van Bennekum A. M., Li L., Piantedosi R., Shamir R., Vogel S., Fisher E. A., Blaner W. S., Harrison E. H. 1999. Carboxyl ester lipase overexpression in rat hepatoma cells and CEL deficiency in mice have no impact on hepatic uptake or metabolism of chylomicron-retinyl ester. Biochemistry. 38: 4150–4156 [DOI] [PubMed] [Google Scholar]
- 18.Harrison E. H., Gad M. Z. 1989. Hydrolysis of retinyl palmitate by enzymes of rat pancreas and liver. Differentiation of bile salt-dependent and bile salt-independent, neutral retinyl ester hydrolases in rat liver. J. Biol. Chem. 264: 17142–17147 [PubMed] [Google Scholar]
- 19.Gad M. Z., Harrison E. H. 1991. Neutral and acid retinyl ester hydrolases associated with rat liver microsomes: relationships to microsomal cholesteryl ester hydrolases. J. Lipid Res. 32: 685–693 [PubMed] [Google Scholar]
- 20.Schindler R., Mentlein R., Feldheim W. 1998. Purification and characterization of retinyl ester hydrolase as a member of the non-specific carboxylesterase supergene family. Eur. J. Biochem. 251: 863–873 [DOI] [PubMed] [Google Scholar]
- 21.Mentlein R., Heymann E. 1987. Hydrolysis of retinyl esters by non-specific carboxylesterases from rat liver endoplasmic reticulum. Biochem. J. 245: 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linke T., Dawson H., Harrison E. H. 2005. Isolation and characterization of a microsomal acid retinyl ester hydrolase. J. Biol. Chem. 280: 23287–23294 [DOI] [PubMed] [Google Scholar]
- 23.Medda S., Swank R. T. 1985. Egasyn, a protein which determines the subcellular distribution of beta-glucuronidase, has esterase activity. J. Biol. Chem. 260: 15802–15808 [PubMed] [Google Scholar]
- 24.Boechzelt H., Karten B., Abuja P. M., Sattler W., Mittelbach M. 1998. Synthesis of 9-oxononanoyl cholesterol by ozonization. J. Lipid Res. 39: 1503–1507 [PubMed] [Google Scholar]
- 25.Barua A. B., Batres R. O., Olson J. A. 1988. Synthesis and metabolism of all-trans-[11–3H]retinyl beta-glucuronide in rats in vivo. Biochem. J. 252: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross A. C.1981. Separation of long-chain fatty acid esters of retinol by high-performance liquid chromatography. Anal. Biochem. 115: 324–330 [DOI] [PubMed] [Google Scholar]
- 27.Barua A. B., Batres R. O., Olson J. A. 1989. Characterization of retinyl beta-glucuronide in human blood. Am. J. Clin. Nutr. 50: 370–374 [DOI] [PubMed] [Google Scholar]
- 28.Troen G., Nilsson A., Norum K. R., Blomhoff R. 1994. Characterization of liver stellate cell retinyl ester storage. Biochem. J. 300: 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U. K.1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 30.Harrison E. H.2000. Lipases and carboxylesterases: possible roles in the hepatic utilization of vitamin A. J. Nutr. 130: 340S–344S [DOI] [PubMed] [Google Scholar]
- 31.Beltramini-Guarini P., Gitzelmann R., Pfister K. 1984. Presence and absence of the microsomal beta-glucuronidase in mice correlates with differences in the processing of the lysosomal enzyme. Eur. J. Cell Biol. 34: 165–170 [PubMed] [Google Scholar]
- 32.McCollum E. V., Davis M. 1913. The necessity of certain lipids in the diet during growth. J. Biol. Chem. 15: 167–175 [Google Scholar]
- 33.Osborne T. B., Mendel L. B. 1913. The relation of growth to the chemical constituent of the diet. J. Biol. Chem. 15: 311–326 [Google Scholar]
- 34.Sanghani S. P., Davis W. I., Dumaual N. G., Mahrenholz A., Bosron W. F. 2002. Identification of microsomal rat liver carboxylesterases and their activity with retinyl palmitate. Eur. J. Biochem. 269: 4387–4398 [DOI] [PubMed] [Google Scholar]
- 35.Akao T., Omura T. 1972. Acetanilide-hydrolyzing esterase of rat liver microsomes. II. Turnover studies. J. Biochem. 72: 1257–1259 [DOI] [PubMed] [Google Scholar]
- 36.Robbi M., Beaufay H. 1994. Cloning and sequencing of rat liver carboxylesterase ES-3 (egasyn). Biochem. Biophys. Res. Commun. 203: 1404–1411 [DOI] [PubMed] [Google Scholar]
- 37.Botting R.2004. Antipyretic therapy. Front. Biosci. 9: 956–966 [DOI] [PubMed] [Google Scholar]
- 38.Tomino S., Paigen K. 1975. Purification and chemical properities of mouse liver lysosomal (L form) beta-glucuronidase. J. Biol. Chem. 250: 8503–8509 [PubMed] [Google Scholar]
- 39.Birkenmeier E. H., Davisson M. T., Beamer W. G., Ganschow R. E., Vogler C. A., Gwynn B., Lyford K. A., Maltais L. M., Wawrzyniak C. J. 1989. Murine mucopolysaccharidosis type VII. Characterization of a mouse with beta-glucuronidase deficiency. J. Clin. Invest. 83: 1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sly W. S., Quinton B. A., McAlister W. H., Rimoin D. L. 1973. Beta glucuronidase deficiency: report of clinical, radiologic, and biochemical features of a new mucopolysaccharidosis. J. Pediatr. 82: 249–257 [DOI] [PubMed] [Google Scholar]
- 41.Whiting J. F., Narciso J. P., Chapman V., Ransil B. J., Swank R. T., Gollan J. L. 1993. Deconjugation of bilirubin-IX alpha glucuronides: a physiologic role of hepatic microsomal beta-glucuronidase. J. Biol. Chem. 268: 23197–23201 [PubMed] [Google Scholar]
- 42.Brunelle F. M., Verbeeck R. K. 1993. Glucuronidation of diflunisal by rat liver microsomes. Effect of microsomal beta-glucuronidase activity. Biochem. Pharmacol. 46: 1953–1958 [DOI] [PubMed] [Google Scholar]
- 43.Sass J. O., Forster A., Bock K. W., Nau H. 1994. Glucuronidation and isomerization of all-trans- and 13-cis-retinoic acid by liver microsomes of phenobarbital- or 3-methylcholanthrene-treated rats. Biochem. Pharmacol. 47: 485–492 [DOI] [PubMed] [Google Scholar]
- 44.Sass J. O., Tzimas G., Nau H. 1994. 9-cis-retinoyl-beta-D-glucuronide is a major metabolite of 9-cis-retinoic acid. Life Sci. 54: PL69–PL74 [DOI] [PubMed] [Google Scholar]
- 45.Genchi G., Wang W., Barua A., Bidlack W. R., Olson J. A. 1996. Formation of beta-glucuronides and of beta-galacturonides of various retinoids catalyzed by induced and noninduced microsomal UDP-glucuronosyltransferases of rat liver. Biochim. Biophys. Acta. 1289: 284–290 [DOI] [PubMed] [Google Scholar]
- 46.Barua A. B., Olson J. A. 1986. Retinoyl beta-glucuronide: an endogenous compound of human blood. Am. J. Clin. Nutr. 43: 481–485 [DOI] [PubMed] [Google Scholar]