Abstract
Hepatic gluconeogenesis is tightly balanced by opposing stimulatory (glucagon) and inhibitory (insulin) signaling pathways. Hepatocyte growth factor (HGF) is a pleiotropic growth factor that mediates diverse biological processes. In this study, we investigated the effect of HGF and its family member, macrophage-stimulating factor (MSP), on hepatic gluconeogenesis in primary hepatocytes. HGF and MSP significantly repressed expression of the key hepatic gluconeogenic enzyme genes, phosphoenolpyruvate carboxykinase (PEPCK), and glucose-6-phosphatase (Glc-6-Pase) and reduced glucose production. HGF and MSP activated small heterodimer partner (SHP) gene promoter and induced SHP mRNA and protein levels, and the effect of HGF and MSP on SHP gene expression was demonstrated to be mediated via activation of the AMP-activated protein kinase (AMPK) signaling pathway. We demonstrated that upstream stimulatory factor-1 (USF-1) specifically mediated HGF effect on SHP gene expression, and inhibition of USF-1 by dominant negative USF-1 significantly abrogated HGF-mediated activation of the SHP promoter. Elucidation of the mechanism showed that USF-1 bound to E-box-1 in the SHP promoter, and HGF increased USF-1 DNA binding on the SHP promoter via AMPK and DNA-dependent protein kinase-mediated pathways. Adenoviral overexpression of USF-1 significantly repressed PEPCK and Glc-6-Pase gene expression and reduced glucose production. Knockdown of endogenous SHP expression significantly reversed this effect. Finally, knockdown of SHP or inhibition of AMPK signaling reversed the ability of HGF to suppress hepatocyte nuclear factor 4α-mediated up-regulation of PEPCK and Glc-6-Pase gene expression along with the HGF- and MSP-mediated suppression of gluconeogenesis. Overall, our results suggest a novel signaling pathway through HGF/AMPK/USF-1/SHP to inhibit hepatic gluconeogenesis.
Glucose homeostasis is tightly regulated by a hormonal network in which glucagon, glucocorticoids, and insulin are the main agents (1, 2). Glycemia is a parameter over which the organism establishes tight control. Insulin negatively regulates transcription of genes involved in hepatic glucose production such as those encoding phosphoenolpyruvate carboxykinase (PEPCK),2 insulin growth factor-binding protein-1 (IGFBP-1), and glucose-6-phosphatase (Glc-6-Pase) via regulation of various transcription factors that bind to the insulin-response unit (1). Hepatic gluconeogenesis is a major contributor to hyperglycemia when insulin secretion is deficient, as in type I diabetes (T1DM), or when there is a deficiency in insulin action, as in type II diabetes (T2DM) (2, 3). Insulin treatment to patients with T1DM and reversing or bypassing the deficient insulin signaling pathway in patients with T2DM is the key to control glucose homeostasis. Therefore, identifying agents or signaling pathways that can inhibit hepatic gluconeogenesis via insulin-independent signaling may provide new therapeutic options to curtail the elevated gluconeogenesis caused by insulin resistance in T2DM.
Hepatocyte growth factor (HGF)/scatter factor was originally identified as a circulating factor that promotes hepatic regeneration after liver injury, displays pleiotropic cellular activities, including angiogenesis, anti-apoptosis, and mitogenesis, in a wide variety of cell types expressing the HGF receptor c-Met (4, 5). The c-Met receptor tyrosine kinase is the receptor for HGF (4). The mature HGF protein binds to its high affinity receptor c-Met, leading to its activation and phosphorylation of multiple serine and tyrosine residue sites (4). Furthermore, HGF levels have been reported to be elevated in obese patients and raised with body mass index (6–10). On the other hand, weight loss after gastroplasty has been shown to be associated with a reduction of HGF plasma levels in obese patients (6). A strong association between elevated serum HGF and metabolic syndrome has also been reported (6–10). HGF stimulates a diverse array of signaling pathways, including Ras, MAPK, and phosphatidylinositol 3-kinase (PI3K) (5). HGF is known to regulate bile acid synthesis in the liver (5). However, the exact role of HGF in hepatic gluconeogenesis remains unclear. Macrophage-stimulating protein (MSP) is another member of the HGF family of growth factors and is also known as hepatocyte growth factor-like protein and scatter factor 2 (11). In addition to stimulation of macrophages, MSP acts on other cell types, including epithelial and hematopoietic cells (11, 12). The MSP receptor is a transmembrane tyrosine kinase RON (in humans) and STK (in mice) (12). Biological activities of MSP/RON are known to be mediated by activation of signal transduction pathways like PI3K, c-Jun N-terminal kinase (JNK), focal adhesion kinase, c-Src, AKT, and MAPK (12). Although both MSP and RON are abundantly expressed in hepatocytes (13), little is known about their role in hepatic metabolic syndromes.
Upstream stimulatory factor (USF)-1 is a transcription factor regulating several genes involved in glucose and lipid metabolism (1, 14). USF-1 binds as a homodimer or heterodimerizes with its highly homologous USF-2 to E-box motifs (CANNTG) of target gene promoters (15). In the liver, expression of the genes coding for glucokinase (GK), liver-type pyruvate kinase (L-PK), fatty-acid synthase, apolipoprotein (apo) A-II, apoA-V, apoC-III, and apoE is up-regulated by USF-1 (1, 13). USF-1 plays an important role in the regulation of genes by insulin as well as glucose (1, 11). Allelic variants of USF1 may confer susceptibility to core features of the metabolic syndrome, such as glucose intolerance and dyslipidemia (11). Reports suggest that HGF phosphorylates and activates USF-1 via MAPK and tyrosine kinase pathways (17). Although USF-1 is known to regulate glycolytic enzyme genes (GK and L-PK) (1, 16), no direct evidence of regulating gluconeogenesis has yet been reported.
Orphan nuclear receptor small heterodimer partner (SHP) (NR0B2) is an atypical member of the nuclear receptor family that lacks a classical DNA-binding domain (DBD) (18–20) and does not have an identified ligand. Most of the studies related to SHP have established it as a transcriptional corepressor inhibiting various transcription factors involved in regulating several metabolic pathways, including bile acid homeostasis and glucose metabolism (19, 20). Studies in human subjects demonstrated mutations in the SHP gene are correlated to early onset diabetes, signifying a critical role of SHP in glucose metabolism (19, 20). The AMP-activated protein kinase (AMPK) is widely regarded to play a major role in metabolic homeostasis. Its activation, upon stress or starvation, is caused by a drop in ATP levels with an increased AMP/ATP ratio (21). In the liver, activation of AMPK leads to an inhibition of gluconeogenic and lipogenic pathways and affects the glucose/insulin-dependent activation of PEPCK, Glc-6-Pase, fatty-acid synthase, and L-PK gene expression (1, 20). Conversely, the knock-out of AMPK triggers a metabolic disturbance associated with high glucose and low insulin levels. However, AMPK is reported to be expressed in the cell nucleus, and AMPK could therefore act directly on transcriptional regulation (1). Recent studies suggest AMPK signaling acts as inducer of SHP gene expression and regulates hepatic gluconeogenesis in animal models (22, 23).
In this study, we demonstrate that HGF, along with MSP, is a novel regulator of PEPCK and Glc-6-Pase gene expression and hepatic glucose production in primary hepatocytes. HGF activates AMPK signaling and USF-1 binding to the SHP gene promoter and increases SHP expression. SHP, in turn, inhibits HNF4α-mediated transcriptional activation and induction of PEPCK and Glc-6-Pase gene expression. From the results of our study, we suggest that HGF/c-Met and MSP/RON signaling may be a potential therapeutic target for modulating hepatic gluconeogenesis and blood glucose levels in diabetes, and we provide a new insight into a novel mechanism for regulation of hepatic gluconeogenesis.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
Recombinant HGF and MSP were from R & D Systems; wortmannin, H89, U0126, 8-bromo-cAMP, and dexamethasone were from Sigma; SB203580, SP600125, and compound C were from Calbiochem; and insulin (Norvolin R) was from Green Cross (Korea). The reporter plasmids pepck-Luc (−490/+72), Glc-6-Pase-Luc (−231/+57), pFR-Luc (Gal4 DBD (UAS)-TK-Luc vector for mammalian one-hybrid assay), and E-box-Luc (22–27) were described previously. A series of human SHP promoter luciferase reporters and mouse Shp promoters were constructed previously (25). The human SHP (−230/+1) wild type and E-box-1 mutant luciferase reporter constructs were subcloned into pGL3 vector using XhoI and HindIII restriction sites. SHP, HNF4α, USF-1, USF-2, and dnUSF-1 plasmids were described previously (22, 23, 26). Gal4 DBD, Gal4 LRH-1, Gal4 scatter factor-1, Gal4 USF-1, and Gal4 USF-1 T153A and pcDNA3-dnAMPKα were described previously (22, 23, 26). A point mutant form of Gal4 USF-1 (Gal4 USF-1 S262A, nucleotides TCT to GCG) was generated by the QuickChange method of site-directed mutagenesis (Stratagene), and all constructs were confirmed by DNA sequencing. siSHP oligonucleotide was described previously (25). pepck, Glc-6-Pase, shp, and gapdh probes for Northern hybridization were described previously (22, 23).
Cell Culture and Transient Transfection Assay
HepG2, H4IIE, and AML12 cells were obtained from the American Type Culture Collection. Maintenance of cell lines and transient transfections were performed as described previously (22). Briefly, cells were transfected with indicated reporter plasmids together with expression vectors encoding various transcription factors or treated with various chemicals. Total cDNA used for each transfection was adjusted to 1 μg/well by adding an appropriate amount of empty vector and cytomegalovirus-β-galactosidase plasmids as an internal control. Cells were harvested 40–48 h post-transfection for luciferase and β-galactosidase assays. The luciferase activity was normalized to β-galactosidase activity and expressed as relative luciferase units (RLU).
Preparation of Recombinant Adenovirus
For ectopic expression of the genes, the adenoviral delivery system was used. Briefly, the cDNA encoding FLAG-USF-1 and HA-HNF4α was cloned into pAdTrack shuttle vector. Recombination of AdTrack-CMV-FLAG-USF-1 and AdTrack-CMV-HA-HNF4α (27) with adenoviral gene carrier vector was performed by transformation into pretransformed AdEasy-BJ21 competent cells. Adenoviruses (Ad) encoding GFP only (Ad-GFP), Ad-dnAMPKα, and Ad-siSHP were described previously (22).
Isolation and Culture of Primary Rat Hepatocytes
Primary hepatocytes were prepared from 200- to 300-g Sprague-Dawley rats by the collagenase perfusion method as described previously (19, 20). Viability of cells was analyzed using trypan blue staining. Cells were maintained in M199 media (Mediatech) overnight for attachment, and chemical treatments were performed as indicated.
Primary Human Hepatocyte (PHH) Culture
PHHs were obtained from the Liver Tissue and Cell Distribution System of the National Institutes of Health (S. Strom, University of Pittsburgh, PA). Hepatocytes were cultured as described previously (5).
RNA Isolation and Analysis
Total RNA was isolated for Northern hybridization using probes for pepck, Glc-6-Pase, shp, and gapdh as described previously (22). Semiquantitative and qPCR analysis in primary rat hepatocytes and PHHs were performed using primers for PEPCK, Glc-6-Pase, SHP, and β-actin as described previously (5, 22). Primers used for detecting l-pk and gk are as follows: l-pk forward 5′-GCTGGGCACTGCCTTCTT and reverse 5′-GTAGCTGAGTGGGGAGGTTGC; gk forward 5′-CTACGTGCGTTGCACCCCAGA and reverse 5′-AGGCCTTGAAGCCCTTGGTCC. The PCR products of l-pk and gk were 295 and 341 bp, respectively.
Western Blot Analysis
Cell lysate preparation and Western blot analysis in primary rat hepatocytes and PHHs, using rabbit monoclonal AMPKα, rabbit monoclonal phospho-AMPKα (Thr-172), rabbit polyclonal ACC, rabbit polyclonal phospho-ACC (Ser-79), rabbit polyclonal USF-1 (C-20), rabbit polyclonal SHP (H-160), and β-tubulin antibodies (Santa Cruz Biotechnology) were described previously (5, 22).
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed according to the manufacturer's protocol (Upstate). Briefly, HepG2 cells were transfected with reporter plasmids, and treatments were performed as indicated. Cells were then fixed with 1% formaldehyde and harvested. Soluble chromatin was immunoprecipitated with rabbit polyclonal USF-1 antibody (C-20, Santa Cruz Biotechnology). After recovering DNA, qPCR was performed using primers encompassing the human SHP promoter (−211/−10) forward 5′-CCGGCCACTTCATTGACT and reverse 5′-TGTTCTGCTGTGGGTG.
Glucose Production Assay
Glucose production from primary rat hepatocytes was measured according to the manufacturer's protocol, using a colorimetric glucose oxidase assay (Sigma). Briefly, after the experimental time period as indicated, the cells were washed three times with phosphate-buffered saline. Then the cells were incubated for 3 h at 37 °C, 5% CO2, in glucose production buffer (glucose-free Dulbecco's modified Eagle's medium, pH 7.4, containing 20 mmol/liter sodium lactate, 1 mmol/liter sodium pyruvate, and 15 mmol/liter HEPES, without phenol red). The glucose assays were performed in triplicate, and the intra-assay coefficient of variation was <5%.
Statistical Analysis
Data are expressed as means ± S.D. Statistical analysis was performed using the Student's t test or analysis of variance analyses followed by Duncan's multiple comparison tests. All experiments were performed at least three times. Differences were considered significant at p < 0.05.
RESULTS
HGF and MSP Repress Hepatic Gluconeogenesis
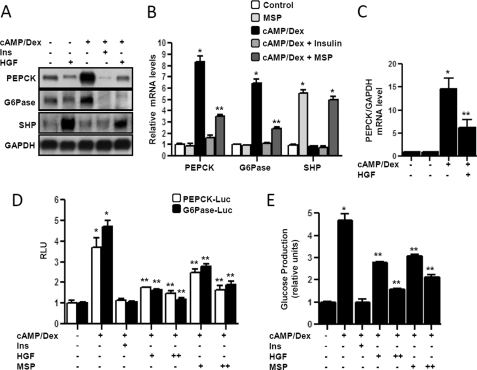
A previous report from our group has demonstrated that HGF repressed bile acid synthesis in PHHs (5). Another study has demonstrated the effect of HGF to be maximal at the concentration of 50 ng/ml in the human hepatoma cell line HepG2 (17). To evaluate the potential role of HGF and MSP on hepatic gluconeogenesis, primary rat hepatocytes (Fig. 1, A and B) and PHHs (Fig. 1C) were exposed to cAMP/Dex treatment. Both HGF and MSP significantly suppressed cAMP/Dex-induced pepck and Glc-6-Pase mRNA expression, and this repression was inversely correlated to the induction of the shp mRNA level (Fig. 1, A and B). Insulin treatment was used as a positive control. HGF was also shown to significantly repress PEPCK mRNA expression in PHHs (Fig. 1C). Next, using transient transfection in AML12 cells with pepck and Glc-6-Pase gene promoters, we demonstrated that HGF and MSP significantly repressed cAMP/Dex-mediated activation of pepck and Glc-6-Pase gene promoters in a dose-dependent manner (Fig. 1D). Consistent with the repression of key gluconeogenic enzyme gene expression, both HGF and MSP treatment dramatically reduced glucose production from gluconeogenesis in primary hepatocytes (Fig. 1E). Taken together, these results clearly indicate a novel regulatory role of HGF and the HGF family member MSP in hepatic gluconeogenesis.
FIGURE 1.
Inhibition of hepatic gluconeogenesis by HGF and MSP in primary hepatocytes. A–C, primary rat hepatocytes (A and B) and primary human hepatocytes (C) were pretreated with HGF (50 ng/ml, A and C), insulin (10 nm, 1 h), and MSP (100 ng/ml, B) for 3 h followed by treatment with cAMP (500 μm) and Dex (100 nm) treatment for 3 h in the continuous presence or absence of HGF and MSP. Total RNA was isolated for Northern hybridization (A) and qPCR analysis (B and C). Data represent means ± S.D. of three individual experiments. *, p < 0.001 and **, p < 0.05 compared with untreated control and cAMP/Dex treated cells, respectively. D, AML12 cells were transfected with pepck and Glc-6-Pase-Luc (200 ng) for 24 h followed by treatment with cAMP (500 μm) and Dex (100 nm) treatment for 3 h in the continuous presence or absence of HGF (50 and 100 ng/ml), MSP (100 and 200 ng/ml), or insulin as mentioned previously. Experiments were done in triplicate, and data are expressed in RLU and as the fold activation relative to the control. Data represent mean ± S.D. of three individual experiments. *, p < 0.05 and **, p < 0.001 compared with untreated control and cAMP/Dex-treated cells, respectively. E, measurement of glucose production. Experiments were performed as described in A and B, using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm). Data represent mean ± S.D. of four individual experiments. *, p < 0.001 and **, p < 0.001 compared with untreated control and cAMP/Dex treated cells, respectively. G6Pase, Glc-6-Pase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
HGF and MSP Induce SHP Gene Expression in Primary Hepatocytes
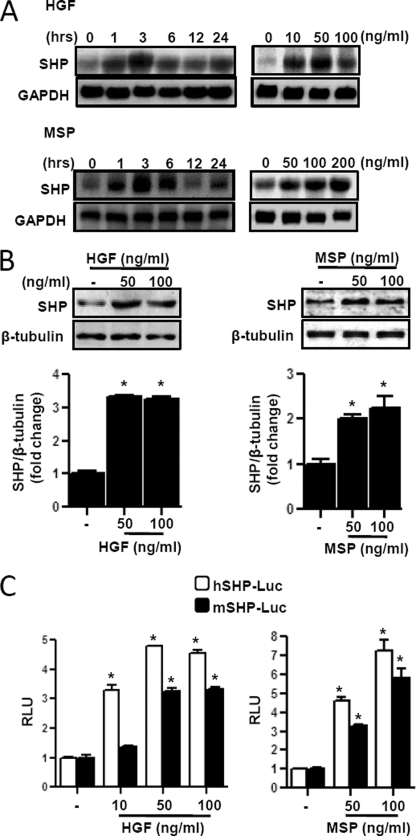
Previous studies from our group demonstrated that SHP is a known repressor of key enzyme genes involved in hepatic gluconeogenesis (22, 23). Therefore, to examine the effect of HGF and MSP, a member of the HGF receptor family, on hepatic gluconeogenesis via SHP, we analyzed the shp gene expression pattern in primary rat hepatocytes following HGF and MSP treatments (Fig. 2). Treatments with HGF and MSP significantly induced shp mRNA expression within 3 h (Fig. 2A) in a time- and dose-dependent manner in primary rat hepatocytes. We tested several other hepatic cell lines (HepG2, AML12, and H4IIE), and HGF was shown to increase the SHP mRNA level consistently in all these tested cell lines (supplemental Fig. 1A). HGF- and MSP-treated primary hepatocytes showed 2–4-fold increase in SHP protein level (Fig. 2B), and dose-dependent treatment of HGF and MSP significantly increased both human and mouse SHP gene promoter (hSHP-Luc and mShp-Luc) activity in HepG2 and AML12 cells, respectively (Fig. 2C). Overall, these results suggest that both HGF and MSP are potent inducers of shp gene expression in primary hepatocytes and various hepatic cell lines.
FIGURE 2.
SHP gene expression is induced by HGF and MSP in primary hepatocytes. Primary hepatocytes were isolated from rats and cultured as described under “Experimental Procedures.” A, cells were treated with HGF or MSP at indicated concentrations or times, and total RNA was isolated for Northern hybridization. shp gene expression was analyzed and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression. The result is representative of three independently performed experiments. B, cells were treated with HGF or MSP at the indicated concentrations for 6 h and harvested for Western blot analysis using the indicated antibodies. The result shown is representative of three independently performed experiments. *, p < 0.001 compared with untreated control. C, HepG2 and AML12 cells were transfected with hSHP-Luc (200 ng) and mShp-Luc (200 ng) respectively. 24 h after transfection, cells were serum-starved for a further 24 h, followed by HGF or MSP treatments at indicated concentrations for 12 h. Experiments were done in triplicate, and data are expressed in RLU, and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001 compared with untreated control.
HGF and MSP Activate AMPK Signaling Pathway and Induce SHP Gene Expression
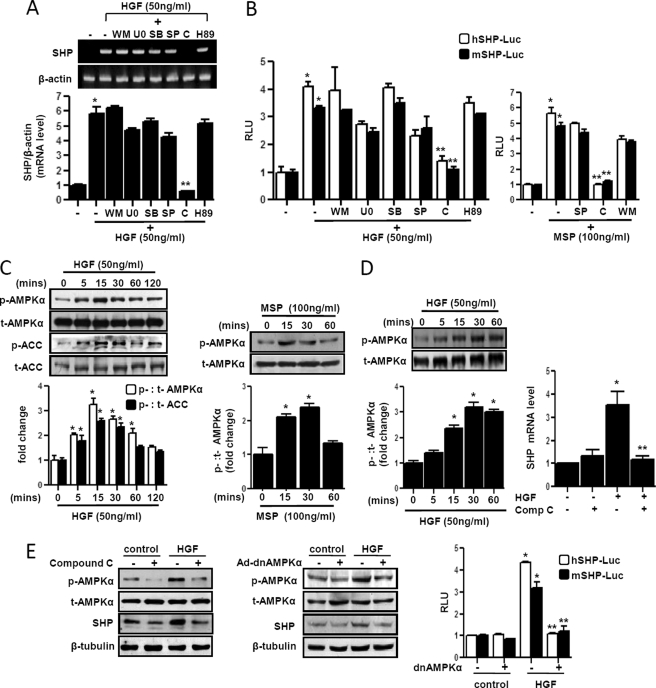
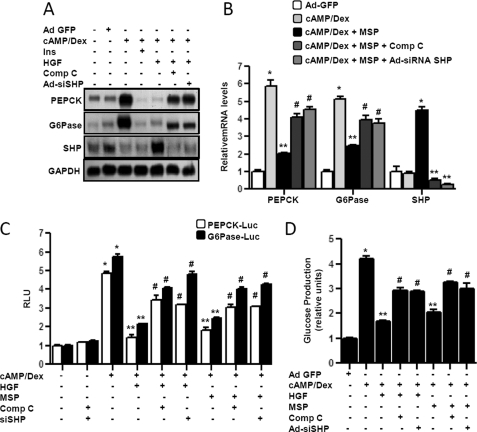
To elucidate the potential signaling pathways involved in HGF-mediated induction of shp gene expression, primary hepatocytes were pretreated with several specific inhibitors of cell signaling pathways followed by HGF treatment (Fig. 3A). Pretreatment of compound C (an AMPK inhibitor) completely abolished HGF-mediated induction of shp mRNA expression. However, there was no significant effect of wortmannin (WM, a PI3K inhibitor), U0126 (an extracellular signal-regulated kinase (ERK) inhibitor), SB203580 (a p38 inhibitor), SP600125 (a JNK inhibitor), and H-89 (protein kinase A inhibitor) on shp mRNA expression. Next, using a transient transfection assay in HepG2 and AML12 cells with hSHP-Luc and mShp-Luc, it was demonstrated that only compound C pretreatment significantly inhibited both HGF- and MSP-mediated increase of both human and mouse SHP promoter activity (Fig. 3B), thereby suggesting that AMPK signaling pathway mediates the HGF and MSP effect on SHP gene expression. To confirm the involvement of AMPK, we performed Western blot analysis to detect phosphorylation of AMPK and its downstream target (ACC) by HGF and MSP in primary hepatocytes (Fig. 3C). HGF rapidly and significantly increased AMPK and ACC phosphorylation within 15 min of treatment (Fig. 3C, left panel). MSP showed similar rapid phosphorylation and activation of AMPK signaling (Fig. 3C, middle panel). The effect of HGF was reconfirmed by detecting increased phosphorylation of the HGF receptor c-Met at the same time point (supplemental Fig. 1B). A similar pattern of AMPK and ACC phosphorylation as well as c-Met phosphorylation by HGF was observed in HepG2 cells (data not shown). Consistently, using PHHs, we demonstrated that HGF activated the AMPK signaling pathway rapidly, and AMPK inhibitor compound C abolished HGF-mediated induction of SHP mRNA expression (Fig. 3D). Finally, to further confirm the role of AMPK in HGF-mediated induction of SHP gene expression, we used adenovirus-mediated overexpression of a dominant negative form of AMPKα (Ad-dnAMPKα) to study the effects of HGF on shp mRNA expression, and SHP gene promoter activity (Fig. 3E) shows that both compound C treatment and Ad-dnAMPKα significantly repressed basal as well as HGF-mediated activation of AMPK. This further resulted in a significant reduction in HGF-mediated induction of Shp protein levels (Fig. 3E, left and middle). Similarly, cotransfection of the dnAMPKα expression vector with SHP gene promoters resulted in a dramatic decrease in HGF-induced SHP promoter activity (right). These results clearly demonstrated that the AMPK signaling pathway was involved in HGF- and MSP-mediated induction of SHP gene expression in hepatocytes.
FIGURE 3.
AMPK mediates HGF- and MSP-mediated induction of SHP gene expression. A, primary rat hepatocytes were pretreated with protein kinase inhibitors wortmannin (WM, 0.1 μm), U0126 (U0, 10 μm), SB203580 (SB, 25 μm), SP600125 (SP, 25 μm), compound C (C, 10 μm) and H-89 (H89, 10 μm) for 1 h followed by treatment with HGF for 3 h. Total RNA was isolated for semiquantitative RT-PCR analysis of shp mRNA expression and was normalized to β-actin expression. Data represent mean ± S.D. of three individual experiments. *, p < 0.05, and **, p < 0.001 compared with untreated control and HGF-treated cells. B, HepG2 and AML12 cells were transfected with hSHP-Luc (200 ng) and mShp-Luc (200 ng), respectively. 24 h after transfection, cells were serum-starved for a further 24 h, followed by pretreatment of inhibitors for 1 h preceding HGF or MSP treatments at indicated concentrations for 12 h. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001, and **, p < 0.001 compared with untreated control and HGF- or MSP-treated cells, respectively. C, primary rat hepatocytes were treated with HGF or MSP at indicated concentration and times for 15 min (right) and harvested for Western blot analysis using indicated antibodies. Result shown is representative of three independently performed experiments. *, p < 0.001 compared with untreated control. D, PHHs were treated with HGF at indicated concentration and for indicated times (left) or treated with HGF for 3 h following pretreatment with compound C (Comp C) (10 μm) (right). Cell lysates were extracted, and Western blot analysis was performed using the indicated antibodies (left). Total RNA was isolated for qPCR analysis SHP mRNA expression and was normalized to β-actin expression (right). Data represent mean ± S.D. of three individual experiments. *, p < 0.05, and **, p < 0.001 compared with untreated control and HGF-treated cells. E, primary rat hepatocytes (left and middle panels) were pretreated with compound C (10 μm) for 1 h or infected with adenovirus dominant negative AMPKα (Ad-dnAMPKα) (50 m.o.i.) for 48 h followed by HGF treatment (50 ng/ml) and harvested for Western blot analysis using indicated antibodies. Result shown is representative of three independently performed experiments. HepG2 and AML12 cells were transfected (right panel) with hSHP-Luc (200 ng) and mShp-Luc (200 ng) respectively, along with dnAMPKα expression vector (200 ng). 24 h after transfection, cells were serum-starved for a further 24 h, followed by HGF treatment (50 ng/ml) for 12 h. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001, and **, p < 0.001 compared with untreated control and HGF treated cells, respectively.
Induction of SHP Gene Expression by HGF Is Specifically Mediated by USF-1
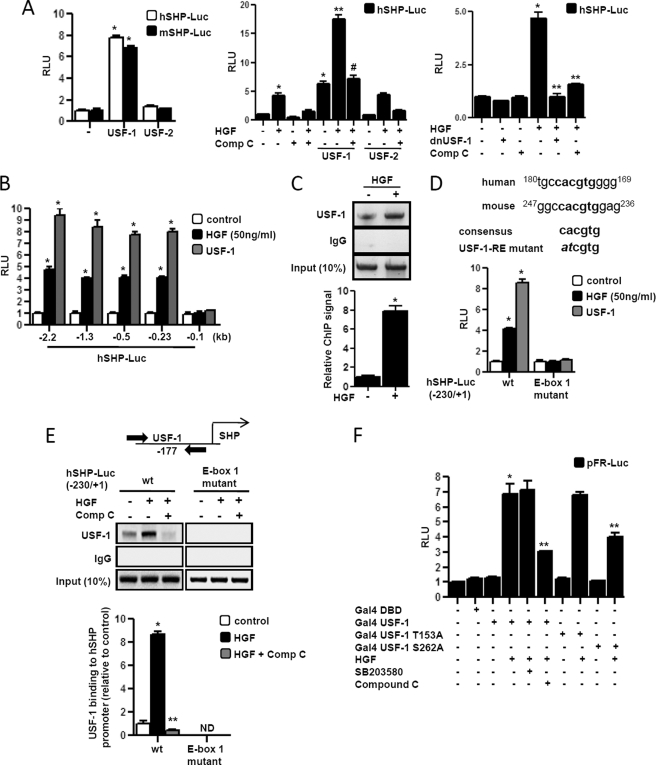
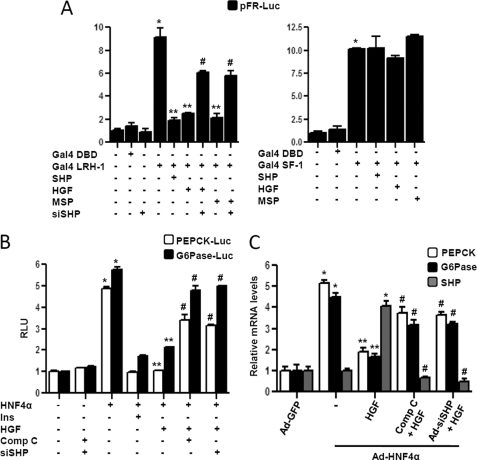
We next attempted to identify the potential transcription factors conferring HGF induction of the SHP gene promoter. A previous report suggests that HGF activates USF-1 binding to other target gene promoters (17). Using transient transfection assay, we tried to evaluate the role of USF-1 in the context of the SHP gene promoter. USF-1 activated human and mouse SHP promoters significantly, whereas USF-2 failed to demonstrate any significant effect on SHP promoters (Fig. 4A, left panel). Treatment of HGF along with USF-1 cotransfection showed significant synergistic activation of the SHP promoter, and pretreatment with compound C dramatically abolished this effect. As expected, USF-2 alone or along with HGF treatment showed no significant change on the SHP gene promoter (Fig. 4A, middle panel). The specificity of HGF synergism on USF-1 transcriptional activity was reconfirmed using E-box reporter construct (supplemental Fig. 2). To verify that both AMPK and USF-1 are downstream of the HGF signaling cascade, dominant negative USF-1 (dnUSF-1) construct was cotransfected with the SHP promoter or compound C was treated prior to HGF treatment. Both dnUSF-1 and compound C significantly abrogated HGF-mediated increase in SHP gene promoter activity (Fig. 4A, right panel), thereby indicating that AMPK and USF-1 mediates HGF effect on SHP gene expression.
FIGURE 4.
USF-1 mediates HGF-mediated induction of SHP gene expression. A, HepG2 and AML12 cells were transfected with pcDNA3-FLAG-USF-1 (400 ng), pcDNA3-FLAG-USF-2 (400 ng), hSHP-Luc (200 ng), mShp-Luc (200 ng), respectively (left), or HepG2 cells were transfected with hSHP-Luc (200 ng) (A middle and right, B and D), pcDNA3-FLAG-USF-1 (400 ng), and pcDNA3-FLAG-USF-2 (400 ng) (middle) or with pcDNA3-dnUSF-1 (400 ng) (right). 24 h post-serum starvation, cells were pretreated with compound C (comp C) (10 μm) for 1 h preceding HGF (50 ng/ml) treatment. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001; **, p < 0.05; #, p < 0.001 compared with untreated control, USF-1 or HGF treated cells and HGF + USF-1 treated cells respectively. B and D, HepG2 cells were transfected with several deletion constructs (B) or E-box 1 mutant construct of hSHP-Luc (200 ng) (D) and USF-1 or treated with HGF as indicated. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001 compared with untreated control. C and E, ChIP assay. HepG2 cells were serum-starved for 24 h followed by HGF treatment for 12 h (C) or HepG2 cells were transfected with hSHP-Luc (−230/+1) (200 ng) wild type (wt) or E-box 1 mutant. Following transfection and serum starvation, cells were pretreated with compound C for 1 h preceding HGF treatment for 12 h. Soluble chromatin was prepared and immunoprecipitated with monoclonal antibody against USF-1 or IgG only as indicated. 10% of the soluble chromatin was used as input. qPCR was performed to determine and quantify the binding of USF-1 to endogenous (C) or transfected (E) hSHP promoter. Data are representative of three individually performed experiments. *, p < 0.001, and **, p < 0.05 compared with untreated control and HGF treated cells respectively. ND, not detectable. F, HepG2 cells were cotransfected with pFR-Luc (200 ng), Gal4 constructs (400 ng each) containing DBD only, and USF-1, USF-1 T153A, and USF-1 S262A as indicated. Post-transfection and serum starvation, cells were pretreated with SB203580 or compound C following HGF treatment as indicated. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001, and **, p < 0.001 compared with untreated control and HGF-treated cells, respectively.
USF-1 binds to E-box sequences (CANNTG) and with specific preference for the nucleotide sequence CACGTG on various gene promoters (15). Previous reports from our group demonstrated the presence of several E-boxes in both human and mouse SHP gene promoters (25). Therefore, to identify the potential sequences conferring USF-1-mediated HGF effect on the SHP promoter, we used a series of deletion constructs of SHP promoters for transient transfection assay. Deletion of the SHP promoter sequence from −2.2 kb to −230 bp had no effect on the promoter activity, and further deletion to −100 bp abolished both HGF and USF-1 activation, suggesting that the region −230 to −100 bp conferred the activation of SHP promoter by HGF or USF-1 (Fig. 4B). Next, we performed ChIP assays to monitor the effect of HGF on USF-1 recruitment to the endogenous SHP gene promoter. Under basal conditions, we could detect USF-1 occupancy on the SHP promoter. However, HGF treatment significantly augmented USF-1 occupancy to the SHP promoter (Fig. 4C). Next, to ascertain the USF-1-responsive region in the SHP gene promoter, we identified an E-box, designated E-box-1, the first and only E-box lying within this region in both human and mouse SHP promoters (Fig. 4D). E-box-1 sequence matched perfectly with the specific USF-1-binding nucleotide sequence (CACGTG). To reconfirm that this E-box-1 mediates HGF and USF-1 activation of SHP, transient transfection assay was performed using wild type and E-box-1 mutant reporters with USF-1 and HGF. This mutant reporter did not respond to either HGF treatment or USF-1 cotransfection (Fig. 4D). Next, we performed ChIP assay to determine the HGF-mediated recruitment of USF-1 to human SHP promoter in HepG2 cells. ChIP assay results demonstrated that USF-1 was present in SHP promoter, and HGF further induced USF-1 binding to SHP chromatin, and pretreatment with compound C drastically abolished that effect. As expected, no binding was observed in E-box-1 mutant SHP promoters (Fig. 4E), therefore suggesting that HGF activates the SHP gene transcription via enhancing USF-1 binding to the promoter, and ChIP assay results provide critical in vivo evidence of the effect of HGF/AMPK/USF-1 signaling cascade resulting in increased SHP gene transcription.
Next, we tried to elucidate the mechanism by which AMPK mediates USF-1 activation by HGF. Previous reports suggest involvement of p38 MAPK and DNA-PK signaling pathways in USF-1 activation (15, 26, 28). Therefore, to address this question, we studied the effects of kinase inhibitors on USF-1 transactivation activity in mammalian one-hybrid assays. Gal4-fused wild type USF-1 (Gal4 USF-1), MAPK-response mutant USF-1 (Gal4 USF-1 T153A), or DNA-PK-response mutant USF-1 (Gal4 USF-1 S262A) constructs were cotransfected with Gal4 reporter pFR-Luc for transfection assays in HepG2 cells pre-treated with HGF and MAPK inhibitor (SB203580) and AMPK inhibitor (compound C). HGF-mediated increase of Gal4 USF-1 transactivity was significantly repressed by compound C treatment but not by SB203580. HGF significantly activated Gal4 USF-1 T153A, comparable with the wild type activation, whereas Gal4 USF-1 S262A activation was significantly but not fully abrogated (Fig. 4F), suggesting that DNA-PK signaling may play a role in mediating AMPK effect on USF-1 activity.
USF-1 Induces SHP Gene Expression to Repress Hepatic Gluconeogenesis
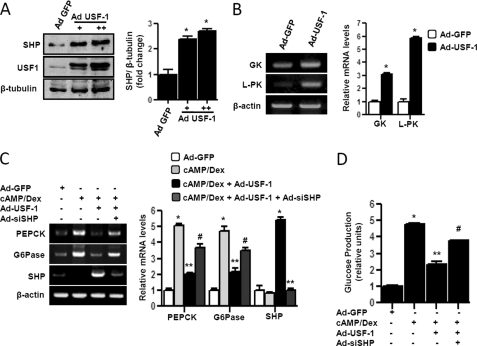
Next, to ascertain the role of USF-1 in regulation of hepatic gluconeogenesis, we overexpressed USF-1 via adenoviral) delivery system in primary rat hepatocytes. Ad USF-1 significantly induced Shp protein level in a dose-dependent manner (Fig. 5A). Similar induction of shp mRNA and protein level was observed in HepG2 cells (data not shown). Previous reports have demonstrated that USF-1 increased glycolysis via activation of key glycolytic enzyme genes, L-PK and GK gene promoters (1, 14, 16). As a positive control, Ad USF-1 treatment was shown to induce l-pk and gk mRNA expression significantly in primary hepatocytes (Fig. 5B). To verify the effect of USF-1 on gluconeogenesis, primary hepatocytes were treated with cAMP/Dex to induce expression of key gluconeogenic enzyme genes pepck and Glc-6-Pase. Ad USF-1 significantly repressed pepck and Glc-6-Pase gene expression with subsequent induction of shp mRNA level, and knockdown of endogenous Shp by adenoviral siRNA specifically targeting SHP (Ad-siSHP) dramatically reversed the inhibitory effects of USF-1 on pepck and Glc-6-Pase mRNA expression (Fig. 5C). Glucose production via gluconeogenesis upon cAMP/Dex treatment in primary hepatocytes was also significantly reduced by Ad USF-1, and as expected, Ad-siSHP significantly diminished the inhibitory effect of USF-1 (Fig. 5D). Overall, these results indicate a novel role of USF-1 in hepatic gluconeogenic program. USF-1 indirectly represses hepatic gluconeogenesis via induction of SHP gene expression.
FIGURE 5.
USF-1 represses gluconeogenesis via induction of SHP gene expression in primary hepatocytes. A and B, primary rat hepatocytes were infected with adenovirus (Ad) GFP (50 m.o.i.) or Ad-USF-1 (+ = 25 m.o.i., ++ = 50 m.o.i.) for 36 h. Cells were harvested for Western blot analysis using indicated antibodies (A), or total RNA was isolated for semi-quantitative RT-PCR analysis (B). Data represent mean ± S.D. of three individual experiments. *, p < 0.001 compared with Ad-GFP-treated cells. C, cells were infected with Ad-GFP, Ad-USF-1, or Ad-siSHP followed by Ad-USF-1 for 36–48 h preceding cAMP (500 μm) and Dex (100 nm) treatment for 3 h. Total RNA was isolated for semiquantitative RT-PCR analysis. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.05, and #, p < 0.001 compared with Ad-GFP-infected cells, cAMP/Dex treatment, and Ad-USF-1 infected cells, respectively. D, measurement of glucose production. Experiments were performed as described in C, using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm). Data represent mean ± S.D. of four individual experiments. *, p < 0.001; **, p < 0.001, and #, p < 0.001 compared with Ad-GFP infected cells, cAMP/Dex treatment, and Ad-USF-1 infected cells, respectively. G6Pase, Glc-6-Pase.
HGF Inhibits Gluconeogenic Gene Expression via SHP-mediated Repression of HNF4α
To elucidate the molecular mechanism by which the HGF/AMPK/SHP pathway represses PEPCK and Glc-6-Pase gene expression, we first confirmed the functional effect of HGF on target transcription factors of SHP. Previous studies have demonstrated that SHP specifically represses nuclear receptor liver receptor homolog-1 (LRH-1) transcriptional activity but not steroidogenic factor-1 (SF-1) transactivity (19, 20). Using the mammalian one-hybrid system (Gal4-fused protein) for transient transfection assay, we found that HGF and MSP significantly and specifically repressed Gal4 LRH-1 activity, and this repressive effect was dramatically reversed by siSHP oligonucleotide (Fig. 6A, left panel). However, consistent with previous reports, HGF family did not show any significant effect on Gal4 scatter factor-1 transactivity (Fig. 6A, right panel). HNF4α is a well known positive regulator of gluconeogenesis (1). Therefore, we investigated the effect of HGF on HNF4α by transfection assay in AML12 cells along with pepck and Glc-6-Pase gene promoters (Fig. 6B) and by adenovirus overexpressing of HNF4α (Ad-HNF4α) in primary hepatocytes to induce pepck and Glc-6-Pase mRNA expression (Fig. 6C). HGF treatment significantly repressed HNF4α-mediated induction of pepck and Glc-6-Pase gene expression, and blocking AMPK signaling by compound C treatment or knockdown of SHP by siRNA oligonucleotide (Fig. 6B) or by Ad-siSHP (Fig. 6C) significantly reversed the repressive effect of HGF on pepck and Glc-6-Pase. These results indicate that repression of pepck and Glc-6-Pase gene expression by HGF is mediated, at least in part, via repression of HNF4α by SHP.
FIGURE 6.
HNF4α is a target of HGF-mediated repression of hepatic gluconeogenesis. A and B, HepG2 cells (A) and AML12 cells (B) were transfected with pFR-Luc, pepck-Luc, Glc-6-Pase-Luc (200 ng each), and expression vectors (400 ng each) as indicated. 24 h after transfection, cells were serum-starved and pretreated with compound C (comp C) (10 μm) for 1 h (B) or treated with HGF (50 ng/ml) and MSP (100 ng/ml) for 12 h. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.001, and #, p < 0.001 compared with untreated control, Gal4-LRH-1- or HNF4α-transfected cells and HGF- or MSP-treated cells, respectively. C, primary hepatocytes were infected with Ad-GFP and Ad-HNF4α or infected with Ad-siSHP for 36–48 h or pretreated with compound C (10 μm) for 1 h preceding HGF treatment for 3 h in the continuing presence of Ad-HNF4α. Total RNA was isolated for qPCR analysis. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.05, and #, p < 0.001 compared with Ad-GFP-infected cells, Ad-HNF4α alone- infected cells and HGF-treated cells, respectively. G6Pase, Glc-6-Pase.
Knockdown of SHP Reversed HGF- and MSP-mediated Repression of Hepatic Gluconeogenesis
To confirm the critical role of AMPK and SHP in conferring the repressive effect of HGF and MSP on hepatic gluconeogenesis in primary hepatocytes, endogenous shp knockdown experiments were performed using Ad-siSHP, and the AMPK signaling pathway was blocked by compound C. Treatment with cAMP/Dex significantly induced pepck and Glc-6-Pase mRNA expression, and this induction was significantly repressed by HGF and MSP treatment (Fig. 7, A and B). Pretreatment with compound C or knockdown of shp dramatically decreased the shp mRNA level, and consequently the HGF- and MSP-mediated repression of cAMP/Dex-induced pepck and Glc-6-Pase mRNA expression was abolished to a significant level. Similarly, using transient transfection assay with pepck and Glc-6-Pase gene promoters, we demonstrated that HGF and MSP repressed cAMP/Dex-mediated increase in promoter transactivation, which was reversed upon blocking the AMPK signaling pathway or by using siSHP oligonucleotides (Fig. 7C). Finally, to verify the effect of knockdown of shp gene expression on HGF- and MSP-mediated repression on gluconeogenesis, we performed glucose production assay from gluconeogenesis in primary hepatocytes (Fig. 7D). HGF and MSP treatments drastically reduced cAMP/Dex-mediated glucose production, and this reduction was reversed by compound C pretreatment or Ad-siSHP, reconfirming the HGF/AMPK/SHP signaling pathway to play a crucial role in regulating hepatic gluconeogenesis in primary hepatocytes.
FIGURE 7.
Knockdown of SHP expression abolished HGF and MSP effects on hepatic gluconeogenesis. A and B, primary rat hepatocytes were infected with Ad-GFP (50 m.o.i.) and Ad-siSHP (50 m.o.i.) for 48 h or pretreated with compound C (10 μm) for 1 h followed by cAMP (500 μm) and Dex (100 nm) treatment for 3 h in the continuous presence or absence of HGF (50 ng/ml), MSP (100 ng/ml), and insulin (10 nm) for 3 h. G6Pase, Glc-6-Pase. Total RNA was isolated for Northern hybridization (A) and qPCR analysis (B). Data represent mean ± S.D. of three individual experiments. *, p < 0.05, **, p < 0.05, and #, p < 0.001 compared with Ad-GFP-infected cells, cAMP/Dex treatment, and HGF- or MSP-treated cells, respectively. C, AML12 cells were transfected with pepck-Luc, Glc-6-Pase-Luc (200 ng each), and siSHP oligonucleotide (20 nm) for 24 h, followed by pretreatment with compound C (10 μm) and treatment with cAMP (500 μm) and Dex (100 nm) for 3 h in the continuous presence or absence of HGF (50 ng/ml), MSP (100 ng/ml), or insulin as mentioned previously. Experiments were done in triplicate, and data are expressed as RLU and as the fold activation relative to the control. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.001, and #, p < 0.001 compared with untreated control, cAMP/Dex-treated cells, and HGF- or MSP-treated cells, respectively. D, measurement of glucose production. Experiments were performed as described in A and B, using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm). Data represent mean ± S.D. of four individual experiments. *, p < 0.001, **, p < 0.001, and #, p < 0.001 compared with Ad-GFP infected cells, cAMP/Dex treatment, and HGF- or MSP-treated cells, respectively.
DISCUSSION
Elevated hepatic gluconeogenesis contributes to the pathogenesis of insulin resistance and diabetes, and targeted inhibition of hepatic gluconeogenesis has emerged as a promising prospect for intervention of type 2 diabetes. Hepatic gluconeogenesis and hepatic glucose production is a tightly regulated product of opposing actions of glucagon, acting via the cAMP-dependent pathways along with glucocorticoids on one side and insulin, acting via the PI3K pathway, on the other hand. In this study we have shown that HGF and the HGF-like protein, MSP, can inhibit hepatic gluconeogenesis through a distinct and novel signaling pathway involving AMPK, USF-1, and the transcriptional corepressor SHP in primary hepatocyte cultures.
In this study we have demonstrated that HGF and the HGF-like protein, MSP, showed significant and dramatic decrease in the expression of key hepatic gluconeogenic genes, PEPCK and Glc-6-Pase, and subsequently led to reduced hepatic glucose production. Although the mechanism of action of HGF and MSP was distinct from insulin, the effects were at a comparably significant and effective level. Previous studies have demonstrated that the hyperglycemic condition due to elevated extracellular glucose concentrations is an independent risk factor for diabetic models leading to development of various cardiovascular complications, and recombinant HGF therapy showed remarkable attenuation of these effects (1–3). Overexpression of HGF in pancreatic β-cells can also prevent streptozotocin-induced onset of diabetes in animal models as well as attenuate β-cell destruction (29). HGF has been known to function as an anti-fibrotic agent in animal models of liver cirrhosis (30). Previous studies on MSP/RON signaling suggest that it activates PI3K signaling and attenuates some macrophage responses in acute inflammation and plays an important role in wound healing phases of tissue injury (11, 12). Both MSP and its receptor, RON, are highly expressed in hepatocytes (13). However, not much is known about the role of MSP/RON signaling in hepatic glucose metabolism, and our study provides evidence for the first time demonstrating that MSP/RON signaling may regulate hepatic glucose metabolism through a signaling pathway different from PI3K signaling. In our study we elucidate the molecular mechanism by which HGF inhibits hepatic gluconeogenesis, utilizing a signaling cascade involving AMPK to activate USF-1 leading to the induction of the transcriptional corepressor SHP. Previous reports from our group demonstrated inhibitory effects of HGF via SHP on accumulation of toxic bile acids and repressed expression of key bile acid metabolism enzyme gene cholesterol 7α-hydroxylase (CYP7A1) in PHHs (5). Therefore, our study suggests that HGF can be a potential therapeutic alternative in treatment of diabetes associated with insulin resistance by improving β-cell function and ameliorating vascular complications, often correlated with diabetes (3), as well as via direct activation of the AMPK signaling pathway to induce SHP gene expression and inhibit PEPCK and Glc-6-Pase gene expression, thereby reducing hepatic glucose production.
Most of the previous studies with HGF and its receptor, c-Met, related to diabetic conditions and its possible ameliorative effects have been reported in the context of the pancreatic β-cell model and in some cases skeletal muscle and adipose tissue. Our results demonstrated a similar negative regulatory effect HGF on gluconeogenesis in the hepatic setting. A previous study using c-Met receptor knock-out model in pancreatic β-cells demonstrated a phenotype with similar early phases of β-cell failure in T2DM along with significant glucose intolerance, although insulin expression remained normal (31). This study concluded that HGF/c-Met signaling is essential for glucose-dependent insulin secretion and genetic alterations in HGF, c-Met, and its downstream signaling pathways may be implicated in the predisposition to develop T2DM (31). Conversely, the HGF/c-Met signaling may provide an attractive pharmacological target for enhancing β-cell function in type 2 diabetes (31). Similarly, various other reports have also demonstrated the efficacy of HGF therapy in ameliorating streptozotocin-induced T1DM as well as diabetic nephropathy (32–36). A previous report also demonstrated ablation of hyperglycemia in diabetic mice model along with preservation of β-cell mass by HGF treatment, thereby suggesting it to function as an insulinotropic factor and a possible therapeutic alternative (35). This information as well as our results demonstrate a significant role of HGF in regulating glucose metabolism not only in the context of the pancreatic β-cell model but also in hepatocytes. However, further research using appropriate animal models are needed to establish the potential therapeutic application of HGF for treating T1DM and T2DM.
AMPK is a well established critical monitor of cellular energy status in all eukaryotes (21). Activation of AMPK signaling has been demonstrated to increase glucose uptake in skeletal muscles, and AMPK plays a major role in regulating various metabolic syndromes (21). In this study we demonstrated that induction of SHP gene expression by HGF and MSP is mediated by AMPK. Previous reports from our group have demonstrated the use of pharmacological activators of AMPK signaling, like anti-diabetic drug metformin (23) or insulin-mimetic compounds like sodium arsenite (22), to induce SHP gene expression and regulate hepatic gluconeogenesis. A recent report suggests that HGF activates AMPK via tumor suppressor kinase LKB1 to function as a critical determinant of hepatocyte proliferation during the liver regeneration stages after partial hepatectomy (37). Here we explored the role of HGF in activating AMPK to inhibit hepatic gluconeogenesis via SHP. Blockade of AMPK signaling attenuates the effect of HGF and MSP in induction of SHP gene expression and consequently on the repression of hepatic glucose production and inhibition of gluconeogenic gene expression. It may be suggested that HGF/c-Met and MSP/RON receptor signaling can be utilized to activate AMPK and SHP, thereby bypassing the ineffective insulin signaling in insulin resistance subjects. A similar approach has been reported using the adiponectin/adiponectin receptor 1 (AdipoR1) signaling pathway to activate AMPK by adenovirally overexpressing AdipoR1 in the leptin receptor-deficient db/db mice model, which exhibits significantly lower expression pattern of AdipoR1 compared with normal mice (38). HGF is implicated to play a significant role in metabolic syndromes, as a previous report on human subjects have shown a direct correlation between elevated serum HGF level and obesity, whereas the plasma HGF level was found to be elevated in fatty liver (6–10). In this context, a previous report demonstrated that plasma insulin levels were dramatically higher in genetically diabetic (db/db) mice compared with normal mice. However, plasma glucose levels remained significantly higher in these animals compared with normal mice. The reason behind this observation was attributed to a drastic decrease in the number of insulin-binding sites in db/db mice (39). Therefore, determination of the expression pattern and receptor sensitivity of c-Met along with the RON receptor in diabetic and obese models may provide with a novel approach in treating hepatic metabolic syndromes.
A previous report demonstrated activation of the E-box-binding transcription factors USF-1 and USF-2 by HGF via MAPK and tyrosine kinase pathways (17). In this study we investigated the role of USF-1 and -2 in mediating HGF effect on the SHP gene promoter. Interestingly, our results suggest that HGF significantly increases USF-1 binding on SHP gene promoter via AMPK signaling. It is consistent with our expression analysis that HGF-mediated induction of SHP mRNA level was significantly decreased only under treatment of the AMPK inhibitor compound C. Our group previously demonstrated the regulation of SHP promoter activity by another E-box-binding transcription factor sterol regulatory element-binding protein (SREBP)-1c (25). USF-1 has been recently demonstrated to be phosphorylated by DNA-PK and insulin signaling (28), and another report suggested that the receptor activator of nuclear factor κB ligand induces osteoclast differentiation via MAPK-dependent activation of USF-1 (26). Using DNA-PK and MAPK mutant USF-1, we verified the kinase signaling required by HGF/AMPK to activate USF-1, because of the absence of any potential AMPK phosphorylation site in USF-1. Our conclusions suggest that DNA-PK may be involved, partially, in mediating AMPK-dependent activation of USF-1, and this result was consistent with a previous report demonstrating positive regulatory effect of p38 MAPK on hepatic gluconeogenesis (40). However, a more detailed elucidation of this pathway is required in the future.
USF-1 protein level is found to be increased in feeding (28) and high glucose conditions (14), correlated to the decrease in hepatic gluconeogenesis under these conditions. Therefore, the induction of SHP gene expression by USF-1 in primary hepatocytes, as demonstrated in this study, may provide a plausible link connecting the decrease of PEPCK and Glc-6-Pase gene expression by SHP to USF-1 expression pattern. Previous studies linking USF-1 to hepatic glucose metabolism demonstrated regulation of key glycolytic enzyme gene expression, GK and L-PK, by USF-1 by binding to the respective gene promoters (1, 14, 16). However, no information was available regarding any direct link between USF-1 and regulation of hepatic gluconeogenesis. Our study demonstrates for the first time that USF-1-mediated induction of transcriptional corepressor SHP leads to substantial inhibitory effect on gluconeogenesis. SHP is a well known transcriptional corepressor of a wide variety of transcription factors regulating various metabolic pathways (19, 20). In this study, we demonstrated that HGF/AMPK/USF-1 signaling cascade induced SHP gene expression, which in turn targets HNF4α, an established positive regulator of PEPCK and Glc-6-Pase gene expression. However, because of its varied repertoire of targets, the possibility of other transcription factors being targeted by SHP, on activation of this signaling cascade, cannot be ruled out and requires further investigation. The most established signaling cascade involving SHP in metabolic regulation is the bile acid/farnesoid X receptor/SHP cascade, which works as a feedback loop to regulate bile acid homeostasis (19, 20). This study provides a novel signaling cascade of HGF/AMPK/USF-1/SHP in regulation of hepatic gluconeogenesis.
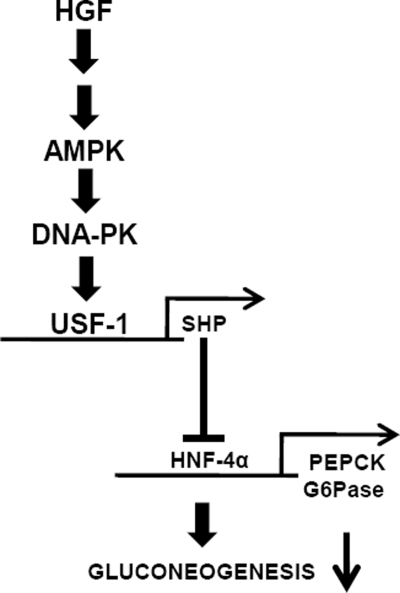
Overall, from these observations, we provide a previously unknown effect of HGF on hepatic gluconeogenesis. HGF activates the AMPK signaling pathway in hepatocytes, which in turn activates the E-box-binding transcription factor USF-1 specifically to bind to the SHP gene promoter. This leads to USF-1-dependent induction of SHP gene expression and subsequently SHP-repressed transcription factor HNF4α and inhibited hepatic gluconeogenesis (Fig. 8).
FIGURE 8.
Schematic diagram of HGF-mediated inhibition of gluconeogenesis in primary hepatocytes. HGF signaling pathway activates AMPK signaling, leading to USF-1 activation via a DNA-PK-dependent pathway. USF-1 indirectly inhibits gluconeogenesis by binding to corepressor SHP gene promoter and induces SHP gene expression, which subsequently inhibits key gluconeogenic enzyme genes PEPCK and Glc-6-Pase (G6Pase) via inhibition of HNF4α and results in decreased gluconeogenesis.
Supplementary Material
Acknowledgments
We thank N. Balachandar and Dr. Seok-Yong Choi for their assistance and helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants DK44442 and DK58379 (to J. Y. L. C.). This work was also supported by Korea Science and Engineering Foundation through the National Research Laboratory Program Grant NRL-ROA-2005-000-10047-0 and by Korea Research Foundation Grant 2006-005-J03003) (to H. S. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- PEPCK
- phosphoenolpyruvate carboxykinase
- HGF
- hepatocyte growth factor
- MSP
- macrophage-stimulating protein
- AMPK
- AMP-activated protein kinase
- USF-1
- upstream stimulatory factor-1
- DNA-PK
- DNA-dependent protein kinase
- SHP
- small heterodimer partner
- Glc-6-Pase
- glucose-6-phosphatase
- HNF4α
- hepatocyte nuclear factor 4α
- L-PK
- liver-type pyruvate kinase
- GK
- glucokinase
- ChIP
- chromatin immunoprecipitation
- m.o.i.
- multiplicity of infection
- RLU
- relative luciferase units
- DBD
- DNA-binding domain
- PHH
- primary human hepatocyte
- MAPK
- mitogen-activated protein kinase
- PI3K
- phosphatidylinositol 3-kinase
- JNK
- c-Jun N-terminal kinase
- ACC
- acetyl-CoA carboxylase
- Dex
- dexamethasone
- dn
- dominant negative
- m
- mouse
- h
- human
- qPCR
- quantitative PCR
- Ad
- adenovirus
- T1DM
- type I diabetes mellitus
- T2DM
- type II diabetes mellitus.
REFERENCES
- 1.Desvergne B., Michalik L., Wahli W. (2006) Physiol. Rev. 86, 465–514 [DOI] [PubMed] [Google Scholar]
- 2.Accili D. (2004) Diabetes 53, 1633–1642 [DOI] [PubMed] [Google Scholar]
- 3.Liu H. Y., Wen G. B., Han J., Hong T., Zhuo D., Liu Z., Cao W. (2008) J. Biol. Chem. 283, 30642–30649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You W. K., McDonald D. M. (2008) BMB Rep. 41, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song K. H., Ellis E., Strom S., Chiang J. Y. (2007) Hepatology 46, 1993–2002 [DOI] [PubMed] [Google Scholar]
- 6.Hiratsuka A., Adachi H., Fujiura Y., Yamagishi S., Hirai Y., Enomoto M., Satoh A., Hino A., Furuki K., Imaizumi T. (2005) J. Clin. Endocrinol. Metab. 90, 2927–2931 [DOI] [PubMed] [Google Scholar]
- 7.Satani K., Konya H., Hamaguchi T., Umehara A., Katsuno T., Ishikawa T., Kohri K., Hasegawa Y., Suehiro A., Kakishita E., Namba M. (2006) Diabet. Med. 23, 617–622 [DOI] [PubMed] [Google Scholar]
- 8.Rehman J., Considine R. V., Bovenkerk J. E., Li J., Slavens C. A., Jones R. M., March K. L. (2003) J. Am. Coll. Cardiol. 41, 1408–1413 [DOI] [PubMed] [Google Scholar]
- 9.Tomiya T., Nagoshi S., Fujiwara K. (1992) Hepatology 15, 1–4 [DOI] [PubMed] [Google Scholar]
- 10.Shiota G., Umeki K., Okano J., Kawasaki H. (1995) J. Med. 26, 295–308 [PubMed] [Google Scholar]
- 11.McElwee K. J., Huth A., Kissling S., Hoffmann R. (2004) J. Invest. Dermatol. 123, 34–40 [DOI] [PubMed] [Google Scholar]
- 12.Danilkovitch A., Leonard E. J. (1999) J. Leukocyte Biol. 65, 345–348 [DOI] [PubMed] [Google Scholar]
- 13.Chen Q., Seol D. W., Carr B., Zarnegar R. (1997) Hepatology 26, 59–66 [DOI] [PubMed] [Google Scholar]
- 14.van Deursen D., Jansen H., Verhoeven A. J. (2008) Diabetologia 51, 2078–2087 [DOI] [PubMed] [Google Scholar]
- 15.Yanai K., Saito T., Hirota K., Kobayashi H., Murakami K., Fukamizu A. (1997) J. Biol. Chem. 272, 30558–30562 [DOI] [PubMed] [Google Scholar]
- 16.Iynedjian P. B. (1998) Biochem. J. 333, 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imagawa S., Fujii S., Dong J., Furumoto T., Kaneko T., Zaman T., Satoh Y., Tsutsui H., Sobel B. E. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 2407–2413 [DOI] [PubMed] [Google Scholar]
- 18.Seol W., Choi H. S., Moore D. D. (1996) Science 272, 1336–1339 [DOI] [PubMed] [Google Scholar]
- 19.Chanda D., Park J. H., Choi H. S. (2008) Endocr. J. 55, 253–268 [DOI] [PubMed] [Google Scholar]
- 20.Lee Y. S., Chanda D., Sim J., Park Y. Y., Choi H. S. (2007) Int. Rev. Cytol. 261, 117–158 [DOI] [PubMed] [Google Scholar]
- 21.Hardie D. G. (2004) J. Cell Sci. 117, 5479–5487 [DOI] [PubMed] [Google Scholar]
- 22.Chanda D., Kim S. J., Lee I. K., Shong M., Choi H. S. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E368–E79 [DOI] [PubMed] [Google Scholar]
- 23.Kim Y. D., Park K. G., Lee Y. S., Park Y. Y., Kim D. K., Nedumaran B., Jang W. G., Cho W. J., Ha J., Lee I. K., Lee C. H., Choi H. S. (2008) Diabetes 57, 306–314 [DOI] [PubMed] [Google Scholar]
- 24.Huang H. P., Liu M., El-Hodiri H. M., Chu K., Jamrich M., Tsai M. J. (2000) Mol. Cell. Biol. 20, 3292–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H. J., Kim J. Y., Park Y. Y., Choi H. S. (2003) Nucleic Acids Res. 31, 6860–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J. H., Kim K., Jin H. M., Youn B. U., Song I., Choi H. S., Kim N. (2008) J. Mol. Biol. 383, 502–511 [DOI] [PubMed] [Google Scholar]
- 27.Xie Y. B., Nedumaran B., Choi H. S. (2009) Nucleic Acids Res. 37, 4100–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong R. H., Chang I., Hudak C. S., Hyun S., Kwan H. Y., Sul H. S. (2009) Cell 136, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park M. K., Kim D. K., Lee H. J. (2003) Exp. Mol. Med. 35, 494–500 [DOI] [PubMed] [Google Scholar]
- 30.Nakamura S., Morishita R., Moriguchi A., Yo Y., Nakamura Y., Hayashi S., Matsumoto K., Matsumoto K., Nakamura T., Higaki J., Ogihara T. (1998) J. Hypertens. 16, 2019–2026 [DOI] [PubMed] [Google Scholar]
- 31.Roccisana J., Reddy V., Vasavada R. C., Gonzalez-Pertusa J. A., Magnuson M. A., Garcia-Ocaña A. (2005) Diabetes 54, 2090–2102 [DOI] [PubMed] [Google Scholar]
- 32.Cruzado J. M., Lloberas N., Torras J., Riera M., Fillat C., Herrero-Fresneda I., Aran J. M., Alperovich G., Vidal A., Grinyó J. M. (2004) Diabetes 53, 1119–1127 [DOI] [PubMed] [Google Scholar]
- 33.Kato N., Nemoto K., Nakanishi K., Morishita R., Kaneda Y., Uenoyama M., Ikeda T., Fujikawa K. (2005) Diabetes 54, 846–854 [DOI] [PubMed] [Google Scholar]
- 34.Dai C., Yang J., Bastacky S., Xia J., Li Y., Liu Y. (2004) J. Am. Soc. Nephrol. 15, 2637–2647 [DOI] [PubMed] [Google Scholar]
- 35.Dai C., Li Y., Yang J., Liu Y. (2003) J. Biol. Chem. 278, 27080–27087 [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Ocaña A., Takane K. K., Syed M. A., Philbrick W. M., Vasavada R. C., Stewart A. F. (2000) J. Biol. Chem. 275, 1226–1232 [DOI] [PubMed] [Google Scholar]
- 37.Vázquez-Chantada M., Ariz U., Varela-Rey M., Embade N., Martínez-Lopez N., Fernández-Ramos D., Gómez-Santos L., Lamas S., Lu S. C., Martínez-Chantar M. L., Mato J. M. (2009) Hepatology 49, 608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamauchi T., Nio Y., Maki T., Kobayashi M., Takazawa T., Iwabu M., Okada-Iwabu M., Kawamoto S., Kubota N., Kubota T., Ito Y., Kamon J., Tsuchida A., Kumagai K., Kozono H., Hada Y., Ogata H., Tokuyama K., Tsunoda M., Ide T., Murakami K., Awazawa M., Takamoto I., Froguel P., Hara K., Tobe K., Nagai R., Ueki K., Kadowaki T. (2007) Nat. Med. 13, 332–339 [DOI] [PubMed] [Google Scholar]
- 39.Kodama H., Fujita M., Yamaguchi I. (1994) Diabetologia 37, 739–744 [DOI] [PubMed] [Google Scholar]
- 40.Cao W., Collins Q. F., Becker T. C., Robidoux J., Lupo E. G., Jr., Xiong Y., Daniel K. W., Floering L., Collins S. (2005) J. Biol. Chem. 280, 42731–42737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.