Abstract
We describe a role for diacylglycerol in the activation of Ras and Rap1 at the phagosomal membrane. During phagocytosis, Ras density was similar on the surface and invaginating areas of the membrane, but activation was detectable only in the latter and in sealed phagosomes. Ras activation was associated with the recruitment of RasGRP3, a diacylglycerol-dependent Ras/Rap1 exchange factor. Recruitment to phagosomes of RasGRP3, which contains a C1 domain, parallels and appears to be due to the formation of diacylglycerol. Accordingly, Ras and Rap1 activation was precluded by antagonists of phospholipase C and of diacylglycerol binding. Ras is dispensable for phagocytosis but controls activation of extracellular signal-regulated kinase, which is partially impeded by diacylglycerol inhibitors. By contrast, cross-activation of complement receptors by stimulation of Fcγ receptors requires Rap1 and involves diacylglycerol. We suggest a role for diacylglycerol-dependent exchange factors in the activation of Ras and Rap1, which govern distinct processes induced by Fcγ receptor-mediated phagocytosis to enhance the innate immune response.
Receptors that interact with the constant region of IgG (FcγR)4 mediate the recognition and elimination of soluble immune complexes and particles coated (opsonized) with immunoglobulins. Clustering of FcγR on the surface of leukocytes upon attachment to multivalent ligands induces their activation and subsequent internalization. Soluble immune complexes are internalized by endocytosis, a clathrin- and ubiquitylation-dependent process (1). In contrast, large, particulate complexes like IgG-coated pathogens are ingested by phagocytosis, a process that is contingent on extensive actin polymerization that drives the extension of pseudopods (2). In parallel with the internalization of the opsonized targets, cross-linking of phagocytic receptors triggers a variety of other responses that are essential components of the innate immune response. These include degranulation, activation of the respiratory burst, and the synthesis and release of multiple inflammatory agents (3, 4).
Like T and B cell receptors, FcγR possesses an immunoreceptor tyrosine-based activation motif that is critical for signal transduction (3, 4). Upon receptor clustering, tyrosyl residues of the immunoreceptor tyrosine-based activation motif are phosphorylated by Src family kinases, thereby generating a docking site for Syk, a tyrosine kinase of the ZAP70 family (3, 4). The recruitment and activation of Syk in turn initiates a cascade of events that include activation of Tec family kinases, Rho- and ARF-family GTPases, phosphatidylinositol 3-kinase, phospholipase Cγ (PLCγ), and a multitude of additional effectors that together remodel the underlying cytoskeleton, culminating in internalization of the bound particle (5, 6).
Phosphoinositide metabolism is thought to be critical for FcγR-induced phagocytosis (7, 8). Highly localized and very dynamic phosphoinositide changes have been observed at sites of phagocytosis: phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) undergoes a transient accumulation at the phagocytic cup, which is rapidly superseded by its complete elimination from the nascent phagosome (7). The secondary disappearance of PtdIns(4,5)P2 is attributable in part to the localized generation of phosphatidylinositol 3,4,5-trisphosphate, which has been reported to accumulate at sites of phagocytosis (9). Activation of PLCγ is also believed to contribute to the acute disappearance of PtdIns(4,5)P2 in nascent phagosomes. Indeed, the generation of diacylglycerol (DAG) and inositol 1,4,5-trisphosphate has been detected by chemical means during FcγR-evoked particle ingestion (10, 11). Moreover, imaging experiments revealed that DAG appears at the time and at the precise site where PtdIns(4,5)P2 is consumed (7).
Two lines of evidence suggest that the DAG generated upon engagement of phagocytic receptors modulates particle engulfment. First, antagonists of PLC severely impair phagocytosis by macrophages (7, 12). This inhibition is not mimicked by preventing the associated [Ca2+] transient, suggesting that DAG, and not inositol 1,4,5-trisphosphate, is the crucial product of the PLC (13). Second, the addition of exogenous DAG or phorbol esters, which mimic the actions of endogenous DAG, augment phagocytosis (14, 15).
Selective recognition of DAG by cellular ligands is generally mediated by specific regions of its target proteins, called C1 domains (16). Proteins bearing C1 domains include, most notably, members of the classical and novel families of protein kinase C (PKC), making them suitable candidates to account for the DAG dependence of phagocytosis. Indeed, PKCα, a classical isoform, and PKCϵ and PKCδ, both novel isoforms, are recruited to phagosomes (12, 15, 17, 18). Although the role of the various PKC isoforms in particle engulfment has been equivocal over the years, Cheeseman et al. (12) convincingly demonstrated that PKCϵ contributes to particle uptake in a PLC- and DAG-dependent manner.
PKCs are not the sole proteins bearing DAG-binding C1 domains. Similar domains are also found in several other proteins, including members of the RasGRP family, chimaerins, and Munc-13 (19–21). One or more of these could contribute to the complex set of responses elicited by FcγR-induced DAG production. The RasGRP proteins are a class of exchange factors for the Ras/Rap family of GTPases (22). There are four RasGRP proteins (RasGRP1 to -4), and emerging evidence has implicated RasGRP1 and RasGRP3 in T and B cell receptor signaling (23–27).
The possible role of DAG-mediated signaling pathways other than PKC in phagocytosis and the subsequent inflammatory response has not been explored. Here, we provide evidence that DAG stimulates Ras and Rap1 at sites of phagocytosis, probably through RasGRPs. Last, the functional consequences of Ras and Rap1 activation were analyzed.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Human IgG and glutathione-agarose beads were from Sigma. PMA, calphostin C, GF109203X, and U73122 were from Calbiochem or Biomol. Latex beads (2.0 and 3.1 μm in diameter) were from Bangs Laboratories. Sheep red blood cells and rabbit anti-sheep red blood cell antibodies were from ICN/Cappel. Anti-sheep red blood cell (RBC) IgM was from Accurate Chemicals. Murine monoclonal antibodies to H-Ras (F235), N-Ras (F155), and RasGRP (199) and rabbit antibodies to K-Ras (C-19) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Monoclonal anti-α-tubulin (B125) was from Sigma. Murine monoclonal H/N-Ras antibodies were from Viromed Biosafety Laboratories. Rabbit antibody to phospho-ERK1/2 (Thr202/Tyr204) was from Cell Signaling. Rat monoclonal anti-mouse LAMP1 (1D4B) was from the Developmental Studies Hybridoma Bank (Iowa City, IA). Mouse anti-T7 antibody was from Novagen. Rabbit anti-RasGRP3 antibodies were previously characterized (26). Polyclonal Rap1 antibodies were from Santa Cruz Biotechnology (Krev-1). Fluorescent or peroxidase-coupled secondary antibodies were from Jackson Laboratories, Pierce, and Molecular Probes.
Cell Culture, DNA Constructs, and Transfection
RAW264.7 macrophages obtained from ATCC were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 5% CO2 at 37 °C. The generation of Rat2 cells stably expressing RasGRP3 was previously described in Ref. 28.
Constructs encoding the untagged dominant negative mutant of H-Ras and H-RasN17, GFP-H-RasN17, T7-tagged N-Ras, and a bacterially expressed RasGTP-binding domain of Raf1 (RBD) fused to GST were gifts from Dr. D. Rotin (Hospital for Sick Children, Toronto, Canada). The generation of GFP-H-Ras, GFP-N-Ras, and GFP-K-Ras, which encode GFP fusion proteins of H-Ras, N-Ras, and K-Ras, respectively, was described in Ref. 29. Construction of the RBD fused to GFP was previously described in Ref. 30. Transfections were accomplished using FuGene 6 (Roche Applied Science) according to the manufacturer's instructions. The Rap1 GTP binding domain of Ral-GDS fused to GST (GST-RalGDS) was generated as described (31).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
RT-PCR was employed to detect mRNA expression of RasGRP1, RasGRP2, and RasGRP3 in RAW264.7 macrophages and murine brain extracts using the Titan RT-PCR kit (Roche Applied Science), as instructed by the manufacturer. mRNA from RAW macrophages and tissue extracts was isolated using the mRNA isolation kit from Roche Applied Science. To amplify RasGRP1, we used two primer sets; amplification of the 5′ region encompassing nucleotides 38–764 was with GRP1U (5′-AGGCTCCGCGGAAACCTTGCCAT-3′) and GRP1D (5′-CACAGGGCAATGGACCTCTCCATG-3′), whereas the 3′ region encompassing nucleotides 841–1310 was with GRP1–841 (5′-AAGTTCATCCATGTGGCTCAGAAAC-3′) and GRP1–1310 (5′-CGGGCATAGGAAAGCTCATAGATTTC-3′). Nucleotides 20–646 of RasGRP2 were amplified using GRP2U (5′-CTGGTTCAAGTGAACAGAAGGTCTGG-3′) and GRP2D (5′-GTTCTGCCAGTTCCATAGGCTCCAAG-3′). The region of RasGRP3 encompassing nucleotides 345–694 was amplified using GRP3U (5′-TTCCTTCTTATGACTGGATGAGG-3′) and GRP3 (5′-GGGCTACATTGATGAACTTTGTG-3′). Amplification of the N-Ras region 250–752 served as a positive control and was performed using the primers NRasU (5′-AGGTGGTGTTGGGAAAAGCGCCCT-3′) and NRasD (5′-GAGTGCCATCGTCACTGCTGTTGAG-3′).
Phagocytosis Assay
Sheep RBCs were opsonized for 1 h using a 1:50 dilution of rabbit anti-sheep RBCs. Latex beads were similarly opsonized using 1 mg/ml human IgG. Phagocytic targets were then washed three times with phosphate-buffered saline. To quantify the phagocytic index in cells expressing the Ras dominant negative allele, H-RasN17, phagocytosis of RBCs was allowed to proceed for 15 min, followed by lysis of adherent extracellular RBCs with water and fixation with 4% paraformaldehyde. Internalized RBCs were identified by bright field microscopy and/or staining with Cy3-conjugated anti-rabbit antibodies. To determine phagosome maturation in the presence of H-RasN17, cells were induced to internalize RBCs, and the phagosomes were allowed to mature for 60 min, followed by fixation and immunostaining for LAMP1. C3bi opsonization and binding assays were performed as described previously (32).
Ras and Rap1 Activation Assay
Affinity precipitation of Ras-GTP and Rap1-GTP was performed as described in Ref. 33. Briefly, GST-RBD (for Ras-GTP) or GST-RalGDS (for Rap1-GTP) was induced in the DH10 Escherichia coli strain using 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. After expression and pelleting, bacteria were washed in HEPES-buffered saline (20 mm HEPES, 150 mm NaCl, pH 7.4) and resuspended in BLB buffer (20 mm HEPES, pH 7.5, 120 mm NaCl, 10% glycerol, 2 mm EDTA, supplemented with a 1:100 dilution of prokaryotic anti-protease mixture). Bacteria were then lysed by two “French press” passes, and the supernatant was cleared by centrifugation. The lysate was supplemented with 0.5% Nonidet P-40 and incubated with glutathione-agarose beads for 30 min. The loaded beads were washed and used to precipitate Ras-GTP or Rap1-GTP within 2 days.
To assay for phagocytosis-dependent activation of Ras or Rap1, macrophages were incubated with opsonized RBCs on ice for 20 min and then warmed to 37 °C for the indicated times. Phagocytosis was arrested by washing with cold phosphate-buffered saline, and macrophage lysates were prepared by the addition of cold MLB buffer (25 mm HEPES, pH 7.5, 150 mm NaCl, 0.25% sodium deoxycholate, 10% glycerol, 1 mm EDTA, and 10 mm MgCl2 added immediately before use) supplemented with a 1:100 dilution of eukaryotic protease inhibitor mixture (Sigma). Cell lysates were then cleared by centrifugation, and the supernatant was incubated with freshly prepared glutathione-agarose beads coupled to GST-RBD or GST-RalGDS for 30 min in the cold. The beads were washed three times with 0.6 ml of MLB buffer, and bound Ras-GTP or Rap1-GTP was then eluted by boiling in twice concentrated Laemmli buffer. Samples were analyzed by SDS-PAGE, blotted onto polyvinylidene difluoride, and subjected to quantitative immunoblotting analysis using monoclonal H/N-Ras antibodies (1:20,000) or polyclonal Rap1 antibodies (1:500).
Immunoblotting
To detect expression of Ras and RasGRP isoforms, RAW, Rat2, Rat2-GRP3 (engineered by retroviral infection to express RasGRP3), and Jurkat cells were lysed in Laemmli buffer. Brain and spleen lysates were prepared by homogenizing tissue in a Dounce homogenizer, followed by the addition of Laemmli buffer and boiling. Lysates were then cleared by centrifugation, and protein concentration was estimated by spotting on Whatman paper and Coommassie Blue staining, using bovine serum albumin as a standard. To detect ERK1/2 activation, RAW cells were lysed in Laemmli buffer immediately after the desired treatment. Twenty to forty micrograms of protein were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Blots were blocked with 5% milk in phosphate-buffered saline or Tris-buffered saline. All primary and secondary antibodies were incubated in 5% milk in phosphate-buffered saline, with the exception of phospho-ERK1/2 (Thr202/Tyr204) antibody, which was in Tris-buffered saline. Rabbit antibodies to RasGRP3, K-Ras, Rap1, and phospho-ERK1/2 were used at 1:250, 1:700, 1:500, and 1:2500, respectively. Monoclonal antibodies to H-Ras, N-Ras, RasGRP1, and tubulin were employed at 1:1000, 1:700, 1:500, and 1:5000, respectively. All horseradish peroxidase-coupled secondary antibodies were used at a 1:5000 dilution.
Immunofluorescence and Confocal Imaging
For immunofluorescence experiments, macrophages were fixed with 4% paraformaldehyde, followed by permeabilization with 0.1% Triton and blocking with 5% donkey serum. Untagged RasN17 was detected using mouse anti-H-Ras (Santa Cruz Biotechnology) at 1:100, T7 epitope-tagged N-Ras was detected with a 1:1000 dilution of mouse anti-T7 antibodies, LAMP1 was detected using undiluted rat anti-mouse LAMP1 supernatant, and endogenous RasGRP3 was stained with a 1:100 dilution of rabbit anti-RasGRP3 antibodies. Staining with anti-phospho-ERK1/2 antibodies was performed as instructed by the supplier and using phosphatase inhibitors. All secondary antibodies were used at 1:1000. Cells were observed with a Zeiss LSM510 scanning laser confocal microscope using the conventional laser lines and filter sets. The method used to monitor fluorescence during the course of phagocytosis in live cells was described (7).
RESULTS
Members of the RasGRP Family Are Expressed in Phagocytic Cells
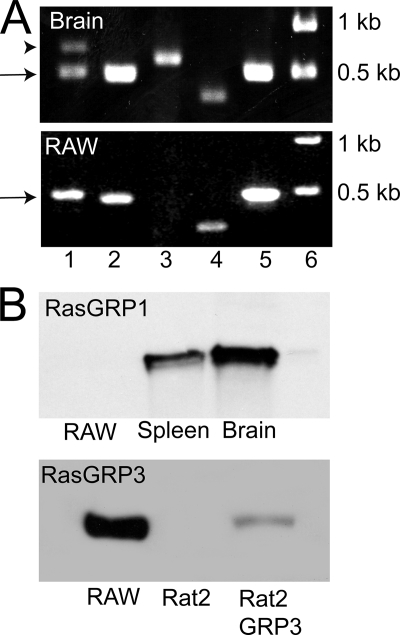
RAW264.7 cells (referred to hereafter as RAW cells) were used for these studies. This murine line has been used extensively for the biochemical and molecular analysis of phagocytosis, because it accurately recapitulates the phagocytic phenotype of monocytes/macrophages and because it is amenable to transfection, albeit with modest efficiency (7, 9, 17). We first analyzed whether RasGRP1, RasGRP2, and/or RasGRP3 are expressed in RAW cells, using RT-PCR. RasGRP4 was not investigated. In accordance with earlier reports, expression of all three RasGRP species was confirmed in mRNA extracted from murine brains, validating the adequacy of the primer sets used (Fig. 1A). The quality of the mRNA extracted from RAW cells was tested by the positive amplification of N-Ras, a widely expressed protein (Fig. 1A, lane 5). As shown in Fig. 1A, mRNA encoding for RasGRP1 and RasGRP3, but not for RasGRP2, was detected in RAW cells (Fig. 1A).
FIGURE 1.
Expression of RasGRP isoforms in macrophages. A, detection of RasGRP isoforms by RT-PCR using as template mRNA extracted from murine brain (top) or from RAW macrophages. Lane 1, RasGRP1 using primers encompassing nucleotides 38–764; lane 2, RasGRP1 using primers encompassing nucleotides 841–1310; lane 3, RasGRP2; lane 4, RasGRP3; lane 5, N-Ras; lane 6, molecular weight markers. The arrowhead and arrow point to the predicted product and an unexpected ∼500-bp product, respectively, amplified using primers encompassing nucleotides 38–764 of RasGRP1. B, detection of RasGRP isoforms by immunoblotting. The top blot contains samples with equivalent protein loads of RAW cells and of murine splenic and brain extracts and was probed with antibodies to RasGRP1. The bottom blot contains samples with equivalent protein loads of RAW cells, Rat2 cells, and Rat2 cells stably transfected with RasGRP3 that were probed with antibodies to RasGRP3.
Western blotting was utilized next to evaluate protein expression of RasGRP1 and RasGRP3 (Fig. 1B). The monoclonal antibody m199 readily detected RasGRP1 in both spleen and brain extracts from mice. However, the same titer of the antibody failed to detect RasGRP1 in RAW cells. This probably implies that the expression level of the protein is low in these cells or, alternatively, that they express a unique splice variant that is not identified by the antibody (see “Discussion”). The suitability of the polyclonal antibody used to detect RasGRP3 was validated using extracts of Rat2 cells that were stably transfected with the protein. Untransfected Rat2 cells, which do not express RasGRP3, were used as negative control (Fig. 1B). As shown in Fig. 1B, RasGRP3 was abundantly expressed in RAW cells. We therefore concentrated our attention on this isoform for the remainder of this study.
RasGRP3 Is Recruited to Phagocytic Cups
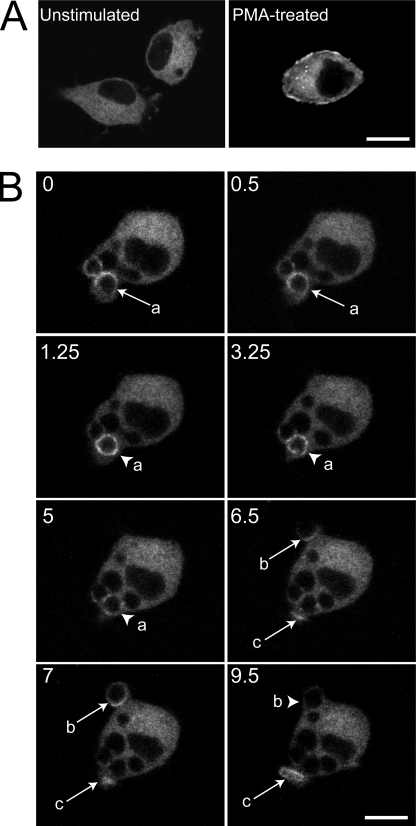
We had reported earlier that DAG accumulates focally at sites of phagosome generation (7). To investigate whether phagosomal DAG is capable of recruiting and potentially activating RasGRP3, RAW macrophages were transfected with a plasmid encoding GFP-tagged RasGRP3. The distribution of RasGRP3-GFP in unstimulated cells was predominantly cytosolic (Fig. 2A), but the addition of a DAG-mimetic phorbol ester (100 nm PMA) induced translocation of a fraction of RasGRP3-GFP to the plasma membrane (Fig. 2A). In a fraction of the cells (∼15%), PMA caused RasGRP3-GFP to accumulate in a juxtanuclear structure, probably the Golgi apparatus (not shown).
FIGURE 2.
RasGRP3 distribution during phagocytosis. RAW cells were transiently transfected with RasGRP3-GFP and analyzed by confocal microscopy. A, resting (untreated) and treated cells with 100 nm PMA for 15 min. B, cells monitored during the engulfment of IgG-opsonized RBCs. The various panels are a temporal series proceeding from left to right and from top to bottom. The numbers indicate the time (min) after acquisition of the first image. Note that RBCs had been added to the cells several min before acquisition of the first image. The arrows indicate a phagocytic cup, and arrowheads point to sealed phagosomes. Selected RBCs undergoing phagocytosis are labeled a–c. Scale bar, 10 μm.
The behavior of RasGRP3 during phagocytosis of IgG-opsonized RBCs was studied next. Little or no redistribution of RasGRP3-GFP was observed at the earliest stages of phagocytosis (e.g. arrow b in Fig. 2B, t = 6.5, points to a phagocytic cup formed within 30 s of RBC attachment to the surface of the macrophage). However, at later stages of phagocytosis, when pseudopod extension was prominent, and particularly when RBC enclosure was completed, the accumulation of RasGRP3 was readily apparent in 50–70% of the phagosomes (e.g. arrowhead a in Fig. 2B, t = 1.25, points to an enclosed RBC), although the extent of recruitment varied between phagocytic cups. RasGRP3-GFP remained on the phagosomal membrane for 1–5 min following closure and then dissociated gradually (e.g. arrowhead a in Fig. 2B, t = 5). Association of endogenous RasGRP3 with the phagosomal membrane was confirmed by immunostaining with the polyclonal antibody described in the legend to Fig. 1 (supplemental Fig. S1A). Although the latter experiments lacked the dynamic information gathered with RasGRP3-GFP, they indicated that the observed association is not the artifactual result of heterologous (over)expression of the exchange factor. Moreover, a yellow fluorescent protein-tagged version of the RasGRP1 isoform expressed in brain was not observed on phagocytic cups or phagosomes, indicating that RasGRP3 is specifically recruited during phagocytosis (supplemental Fig. S1B).
Ras Is Activated during Phagocytosis
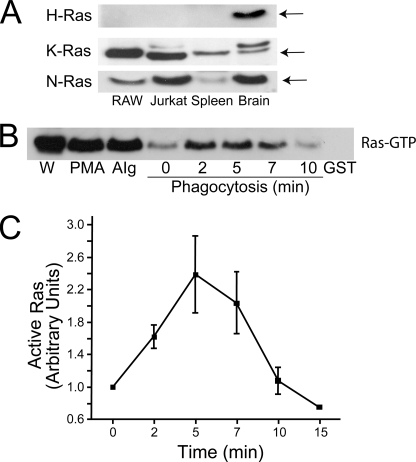
The recruitment of RasGRP3 to sites of phagocytosis suggested that Ras may be activated during FcγR-mediated phagocytosis, at least in part by a DAG-dependent process. To test this hypothesis, we first determined which isoforms of Ras were expressed in RAW cells by immunoblotting whole cell lysates. As illustrated in Fig. 3A, N-Ras and K-Ras were clearly present in RAW macrophages. In contrast, H-Ras was not detected in either RAW cells, in other leukocytes like Jurkat lymphocytes or in murine spleen extracts, but was observed in the brain, confirming the reactivity of the antibody used (Fig. 3A). Therefore, N-Ras and K-Ras became the focus of our subsequent studies in RAW cells.
FIGURE 3.
Expression of endogenous Ras isoforms in macrophages and activation during phagocytosis. A, detection of Ras isoforms by immunoblotting. Blots contain samples with equivalent protein loads of RAW and Jurkat cells and of murine splenic and brain extracts and were probed with antibodies to H-Ras, K-Ras, and N-Ras (from top to bottom, as indicated). The arrows point to Ras. B, Ras activation in RAW cells was assessed using the RBD-GST pull-down assay described under “Experimental Procedures.” As a control, beads coated with only GST were mixed with activated lysate (GST). The lane labeled W is a fraction of whole cell lysate. Samples were stimulated by PMA (100 nm) by the addition of aggregated IgG (AIg) to simultaneously stimulate all FcγR or by exposure to IgG-coated RBCs (Phagocytosis) for the indicated time (min). C, quantification of the course of activation of Ras during phagocytosis, from four experiments like that illustrated in B. Quantification was performed by densitometry of scanned films using Image J software. To enable comparison between experiments, all values were normalized to the intensity of the unstimulated (t = 0) cells. Data are means ± S.E.
We employed a GST fusion of RBD to assess the activation of Ras during FcγR-mediated phagocytosis. This GST-RBD fusion protein selectively precipitates the activated form of Ras, which was then quantified by immunoblotting (33). The effectiveness of the method was initially confirmed by stimulating the cells with PMA, a well established activator of Ras (26) (Fig. 3B). Two different approaches were used to analyze the effect of Fcγ receptor activation on Ras. First, the receptors were cross-linked with aggregated IgG to produce a global and synchronous stimulation. This manipulation induced a marked activation of Ras (cf. stimulated lane with unstimulated control, labeled 0 in Fig. 3B). Second, Fcγ receptors were activated focally by exposure to IgG-opsonized RBCs. When the onset of phagocytosis was synchronized by preincubating RAW cells with the IgG-opsonized RBCs in the cold, a rapid and transient activation of Ras was observed, which was detectable within 2 min, peaked after ∼5 min and was abated by 10 min (Fig. 3, B and C). A similar transient activation of Ras had been reported earlier following antibody-mediated cross-linking of Fc receptors in U937 cells (34).
Distribution of N-Ras during Phagocytosis
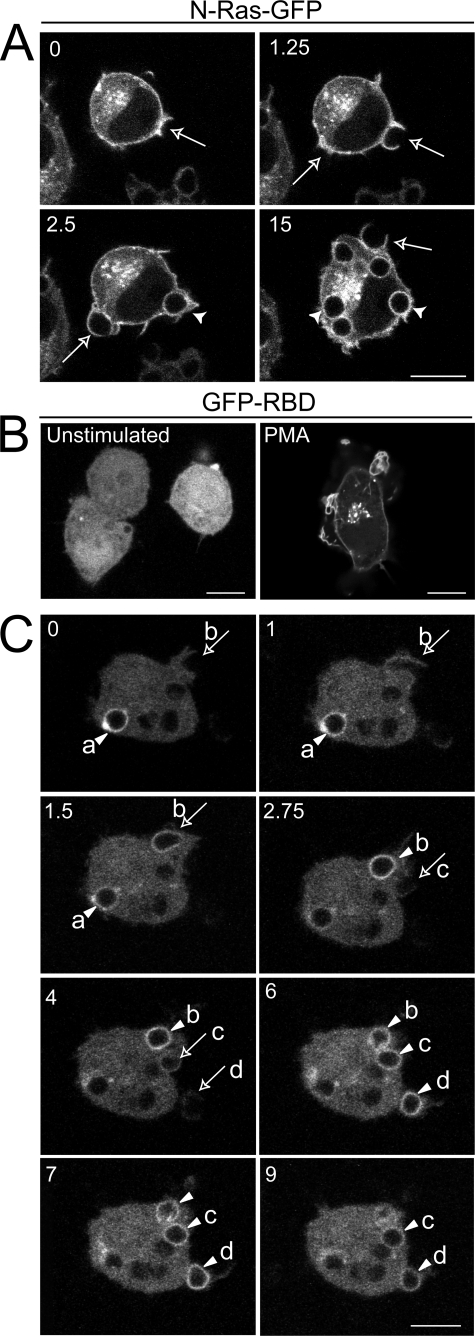
Like Ras, small GTPases of the Rho family are also activated during phagocytosis (35). Rac and Cdc42, which are activated upon cross-linking of FcγR, accumulate at sites of phagocytosis (36). To analyze whether Ras is also recruited to the phagocytic cup, we transfected RAW cells with either GFP-tagged N-Ras or K-Ras constructs because the endogenous Ras isoforms were not detectable by immunostaining with available antibodies. Expression of the fluorescent proteins also enabled us to perform dynamic studies in living cells. As reported in other systems (37), GFP-N-Ras was enriched in the plasma membrane but was also present in a juxtanuclear compartment, probably the Golgi complex (Fig. 4A). GFP-K-Ras was similarly found in the plasmalemma with a small fraction in diffuse vesicular structures, possibly endosomes (38, 39) (see Ref. 40 for illustration). Expression of the GFP-tagged proteins had no discernible effect on the kinetics or efficiency of phagocytosis. Neither N-Ras (Fig. 4A) nor K-Ras (not shown; see Ref. 40) displayed a propensity to accumulate in phagocytic cups, compared with their density in unengaged regions of the surface membrane. Notably, GFP-N-Ras remained associated with the phagosomes for at least 20 min, although N-Ras activation is largely terminated after 10 min. By comparison, GFP-K-Ras was depleted from the phagosomal membrane following RBC engulfment, an effect attributed recently to changes in the surface charge of the membrane upon phagocytosis (40). These observations suggest that activation of Ras during phagocytosis does not involve its preferential recruitment to the nascent phagosome; nor is deactivation dependent on its detachment from the phagosomal membrane.
FIGURE 4.
Dynamics of Ras and Ras-GTP during phagocytosis. A, RAW cells were transiently transfected with a GFP fusion protein of N-Ras. Phagocytosis was initiated by exposure of the transfected cells to IgG-opsonized RBCs. Fluorescence was analyzed by confocal laser microscopy. The numbers indicate the time (min) after acquisition of the first image. Note that RBCs had been added to the cells before acquisition of the first image. Images are representative of five experiments. B and C, RAW macrophages were co-transfected with GFP-RBD and T7-N-Ras and analyzed by laser confocal microscopy. B, resting (untreated) and cells treated with 100 nm PMA; C, cells monitored during the engulfment of IgG-opsonized RBCs. The various panels are a temporal series proceeding from left to right and top to bottom. The numbers indicate the time (min) after acquisition of the first image. Note that RBCs had been added to the cells several min before acquisition of the first image. The arrows point to phagocytic cups, and the arrowheads point to sealed phagosomes. Selected RBCs undergoing phagocytosis are labeled a–d. Scale bar, 10 μm.
Localized Activation of Ras at the Phagocytic Cup
Because RasGRP3 was recruited to the phagosomal cup, we hypothesized that this leads to localized activation of K-Ras and N-Ras. To this end, we used a GFP fusion protein of the Raf-1 RBD domain (GFP-RBD), which was used successfully to probe the subcellular dynamics of RasGTP (30). RAW cells were co-transfected with GFP-RBD and N-Ras. Overexpression of Ras was required, since the endogenous levels of RasGTP were insufficient for detection by this method, as seen in other systems (30). As shown in Fig. 4B, in unstimulated cells, GFP-RBD was mostly cytosolic, with no obvious association with the plasma membrane. Treatment of cells with 100 nm PMA induced migration of GFP-RBD to a juxtanuclear location, probably the Golgi complex, and to a lesser extent to the plasma membrane (Fig. 4B).
The distribution of GFP-RBD was next studied during the course of phagocytosis. Most frequently, little or no accumulation of RasGTP was detectable at the earliest stages of phagosomal cup formation (e.g. arrow b in Fig. 4C, t = 0 and 1 min; arrow c, t = 2.75 min; and arrow d, t = 4 min). By contrast, focal accumulation of GFP-RBD was discernible on the advancing pseudopods when they had nearly surrounded the RBCs (e.g. arrow b in Fig. 4C, t = 1.5 min). However, the formation of RasGTP was most obvious on the membrane of the phagosomes immediately after closure (e.g. arrowhead a in Fig. 4C, t = 0 min and t = 1 min; arrow b, t = 2.75 min; and arrowheads c and d, t = 6 and 7 min). Activated Ras was detectable on newly formed phagosomes for 4–12 min after sealing (e.g. arrowheads in Fig. 4C, t = 9 min), waning thereafter. This transient pattern of RasGTP accumulation on the phagosomal membrane parallels the kinetics of Ras activation determined by biochemical means following synchronized phagocytosis by a population of macrophages (Fig. 3C). These observations suggest that the Ras activation detected in the cell lysates originated largely from the phagosomal membrane. They also confirm the notion that intracellular organelles, such as the Golgi complex, the endoplasmic reticulum (30), and, in our case, the phagosome, can serve as platforms for focal Ras signaling.
Contribution of DAG to the Activation of Ras
Taken together, the results presented above imply that RasGRP3 plays a role in the activation of Ras at the phagosomal membrane. Not only is the exchange factor probably recruited to the nascent phagosomes by the local generation of DAG, but the course of recruitment of RasGRP3 parallels the formation of RasGTP on the phagocytic vacuole (cf. Figs. 2 and 4). However, Ras is activated by a variety of exchange factors, some of which may also be stimulated by phagocytic receptors (see “Discussion”). We therefore performed studies to evaluate the contribution of DAG and RasGRP3 to the observed stimulation. We first examined the sensitivity of Ras activation to the PLC antagonist, U73122, which was previously shown to effectively inhibit PLC activity in macrophages (7). As illustrated in Fig. 5A, the activation of Ras normally seen in otherwise untreated cells performing phagocytosis was eliminated by pretreatment of the cells with 5 or 10 μm U73122. In addition, calphostin C, which impairs binding of DAG to its protein effectors (21), also precluded Ras activation during phagocytosis (Fig. 5A). It is unlikely that calphostin C produced this effect by interfering with the binding of DAG to classical or novel PKC isoforms, since the addition of GF109203X, a general antagonist of PKC activity, did not affect Ras stimulation during ingestion (Fig. 5A). Thus, Ras stimulation was suppressed not only when the formation of DAG was prevented by inhibition of PLC but also when the interaction of DAG with its target molecules was impaired. RasGRP3, one such DAG effector, is most likely involved in Ras stimulation.
FIGURE 5.
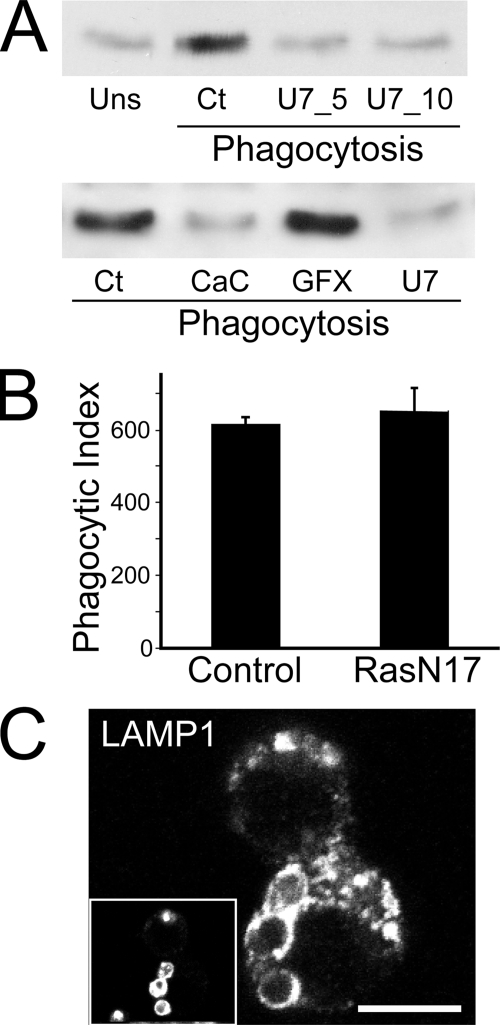
Diacylglycerol-dependent activation of Ras and its involvement in phagosome formation and maturation. A, diacylglycerol-dependent Ras activation in RAW cells. Ras activation was probed using the RBD-GST pull-down assay described under “Experimental Procedures” and in the legend to Fig. 3. Cells were either unstimulated (Uns) or allowed to undergo phagocytosis of IgG-coated RBCs. Additionally, cells undergoing phagocytosis were either untreated (Ct), pre-exposed to 5 μm (U7_5) or 10 μm (U7_10, U7) U73122 for 20 min, or pretreated for 30 min with 0.5 μm calphostin C (CaC) or 10 μm GF109203X (GFX). Phagocytosis was allowed to proceed for 5–7 min prior to cell lysis. Blots are representative of four similar experiments. B, quantification of phagocytosis in control cells and in cells expressing the dominant negative allele H-RasN17. The phagocytic index (the number of RBCs internalized/100 macrophages) was calculated in three similar experiments. Data are means ± S.E. C, cells expressing H-RasN17 were allowed to internalize IgG-coated RBCs and chased for 1 h. Cells were fixed and stained for LAMP-1 to verify phagosome maturation. The inset shows internalized IgG-coated RBCs. Scale bar, 10 μm.
Investigation of Ras Signaling in Phagocytosis and Phagosome Maturation
Activation of Ras upon FcγR activation suggested that the GTPase may contribute to signaling phagocytosis. We therefore tested if signals emanating from active Ras were required for phagosome formation or maturation in RAW cells, by expressing an untagged dominant negative allele, H-RasN17. Expression of RasN17 was confirmed by immunostaining with anti-H-Ras. We employed H-RasN17 because it was previously shown to impede signaling from all three Ras isoforms, unlike N-RasN17 and K-RasN17, which specifically inhibit N-Ras or K-Ras, respectively (41). We monitored and compared the phagocytic index of control cells that did not express H-RasN17 and of cells expressing the Ras dominant negative construct. We found that H-RasN17 expression had no discernible effect on the ability of RAW cells to engulf IgG-opsonized RBCs (Fig. 5B). We next investigated whether inhibition of Ras altered the ability of formed phagosomes to mature. Maturation was assessed by the acquisition of the late endosome/lysosome marker LAMP-1 (Fig. 5C). As with phagocytosis, expression of RasN17 was without effect on the ability of phagosomes to acquire LAMP-1. Together, these results suggest that Ras does not directly regulate phagocytosis or phagosome maturation.
ERK Activation during Phagocytosis Requires Ras and DAG-mediated Signaling
Engagement of FcγRs elicits an inflammatory response consisting of induction of expression and secretion of cytokines, such as TNFα and IL-1 (42, 43). Expression of TNFα in leukocytes was shown to depend on FcγR-induced activation of the mitogen-activated protein kinases, ERK1 and -2 (42, 43). Because these kinases can be activated by Raf, a downstream effector of Ras, we examined whether Ras and diacylglycerol-dependent signaling influence ERK activity during phagocytosis.
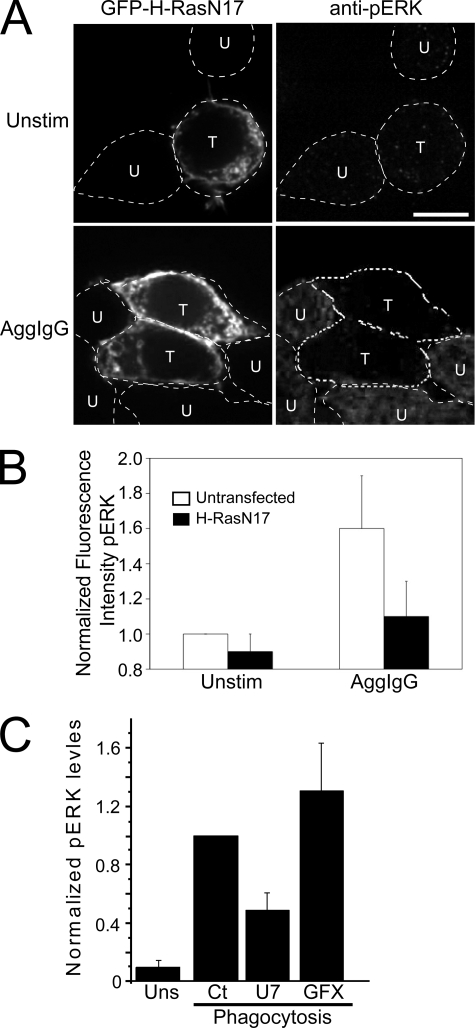
We first investigated whether Ras was required for ERK activation during phagocytosis by immunostaining cells with an antibody that detects ERK phosphorylated at positions Thr202/Tyr204 (pERK). To this end, cells were transfected with GFP-H-RasN17, and the transfectants were identified by their fluorescence. In unstimulated cells, pERK was perceptible near background levels, whether or not cells expressed H-RasN17 (Fig. 6A, top). In contrast, the addition of aggregated IgG to cells elicited phosphorylation of the ERK proteins (Fig. 6A, untransfected cells). Of note, the presence of H-RasN17 in cells severely curtailed the formation of pERK following stimulation with aggregated IgG (Fig. 6A, transfected cells in the lower panels). Quantification of experiments like those in Figs. 6A indicated that H-RasN17 significantly inhibited pERK formation in cells activated by aggregated IgG (Fig. 6B).
FIGURE 6.
The role of Ras and DAG in ERK activation during phagocytosis. A, RAW cells were transfected with GFP-H-RasN17 and immunostained for pERK after the indicated treatments. In the top panels, cells were untreated. In the bottom panels, cells were treated with 10 mg/ml aggregated IgG (AggIgG) for 10 min. The dashed lines outline cells. Cells transfected with GFP-H-RasN17 are denoted by a T, whereas those that remained untransfected are indicated with a U. Scale bar, 10 μm. B, quantification of pERK fluorescence using ImageJ, from experiments like those in A. To facilitate comparison, the signal was normalized to unstimulated, untransfected cells. Data are means of three experiments, and S.E. is shown. C, quantification of ERK1/2 phosphorylation by immunoblotting in unstimulated cells (Uns) and in cells exposed to IgG-opsonized RBCs for 10 min (Phagocytosis). Cells undergoing phagocytosis were untreated (Ct) or had been pretreated with U73122 (U7) or GF109203X (GFX). The pERK content of the cells was determined as described under “Experimental Procedures” and is normalized to that of control phagocytosis (Ct). Equal loading was ensured or corrected for by stripping the blots and probing for cellular tubulin. Data are means ± S.E. of six experiments.
We next used immunoblotting to study whether DAG was needed for ERK activation. Whole cell lysates of cells treated under the indicated conditions were blotted with antibodies to pERK and tubulin, the latter used to normalize the pERK signal. Results from three such experiments were quantified, and the results are summarized in Fig. 6C. As observed by immunofluorescence, FcγR-mediated phagocytosis triggered phosphorylation of ERK (Fig. 6C). Importantly, preventing DAG synthesis by treatment with the PLC antagonist, U73122, decreased pERK levels. This effect was not due to impairment of PKC activation, since inhibition of this family of kinases with GF109203X in fact exaggerated the stimulation of ERK observed during phagocytosis (Fig. 6C). Together, these findings implicate the DAG-Ras signaling axis in the regulation of ERK1/2 activity elicited by FcγR engagement.
Rap1 GTPase Is Activated by Diacylglycerol and Modulates Cross-activation of the Complement Receptor CR3 by Fcγ Receptors
In addition to activating Ras, RasGRP3 also serves as an exchange factor for Rap1 (22), another member of the Ras superfamily of GTPases. Rap1 regulates phagocytosis in Dictyostelium (44) and is required for complement receptor-mediated phagocytosis (45). We previously demonstrated that engagement of FcγR cross-activates the complement receptor, CR3, a member of the integrin family, enabling cells to recognize and bind complement-opsonized particles (32). Moreover, this cross-activation requires a DAG-dependent signaling pathway (32). Therefore, we speculated that Rap1 may control FcγR-mediated cross-activation of CR3 receptors in a DAG-dependent manner.
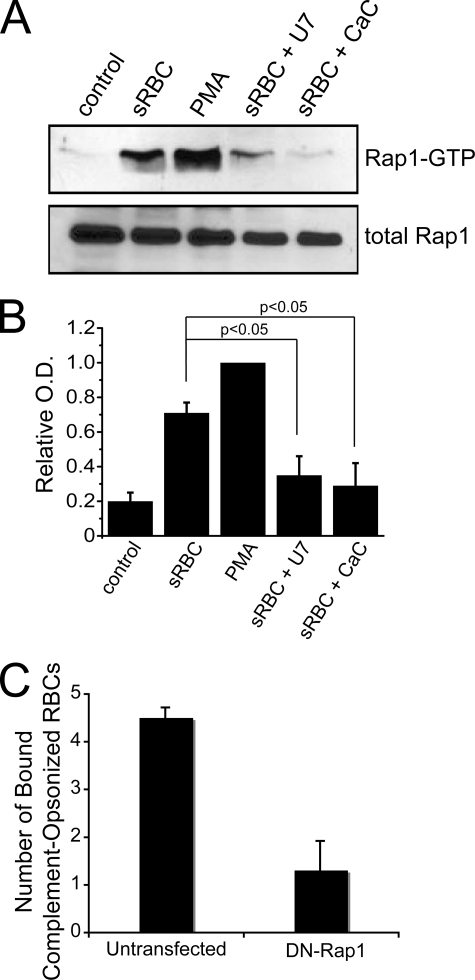
To test this hypothesis, we measured Rap1 activation by affinity precipitation with a GST fusion of the Rap1/GTP-binding domain of RalGDS (Fig. 7A). As expected, PMA, a DAG-mimetic compound, strongly stimulated Rap1 activation. Similarly, FcγR-mediated phagocytosis of IgG-opsonized RBCs promoted Rap1 activation (Fig. 7A). Importantly, the PLC inhibitor U73122 and the DAG antagonist, calphostin C, potently blocked Rap1 stimulation after Fcγ receptor engagement (Fig. 7, A and B). Data of three similar experiments are quantified in Fig. 7B. Together, the data suggest that Rap1 activation during FcγR-mediated phagocytosis requires DAG.
FIGURE 7.
Role of DAG-activated Rap1 in cross-activation of complement receptors. A, Rap1 activation in RAW cells. Activation was probed using a GST fusion of the Rap1-GTP-binding domain of Ral1 in a pull-down assay described under “Experimental Procedures.” Cells were either unstimulated (Control), treated with PMA, or allowed to undergo 5-min phagocytosis of IgG-opsonized sheep RBCs either without (sRBC) or with treatment with U73122 (U7) or calphostin C (CaC). Loading was controlled by immunoblotting for total Rap1. B, quantification of Rap1 activation. Quantification was performed by densitometry of scanned films using Image J software. To enable comparison between experiments, all values were normalized to the intensity of PMA-treated cells. Data are means ± S.E. *, p < 0.05 compared with untreated cells undergoing phagocytosis using Student's t test. C, Rap1 requirement for CR3 cross-activation by Fcγ receptors. Cells were transfected with or without dominant negative Rap1 (DN-Rap1) and activated with aggregated IgG. Subsequently, the number of complement-opsonized RBCs bound to the macrophages was quantified after 30 min, as described in Ref. 32. Data shown are means ± S.E. from three different experiments.
We then tested whether Rap1 played a direct role in the cross-activation of CR3 receptors induced by FcγR clustering. To this effect, we cross-linked FcγRs with aggregated IgG and followed the ability of cells to adhere to complement-opsonized sheep RBCs in the presence or absence of a dominant negative allele of Rap1 (Fig. 7C). Notably, we found a striking inhibitory effect in cells that expressed the dominant negative Rap1. Although cells that expressed only the endogenous Rap1 bound on average 4.4 ± 0.2 complement-opsonized RBCs/cell, macrophages expressing dominant negative Rap1 bound only 1.3 ± 0.6 complement-opsonized RBCs/cell (Fig. 7C). This result strongly suggests that Rap1 plays an integral role in the cross-activation of complement receptors during FcγR engagement (Fig. 8).
FIGURE 8.
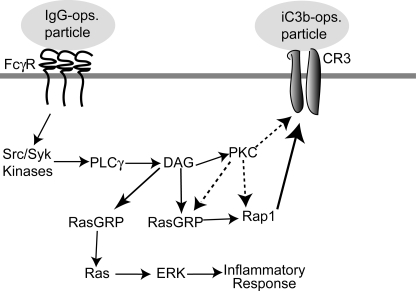
A model for DAG-dependent activation of Ras and Rap1. FcγR clustering induces activation of Src/Syk kinases, which triggers PLCγ and the subsequent release of DAG. We envision the DAG-mediated signal ramifying into the recruitment and activation of PKCs and RasGRPs. Stimulated RasGRPs then convert Ras and Rap1 to their active forms. In our model, Ras activation is independent of PKC and modulates ERK activity and the subsequent inflammatory response, although other signals may contribute to ERK regulation. Rap1, in turn, governs cross-activation of complement and possibly other receptors. However, since cross-activation requires PKC (32), it is unclear whether Rap1 activation needs PKC or if PKC and Rap1 act as parallel but complementary and indispensable activities in this cross-activation.
DISCUSSION
RasGRPs were initially identified in neuronal tissues (46), but members of this family of Ras nucleotide exchange factors were subsequently shown to be expressed and functionally relevant in immune cells (23, 25, 26, 47). Indeed, interference with DAG metabolism in T cells alters Ras signaling, presumably by modulation of RasGRP activity (48, 49). Moreover, ablation of genes encoding members of the RasGRP family results in abnormal lymphocyte development and function and impaired activation of the Ras-ERK signaling axis following stimulation of immune receptors (24, 23, 49).
The experiments described here indicate that RasGRP exchangers are also expressed in macrophages and are important during the course of the innate immune response. RasGRP1 and -3 were identified in RAW cells by RT-PCR, but only RasGRP3 was detectable by immunoblotting. This may indicate that the translation of RasGRP1 is inefficient or that the protein is not stable. However, it is conceivable that macrophages express a splice variant that does not contain the epitope recognized by the monoclonal antibody used. In accordance with this interpretation, we found that two separate products were amplified when using brain mRNA and primers encompassing nucleotides 38–764 of RasGRP1. The larger of these products was predicted on the basis of the published sequence, but the smaller one was unexpected. Of note, only the latter was detectable in RAW cells. The smaller product is unlikely to be an artifact, since RasGRP1 was clearly detected in both brain and RAW cells when using a different primer set (Fig. 1). Therefore, the expression of RasGRP1 in macrophages remains uncertain, and the possible existence of splice variants of this protein needs to be explored in more detail.
Ras is activated in cells stimulated by IgG-opsonized particles, at least in part, by a DAG-mediated process that does not appear to depend on PKC (Fig. 8). Three important findings indicate that RasGRP3 contributes to this response. First, this exchange factor is abundantly expressed in macrophages (Fig. 1) and was found to associate with nascent phagosomes (Fig. 2). Second, RasGRP3 recruitment coincided with the stimulation of Ras. Third, Ras stimulation was abrogated by DAG antagonists, which presumably prevented DAG binding to the C1 domain of RasGRP3 (Fig. 5A). Our results suggest that RasGRP3 may activate Ras independently of PKC signaling during FcγR-mediated phagocytosis in macrophages. This is in contrast to evidence suggesting that PKC-mediated phosphorylation of RasGRP3 is required for full activation in B cells (50, 51). Possibly, non-PKC protein kinases can activate RasGRP3 in RAW cells during phagocytosis.
It is important to note that although our results implicate RasGRP3 in the activation of Ras, they do not exclude other stimulatory pathways. To the extent that FcγR signaling is initiated by tyrosine phosphorylation, one might anticipate that the canonical SOS pathway would contribute also to the activation of Ras during phagocytosis. In fact, we can readily detect recruitment to forming phagosomes of Grb2 (supplemental Fig. S2), an adaptor protein that links SOS with phosphotyrosine moieties of activated receptor complexes. We were unable to express full-length SOS in macrophages, but a more stable construct containing its catalytic domain was found to associate with nascent phagosomes very weakly (supplemental Fig. S2). Nevertheless, based on the involvement of SOS in TCR, BCR, and FcμR signaling, SOS probably contributes also to the localized stimulation of Ras during phagocytosis (Fig. 4C). A similar co-activation of SOS and RasGRP is thought to occur in TCR signaling (52, 53). In the case of FcγR-mediated phagocytosis by macrophages, however, the contribution of SOS appears to be secondary, to the extent that the activation of Ras was largely abrogated by DAG antagonists. Similarly, Ras/ERK activation following BCR stimulation is also thought to occur predominantly through RasGRP1/RasGRP3, rather than SOS (25).
A predominant role for DAG and RasGRP3 is also consistent with the observed kinetics of Ras activation. Accumulation of RasGTP was conspicuously absent at the earliest stages of pseudopod formation and extension and was apparent only later, becoming most obvious shortly after phagosome sealing. This parallels closely the course of accumulation of DAG (7) and of RasGRP3 on phagosomes but is distinct from the kinetics of association and detachment of Grb2 and of the fragment of SOS described above.
Useful information can also be derived comparing the time of residence of Ras on phagosomes with the course of its activation. N-Ras remained associated with phagosomes for an extended time (>20 min), which clearly exceeded the period of association of the GFP-RBD probe, an index of RasGTP formation. Because the intrinsic GTPase activity of Ras is low, termination of the stimulatory signal alone cannot readily explain the inactivation. Instead, these observations suggest that a RasGAP is responsible for the elimination of RasGTP. In this context, it is noteworthy that IgG-opsonized particles engage not only activating FcγRs but also the inhibitory isoform FcγRIIB. Cross-linking of these receptors induces the recruitment of p62dok, a RasGAP-binding protein (54). The ensuing recruitment of RasGAP could readily explain the observed rapid termination of Ras signaling, despite the continued presence of N-Ras on the phagosomal membrane.
Particle internalization and phagosome maturation are clearly independent of Ras. There is precedent for FcγR-mediated activation of signaling pathways that do not appear to control particle internalization. For example, activation of phosphatidylinositol 3-kinase and of ERK in monocytes (55, 56), elevation of cytosolic [Ca+2] in macrophages (57), and stimulation of receptor ubiquitylation (1) are all well documented to occur after FcγR-receptor engagement but they are all dispensable for phagocytosis itself. These findings are not necessarily superfluous or paradoxical, because, in parallel with particle engulfment, FcγRs trigger an array of ancillary reactions. Notable among these are the respiratory burst, the generation of nitric oxide, and the synthesis and secretion of a variety of inflammatory mediators (3, 4). FcγR engagement leads to ERK- and/or NFκB-dependent expression of inflammatory gene products (42, 43, 55, 58). In this study, we found that FcγR-induced stimulation of ERK requires Ras and is in part dependent on DAG signaling. Because ERK lies upstream of the pathway leading to the activation of NFκB and AP-1, which in turn control the expression of tumor necrosis factor α and other cytokines in myeloid cells (42, 43, 55, 58), we propose that the activation of Ras by FcγR is a component of the inflammatory response. It will be of interest to determine whether Ras fails to be activated by other types of phagocytic stimuli, such as complement-opsonized particles, which are not inflammatory.
RasGRP3 increases the rate of nucleotide exchange not only of Ras but also Rap GTPases (22). Rap proteins have been detected in the endo/phagocytic compartment (59) and have been shown to play a role in phagocytosis in Dictyostelium (44). In mammalian phagocytes, they may contribute to phagocytosis by mediating the activation of integrins, as shown in lymphoid cells (60, 61). Indeed, stimulation of FcγR in macrophages activates the complement receptor CR3, an integrin, and this response involves PLC (32). Strikingly, in cells stimulated by IgG-opsonized RBCs, we detected activation of Rap1 by a process that required PLC activity and DAG. Cross-stimulation of CR3 integrins also involves PKC (32). This implies that either Rap1 activation requires two different targets of DAG, namely RasGRP3 and PKC, or that integrin stimulation is accomplished through two parallel DAG signals, one consisting of the RasGRP3-Rap1 axis and the other involving PKC (Fig. 8). Moreover, one might have predicted that a dominant negative mutant of Ras would also block activation of Rap1 by RasGRP3, in as much as the dominant negative protein acts by sequestering the exchange factor in an inactive complex. However, H-RasN17 may only block RasGRP3 activation of Ras in membrane microdomains that serve as Ras signaling platforms, whereas the dominant negative Rap1 protein might selectively block Rap1 signaling in Rap1-specific microdomains.
In summary, we provide evidence for the presence, activation, and functional role of RasGRPs, a family of DAG-dependent exchange factors for Ras and Rap1, during FcγR-mediated signaling in macrophages. Because of their likely role in the activation of gene transcription, RasGRP3 and other members of the RasGRP family may become a target for the development of anti-inflammatory agents and for the design of strategies to manage immune-mediated diseases like arthritis and lupus.
Supplementary Material
Acknowledgments
We thank Donna Mojdami and Analyn Yu for help performing some of the experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants GM55279 and CA116034 (to the laboratory of M. R. P.). This work was also supported by Canadian Institutes of Health Research (CIHR) grant 7075. Work in the laboratory of R. E. H. was supported by CIHR.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Supported by the Natural Sciences and Engineering Research Council of Canada.
Holder of the Pitblado Chair in Cell Biology.
- FcγR
- Fcγ receptor
- DAG
- diacylglycerol
- ERK
- extracellular signal-regulated kinase
- pERK
- phosphorylated ERK
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PKC
- protein kinase C
- PLC
- phospholipase C
- PMA
- phorbol 12-myristate 13-acetate
- RBC
- red blood cell
- RBD
- RasGTP-binding domain of Raf1
- RT
- reverse transcription.
REFERENCES
- 1.Booth J. W., Kim M. K., Jankowski A., Schreiber A. D., Grinstein S. (2002) EMBO J. 21, 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen L. A., Aderem A. (1996) Curr. Opin. Immunol. 8, 36–40 [DOI] [PubMed] [Google Scholar]
- 3.Daëron M. (1997) Annu. Rev. Immunol. 15, 203–234 [DOI] [PubMed] [Google Scholar]
- 4.Gessner J. E., Heiken H., Tamm A., Schmidt R. E. (1998) Ann. Hematol. 76, 231–248 [DOI] [PubMed] [Google Scholar]
- 5.Castellano F., Chavrier P., Caron E. (2001) Semin. Immunol. 13, 347–355 [DOI] [PubMed] [Google Scholar]
- 6.Greenberg S., Grinstein S. (2002) Curr. Opin. Immunol. 14, 136–145 [DOI] [PubMed] [Google Scholar]
- 7.Botelho R. J., Teruel M., Dierckman R., Anderson R., Wells A., York J. D., Meyer T., Grinstein S. (2000) J. Cell Biol. 151, 1353–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox D., Tseng C. C., Bjekic G., Greenberg S. (1999) J. Biol. Chem. 274, 1240–1247 [DOI] [PubMed] [Google Scholar]
- 9.Marshall J. G., Booth J. W., Stambolic V., Mak T., Balla T., Schreiber A. D., Meyer T., Grinstein S. (2001) J. Cell Biol. 153, 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Della Bianca V., Grzeskowiak M., Dusi S., Rossi F. (1993) J. Leukocyte Biol. 53, 427–438 [DOI] [PubMed] [Google Scholar]
- 11.Della Bianca V., Grzeskowiak M., Rossi F. (1990) J. Immunol. 144, 1411–1417 [PubMed] [Google Scholar]
- 12.Cheeseman K. L., Ueyama T., Michaud T. M., Kashiwagi K., Wang D., Flax L. A., Shirai Y., Loegering D. J., Saito N., Lennartz M. R. (2006) Mol. Biol. Cell 17, 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg S., el Khoury J., di Virgilio F., Kaplan E. M., Silverstein S. C. (1991) J. Cell Biol. 113, 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karimi K., Gemmill T. R., Lennartz M. R. (1999) J. Leukocyte Biol. 65, 854–862 [DOI] [PubMed] [Google Scholar]
- 15.Larsen E. C., DiGennaro J. A., Saito N., Mehta S., Loegering D. J., Mazurkiewicz J. E., Lennartz M. R. (2000) J. Immunol. 165, 2809–2817 [DOI] [PubMed] [Google Scholar]
- 16.Oancea E., Teruel M. N., Quest A. F., Meyer T. (1998) J. Cell Biol. 140, 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen E. C., Ueyama T., Brannock P. M., Shirai Y., Saito N., Larsson C., Loegering D., Weber P. B., Lennartz M. R. (2002) J. Cell Biol. 159, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brumell J. H., Howard J. C., Craig K., Grinstein S., Schreiber A. D., Tyers M. (1999) J. Immunol. 163, 3388–3395 [PubMed] [Google Scholar]
- 19.Kazanietz M. G., Lewin N. E., Bruns J. D., Blumberg P. M. (1995) J. Biol. Chem. 270, 10777–10783 [DOI] [PubMed] [Google Scholar]
- 20.Areces L. B., Kazanietz M. G., Blumberg P. M. (1994) J. Biol. Chem. 269, 19553–19558 [PubMed] [Google Scholar]
- 21.Lorenzo P. S., Beheshti M., Pettit G. R., Stone J. C., Blumberg P. M. (2000) Mol. Pharmacol. 57, 840–846 [PubMed] [Google Scholar]
- 22.Yamashita S., Mochizuki N., Ohba Y., Tobiume M., Okada Y., Sawa H., Nagashima K., Matsuda M. (2000) J. Biol. Chem. 275, 25488–25493 [DOI] [PubMed] [Google Scholar]
- 23.Ebinu J. O., Stang S. L., Teixeira C., Bottorff D. A., Hooton J., Blumberg P. M., Barry M., Bleakley R. C., Ostergaard H. L., Stone J. C. (2000) Blood 95, 3199–3203 [PubMed] [Google Scholar]
- 24.Dower N. A., Stang S. L., Bottorff D. A., Ebinu J. O., Dickie P., Ostergaard H. L., Stone J. C. (2000) Nat. Immunol. 1, 317–321 [DOI] [PubMed] [Google Scholar]
- 25.Oh-hora M., Johmura S., Hashimoto A., Hikida M., Kurosaki T. (2003) J. Exp. Med. 198, 1841–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teixeira C., Stang S. L., Zheng Y., Beswick N. S., Stone J. C. (2003) Blood 102, 1414–1420 [DOI] [PubMed] [Google Scholar]
- 27.Bivona T. G., Pérez De Castro I., Ahearn I. M., Grana T. M., Chiu V. K., Lockyer P. J., Cullen P. J., Pellicer A., Cox A. D., Philips M. R. (2003) Nature 424, 694–698 [DOI] [PubMed] [Google Scholar]
- 28.Lorenzo P. S., Kung J. W., Bottorff D. A., Garfield S. H., Stone J. C., Blumberg P. M. (2001) Cancer Res. 61, 943–949 [PubMed] [Google Scholar]
- 29.Prior I. A., Harding A., Yan J., Sluimer J., Parton R. G., Hancock J. F. (2001) Nat. Cell Biol. 3, 368–375 [DOI] [PubMed] [Google Scholar]
- 30.Chiu V. K., Bivona T., Hach A., Sajous J. B., Silletti J., Wiener H., Johnson R. L., 2nd, Cox A. D., Philips M. R. (2002) Nat. Cell Biol. 4, 343–350 [DOI] [PubMed] [Google Scholar]
- 31.McLeod S. J., Ingham R. J., Bos J. L., Kurosaki T., Gold M. R. (1998) J. Biol. Chem. 273, 29218–29223 [DOI] [PubMed] [Google Scholar]
- 32.Jongstra-Bilen J., Harrison R., Grinstein S. (2003) J. Biol. Chem. 278, 45720–45729 [DOI] [PubMed] [Google Scholar]
- 33.de Rooij J., Bos J. L. (1997) Oncogene 14, 623–625 [DOI] [PubMed] [Google Scholar]
- 34.Park R. K., Erdreich-Epstein A., Liu M., Izadi K. D., Durden D. L. (1999) J. Immunol. 163, 6023–6034 [PubMed] [Google Scholar]
- 35.Patel J. C., Hall A., Caron E. (2002) Mol. Biol. Cell 13, 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massol P., Montcourrier P., Guillemot J. C., Chavrier P. (1998) EMBO J. 17, 6219–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choy E., Chiu V. K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I. E., Philips M. R. (1999) Cell 98, 69–80 [DOI] [PubMed] [Google Scholar]
- 38.Bivona T. G., Philips M. R. (2003) Curr. Opin. Cell Biol. 15, 136–142 [DOI] [PubMed] [Google Scholar]
- 39.Prior I. A., Hancock J. F. (2001) J. Cell Sci. 114, 1603–1608 [DOI] [PubMed] [Google Scholar]
- 40.Yeung T., Terebiznik M., Yu L., Silvius J., Abidi W. M., Philips M., Levine T., Kapus A., Grinstein S. (2006) Science 313, 347–351 [DOI] [PubMed] [Google Scholar]
- 41.Matallanas D., Arozarena I., Berciano M. T., Aaronson D. S., Pellicer A., Lafarga M., Crespo P. (2003) J. Biol. Chem. 278, 4572–4581 [DOI] [PubMed] [Google Scholar]
- 42.Rose D. M., Winston B. W., Chan E. D., Riches D. W., Gerwins P., Johnson G. L., Henson P. M. (1997) J. Immunol. 158, 3433–3438 [PubMed] [Google Scholar]
- 43.Trotta R., Kanakaraj P., Perussia B. (1996) J. Exp. Med. 184, 1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seastone D. J., Zhang L., Buczynski G., Rebstein P., Weeks G., Spiegelman G., Cardelli J. (1999) Mol. Biol. Cell 10, 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caron E., Self A. J., Hall A. (2000) Curr. Biol. 10, 974–978 [DOI] [PubMed] [Google Scholar]
- 46.Ebinu J. O., Bottorff D. A., Chan E. Y., Stang S. L., Dunn R. J., Stone J. C. (1998) Science 280, 1082–1086 [DOI] [PubMed] [Google Scholar]
- 47.Yang Y., Li L., Wong G. W., Krilis S. A., Madhusudhan M. S., Sali A., Stevens R. L. (2002) J. Biol. Chem. 277, 25756–25774 [DOI] [PubMed] [Google Scholar]
- 48.Topham M. K., Prescott S. M. (2001) J. Cell Biol. 152, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong X. P., Hainey E. A., Olenchock B. A., Zhao H., Topham M. K., Koretzky G. A. (2002) J. Biol. Chem. 277, 31089–31098 [DOI] [PubMed] [Google Scholar]
- 50.Aiba Y., Oh-hora M., Kiyonaka S., Kimura Y., Hijikata A., Mori Y., Kurosaki T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16612–16617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Y., Liu H., Coughlin J., Zheng J., Li L., Stone J. C. (2005) Blood 105, 3648–3654 [DOI] [PubMed] [Google Scholar]
- 52.Gong Q., Cheng A. M., Akk A. M., Alberola-Ila J., Gong G., Pawson T., Chan A. C. (2001) Nat. Immunol. 2, 29–36 [DOI] [PubMed] [Google Scholar]
- 53.Caloca M. J., Zugaza J. L., Matallanas D., Crespo P., Bustelo X. R. (2003) EMBO J. 22, 3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamanashi Y., Tamura T., Kanamori T., Yamane H., Nariuchi H., Yamamoto T., Baltimore D. (2000) Genes Dev. 14, 11–16 [PMC free article] [PubMed] [Google Scholar]
- 55.García-García E., Sánchez-Mejorada G., Rosales C. (2001) J. Leukocyte Biol. 70, 649–658 [PubMed] [Google Scholar]
- 56.Karimi K., Lennartz M. R. (1998) Inflammation 22, 67–82 [DOI] [PubMed] [Google Scholar]
- 57.Di Virgilio F., Meyer B. C., Greenberg S., Silverstein S. C. (1988) J. Cell Biol. 106, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drechsler Y., Chavan S., Catalano D., Mandrekar P., Szabo G. (2002) J. Leukocyte Biol. 72, 657–667 [PubMed] [Google Scholar]
- 59.Pizon V., Desjardins M., Bucci C., Parton R. G., Zerial M. (1994) J. Cell Sci. 107, 1661–1670 [DOI] [PubMed] [Google Scholar]
- 60.Reedquist K. A., Ross E., Koop E. A., Wolthuis R. M., Zwartkruis F. J., van Kooyk Y., Salmon M., Buckley C. D., Bos J. L. (2000) J. Cell Biol. 148, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bivona T. G., Wiener H. H., Ahearn I. M., Silletti J., Chiu V. K., Philips M. R. (2004) J. Cell Biol. 164, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.