Abstract
The small leucine-rich repeat proteins, fibromodulin and osteoadherin, have N-terminal extensions with a variable number of O-sulfated tyrosine residues. This modification combined with a number of aspartic and glutamic acid residues results in a highly negatively charged domain of less than 30 amino acids. We hypothesized that this domain shares functional properties with heparin regarding binding to proteins and polypeptides containing clusters of basic amino acids. Two other family members, PRELP and chondroadherin, have distinctly different clusters of basic amino acids in their N and C termini, respectively, and PRELP is known to bind to heparin via this domain. Another heparin-binding protein is the cytokine Oncostatin M, with a different cluster of basic amino acids in its C terminus. We used polypeptides representing these basic domains in solid phase assays and demonstrate interactions with the negatively charged N-terminal domain of fibromodulin and full-length osteoadherin. The tyrosine sulfate domains also bound heparin-binding proteins such as basic fibroblast growth factor-2, thrombospondin I, MMP13, the NC4 domain of collagen IX, and interleukin-10. Fibronectin with large heparin-binding domains did not bind, neither did CILP containing a heparin-binding thrombospondin type I motif without clustered basic amino acids. Affinity depends on the number and position of the sulfated tyrosine residues shown by different binding properties of 10-kDa fragments subfractionated by ion-exchange chromatography. These interactions may sequester growth factors, cytokines, and matrix metalloproteinases in the extracellular matrix as well as contribute to its organization.
The integrity of the extracellular matrix depends on a multitude of interactions between molecular constituents leading to the formation of major macromolecular assemblies important for tissue functions. A major component in most types of extracellular matrix is the network of fibrillar structures primarily composed of collagen I in fibrous tissues and bone, whereas cartilage contains the similar collagen II.
These collagen fibrils contain a number of associated molecules, often bound to their surface. One such molecule is the distinct collagen IX, containing three triple helical domains each surrounded by non-triple helical domains. Another set of molecules binding to triple helical collagen is the members of the small leucine-rich repeat protein family, such as fibromodulin (1), lumican (2), decorin (3), biglycan (4), PRELP (5), chondroadherin (6), and possibly osteoadherin. The typical LRR3 protein contains 10–11 repeats of some 25 amino acids with leucine residues at conserved locations. This domain represents a common denominator for the family and contains structures providing for interaction with, e.g. triple helical collagen (7–9). The LRR proteins contain an extension at either the N- or C-terminal end or, in a few cases, at both ends. These extensions may contribute to a second function exemplified by PRELP, where the N-terminal with a stretch of clustered arginine residues provides binding to heparin/heparan sulfate containing optimally five or more disaccharides with three sulfate groups each (10). In decorin and biglycan, the N-terminal extension have substituents of glycosaminoglycan chains of dermatan/chondroitin sulfate that can contribute to collagen binding (11) as well as provide putative self interactions with a similar chain on another molecule. In particular, it has been shown that decorin and biglycan will bind via their protein core to the N-terminal globular domain of collagen VI (4) and direct the formation of the collagen VI-beaded filament network, provided that the glycosaminoglycan chains are intact.
There are a number of proteins known to interact with heparin. Whereas heparin is not present in the extracellular matrix, these proteins may bind to stretches within the heparan sulfate chains enriched in disaccharides having high sulfate content. The heparan sulfate is found particularly as a component of cell surface proteoglycans such as glypicans (12) and syndecans (13) and of the extracellular matrix proteoglycans perlecan (14) and agrin (15). Important ligands for these chains are growth factors exemplified by members of the FGF family. Other molecules that bind to heparan sulfate include fibronectin, having two such domains with molecular weights of around 20,000 (16). Also several members of the metalloproteinase family contain heparin-binding motifs as do many cytokines.
The most common heparin-binding sequence contains clusters of basic amino acids, often with additional proline residues. PRELP and chondroadherin as well as the other proteins mentioned represent examples having such sequences. A different type of motif, first found in thrombospondin I, contains consecutive repeats of a WXZ sequence, where the tryptophan may be mannosylated (17, 18). This is referred to as the thrombospondin type I motif heparin-binding structure. Thrombospondin I in addition contains a heparin-binding basic cluster of amino acids (19). CILP on the other hand only contains the thrombospondin type 1 motif. These domains can bind to heparin with high affinity, an interaction that can be disrupted by high salt.
A very different type of extension is found in the N-terminal part of fibromodulin and osteoadherin. These proteins contain a number of tyrosine residues, which may and often do, carry a sulfate group. Thus, fibromodulin contains up to nine such residues and osteoadherin as many as eight, where six are located in the N-terminal and two in the C-terminal extension (20). Any given preparation contains molecules within the same species with a range of levels of O-sulfate-substituted tyrosine. The functional significances of these domains have been unknown. We now show that these domains can mimic heparin in several interactions.
EXPERIMENTAL PROCEDURES
Synthetic Heparin-binding Peptides
Three peptides corresponding to the heparin-binding motif of the N-terminal sequence of PRELP (QPTRRPRPGTGPGRRPRPRPRPTPC, 2847.3 Da) referred to as PRELP-QPTR, the C-terminal sequence of chondroadherin (CKFPTKRSKKAGRH, 1644 Da) referred to as CHAD-CKF, and the basic sequence close to the C-terminal of Oncostatin M (CLRKGVRRTRPSRKGKRLMTRG, 2613.2 Da) referred to as OSM-CLRK, were commercially synthesized by Schafer-N, Denmark. Two of the peptides were also synthesized containing a C-terminal biotin in the case of the PRELP peptide (PRELP-QPTR-biotin) and an N-terminal biotin in the case of the chondroadherin peptide (CHAD-CKF-biotin).
Binding of the Heparin-binding Peptides PRELP-QPTR, CHAD-CKF, and OSM-CLRK to Heparin
The synthetic peptides PRELP-QPTR, OSM-CLRK, and CHAD-CKF at 1 mg/ml in PBS (5 mm sodium phosphate, 0.15 m NaCl, pH 7.4) were applied onto a 1-ml HiTrap heparin column (Amersham Biosciences) equilibrated in PBS. Bound peptides were eluted with a 40-ml salt gradient of 0.15 to 2 m NaCl in 5 mm NaH2PO4, pH 7.4. Absorbance at 215 nm was recorded, and fractions of 0.6 ml were collected. The identity and content of peptides in fractions corresponding to the peaks were analyzed by MALDI-TOF MS as described below.
Generation and Purification of the N-terminal Part of Bovine Fibromodulin (Gln19–Lys98) the 10-kDa Tyrosine Sulfate Fragment
This fragment was prepared by digestion of fibromodulin purified from bovine cartilage (21) with endoproteinase Lys-C (Wako Chemicals GmbH, Germany) as described (22). Digestion was stopped by adding 6 m Urea, 10 mm Tris, pH 7.6, to adjust the solution to conditions of the equilibration buffer for subsequent anion exchange chromatography. The sample was applied onto a 1-ml Mono Q column (Amersham Biosciences) attached to an Äkta high-performance liquid chromatography system (Amersham Biosciences) and eluted with a 10-ml salt gradient from 0 to 0.22 m NaCl followed by a 30-ml salt gradient from 0.22 to 1.5 m NaCl in the Urea-Tris buffer. Fractions of 1 ml were collected. Identification was done by desalting aliquots on C18 stage tips (EmporeTM extraction disk C18, 3M, Minneapolis, MN) eluted with 50% acetonitrile onto an anchorchip target plate for MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) containing spots of 2,5-dihydroxybenzoic acid matrix. Mass spectrometry was performed with a Bruker reflex III MALDI-TOF MS to identify peptides present.
Six pools were defined from the chromatogram (fig 5) and desalted separately on a column of Sephasil C8 (Amersham Biosciences) attached to a SMARTTM micro-high-performance liquid chromatography system (Amersham Biosciences). Bound polypeptides were eluted and fractionated (100 μl/fraction) by a gradient of 0 to 100% acetonitrile in 0.1% trifluoroacetic acid. These fractions were analyzed by MALDI-TOF MS to identify polypeptides present. Defined fractions were pooled and freeze dried. Pools 4–6 were dissolved in PBS and stored at −20 °C. Pools 1–3 were dissolved in 6 m Urea, 20 mm Bis-Tris, pH 5.5, and purified further by MiniQ (Amersham Biosciences) anion-exchange chromatography using the SMART system. Eluted fractions were analyzed as described above with MALDI-TOF MS. The major portion (3 of 4) of the selected fractions was desalted on Sephasil C8 as described. After analyses of these fractions by MALDI-TOF mass spectrometry, selected fractions were pooled, freeze dried, and re-dissolved in PBS.
FIGURE 5.
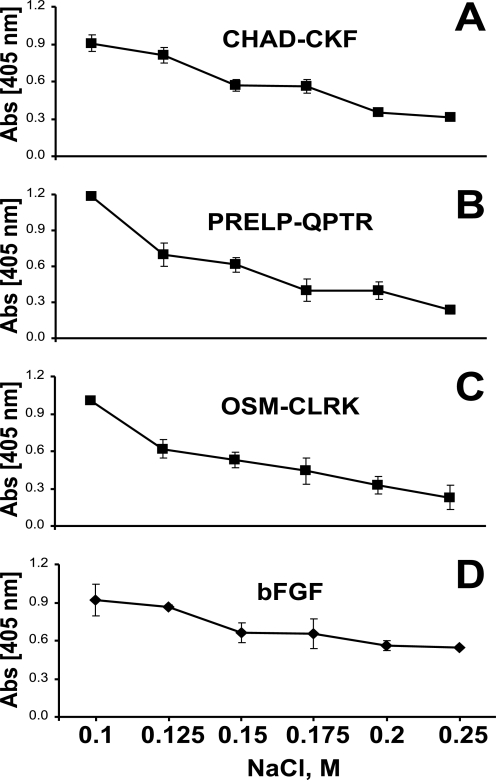
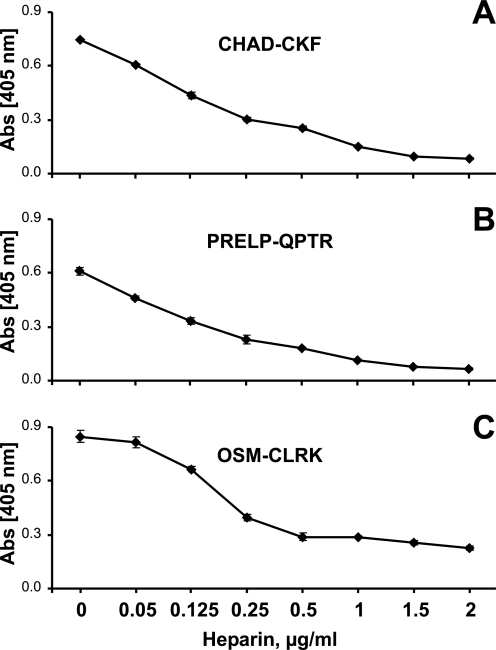
Interaction of the tyrosine sulfate-rich domains of fibromodulin with heparin-binding motifs of chondroadherin, PRELP, Oncostatin M, and bFGF in presence of sodium chloride. The N-terminal of fibromodulin was coated onto a plastic surface overnight, and the remaining sites were blocked with ovalbumin. The three heparin-binding peptides CHAD-CKF (A), PRELP-QPTR (B), OSM-CLRK (C), and basic-FGF (D) were added to coated wells in the presence of sodium chloride at different concentrations. Any bound peptide was detected with alkaline phosphate-conjugated streptavidin and any bound protein with specific antiserum.
Generation and Purification of the Bovine Fibromodulin (Asn33–Arg74) 5-kDa Tyrosine Sulfate Fragment
Bovine fibromodulin was digested in TBS (25 mm Tris, 0.15 m NaCl, pH 7.4) overnight at 37 °C with trypsin (Sigma-Aldrich). Trypsin activity was then inhibited by adding Nα-p-tosyl-l-lysine chloromethyl ketone-HCl (Sigma-Aldrich), and the sample was further incubated for 30 min at 37 °C. To remove keratan sulfate, interfering with subsequent purification, fibromodulin was de-glycosylated by adding N-glycosidase F (Roche Applied Science) and incubated overnight at 37 °C as described (22). This enzyme activity was inhibited by adding 4 volumes of 6 m Urea, 10 mm Tris, pH 7.6. Purification and desalting of this 5-kDa fibromodulin fragment rich in tyrosine sulfate was performed using the conditions described for the 10-kDa fragment for separations on Mono Q and Sephasil C8.
Expression and Purification of the N-terminal Domain of Bovine Fibromodulin in E. coli
The plasmid pET27b(+) (Novagen) with cDNA from bovine fibromodulin (8) was used as a template to amplify the N-terminal of fibromodulin with the primer (5′-AATCCATGGGGCAATATGAGGAAGAC- 3′ and 5′-AATGCTAGCGGGCAGGTACTTGAGATTG-3′) corresponding to nucleotides 116–361 with flanking NcoI and NheI sites. The generated fragment was ligated into the PCRscript Amp sk(+) vector and cloned into XL10-Gold ultracompetent cells according to the manual of the manufacturer (Qiagen). One positive clone was digested with the enzymes NcoI and NheI and subcloned into the vector pET-27b(+). The construct was amplified in JM 110 E. coli (Stratagene) followed by transformation into the protein expression system Rosetta GamiTM 2 E. coli (Novagen). The transfected cells were grown in 2xTY (1.6% tryptone, 1% yeast extract, 0.5% NaCl) medium with kanamycin sulfate (50 μg/ml) added (Sigma-Aldrich). After isopropyl 1-thio-β-d-galactopyranoside induction the expressed N-terminal of fibromodulin were extracted from the periplasm according to the manual of the manufacturer.
Extracted fragment of the N-terminal of fibromodulin was purified by Ni2+ affinity to a HisTrapTM HP column (Amersham Biosciences). Unbound sample was removed by washing with 10 column volumes of 50 mm Tris, pH 8.0, 0.5 m NaCl. Bound material was eluted with 10 column volumes of a 0 to 0.5 m gradient of imidazole in wash buffer.
Fractions were analyzed by SDS-PAGE Tris/Tricine. Bands of interest were cut out from gel and analyzed for fragment present by in-gel trypsin digestion and MALDI-TOF MS (23).
Fractions containing the N-terminal fragment of fibromodulin were pooled, diluted, and dialyzed into 25 mm Bis-Tris, pH 6.0, and applied onto a 1-ml anion exchange Mono Q column attached to an Äkta high-performance liquid chromatography system. Bound fragments were eluted by a 20-ml salt gradient from 0 to 1.0 m NaCl in Bis-Tris buffer. Fractions were analyzed by SDS-PAGE Tris/Tricine. Fractions collected (1 ml) containing the fragment were pooled and diluted four times with 25 mm Tris, pH 7.4, and enzymatically digested with endoproteinase Lys-C overnight at 37 °C to remove the His tag.
The digest was applied onto a reversed-phase column Source 5, and bound fragments were eluted and fractionated by a 20-column volume gradient of 0–100% acetonitrile in 0.1% trifluoroacetic acid (0.65 ml). The fractions were analyzed by MALDI-TOF MS to identify polypeptides present. Defined fractions were pooled, freeze dried, and dissolved in PBS.
A portion of the purified fragment was enzymatically cleaved with trypsin overnight at 37 °C and applied onto the Source 5 reversed-phase column as described above. Fragments present were identified by MALDI-TOF MS and the polypeptide corresponding to the non-sulfated 5-kDa fragment was pooled, freeze dried, and dissolved in PBS.
Expression and Purification of the NC4 Domain of Human Collagen Type IX in E. coli
A vector containing an E. coli codon bias optimized cDNA sequence coding for the NC4 domain of collagen IX was used as template to amplify the sequence with the primer forward 5′-CACCTCTGCGGCGGTGAAACGT-3′ and reverse 5′-CCTTATTACTGGCTCGGGGTAA-3′. The generated PCR fragment was ligated into the vector pET200/D-TOPO (Invitrogen) and amplified in TOP10 E. coli (Invitrogen). The sequence identity was confirmed, and the sequence was cloned into the expression vector pAM 104-2 and transformed into the Rosetta DE3TM E. coli (Novagen) expression system. A single colony was inoculated in Luria Broth with kanamycin sulfate (50 μg/ml), and expression of the protein was induced by isopropyl 1-thio-β-d-galactopyranoside according to the manual of the manufacturer. Generated NC4 domain of collagen IX were extracted from bacteria under native conditions (24) and purified by Ni2+ affinity using a HisTrapTM HP column attached to an ÄKTA high-performance liquid chromatography system as described above.
Biotinylation of the N-terminal Fibromodulin 10-kDa Lys-C (Gln19–Lys98) and the 5-kDa (Asn33–Arg74) Fragments Generated by Trypsin
The purified N-terminal tyrosine sulfate-rich fragments generated by trypsin (5 kDa) and Lys-C (10 kDa), respectively (1 mg/ml in PBS), were mixed with EZ-link-sulfo-NHS biotin, 1 mg/ml (Pierce), and incubated for 1 h at 4 °C. Excess biotin was removed by dialysis (membrane cut-off, 3500 Da), precoated with 1 mg/ml ovalbumin (Sigma-Aldrich), in PBS, for 1 h at 4 °C. The fragments were checked for derivatization with a single biotin by MALDI-TOF MS as described above.
Biotinylation of the Non-sulfated 5-kDa (Asn33–Arg74) Fragments Generated by Trypsin
Purified fragments of non-sulfated 5 kDa from the source 5 column purification were mixed with a 10-fold excess of EZ-link-sulfo-NHS biotin and incubated for 1 h at room temperature, desalted on a Source 5 column, and checked for derivatization as described above.
Biotinylation of the Oncostatin M Peptide
The peptide from the Oncostatin M heparin binding sequence (1 μg/μl in PBS) was mixed with 25 mm EZ link maleimide PEO2 biotin (Pierce) and incubated for 2 h in the dark at room temperature. Excess biotin was removed by using the desalting column NAP 5 (Amersham Biosciences) conditioned with bovine serum albumin (1 mg/ml in PBS). The peptide was checked for derivatization with biotin by MALDI-TOF MS as described above.
125I Radioiodination of Interleukin 10
Radioiodination of IL-10 (5 μg) was performed by using IODO-GEN® precoated iodination tubes (Pierce) according to the manual from the manufacturer. In brief, 100 μl of PBS and 0.5 mCi of Na125I (NEZ033A005MC, PerkinElmer Life Sciences) was added to the IODO-GEN® tube and incubated for 6 min. The solution was transferred to a vial containing IL-10 (0.1 μg/ml) in PBS and incubated for another 9 min. A PD10 column (Amersham Biosciences) was blocked for nonspecific adsorption with bovine serum albumin (2 mg/ml PBS) and equilibrated in PBS. The iodinated sample was applied for desalting, and fractions were collected. Fractions containing desalted 125I-IL-10 were pooled and used in the binding assay.
Interaction of Proteins Containing Heparin-binding Motifs with Fibromodulin and Osteoadherin
The bovine Lys-C-generated N-terminal 10-kDa tyrosine sulfate-rich fragments of fibromodulin and recombinant human osteoadherin, respectively, were coated (2 μg/ml in PBS) onto 96-well microtiter plates (MaxiSorb C96, Nunc, Denmark) overnight at room temperature. Plates were washed with PBS, and remaining sites were blocked with bovine serum albumin (Serva, Heidelberg, Germany) at 2 mg/ml in PBS for 1 h at room temperature. Plates were then extensively washed with PBS, 0.05% Tween 20. Ligands, such as human recombinant PRELP (10), human recombinant PRELP truncated to not contain the heparin-binding domain (10), human fibronectin (purified by use of heparin-Sepharose and gelatin-Sepharose according to routine procedures), bovine recombinant FGF-2 (Upstate, Dundee, UK), bovine antithrombin III (Sigma-Aldrich), bovine thrombospondin I (prepared in the laboratory with purity verified by SDS-PAGE and identity confirmed by antibody reactivity, data not shown), 125I-labeled IL-10 (ProSpec-Tany TechnoGene Ltd., Israel), NC4 domain of collagen IX, and recombinant CILP-1 (25, 26) expressed in 293 EBNA cells without tag (a kind gift from Dr. P. Lorenzo, Lund University) were added (2 μg/ml in PBS, 0.05% Tween 20). All ligands were allowed to interact for 1 h at room temperature. Unbound proteins were removed by extensive washing followed by adding the specific antiserum for each ligand diluted in PBS, 0.05% Tween 20. After 1 h at room temperature bound antibodies were detected with an alkaline phosphatase-conjugated swine anti-rabbit antibody (Dako A/S, Denmark), except when the primary antibody was a monoclonal in the case of FGF-2 (Upstate, Dundee, UK), thrombospondin I (a kind gift from Prof. Deane Mosher, University of Wisconsin), and antithrombin III (Sigma-Aldrich), where a rabbit anti-mouse-alkaline phosphatase-conjugated antibody (Dako A/S, Denmark) was used. Wells with IL-10 were analyzed for radioactivity using a gamma counter. Enzyme activity was measured using 4-nitrophenyl phosphate disodium salt (Serva) as the substrate (1 mg/ml in 1 m diethanolamine, 0.5 mm MgCl2, 3 mm sodium azide, pH 9.8), and monitoring absorbance at 405 nm using the ASYS Hitech Expert 96 microtiter plate reader. All samples were analyzed in triplicates.
In some wells remaining sites were blocked with ovalbumin (1 mg/ml TBS, 0.05% Tween 20, Sigma-Aldrich) for 1 h at 4 °C and extensively washed with TBS, 0.05% Tween 20. The ligand MMP13 (a kind gift from Dr. Peter Mitchell, Pfizer, Groton) was added to these wells at 5 μg/ml in TBS, 0.05% Tween 20, 5 mm CaCl2, 5 mm ZnCl2 and allowed to adhere for 1 h at 4 °C. Unbound proteins were removed by extensive washing with TBS, 0.05% Tween 20, followed by adding specific antibodies to the ligand in TBS, 0.05% Tween 20 for 1 h at 4 °C. Bound antibodies were detected by a rabbit anti-mouse-alkaline phosphatase-conjugated antibody (Dako A/S, Denmark). Enzyme activity was measured as described above.
Interaction of the Tyrosine Sulfate-rich Domains of Fibromodulin and Intact Osteoadherin with Heparin-binding Motifs of Chondroadherin, PRELP, and Oncostatin M
The peptides, C-terminal CHAD-CKF, N-terminal PRELP-QPTR, and C-terminal OSM-CLRK, respectively, were coated onto microtiter plates (Nunc C96, 20 μm in PBS) overnight at room temperature. In other experiments the 10-kDa tyrosine sulfate-rich fragment generated from bovine fibromodulin or intact osteoadherin (2 μg/ml PBS) were similarly coated overnight at room temperature. Plates were washed with PBS and remaining sites were blocked with ovalbumin (1 mg/ml in PBS) for 1 h at room temperature, and extensively washed with PBS, 0.05% Tween 20. Ligands, i.e. the biotin-tagged fibromodulin 10-kDa fragment, the biotin-tagged fibromodulin 5-kDa fragment, or the biotinylated bacterially expressed non-sulfated 5-kDa fragment were added (2 μg/ml in PBS, 0.05% Tween 20) to plates coated with peptides. Alternatively the biotin-tagged CHAD-CKF peptide, biotin-tagged N-terminal PRELP-QPTR peptide- or biotin-tagged OSM-CLRK peptide (2 μg/ml PBS, 0.05% Tween 20) were added to the surfaces coated with fibromodulin fragments or intact osteoadherin. All ligands were allowed to interact for 1 h at room temperature. Unbound peptides were removed by extensive wash with PBS, 0.05% Tween 20 followed by incubation with alkaline phosphatase-conjugated streptavidin (Roche Applied Science) for 1 h at room temperature. After wash, plates were incubated with substrate for 1 h, and enzyme activity was measured, as described above. All samples were analyzed in triplicates.
Interaction of the Tyrosine Sulfate-rich Domains of Fibromodulin with Heparin-binding Motifs of Chondroadherin, PRELP, and Oncostatin M in the Presence of Different Concentrations of Sodium Chloride
The N-terminal fragment of fibromodulin was coated onto microtiter plates (Nunc C96, 2 μg/ml) in PBS overnight at room temperature. Plates were washed with PBS, and remaining sites were blocked with ovalbumin (1 mg/ml in PBS) for 1 h at room temperature followed by washing with PBS. The biotinylated peptides, C-terminal CHAD-CKF, N-terminal PRELP-QPTR, C-terminal OSM-CLRK, and FGF-2, respectively, were added (2 μg/ml) in 6 m Urea, 16 mm NaH2PO4, 0.05% Tween 20, pH 7.4, with sodium chloride present at different concentrations (0.1, 0.125, 0.15, 0.175, 0.2, and 0.25 m) and incubated for 1 h at room temperature. Unbound sample was removed by washing with 16 mm NaH2PO4, 0.1 m NaCl, 0.05% Tween 20, pH 7.4. The presence of biotinylated peptides was detected by alkaline phosphatase-conjugated streptavidin and the presence of protein was detected by a specific antiserum in 16 mm NaH2PO4, 0.1 m NaCl, 0.05% Tween 20, pH 7.4, as described above. All samples were analyzed in triplicates.
Competition between Binding of the Tyrosine Sulfate-rich Domain of Fibromodulin and Heparin to the Heparin-binding Motifs of Chondroadherin, PRELP, and Oncostatin M
The heparin-binding peptides, C-terminal CHAD-CKF, N-terminal PRELP-QPTR, and C-terminal OSM-CLRK, respectively, were coated onto microtiter plates (Nunc C96, 20 μm in PBS) overnight at room temperature. Plates were washed with PBS, and remaining sites were blocked with ovalbumin (1 mg/ml in PBS) for 1 h at room temperature, followed by extensive wash with PBS, 0.05% Tween 20. The N-terminal fragment of fibromodulin was added (2 μg/ml in PBS, 0.05% Tween 20) in the presence of heparin at different concentrations (0.06, 0.12, 0.25, 0.50, 1.0, 1.5, and 2.0 μg/ml) for 1 h at room temperature. Unbound fragments were removed by washing with PBS, 0.05% Tween 20 followed by incubation with an antibody specific for fibromodulin at room temperature for 1 h. After washing, bound antibodies were detected by a swine anti rabbit alkaline phosphatase-conjugated antibody. Enzyme activity was measured as described above. All samples were analyzed in triplicates.
Interactions of the Heparin-binding Peptides with Sub-fractions of the Tyrosine Sulfate-rich Domain Differing in Charge Density
Immunopure streptavidin (Pierce), 0.2 μg/ml in PBS, was coated overnight at room temperature onto MaxiSorb immunoplates (Nunc C96). Unbound protein was removed by washing the plate with PBS, and remaining sites were blocked with ovalbumin (1 mg/ml in PBS) for 1 h at room temperature. After wash with PBS, biotin-tagged CHAD-CKF and PRELP-QPTR, respectively, were incubated with the coated wells for 1 h at room temperature in PBS, 0.05% Tween 20. Wells were washed, and pools (1–5) of the fractionated fibromodulin 10-kDa Lys-C-generated fragment were added at concentrations of 0.2, 1, and 2 μg/ml in PBS, 0.05% Tween 20. After incubation for 1 h at room temperature, the plate was probed with primary antibody to fibromodulin, secondary antibody enzyme conjugate and substrate with washes between incubations, as described above. All analyses were made in triplicates.
Binding of MMP13 to Fractions of the Tyrosine Sulfate Domain of Fibromodulin with Different Sulfation and Charge Properties as Well as to Intact Osteoadherin
MMP13 (10 μg/ml in 50 mm Tris pH 8.0, a kind gift from Dr. Peter Mitchell, Pfizer) was incubated overnight at 4 °C in MaxiSorb immunoplates (Nunc, C96). Non-coated sites were blocked with ovalbumin (1 mg/ml in TBS) for 1 h at 4 °C. Osteoadherin (2 μg/ml), the fibromodulin biotinylated 5- or 10-kDa tyrosine sulfate-rich fragments (2 μg/ml) were added to coated wells in TBS, 0.05% Tween 20, 5 mm CaCl2, 5 mm ZnCl2 1 h at 4 °C. In separate experiment sub-fractions pool 1–5 (at 0.2, 1, and 2 μg/ml) from the Mono Q separation of the 10-kDa fragment were similarly analyzed. Unbound ligands were removed by washing followed by adding diluted antiserum against the specific protein. Bound antibodies were detected using alkaline phosphatase-conjugated swine anti-rabbit IgG as described above. All samples were analyzed in triplicates.
Expression and Purification of Human Osteoadherin
A cDNA fragment of rat osteoadherin (0.5 kb) was used to screen a lambda ZapII human osteoblast library (a kind gift from Dr. Larry Fischer, Institute of Craniofacial and Dental Research, NIH) with the ZAP ExpressionTM Vector kit from Stratagene. Approximately 840,000 plaque-forming recombinants were screened. Eight positive clones were found and re-screened according to the manufacturer. DNA sequencing reactions were performed with the dye terminator procedure (Applied Biosystems) and analyzed on an ABI PRISM 310 Genetic Analyzer. Assembly and analysis of the result were performed with the Lasergene DNA Star software package. One clone was found representing the cDNA sequence coding for full-length human osteoadherin. Full-length osteoadherin was amplified with primers (5′-GCCAAGCTTCAATATGAAACTTATC-3′ and 5′-GCTGGATCCCTATTCTTGATTTTC-3′) corresponding to nucleotides 161–1363 with flanking HindIII and BamHI sites. The fragment generated and a pCEP4-BM40-hisEK expression vector (10) were digested with HindIII and BamHI and ligated. Full-length Osteoadherin in the pCEP4-BM40-hisEK expression vector was transfected into 293-EBNA cells with FuGENE 6 Transfection Reagent (Roche Applied Science, Basel, Switzerland). The cells were grown in 293 SFM II medium (Invitrogen) in the presence of hygromycin B (260 μg/ml, Duchefa Biochemicals). Ethylene glycol, 20%, was added to conditioned medium, which was concentrated and transferred into equilibration buffer (50 mm Tris, pH 8.0, 0.5 m NaCl, 20% ethylene glycol, 10 mm imidazole) by dialysis. The sample was filtered and applied onto a 1-ml HiTrapTM Chelating HP column (Amersham Biosciences) primed with Ni2+. Unbound proteins were removed by washing, and captured proteins were eluted with 20 column volumes of a linear 0.1–1 m imidazole gradient in equilibration buffer. Fractions containing eluted proteins were dialyzed against 50 mm Tris, pH 8.0, 10 mm NaCl, 20% ethylene glycol, and applied onto a 1-ml Mono Q column. The sample was eluted in 1-ml fractions with a salt gradient (10 mm NaCl to 1 m NaCl) and analyzed by SDS-polyacrylamide gel electrophoresis using 4–16% gradient gels according to the procedure of Laemmli (27). Bands of interest were cut out and analyzed for proteins present by in-gel trypsin digestion and MALDI-TOF MS (23). Fractions containing pure osteoadherin were stored at 4 °C.
RESULTS
The highly anionic, N-terminal tyrosine sulfate domain of fibromodulin was considered to have the potential to mimic heparin in interactions. Therefore we decided to test whether peptides representing three variants of heparin binding motifs could bind to this domain. We selected the domain from the three proteins PRELP, chondroadherin, and Oncostatin M because they contain basic amino acids in different patterns.
Heparin Binding of Peptides
In initial experiments the binding of the three peptides to heparin was evaluated using elution with increasing salt as an indicator of affinity (Fig. 1).
FIGURE 1.
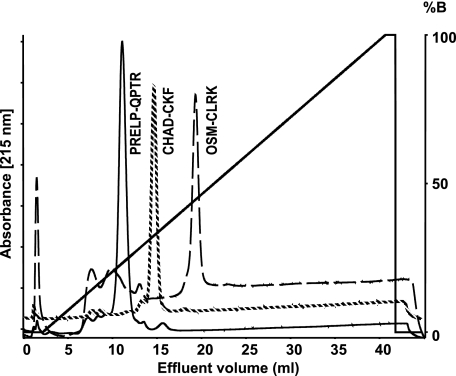
Binding of peptides representing the putative heparin binding motifs of chondroadherin, PRELP, and Oncostatin M to heparin. The peptides from PRELP (PRELP-QPTR, solid line), chondroadherin (CHAD-CKF, dotted line), and Oncostatin M (OSM-CLRK, dashed line) were chromatographed separately on a 1-ml heparin-Sepharose column to verify affinity to heparin. Elution was performed with a salt gradient of 0.15 to 2 m NaCl. The identity of the peptide in the respective peak was confirmed by MALDI-TOF MS.
The respective peptide was chromatographed on heparin-Sepharose and eluted with a gradient of NaCl. Interestingly, the PRELP peptide with only clusters of arginine residues was eluted at 0.61 m NaCl before the shorter chondroadherin peptide containing also doublet lysine residues eluting at 0.8 m NaCl. The longer peptide of Oncostatin M also containing a mix of arginine and lysine residues bound most strongly eluting at 1.0 m NaCl (Fig. 1).
Interactions of Heparin-binding Domain Peptides of PRELP, Chondroadherin, and Oncostatin M with the Isolated Tyrosine Sulfate Domain of Fibromodulin
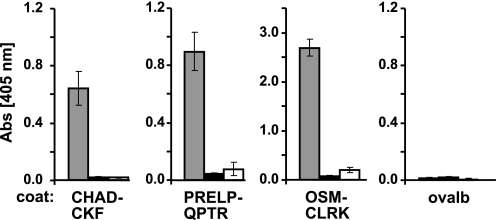
Binding of the heparin-binding peptides to the anionic tyrosine sulfate domain isolated after digestion with Lys-C and purified by anion-exchange chromatography (pooled fractions 1–6, see Fig. 8) was tested in solid phase assays (Fig. 2). The peptides were coated onto the plastic surface, and the ability of biotin-tagged 10-kDa fragment to bind was tested (Fig. 2, left part). Indeed all the peptides bound to the tyrosine sulfate domain. Binding was also observed when the 10-kDa fragment was coated onto the plastic surface, and peptides tagged with biotin in one end were applied (Fig. 2, right part).
FIGURE 8.
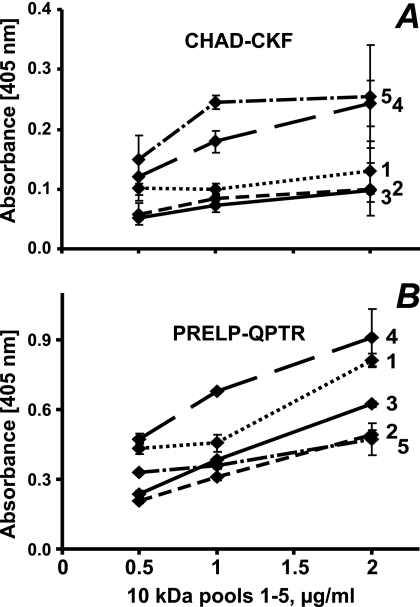
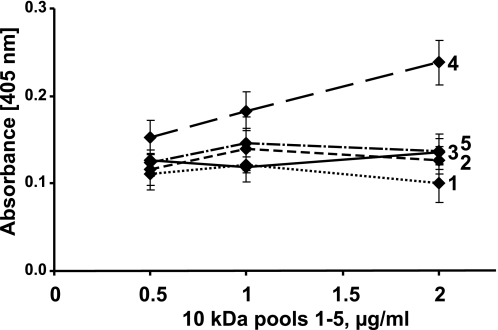
Interaction of heparin-binding domains of PRELP (PRELP-QPTR) and chondroadherin (CHAD-CKF) with subfractions of the tyrosine sulfate rich N-terminal fragment of fibromodulin. To investigate whether the degree and distribution of O-sulfate in the tyrosine-rich N-terminal fragment of fibromodulin influence the binding to different heparin-binding motifs, the biotinylated peptide CHAD-CKF (A) and biotinylated PRELP-QPTR (B) were added onto streptavidin-coated plastic surface, and the tyrosine sulfate-rich N-terminal fragment of fibromodulin, pools 1–5 (1, dotted line; 2, line with short dashes; 3, solid line; 4, line with large dashes; and 5, line with dashes and dots) from the Mono Q separation in Fig. 5, were added at different dilutions. Bound fragments were detected using a polyclonal antibody against full-length fibromodulin.
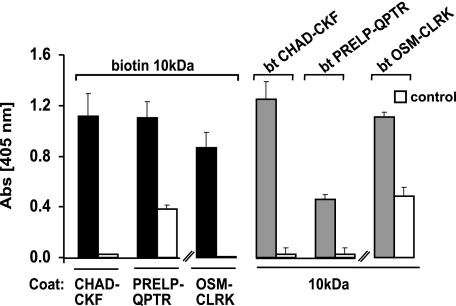
FIGURE 2.
Interaction of the 10-kDa tyrosine sulfate-rich fragment of fibromodulin with CHAD-CKF, OSM-CLRK, and PRELP-QPTR. To demonstrate that the tyrosine sulfate-rich domain can bind to heparin-binding protein motifs, three synthetic peptides representing the heparin-binding motif in chondroadherin (CHAD-CKF), Oncostatin M (OSM-CLRK), and PRELP (PRELP-QPTR) were used. The peptides (left part) or alternatively the tyrosine sulfate-rich N-terminal fragment of fibromodulin (right part) were coated onto a plastic surface overnight and remaining sites were blocked with ovalbumin. Biotin-tagged tyrosine sulfate-rich N-terminal fragment of fibromodulin (left part) or either of biotin-tagged CHAD-CKF, biotin-tagged OSM-CLRK, or PRELP-QPTR peptides (right part), were added to the respective coated wells, and bound biotinylated ligands were detected with alkaline phosphate-conjugated streptavidin. Controls were performed with only a blocking agent present on the plastic surface.
To verify that the binding occurred via the tyrosine sulfate domain proper, a 5-kDa fragment generated by trypsin digestion of the larger 10-kDa piece was used. This fragment is devoid of the cysteine loop structure and lacks only the one most N-terminal tyrosine residue. It was found to efficiently bind to the peptides from chondroadherin, PRELP, and Oncostatin M, respectively, coated to the microtiter plate surface (Fig. 3).
FIGURE 3.
Interaction of the trypsin generated N-terminal 5-kDa sulfated fragment of fibromodulin and its non-sulfated variant expressed in bacteria with heparin-binding peptides of chondroadherin, PRELP, and Oncostatin M. The three heparin-binding peptides CHAD-CKF, PRELP-QPTR, and OSM-CLRK were coated onto a plastic surface overnight, and the remaining sites were blocked with ovalbumin. Biotinylated sulfated (gray columns) or non-sulfated (■) N-terminal fragments of fibromodulin were added to the wells. Bound fragments were detected by alkaline phosphates conjugated streptavidin. Controls (□) were performed with only a blocking agent present on the plastic surface.
To verify that binding primarily involved one or more sulfated tyrosine residues, the N-terminal domain of fibromodulin was expressed in E. coli, which do not generate post-translation modifications such as sulfation. This fragment and control sulfated 5-kDa fragment were tested for binding to the three basic heparin-binding peptides CHAD-CKF, PRELP-QPTR, and OSM-CLRK coated onto a plastic surface. Indeed the non-sulfated fragment showed no significant binding compared with the sulfated fragment (Fig. 3).
Heparin was used in a competition assay to verify the hypothesis that heparin and the N-terminal of fibromodulin compete for the same binding site of the cationic peptides. The three heparin-binding peptides were coated onto a plastic surface, and the 10-kDa fragment was added together with different concentrations of heparin. Indeed by adding heparin in the assay the binding of the 10-kDa fragment of fibromodulin to the peptides was inhibited in a dose-dependent manner (Fig. 4).
FIGURE 4.
Competition between heparin and the N-terminal fragment of fibromodulin in binding to the three different heparin-binding peptides. The three heparin-binding peptides CHAD-CKF (A), PRELP-QPTR (B), and OSM-CLRK (C) were coated onto a plastic surface overnight, and the remaining sites were blocked with ovalbumin. The Lys-C-generated fragment of the fibromodulin N-terminal were added to coated wells in the presence of different concentrations of heparin. Bound fibromodulin fragment was detected with an antibody against fibromodulin.
Interaction was also studied in the presence of sodium chloride to evaluate the dependence on ionic strength of the binding between the molecules. In initial experiments the interaction was inhibited at low salt concentration using 5 mm sodium phosphate buffer, pH 7.4 (data not shown). Binding was evaluated further by increasing the phosphate concentration and decreasing the initial sodium chloride concentration (to a more physiological like condition) and also including Urea to focus on ionic interactions. Binding of the three basic peptides and bFGF to the coated 10-kDa fragment was gradually abolished with increasing ionic strength (Fig. 5, A–D). Urea present would prevent any self-interaction of peptides at increasing salt concentration.
Interactions of Proteins Containing Heparin-binding Motifs with the Fibromodulin Tyrosine Sulfate Domain
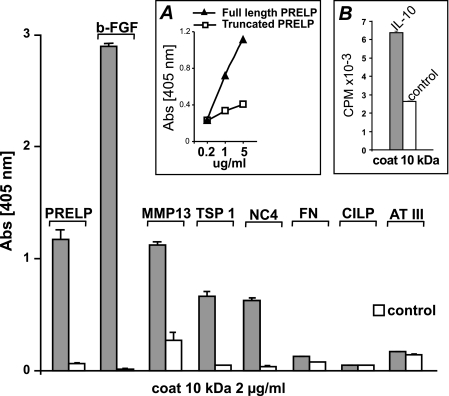
The N-terminal, 10 kDa tyrosine sulfate domain fragment of fibromodulin, was isolated after digestion of the protein with Lys-C followed by ion exchange chromatography (combined pools 1–6 Fig. 7). The combined pool of all 10-kDa fragments was coated onto polystyrene microtiter plates and used to test interactions with the heparin-binding proteins PRELP, FGF-2, MMP13, fibronectin, CILP-1, TSP-1, NC4 domain of collagen IX and antithrombin III in solid phase assay, Fig. 6. Most of the proteins showed binding. The exceptions were fibronectin that contains two much larger heparin-binding domains than the other proteins, which would favor strong binding to the long heparin polymer. CILP-1 only contains a thrombospondin motif domain of different character with the heparin binding WSP repeat structure. This apparently does not bind to the tyrosine sulfate domain. Thrombospondin also contains this motif as well as a second heparin binding structure of a cluster of basic amino acids that apparently promotes binding. Antithrombin III with a unique requirement for a 3-sulfate in the heparin did not bind to the 10-kDa tyrosine sulfate domain (Fig. 6).
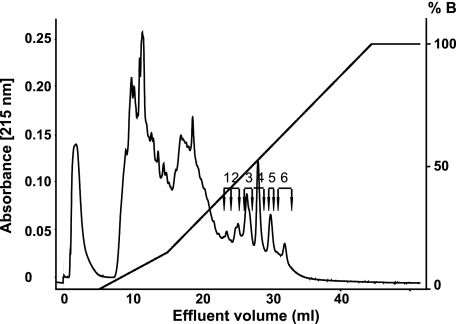
FIGURE 7.
Fractionation of an Lys-C digest of fibromodulin on Mono Q. Bovine fibromodulin, cleaved by the enzyme endoproteinase Lys-C, was applied onto a 1-ml Mono Q column and eluted with a salt gradient (0 to 1.5 m NaCl). The tyrosine sulfate-rich 10-kDa fragments were fractionated into six individual, peaks containing fragments differing in sulfation. Fractions corresponding to the peaks were divided into six pools as indicated in the graph and used in further experiments. For some experiments an equal aliquot of each fraction was pooled and used as the total 10-kDa fragment preparation. Note that the peaks are not all symmetrical, indicating heterogeneity.
FIGURE 6.
Interaction of heparin-binding motifs with fibromodulin. The isolated N-terminal 10-kDa tyrosine sulfate fragment of fibromodulin was coated onto microtiter plates overnight, and unbound sites were blocked with bovine serum albumin except for wells to contain MMP13 as a ligand. These were blocked with ovalbumin. Proteins with known heparin binding properties, i.e. bovine PRELP, bovine FGF-2 human MMP13, human fibronectin, bovine CILP, bovine thrombospondin I, recombinant collagen IX NC-4 domain, bovine antithrombin III as well as non-heparin binding truncated bovine PRELP (A, inset) lacking the entire N-terminal were allowed to interact, and remaining proteins were detected with specific antibodies. [125I]IL10 was also allowed to interact, and bound radioactivity after washing was measured in a gamma counter (B). Controls were performed with only a blocking agent present.
In corroborating experiments, we could show that PRELP as one example of an intact protein did bind to the intact fibromodulin. When the N-terminal heparin-binding structure of the PRELP was truncated, the protein no longer showed binding to fibromodulin corroborating that binding depended on the two implicated domains (Fig. 6A, inset).
In a separate protocol (all antibodies from the supplier of the IL-10 were inactive in several assay protocols with the factor that we proved to represent IL-10 only by MS) the heparin-binding cytokine IL-10 was shown to bind to the tyrosine sulfate domain (Fig. 6B, inset).
Interaction of Various Heparin-binding Peptides/Proteins with Sub-fractions of the Tyrosine Sulfate Domain of Fibromodulin with Different Anionic Charge
Previous analysis of the 10-kDa fibromodulin fragment by mass spectrometry indicated that there was pronounced variability in degree of tyrosine sulfation. The 10-kDa fragment of fibromodulin was therefore fractionated into differently charged fractions by chromatography on Mono Q. Several distinct peaks were apparent, eluting at ionic strengths from 0.6 to 1 m NaCl (Fig. 8). These peaks having the same protein core, as observed by mass spectrometry, must represent fragments containing different numbers and locations of tyrosine sulfate residues. Further attempts to sub-fractionate the material in some of the peaks demonstrated inhomogeneity with partly resolved peaks eluting from the Mini Q (data not shown), possibly a result of different clustering of the sulfate groups. Because separation was not sufficient to provide homogeneous materials, further experiments made use of the fractions pooled as depicted in Fig. 7.
To document dependence on the levels of tyrosine sulfation for binding of the heparin-binding peptides from chondroadherin and PRELP, respectively, these were tagged with biotin in their respective N and C termini representing the internal end. These peptides were then immobilized to streptavidin-coated surfaces. The binding of fragments representing the different pools of fibromodulin was studied at different concentrations for the chondroadherin peptide (Fig. 8A) and for the PRELP peptide (Fig. 8B). Both peptides were shown to bind to material in fractions of 10 kDa, with the exception of those eluted at lower salt, which shows poor or no binding to the chondroadherin peptide (Fig. 8A). The slope of the dilution curves differed, indicating different affinities for the materials in the fractions. Those eluted at high salt (fractions 4 and 5) showed a steeper slope and higher binding apparently reaching plateau level for fraction 5. The fact that several of the binding curves were not linear may reflect a different pattern of sulfation, although the number of sulfates may be similar for all those molecules in the particular sub-fraction. The PRELP peptide with a different type of basic amino acid clusters showed somewhat different binding characteristics (Fig. 8B). In this case the 10-kDa fraction requiring the highest salt for elution from the ion exchanger showed the poorest binding, and in general the curves where somewhat differently ordered.
The material eluting at highest salt from the Mono Q showed no binding of either peptide. A further investigation into the presence of this fragment by SDS-PAGE and mass spectrometry of the remaining material indicated that it had been lost, probably a result of its low abundance. All the other fractions were added at the same molar absorbance.
Binding of MMP13 to the Fractions of the Tyrosine Sulfate Domain from Fibromodulin with Different Charge Properties
MMP13 contains a large number of basic amino acids. It appears however, that heparin binding may depend on their three-dimensional arrangement of MMP13 (28). We, therefore, decided to test the binding of this protein to the differently charged tyrosine sulfate domain fractions from the Mono Q separation. Binding was tested in presence of Zn2+ and Ca2+ ions. Interestingly, fraction 4 showed the most prominent binding (Fig. 9), whereas the other fractions did not exhibit proper concentration dependence and may therefore not bind significantly. In further experiments we tested binding of the 5-kDa fragment only containing the tyrosine sulfate domain, albeit also containing a previously described cleavage site of MMP13. Indeed, this fragment did bind the MMP13 (Fig. 10) and could be cleaved by the enzyme (data not shown).
FIGURE 9.
Interaction of MMP-13 with subfractions of the tyrosine sulfate-rich N-terminal fragment of fibromodulin. MMP13 was coated onto a plastic surface overnight and unspecific sites were blocked with ovalbumin. The Lys-C-generated tyrosine sulfate-rich N-terminal fragment of fibromodulin, pools 1–5 (1, dotted line; 2, line with short dashes; 3, solid line; 4, line with large dashes; and 5, line with dashes and dots) from the Mono Q separation in Fig. 7, were added to the coated wells. Bound fragments were detected with a polyclonal antibody against full-length fibromodulin.
FIGURE 10.
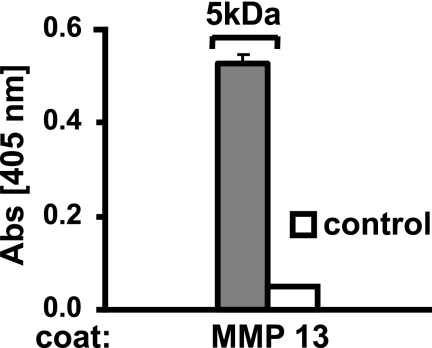
Interaction of MMP-13 with the “small” tyrosine sulfate 5-kDa N-terminal fragment of fibromodulin generated by trypsin digestion. MMP13 was coated overnight onto a plastic surface, and the remaining sites were blocked with ovalbumin. The biotinylated 5-kDa N-terminal fragment of fibromodulin was allowed to interact. Bound fragment were detected with streptavidin-AP. Control were performed with no coated protein present.
Interaction of Osteoadherin with Heparin-binding Proteins
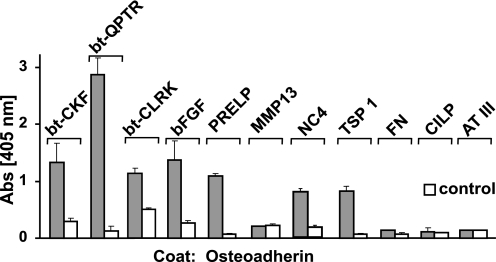
Another protein with a prominent tyrosine sulfate domain in its N-terminal part is osteoadherin. The clustering of sulfate residues in this domain is somewhat different compared with fibromodulin, and there are maximally only six sulfate residues. We therefore investigated whether also this protein could bind to the heparin-binding structures previously tested, including the peptides from PRELP, chondroadherin, and Oncostatin M as well as the intact proteins of PRELP, FGF-2, MMP13, fibronectin, CILP, TSP-1, and antithrombin III. Despite the differences in the tyrosine sulfate domain binding to osteoadherin was the same as that to the fibromodulin tyrosine sulfate domain, with the interesting exception that MMP-13 showed no binding to the former (Fig. 11).
FIGURE 11.
Interaction of osteoadherin with heparin-binding motifs. Osteoadherin was coated overnight onto a plastic surface and unspecific sites blocked with ovalbumin (in the case of PRELP-QPTR, CHAD-CKF, OSM-CLRK, and MMP13 adhesives) or bovine serum albumin (in the case of bovine PRELP, FGF-2, collagen IX NC 4 domain, human fibronectin, human CILP, bovine thrombospondin, and antithrombin III adhesives). Candidate ligands, i.e. the heparin-binding peptide CHAD-CKF and PRELP-QPTR and OSM-CLRK, as well as bovine PRELP, bovine FGF-2, human MMP13, collagen IX NC4 domain, human fibronectin, human CILP, bovine thrombospondin I, and bovine antithrombin III were allowed to interact with the coated protein. Bound peptides were detected with streptavidin-AP and bound proteins with primary antibodies specific to the respective protein and a secondary antibody-AP enzyme conjugate. All controls were made with only the blocking protein coat on the plastic surface.
DISCUSSION
Over the years studies of tyrosine sulfation in proteins have been limited, hampered by lack of technology to identify sulfate substitution of the tyrosine residues. However, mass spectrometry, using special modes, provides a possibility to identify the presence of sulfated tyrosine residues. By using such technology, we identified a short polypeptide domain in the N terminus of the LRR protein fibromodulin, which contains a variable number of sulfated tyrosine residues (20). Another member of the same family, OSAD, contains a similar N-terminal domain, albeit the tyrosine residues putatively carrying sulfate are distributed in a different manner and are somewhat fewer (20).
In view of the many sulfate residues over a short peptide stretch, the charge density is high and can be compared with that of the extremely highly charged heparin/heparan sulfate and other molecules among the glycosaminoglycans. The charge density is further enhanced by a similar number of acidic amino acids contributing carboxyl groups, not unlike the situation in the glycosaminoglycans. Because heparin interacts with a number of biologically active molecules and regulates their functions, it became relevant to search for an analogous function of the tyrosine sulfate domain of these LRR proteins.
In initial experiments we could indeed show that the small domain of fibromodulin containing the tyrosine sulfate residues was able to bind other proteins known to interact with heparin. In the case of PRELP, Oncostatin M, and chondroadherin, peptides corresponding to the heparin-binding domain were synthesized and shown to bind selectively to the tyrosine sulfate domain preparation. In further support of the similar nature of the interactions between peptides and the tyrosine sulfate domain with those between peptides and heparin, we could show that the two polyanionic compounds compete for binding of the peptides.
The specificity for the interaction varied between subfractions of the domain with different levels of charge. In general those fragments having a higher charge density and therefore more extensively retained on an anion-exchange resin, showed a more pronounced interaction with the heparin-binding peptides. At the same time, even the fractions with apparent similar charge density showed binding characteristics indicating that interaction was influenced by the variability in the positioning of the sulfate groups along the stretch of tyrosine residues.
Interactions between the sulfate domain of fibromodulin and the heparin binding molecules were influenced by addition of salt. Increasing salt concentration in the assay gradually decreased binding of the peptides, demonstrating the dependence of ionic interactions. In assays using a non-sulfated N-terminal fragment of fibromodulin the binding to the heparin-binding peptides was abolished, demonstrating the importance of the sulfate residues.
The much longer heparin-binding domains of fibronectin appeared not to be able to bind to the tyrosine sulfate-rich structure. A possible explanation is that the clustering of basic amino acids in fibronectin is different and does not provide for a tight binding to this peptide domain that is many times shorter than heparin.
MMP13, like many other components and enzymes present in the extracellular matrix, can bind to heparin. We could show that the enzyme did bind via ionic interactions to the tyrosine sulfate domain of fibromodulin. Upon binding, the enzyme could cleave the polypeptide at a site previously identified (22). Whether this binding is a prerequisite for cleavage we do not know. It is interesting that fibromodulin has been shown to be present in the tissue bound along the collagen fibers, at the gap region (29, 30). It is likely that the domain carrying the tyrosine sulfates is extending away from the fiber, because MMP13 added to tissue explants will cleave the protein such as to induce initial release of the most part of the tyrosine sulfate domain (22). At the same time, the part of fibromodulin containing the leucine-rich repeat region is retained in the tissue. It is possible that the protein bound to the collagen fiber surface has a role in directing the enzyme to appropriate cleavage on the collagen being its major substrate in the tissue.
Osteoadherin is a close relative to fibromodulin within the leucine-rich repeat protein family. The two proteins differ in the distribution of the sulfate residues in their tyrosine-rich N-terminal domains. However, they showed similarities in binding of the heparin-binding proteins. Notably, binding to MMP13 was found to differ, such that osteoadherin did not show detectable interaction. Also, for the shorter heparin-binding peptide of chondroadherin there may be a small, barely detectable difference in binding selectivity.
It is known that the extracellular matrix of, e.g. cartilage, contains pro-MMPs, which can be activated (6, 31). In view of the findings in the present work it is possible that anionic domains with tyrosine sulfate residues may serve to sequester these pro-enzymes to specific structures in the tissue. Upon activation they may cleave substrates locally, e.g. fibromodulin and osteoadherin, opening up for faster processing.
FGF-2, as a representative of the heparin binding growth factors, did bind to the tyrosine sulfate domains. This may result in sequestering of the growth factor in the matrix to be released upon degradation of the structures containing fibromodulin. These released factors may in turn trigger a repair response by the cells.
The LRR proteins studied that contain a heparin-binding domain, as well as those containing tyrosine sulfate domains, do bind to collagen via their LRR regions. In the case of PRELP, it has been shown that the protein bound onto the collagen can still bind to perlecan via its heparan sulfate chains (10). It is therefore possible that the fibromodulin or osteoadherin molecules bound at the surface of one collagen fibril may cross-link to the heparin-binding motif of PRELP or chondroadherin sitting on another collagen fibril. These interactions depend on the presence of the tyrosine sulfate-rich domain capable of binding to the domain of clustered basic amino acids on another collagen bound molecule. For this role it is essential that the proteins contain two functional domains such that they can bind to collagen at the same time as interacting with another partner molecule.
Collagen IX contains an N-terminal part consisting of the NC4 and collagen 3 domains that protrudes out from the collagen fiber (32). The NC-4 domain is capable of binding to heparin (33). We now show that this domain also binds to the tyrosine sulfate domain of fibromodulin.
In conclusion, the properties of the tyrosine sulfate-rich domains have the potential as important contributors to the interactions essential to tissue integrity as well as in sequestering factors in the matrix, which in tissue injury may have roles in repair responses.
This work was supported by The Swedish Research Council, King Gustaf V's 80-year jubilee fund, the Österlund Foundation, the Kock Foundation, the Magnus Bergvall Foundation, and Anamar Medical. Mass spectrometers were purchased through grants from the IngaBritt and Arne Lundbergs Research Foundation.
- LRR
- leucine-rich repeat
- FGF
- fibroblast growth factor
- PBS
- phosphate-buffered saline
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- MS
- mass spectrometry
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Hedbom E., Heinegård D. (1989) J. Biol. Chem. 264, 6898–6905 [PubMed] [Google Scholar]
- 2.Blochberger T. C., Vergnes J. P., Hempel J., Hassell J. R. (1992) J. Biol. Chem. 267, 347–352 [PubMed] [Google Scholar]
- 3.Vogel K. G., Paulsson M., Heinegård D. (1984) Biochem. J. 223, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiberg C., Hedbom E., Khairullina A., Lamandé S. R., Oldberg Å., Timpl R., Mörgelin M., Heinegård D. (2001) J. Biol. Chem. 276, 18947–18952 [DOI] [PubMed] [Google Scholar]
- 5.Bengtsson E., Mörgelin M., Sasaki T., Timpl R., Heinegård D., Aspberg A. (2002) J. Biol. Chem. 277, 15061–15068 [DOI] [PubMed] [Google Scholar]
- 6.Månsson B., Wenglén C., Mörgelin M., Saxne T., Heinegård D. (2001) J. Biol. Chem. 276, 32883–32888 [DOI] [PubMed] [Google Scholar]
- 7.Kalamajski S., Aspberg A., Oldberg Å. (2007) J. Biol. Chem. 282, 16062–16067 [DOI] [PubMed] [Google Scholar]
- 8.Kalamajski S., Oldberg Å. (2007) J. Biol. Chem. 282, 26740–26745 [DOI] [PubMed] [Google Scholar]
- 9.Svensson L., Heinegård D., Oldberg Å. (1995) J. Biol. Chem. 270, 20712–20716 [DOI] [PubMed] [Google Scholar]
- 10.Bengtsson E., Aspberg A., Heinegård D., Sommarin Y., Spillmann D. (2000) J. Biol. Chem. 275, 40695–40702 [DOI] [PubMed] [Google Scholar]
- 11.Scott J. E., Orford C. R. (1981) Biochem. J. 197, 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R. L., Lander A. D. (2001) J. Biol. Chem. 276, 7507–7517 [DOI] [PubMed] [Google Scholar]
- 13.Rapraeger A., Jalkanen M., Endo E., Koda J., Bernfield M. (1985) J. Biol. Chem. 260, 11046–11052 [PubMed] [Google Scholar]
- 14.Hassell J. R., Robey P. G., Barrach H. J., Wilczek J., Rennard S. I., Martin G. R. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 4494–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsen G., Halfter W., Kröger S., Cole G. J. (1995) J. Biol. Chem. 270, 3392–3399 [DOI] [PubMed] [Google Scholar]
- 16.Mostafavi-Pour Z., Askari J. A., Whittard J. D., Humphries M. J. (2001) Matrix Biol. 20, 63–73 [DOI] [PubMed] [Google Scholar]
- 17.Hofsteenge J., Huwiler K. G., Macek B., Hess D., Lawler J., Mosher D. F., Peter-Katalinic J. (2001) J. Biol. Chem. 276, 6485–6498 [DOI] [PubMed] [Google Scholar]
- 18.Guo N. H., Krutzsch H. C., Nègre E., Vogel T., Blake D. A., Roberts D. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3040–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixit V. M., Grant G. A., Santoro S. A., Frazier W. A. (1984) J. Biol. Chem. 259, 10100–10105 [PubMed] [Google Scholar]
- 20.Önnerfjord P., Heathfield T. F., Heinegård D. (2004) J. Biol. Chem. 279, 26–33 [DOI] [PubMed] [Google Scholar]
- 21.Heinegård D., Larsson T., Sommarin Y., Franzén A., Paulsson M., Hedbom E. (1986) J. Biol. Chem. 261, 13866–13872 [PubMed] [Google Scholar]
- 22.Heathfield T. F., Önnerfjord P., Dahlberg L., Heinegård D. (2004) J. Biol. Chem. 279, 6286–6295 [DOI] [PubMed] [Google Scholar]
- 23.Lorenzo P., Aspberg A., Önnerfjord P., Bayliss M. T., Neame P. J., Heinegård D. (2001) J. Biol. Chem. 276, 12201–12211 [DOI] [PubMed] [Google Scholar]
- 24.Frangioni J. V., Neel B. G. (1993) Anal. Biochem. 210, 179–187 [DOI] [PubMed] [Google Scholar]
- 25.Lorenzo P., Bayliss M. T., Heinegård D. (1998) J. Biol. Chem. 273, 23463–23468 [DOI] [PubMed] [Google Scholar]
- 26.Lorenzo P., Neame P., Sommarin Y., Heinegård D. (1998) J. Biol. Chem. 273, 23469–23475 [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28.Gomis-Rüth F. X., Gohlke U., Betz M., Knäuper V., Murphy G., López-Otín C., Bode W. (1996) J. Mol. Biol. 264, 556–566 [DOI] [PubMed] [Google Scholar]
- 29.Hedbom E., Heinegård D. (1993) J. Biol. Chem. 268, 27307–27312 [PubMed] [Google Scholar]
- 30.Hedlund H., Mengarelli-Widholm S., Heinegård D., Reinholt F. P., Svensson O. (1994) Matrix Biol. 14, 227–232 [DOI] [PubMed] [Google Scholar]
- 31.Ganu V., Goldberg R., Peppard J., Rediske J., Melton R., Hu S. I., Wang W., Duvander C., Heinegård D. (1998) Arthritis Rheum. 41, 2143–2151 [DOI] [PubMed] [Google Scholar]
- 32.Vaughan L., Mendler M., Huber S., Bruckner P., Winterhalter K. H., Irwin M. I., Mayne R. (1988) J. Cell Biol. 106, 991–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pihlajamaa T., Lankinen H., Ylöstalo J., Valmu L., Jäälinoja J., Zaucke F., Spitznagel L., Gösling S., Puustinen A., Mörgelin M., Peränen J., Maurer P., Ala-Kokko L., Kilpelaïnen I. (2004) J. Biol. Chem. 279, 24265–24273 [DOI] [PubMed] [Google Scholar]