Abstract
δ-Catenin was first identified because of its interaction with presenilin-1, and its aberrant expression has been reported in various human tumors and in patients with Cri-du-Chat syndrome, a form of mental retardation. However, the mechanism whereby δ-catenin is regulated in cells has not been fully elucidated. We investigated the possibility that glycogen-synthase kinase-3 (GSK-3) phosphorylates δ-catenin and thus affects its stability. Initially, we found that the level of δ-catenin was greater and the half-life of δ-catenin was longer in GSK-3β−/− fibroblasts than those in GSK-3β+/+ fibroblasts. Furthermore, four different approaches designed to specifically inhibit GSK-3 activity, i.e. GSK-3-specific chemical inhibitors, Wnt-3a conditioned media, small interfering RNAs, and GSK-3α and -3β kinase dead constructs, consistently showed that the levels of endogenous δ-catenin in CWR22Rv-1 prostate carcinoma cells and primary cortical neurons were increased by inhibiting GSK-3 activity. In addition, it was found that both GSK-3α and -3β interact with and phosphorylate δ-catenin. The phosphorylation of ΔC207-δ-catenin (lacking 207 C-terminal residues) and T1078A δ-catenin by GSK-3 was noticeably reduced compared with that of wild type δ-catenin, and the data from liquid chromatography-tandem mass spectrometry analyses suggest that the Thr1078 residue of δ-catenin is one of the GSK-3 phosphorylation sites. Treatment with MG132 or ALLN, specific inhibitors of proteosome-dependent proteolysis, increased δ-catenin levels and caused an accumulation of ubiquitinated δ-catenin. It was also found that GSK-3 triggers the ubiquitination of δ-catenin. These results suggest that GSK-3 interacts with and phosphorylates δ-catenin and thereby negatively affects its stability by enabling its ubiquitination/proteosome-mediated proteolysis.
δ-Catenin was first identified as a molecule that interacts with presenilin-1 (PS-1)2 by yeast two-hybrid assay (1) and was found to belong to the p120-catenin subfamily of armadillo proteins, which characteristically contain 10 Arm repeats (2). In addition to its interaction with PS-1 and its abundant expression in brain (3, 4), several lines of evidence indicate that δ-catenin may play a pivotal role in cognitive function. First, the hemizygous loss of δ-catenin is known to be closely correlated with Cri-du-Chat syndrome, a severe form of mental retardation in humans (5). Second, severe learning deficits and abnormal synaptic plasticity were found in δ-catenin-deficient mice (6). Moreover, in δ-catenin−/− mice, paired pulse facilitation (a form of short term plasticity) was found to be reduced, and long term potentiation, which is related to the forming and storage mechanisms of memory, was deficient (7, 8). Third, δ-catenin interacting molecules, such as PSs (1, 9), cadherins (10), S-SCAM (2), and PSD-95 (11), have been shown to play important roles in modulating synaptic plasticity. However, even though the maintenance of an adequate δ-catenin level is known to be critical for normal brain function, few studies have been undertaken to identify the factors that regulate δ-catenin stability in cells. We have previously demonstrated that PS-1 inhibits δ-catenin-induced cellular branching and promotes δ-catenin processing and turnover (12).
Because of structural similarities among β-catenin, p120-catenin, and δ-catenin and to their shared binding partners (i.e. PS-1 (1, 9) and cadherins (10)), glycogen-synthase kinase-3 (GSK-3) drew our attention as a potential candidate effector of δ-catenin stability in cells. GSK-3 is a serine/threonine kinase and has two highly homologous forms, GSK-3α and GSK-3β, in mammals (13). Although GSK-3α and GSK-3β have similar structures, they differ in mass (GSK-3α (51 kDa) and GSK-3β (47 kDa) (13)) and to some extent in function (14). GSK-3 is a well established inhibitor of Wnt signaling. Moreover, it is known to phosphorylate β-catenin, which results in its degradation via ubiquitination/proteosome-dependent proteolysis (15). GSK-3 is ubiquitously distributed in the human body, but it is particularly abundant in brain (13), and it is interesting that δ-catenin is also abundant in the nervous system (4) and that GSK-3 participates in the progression of Alzheimer disease (16). The majority of GSK-3 substrates have the consensus sequence (Ser/Thr)-Xaa-Xaa-Xaa-(Ser/Thr) (17). Interestingly, we found that δ-catenin has several putative phosphorylation sites targeted by GSK-3, which suggests that δ-catenin can be regulated by GSK-3 in the same way as β-catenin.
In this report, we demonstrate that both GSK-3α and -3β interact with and phosphorylate δ-catenin and that this leads to its subsequent ubiquitination and degradation via proteosome-dependent proteolysis. Our results strongly suggest that GSK-3 is a key regulator of δ-catenin stability in cells.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
The construction of wild type (WT)-, ΔC207-, ΔN85–325-δ-catenin in pEGFP-C1 has been previously described (18). ΔN85–325/ΔC207- and ΔN19–1153-δ-catenin were constructed using two subcloning steps. The δ-catenin T1078A and T337A mutants, in which both the Thr1078 and Thr337 residues were substituted to Ala, were generated by site-directed mutagenesis. The HA-tagged GSK-3β wild type (hereafter GSK-3β WT) and kinase dead (hereafter GSK-3β KD) constructs were kindly donated by Kang-Yeol Choi (Yonsei University, Seoul, Korea). We generated HA- tagged GSK-3α wild type (hereafter GSK-3α WT) and kinase dead (hereafter GSK-3α KD) constructs in a cytomegalovirus promoter-derived mammalian expression vector (pCS4–3Myc, -3HA) by PCR amplification. siRNA for control (catalog number 6201), GSK-3α (catalog number 6312), and GSK-3α/β (catalog number 6301) were purchased from Cell Signaling Technology, Inc.
The antibodies used were as follows: anti-δ-catenin (BD Bioscience and Upstate Biotechnology); anti-β-catenin (Sigma); anti-GFP (Clontech, Abcam, and Sigma); anti-β-tubulin (Sigma); anti-actin (Santa Cruz Biotechnology); and anti-GSK-3 (α + β) (Abcam). The anti-HA antibody was obtained using 12CA5 hybridoma cells.
Cell Culture, Transfection, and Cycloheximide Treatment
The wild type mouse embryonic fibroblast (MEF) and HEK293 or CWR22Rv-1 human prostate cell lines were maintained in DMEM or in RPMI supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a 5% CO2 atmosphere. The cells were transfected using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions or using the calcium phosphate method. siRNA was transfected by Lipofectamine Plus reagent. To determine protein stability, the cells were treated with cycloheximide (CHX, 40 μg/ml; Sigma) and then harvested at different times, as indicated in the figure.
Pulse-Chase
The cells were maintained in methionine/cysteine-free DMEM (Sigma) supplemented with 1% fetal bovine serum and 0.04 m l-glutamine (Sigma) overnight. After the cells were treated with new conditioned DMEM supplemented with 150 μCi of 35S (Agenbio) for 1 h, the cells were washed with phosphate-buffered saline once, maintained in DMEM, and harvested at 0, 4, 6, 8, 12, 14, and 16 h with lysis buffer. The cell extracts were immunoprecipitated, resolved by SDS-PAGE, and exposed to film at −80 °C for 3 days.
Immunoblotting and Immunoprecipitation
Transfected cells were harvested with lysis buffer (10% glycerol, 25 mm HEPES, 150 mm NaCl, 1 mm EDTA, 25 mm NaF, 1 mm Na3VO4, 1% Nonidet P-40, 0.2 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture). Equal amounts of protein samples were solubilized and boiled with SDS sample buffer for 2 min and then separated by SDS-PAGE. Proteins were run on appropriate gels, transferred to hydrophobic polyvinylidene difluoride membranes (Millipore), developed with ECL Western blotting detection reagents (Millipore), and detected with x-ray film (Agfa) or LAS-4000 (Fujifilm). Immunoprecipitation was performed as previously described (19).
In Vitro Kinase Assays
δ-Catenin was transiently transfected into HEK293 cells, and δ-catenin proteins were collected by immunoprecipitation using anti-δ-catenin antibody and protein G-Sepharose (GE Healthcare). The immune complexes so obtained were washed three times with lysis buffer and once with Tris-buffered saline (pH 7.4) and then incubated for 30 min at 30 °C in a 20-μl reaction mixture supplemented with recombinant GSK-3β protein (New England Biolabs) and 10 μCi of [γ-32P]ATP (BMS). The reactions were stopped by adding SDS sample buffer. The phosphorylation status of δ-catenin was determined by autoradiography.
Cortical Neuron Culture
Cultured cortical neurons were prepared from embryonic day 18 fetal Sprague-Dawley rats and were plated on poly-d-lysine-coated five 18-mm glass coverslips at a density of 200,000 cells/60-mm dish. The cultures were grown in neurobasal medium (Invitrogen) supplemented with 2% B-27 and 0.5 mm l-glutamine.
Phosphoprotein Analysis by LC-MS/MS
δ-Catenin was transfected into both GSK-3β+/+ and GSK-3β−/− fibroblasts, and δ-catenin proteins were purified by immunoprecipitation using anti-δ-catenin antibody and subjected to SDS-PAGE. For the detection of phosphoproteins, the gels were stained using a Pro-Q Diamond phosphoprotein gel stain kit (Molecular Probes) according to the manufacturer's protocol. After fluorescent scanning, the gels were stained again using a modified CBB-G250 method for visualization of the total purified proteins and then subjected to in-gel digestion. The gel pieces were destained with 30% EtOH for 1 h. In-gel digestion was performed in three steps following the protocol modified by Russell et al. (20). After trypsin digestion, phosphopeptides were enriched using phosphopeptide enrichment spin column (Clontech Laboratory) or TiO2 (Glygen Corp.) according to the manufacturer's protocol.
Ultra performance liquid chromatography-MS/MS analyses were performed at Korea Basic Science Institute using a nanoAcquity ultra performance liquid chromatography system (Waters, Milford, MA) in combination with a Synapt high definition mass spectrometry mass spectrometer (Waters Micromass, Manchester, UK) and performed at Kyungpook National University using Q-TOF Premier (Waters Micromass). The column used was a 150-mm 75-μm BEH C18 column packed with 1.7-μm particles with a pore width of 130 Å. The LC was coupled with the mass spectrometer using a Pico Tip sprayer (Waters) operated using PicoTips (New Objective, Woburn, MA) with an inner diameter of 10 μm. Ultra performance liquid chromatography-MS/MS analyses were carried out at a flow rate of 400 nl/min and a column temperature of 30 °C. The samples were loaded directly on the analytical column. The gradient condition of mobile phase A and B, 3–40% mobile phase B over 110 min, 40–70% mobile phase B over 30 min at a flow rate of 300 nl/min, followed by a 10-min rinse of 80% of mobile phase B. This resulted in a total run time of 150 min. The mass spectrometer was operated in positive ion mode with a capillary voltage of 2400 V and a cone voltage of 40 V. The data were acquired in data dependent acquisition mode or MSE mode. For data dependent acquisition mode, one survey scan of 1 s was carried out followed by up to three MS/MS scans (of 1 s) of each of the three most intense precursor ions. Only multiply charged ions were allowed for fragmentation. MS/MS spectra were processed using MassLynx 4.1 to peak lists and searched against the protein data base containing protein sequence using MASCOT 2.2.1. MS/MS spectra obtained by MSE mode are analyzed by PLGS2.3. These searches were performed using enzyme option with trypsin at a MS tolerance of 1.2 Da and a MS/MS tolerance of 0.2 Da including the following variable modifications: carbamidomethyl-Cys, oxidized Met, phosphorylation at Ser, Thr, and Tyr.
RESULTS
GSK-3 Negatively Affects the Stability of δ-Catenin
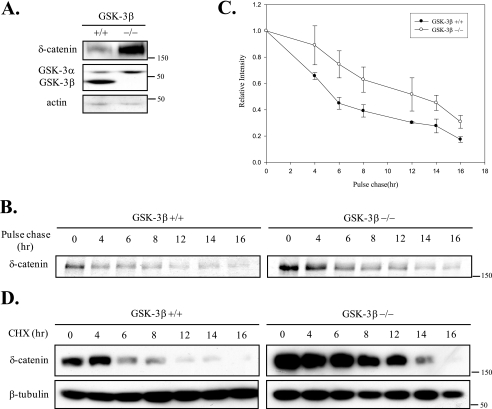
Most substrates of GSK-3 possess the recognition sequence (S/T)-Xaa-Xaa-Xaa-(S/T) (17). Our protein motif analysis revealed that δ-catenin contains several putative GSK-3 phosphorylation sequences. Because GSK-3 is a well established regulator of β-catenin (15) and p120-catenin (21) (prototypes of Arm family proteins), we decided to test our hypothesis that GSK-3 affects the stability of δ-catenin. The expression level of δ-catenin was found to be ∼2.6 times higher in GSK-3β−/− fibroblasts than in GSK-3β+/+ fibroblasts (Fig. 1A). Experiments of pulse-chase showed that the half-life of δ-catenin in GSK-3β−/− fibroblasts (12.3 h) was noticeably longer than that in GSK-3β+/+ fibroblasts (5.4 h) (Fig. 1, B and C). Treatment with cycloheximide (a protein synthesis inhibitor) also showed that the half-life of δ-catenin in GSK-3β−/− fibroblasts (10.2 h) was longer than that in GSK-3β+/+ fibroblasts (5.6 h) (Fig. 1D). These results indicated that GSK-3 does affect δ-catenin stability.
FIGURE 1.
GSK-3β affects the level of δ-catenin. A, δ-catenin was transfected into GSK-3β+/+ fibroblast cells and GSK-3β−/− fibroblast cells. δ-Catenin, GSK-3α and -3β, and actin proteins were detected using its specific antibody. B, GSK-3β+/+ and GSK-3β−/− fibroblast cells were transfected with δ-catenin. The cells were treated with methionine/cysteine-free DMEM (Sigma) containing 1% fetal bovine serum and 0.04 m l-glutamine (Sigma) overnight. After cells were treated with new conditioned medium containing 150 μCi of 35S for 1 h, the cells were harvested at indicated time. The extracts were immunoprecipitated with anti-δ-catenin. C, graph represented the relative intensities of δ-catenin. Band intensities at the various time points in B were measured using TINA 2.09 software program (Raytest). D, after GSK-3β+/+ fibroblast cells and GSK-3β−/− fibroblast cells had been transfected with δ-catenin, the cells were treated with cycloheximide (40 μg/ml) and harvested at the times indicated. β-Tubulin was used as a loading control.
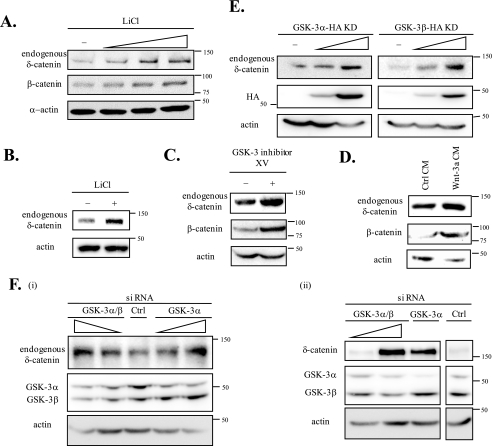
To confirm that GSK-3 negatively affects the level of δ-catenin, we used four different approaches that are known to specifically inhibit GSK-3 activity: 1) treatments with a GSK-3-specific chemical inhibitor, including LiCl and GSK-3 inhibitor XV; 2) Wnt-3a conditioned media; 3) the overexpression of GSK-3α KD or GSK-3β KD; and 4) siRNA specific for either GSK-3α/β or GSK-3α. First, we examined the effects of LiCl on endogenous δ-catenin levels. In CWR22Rv-1 cells (a prostate cancer cell line expressing δ-catenin (22)) endogenous δ-catenin levels were increased by treatment with LiCl (up to 30 mm). We used β-catenin, the expression of which is known to be affected by GSK-3 (15), as a positive control (Fig. 2A). Levels of endogenous δ-catenin in primary cortical neurons were increased by LiCl treatment (Fig. 2B). We also identified that levels of endogenous δ-catenin were increased by GSK-3 inhibitor XV in CWR22Rv-1 cells (Fig. 2C). Second, we examined the effects of Wnt-3a conditioned media on levels of endogenous δ-catenin in CWR22Rv-1 cells. The results obtained revealed that culture with Wnt-3a conditioned media increased levels of endogenous δ-catenin and of β-catenin compared with culture in control conditioned media (Fig. 2D). Moreover, the overexpression of either GSK-3α KD or GSK-3β KD induced dose-dependent increases in endogenous δ-catenin levels (Fig. 2E). The transfection with siRNA knocking down either GSK-3α/β or GSK-3α significantly increased the level of exogenous δ-catenin in GSK-3β+/+ fibroblasts and endogenous δ-catenin in CWR22Rv-1 cells as well (Fig. 2F). All of the above results indicate that GSK-3 negatively regulates δ-catenin levels.
FIGURE 2.
GSK-3 negatively affects δ-catenin levels. A, CWR22Rv-1 cells were treated with different concentrations of LiCl, a specific inhibitor of GSK-3 (10, 20, and 30 mm) for 12 h. The cells were then harvested, and the endogenous δ-catenin was detected using anti-δ-catenin antibody. β-Catenin was used as positive control (Ctrl) to determine the effect of LiCl, and α-actin was used as a loading control. B, primary cortical neurons were treated with 1 mm of LiCl for 12 h. Endogenous δ-catenin was detected using anti-δ-catenin antibody. Actin was used as a loading control. C, CWR22Rv-1 cells were treated with GSK-3 inhibitor XV (0.6 nm; Calbiochem) for 16 h. β-Catenin was used as a positive control, and actin was used as a loading control. D, CWR22Rv-1 cells were treated with Wnt-3a or control conditioned media for 8 h. Endogenous δ-catenin was detected using anti-δ-catenin antibody. β-Catenin was used as a positive control, and actin was used as a loading control. E, CWR22Rv-1 cells were transfected with two different doses of the GSK-3α and -3β KD tagged with HA. GSK-3α and -3β KD were detected with anti-HA and actin as a loading control. F, CWR22Rv-1 cells were transfected with either siRNA for GSK-3α/β or GSK-3α (panel i) and GSK-3β+/+ cells were transfected with δ-catenin together with either siRNA for GSK-3α/β or GSK-3α (panel ii). Control siRNA was also transfected and used as a negative control. Endogenous (panel i) or exogenous δ-catenin (panel ii) was detected with anti-δ-catenin antibody, and GSK-3α/β was detected with anti-GSK-3α/β antibody.
Both GSK-3α and -3β Interact with and Phosphorylate δ-Catenin, and Thr1078 of δ-Catenin Is One of Multiple Phosphorylation Sites by GSK-3
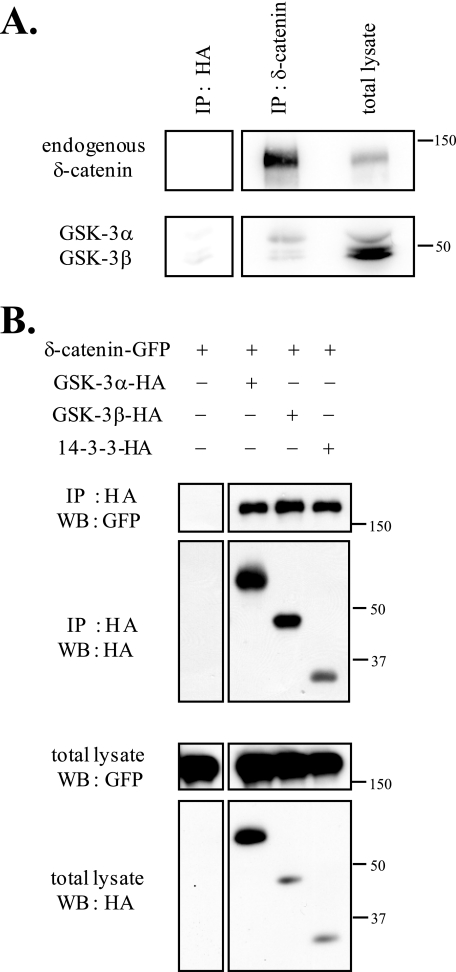
To demonstrate that GSK-3α and GSK-3β interact with endogenous and exogenous δ-catenin, we performed immunoprecipitation assays (Fig. 3). Purified immunocomplex of 1 day post-natal mouse brain lysate with anti-δ-catenin antibody was used for immunoprecipitation (Fig. 3A). As shown in Fig. 3A, both GSK-3α and -3β interacted well with endogenous δ-catenin. For the immunoprecipitation assay of exogenous δ-catenin, 14-3-3 (a known binding partner of δ-catenin) was used as a positive binding control (23). We used an anti-HA antibody (Fig. 3B, top two panels) to immunoprecipitate GSK-3α-HA or GSK-3β-HA immunocomplexes, and immunoprecipitated proteins were immunoblotted using the same antibody (Fig. 3B, bottom two panels). As shown in Fig. 3B, both GSK-3α and -3β interacted well with exogenous δ-catenin.
FIGURE 3.
δ-Catenin interacts with GSK-3α or GSK-3β. A, purified immunocomplex of 1 day post-natal mouse brain lysate with anti-δ-catenin antibody was used for immunoprecipitation (IP). Endogenous δ-Catenin was detected using anti-δ-catenin antibody, and endogenous GSK-3α and -3β proteins were detected using anti-GSK-3 (α + β) antibody. HA antibody was used as a negative control. B, δ-catenin-GFP was co-transfected with GSK-3α, GSK-3β, or 14-3-3 into HEK293 cells to determine whether it interacts with GSK-3α and GSK-3β. The interaction between δ-catenin and 14-3-3 was used as a positive control. Immunoprecipitations were performed using HA antibody, and immunoprecipitated proteins were immunoblotted with anti-GFP antibody or HA antibody. WB, Western blot.
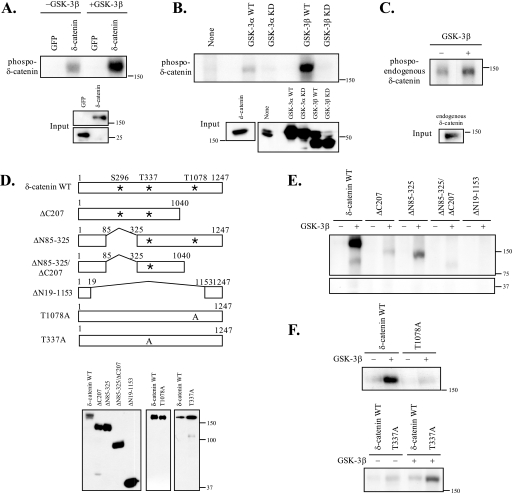
Next, we performed in vitro kinase assays using two different approaches. First, we used a purified GSK-3β enzyme and [γ-32P]ATP. We immunoprecipitated proteins with specific antibodies (anti-GFP or -δ-catenin antibody), divided them equally into three tubes, and used two tubes for GSK-3 kinase assays (one tube without GSK-3β enzyme and one tube with GSK-3β) and one tube for Western blotting to determine its relative loading. As shown in Fig. 4A, incubation with GSK-3β purified enzymes significantly increased the phosphorylation of δ-catenin, but not of GFP. The low levels of phosphorylated δ-catenin observed in non-GSK-3β-incubated samples may have been due to low levels of kinases co-immunoprecipitated with δ-catenin (Fig. 4A). Second, we performed in vitro kinase assays using GSK-3 immunocomplexes and [γ-32P]ATP as shown in Fig. 4B. We demonstrated that the GSK-3β WT noticeably increased the phosphorylated δ-catenin, whereas the GSK-3β KD did not. In addition, we found that GSK-3α phosphorylates δ-catenin as well (Fig. 4B). We also confirmed phosphorylation of endogenous δ-catenin by in in vitro kinase assays using 1 day post-natal mouse brain lysates (Fig. 4C).
FIGURE 4.
GSK-3 phosphorylates δ-catenin, and the Thr1078 residue of δ-catenin is one of multiple phosphorylation sites by GSK-3. In vitro kinase assays were performed as described under “Experimental Procedures.” A and E, GFP protein was used as a negative control. Full-length (A) or deletion constructs of δ-catenin (ΔC207, ΔN85–325, ΔN85–325/C207, and ΔN19–1153) (E) were transiently transfected into HEK293 cells. The level of each protein used for kinase assays was represented by Western blot (D). B, GSK-3α WT/KD, GSK-3β WT/KD, or δ-catenin were transfected into HEK293 cells. Each immunocomplex was formed by immunoprecipitating with anti-HA antibody or anti-δ-catenin antibody and protein G-Sepharose. HEK293 cell lysate (indicated as none) was used as a negative control. After mixing δ-catenin immunocomplex and the immunocomplexes of GSK-3α WT/KD or GSK-3β WT/KD, the mixed immunocomplexes were incubated for 30 min at 30 °C in a 20-μl reaction mixture with 10 μCi of [γ-32P]ATP. C, purified immunocomplex of 1 day post-natal mouse brain lysate with anti-δ-catenin antibody was used as substrates for in vitro kinase assays. D, a schematic diagram of δ-catenin constructs. F, T1078A-δ-catenin, T337A-δ-catenin, or δ-catenin WT were transiently transfected into HEK293 cells, and δ-catenin immunocomplexes were used for GSK-3 kinase assays as described under “Experimental Procedures.”
Our protein motif scan analysis for putative phosphorylation sites on δ-catenin by GSK-3 revealed that Ser296, Thr337, and Thr1078 on mouse δ-catenin are putative sites based on medium stringency searches. To map the GSK-3 phosphorylation site on δ-catenin, we used four different deletion δ-catenin constructs containing different numbers of putative GSK-3 phosphorylation sites and two different point-mutated δ-catenin constructs, as illustrated in Fig. 4D. As shown in Fig. 4E, ΔN19–1153-δ-catenin, containing none of these putative GSK-3 phosphorylation sites, showed no phosphorylation. ΔN85–325-δ-catenin, lacking one putative Ser296 site, showed a reduced level of δ-catenin phosphorylation by GSK-3. However, the ΔC207-δ-catenin deletion mutant, lacking a putative Thr1078 GSK-3 phosphorylation site, showed a greater reduction in phosphorylation, thus suggesting that Thr1078 can be a major phosphorylation site. ΔN85–325/ΔC207-δ-catenin, containing only one Thr337 putative GSK-3 phosphorylation site, showed low but detectable levels of phosphorylated δ-catenin. Thus, even though Thr1078 of δ-catenin can be a major phosphorylation site by GSK-3, our results also suggested that GSK-3 phosphorylate multiple sites in δ-catenin, as it does to other substrates. To directly demonstrate that Thr1078 of δ-catenin is one of phosphorylation sites by GSK-3, a δ-catenin T1078A mutant, in which the Thr1078 residue was substituted to Ala, was generated by site-directed mutagenesis. As shown in Fig. 4F, the δ-catenin T1078A mutant showed a significant reduction in phosphorylation by GSK-3 compared with that of wild type δ-catenin, demonstrating that Thr1078 of δ-catenin is indeed one of multiple phosphorylation sites by GSK-3. In contrast, the δ-catenin T337A mutant showed no reduction in phosphorylation by GSK-3 compared with that of wild type δ-catenin, ruling out the possibility that Thr-337 of δ-catenin is one of the phosphorylation sites by GSK-3 (Fig. 4F).
To investigate the phosphorylation status of putative site(s) on δ-catenin in GSK-3β+/+ and GSK-3β−/− fibroblasts, we performed LC-MS/MS analyses with tryptic peptide fragment of δ-catenin obtained from above fibroblasts. The position of phosphorylated residue in the tryptic fragment, which was detected in GSK-3β+/+ and/or GSK-3β−/− fibroblasts, was summarized in Table 1. Among three putative GSK-3 phosphorylation sites on δ-catenin, Thr1078 residue was phosphorylated only in GSK-3β+/+ but not in GSK-3β−/− fibroblasts (Table 1 and supplemental Fig. S1). Ser296 was phosphorylated in both GSK-3β+/+ and GSK-3β−/− fibroblasts. Thr337 was not phosphorylated in GSK-3β+/+ and GSK-3β−/− fibroblasts. As shown in Table 1, five Thr and seven Ser residues were constitutively phosphorylated in both GSK-3β+/+ and GSK-3β−/− fibroblasts. The phosphorylation of Tyr was relatively rare, and only Tyr954 was constitutively phosphorylated in both cell lines. Interestingly, Ser769 on δ-catenin was phosphorylated in GSK-3β+/+ fibroblasts but not in GSK-3β−/− fibroblasts.
TABLE 1.
Ser/Thr phosphorylation sites in mouse δ-catenin
List of the tryptic peptide fragments recovered from mouse δ-catenin in GSK-3β+/+ and GSK-3β−/− fibroblasts. + indicates specific phosphorylation of Ser/Thr site(s), whereas − indicates that the Ser/Thr site(s) were not phosphorylated. Amino acid numbers are based on the mouse δ-catenin sequence.
| Position | Tryptic fragment | GSK-3β+/+ fibroblasts | GSK-3β−/− fibroblasts |
|---|---|---|---|
| Thr66 | LT*RELEAER | + | + |
| Ser192, Thr193 | S*T*QAR | + | + |
| Ser216 | AGHLAGS*EPAPPPPPPR | + | + |
| Ser295, Ser296 | GS*S*PKQ | + | + |
| Thr337 | VTSPPTVQST*ISSSPIHQLSSLI QTYATLSPTK | − | − |
| Thr645, Thr646 | LLRKT*T*DLEI | + | + |
| Thr713 | NAT*GCLR | + | + |
| Ser769 | NLS*YR | + | − |
| Ser1018 | LVGISKS*KGDK | + | + |
| Ser1046 | S*LYK | + | + |
| Thr1078 | T*PSISPVRVSPNNR | + | − |
| Ser1087 | VS*PNNRSASAPASPREMISLK | − | + |
| Ser1094 | VSPNNRSAS*APASPREMISLK | + | + |
δ-Catenin Undergoes GSK-3-mediated Ubiquitination and Subsequent Proteosome-mediated Degradation
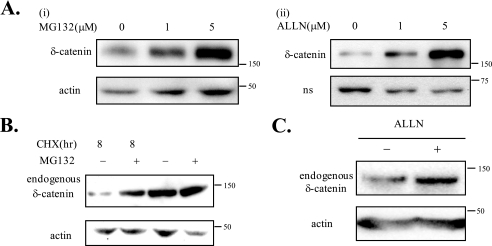
We decided to test the possibility that δ-catenin phosphorylation by GSK-3 triggers its degradation via an ubiquitination/proteosome-dependent pathway. To determine whether δ-catenin is degraded in a proteosome-dependent manner, we used three different methods. As shown in Fig. 5A, treatments with either MG132 or ALLN (specific inhibitors of proteosome-dependent proteolysis) significantly enhanced exogenous δ-catenin levels in MEFs in a dose-dependent manner. After treatments with CHX, a specific protein synthesis inhibitor, for 8 h, the level of δ-catenin was hardly detectable, but we could observe that the level of δ-catenin was increased by treatments with MG132 (Fig. 5B, first lane versus second lane). When cells were not treated with CHX, we could observe the same pattern (Fig. 5B, third lane versus fourth lane). The effects of ALLN were confirmed on endogenous δ-catenin in CWR22Rv-1 cells (Fig. 5C). These results consistently demonstrate that δ-catenin undergoes proteosome-dependent proteolysis.
FIGURE 5.
δ-Catenin undergoes the proteosomal degradation. A, to determine the effects of MG132 or ALLN on exogenous δ-catenin, MEF cells were transfected with δ-catenin and treated with different concentrations of MG132 or ALLN for 4 h prior to lysis (upper panels). δ-Catenin-GFP proteins were detected by anti-δ-catenin (panel i) or anti-GFP (panel ii) antibody. Actin (panel i) and nonspecific band (indicated as ns) of anti-GFP (panel ii) were used as a loading control. B, to determine the effects of MG132 on endogenous δ-catenin, CWR22Rv-1 cells were treated with CHX for 8 h (first lane) or with MG132 for 2 h followed by CHX for 8 h (second lane) left with no treatments (third lane) or with MG132 alone for 2 h (fourth lane) prior to lysis. Proteins of δ-catenin were detected by anti-δ-catenin antibody, and actin was used as a loading control. C, to determine the effects of ALLN on endogenous δ-catenin, CWR22Rv-1 cells were treated with ALLN for 4 h prior to lysis. Actin was used as a loading control.
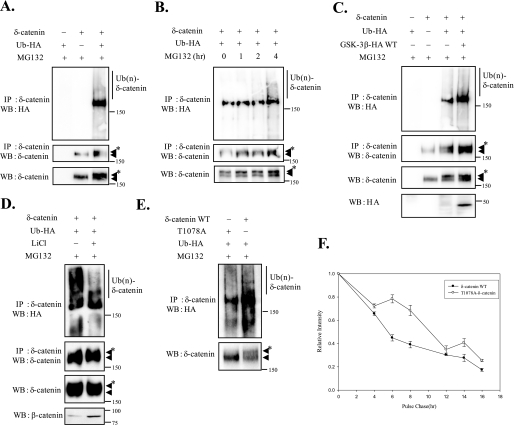
To test that δ-catenin can be ubiquitinated, we transfected MEF cells with δ-catenin together with or without HA-tagged Ub and treated them with MG132. Immunoprecipitation assays followed by Western blotting analysis detected a ladder of HA-marked δ-catenin (Fig. 6A). Moreover, these ubiquitinated δ-catenin bands were multiple and smeared, which is a characteristic of the banding pattern of poly-ubiquitinated proteins. Moreover, when we treated cells with MG132 for different times, we observed increases in the levels of total δ-catenin (Fig. 6B, bottom panel) and concomitant accumulation of ubiquitinated δ-catenin (Fig. 6B, top panel), suggesting that ubiquitinated δ-catenin undergoes subsequent proteosome-dependent degradation.
FIGURE 6.
δ-Catenin undergoes GSK-3-mediated ubiquitination and proteosome-dependent degradation. A, MEF cells were transfected with Ub-HA alone, δ-catenin alone, or Ub-HA+δ-catenin as indicated. The cells were treated with MG132 for 4 h prior to lysis and then anti-δ-catenin immunoprecipitations (IP) followed by anti-HA or anti-δ-catenin Western analyses were carried out on the extracts (top and middle panels). Anti-δ-catenin Western blot (WB) analysis was also performed on cell extracts (bottom panel). B, MEF cells were transfected with δ-catenin and Ub-HA. The cells were treated with MG132 for different times, as indicated. The extracts were subjected to immunoprecipitation and Western analysis with anti-HA or anti-δ-catenin antibody (top and middle panels). For lysates Western analysis was also performed with anti-δ-catenin (bottom panel). C, MEF cells were transfected Ub-HA alone, δ-catenin alone, and co-transfected with δ-catenin, Ub-HA, and with or without GSK-3β WT. Extracts of adjusted amounts of δ-catenin in these two groups to be equal were immunoprecipitated by anti-δ-catenin antibody, and Western analyses were performed by anti-HA or anti-δ-catenin antibody (top and second panels). Anti-δ-catenin antibody was used for detecting δ-catenin and ubiquitinated δ-catenin on cell extracts (third panel). Anti-HA antibody was used for detecting GSK-3β WT (bottom panel). D, MEF cells were co-transfected with δ-catenin and Ub-HA. The cells were treated with or without LiCl for 4 h and then treated with MG132 for 2 h. Immunoprecipitations were performed with anti-δ-catenin, and Western analyses were performed with anti-HA or anti-δ-catenin (top and second panels). Anti-δ-catenin Western blot analysis was also performed on cell extracts (third panel). β-Catenin was used for confirming effects of LiCl on cells (bottom panel). E, MEF cells were transfected with Ub-HA and with either T1078-δ-catenin or δ-catenin WT. The cells were treated with MG132 for 4 h prior to lysis. Immunoprecipitations were performed with anti-δ-catenin, and Western analyses were performed with anti-HA (top panel). Anti-δ-catenin Western blot analysis was also performed on cell extracts (bottom panel). ◀ indicates δ-catenin and ◀* indicates ubiquitinated δ-catenin. F, GSK-3β+/+ fibroblasts were transfected with either wild type or T1078A δ-catenin. Pulse-chase experiments were performed as described in the legend to Fig. 1. F, graph represented the relative intensities of δ-catenin. Band intensities at the various time points were measured using TINA 2.09 software program (Raytest).
To test the possibility that δ-catenin phosphorylation by GSK-3 affects its ubiquitination, we transfected δ-catenin and HA-Ub together with GSK-3β wild type and compared ubiquitinated δ-catenin levels in these two groups (Fig. 6C). Because GSK-3 negatively regulates the stability of δ-catenin, we adjusted amounts of δ-catenin in these two groups to be equal to judge the level of ubiquitinated δ-catenin in the same amounts of stable δ-catenin. As was expected, the overexpression of GSK-3β WT induced an increase in the level of ubiquitinated δ-catenin. A decrease in the level of ubiquitinated δ-catenin was also detected after treatment of LiCl with an increase in total δ-catenin levels (Fig. 6D). We have also compared the ubiquitination levels between wild type and T1078A δ-catenin. As shown in Fig. 6E, wild type δ-catenin showed more ubiquitination (Ub(n)- δ-catenin in the top panel; arrowhead with an asterisk in the bottom panel) compared with the T1078A δ-catenin mutant. These results indicate that the GSK-3-mediated phosphorylation of δ-catenin triggers its ubiquitination and subsequent proteosome-dependent degradation. Pulse-chase experiment revealed that half-life of δ-catenin T1078A mutant (10.4 h) was longer than wild type δ-catenin (5.6 h) as expected (Fig. 6F).
DISCUSSION
δ-Catenin is mainly expressed in brain and has been implicated in the regulation of synaptic plasticity and cognitive function via its interaction with Cadherins, PS-1, PSD-95, and GRIP (24). The aberrant expression of δ-catenin has been reported in patients with Cri-du-Chat syndrome, a form of mental retardation (5). Furthermore, severe learning deficits and abnormal synaptic plasticity were observed in δ-catenin-deficient mice (6). In adult humans, the overexpression of δ-catenin has been reported in various tumors (22), including those of breast and prostate carcinomas, which suggests that levels of δ-catenin should be tightly regulated in cells in terms of both location and amount. However, few attempts have been made to identify the factor(s) that affect(s) the stability of δ-catenin in cells. In the present study, we found that GSK-3 is a strong negative regulator of δ-catenin and that GSK-3 interacts with and phosphorylates δ-catenin triggering its ubiquitination and subsequent proteosome-dependent degradation.
In an attempt to identify the regulators of δ-catenin, GSK-3 was chosen for several reasons. Most importantly p120-catenin and β-catenin, which are two major proteins in the Arm repeat protein family, have been reported as GSK-3 substrates (15, 21), and δ-catenin shares many of the characteristics of p120-catenin. Most of the binding partners of p120-catenin, including cadherins, PS-1, and Kaiso (25), have also been reported to bind δ-catenin (1, 9, 10). β-Catenin also binds PS-1 and E-cadherin but utilizes binding domains that differ from those used by p120-catenin or δ-catenin (10, 26). Moreover, GSK-3 is known to phosphorylate four different Ser/Thr residues on β-catenin at its N terminus (27) and to phosphorylate the Ser252 and Thr310 residues of p120-catenin (in vitro but not in vivo) (21), indicating that the central Arm repeat domains may not be a common target for phosphorylation by GSK-3. These similar binding characteristics of Arm repeat family proteins placed GSK-3 on top of our list as a potential regulator of δ-catenin. Another intriguing hint emerged from computerized analyses of protein motif scans of δ-catenin. Substrates of GSK-3 have the (S/T)-Xaa-Xaa-Xaa-(S/T) consensus sequence, and analysis revealed that mouse δ-catenin contains three putative GSK-3 phosphorylation sites (Ser296, Thr337, and Thr1078) based on medium stringency searches. Therefore, we decided to test our hypothesis that δ-catenin is regulated by GSK-3.
The half-life of δ-catenin in GSK-3β−/− fibroblasts was found to be almost twice that in GSK-3β+/+ fibroblasts, suggesting that GSK-3 may indeed negatively regulate the stability of δ-catenin. The results of the four different approaches used to inhibit the activity of GSK-3 also supported our hypothesis. Furthermore, the interaction between δ-catenin and GSK-3α or GSK-3β was proven by performing immunoprecipitation assays, and the phosphorylation of δ-catenin by GSK-3 was demonstrated by in vitro GSK-3 kinase assays. Moreover, a computer motif scan and an in vitro kinase assay using various deletion constructs of δ-catenin showed that Thr1078 can be a major phosphorylation site by GSK-3. However, both ΔC207-δ-catenin deletion mutant (lacking the putative Thr1078 GSK-3 phosphorylation site) and T1078A site-directed δ-catenin mutant showed low but detectable levels of phosphorylated δ-catenin. This suggests that multiple phosphorylation sites on δ-catenin are targeted by GSK-3, as is the case for other GSK-3 substrates such as β-catenin.
Using LC-MS/MS analyses, we compared the phosphorylation site(s) in GSK-3β+/+ versus GSK-3β−/− fibroblasts. Among 865 tryptic peptide sequences commonly recovered from both GSK-3β+/+ and GSK-3β−/− fibroblasts, five Thr, seven Ser, and one Tyr residues were constitutively phosphorylated in both cell lines. Among three putative phosphorylation sites on δ-catenin by GSK-3, we ruled out Thr337 as a GSK-3 target site because T337A δ-catenin mutant showed significant phosphorylation by GSK-3β (Fig. 4E). Ser296 on δ-catenin was constitutively phosphorylated in both GSK-3β+/+ and GSK-3β−/− fibroblasts (Table 1). However, we cannot rule out the possibility that Ser296 on δ-catenin can be still one of the multiphosphorylation sites by GSK-3 for two reasons. First, there is still active GSK-3α in GSK-3β−/− fibroblasts. Second, ΔN85–325-δ-catenin, lacking one putative Ser296 site, showed a reduced level of δ-catenin phosphorylation by GSK-3. Our biochemical data strongly suggested that Thr1078 can be a major phosphorylation site by GSK-3. The Thr1078 residue on δ-catenin was not phosphorylated in GSK-3β−/− fibroblasts. There was a report by others showing the phosphorylation of Thr1078 on δ-catenin in PSD preparations from murine brain (28), suggesting that Thr1078 on δ-catenin can be indeed phosphorylated. In addition to this, we provide evidence that Thr1078 residue on δ-catenin is a direct phosphorylation site by GSK-3. Interestingly, Ser769 on δ-catenin was phosphorylated in GSK-3β+/+ fibroblasts but not in GSK-3β−/− fibroblasts, suggesting that Ser769 can be one of the target phosphorylation sites by GSK-3β. Because many substrates of GSK-3, including β-catenin and p120-catenin, contain multiple phosphorylation sites, future experiments to pinpoint the additional phosphorylation sites on δ-catenin by GSK-3 are required.
Recently, Abu-Elneel et al. (29) reported the constitutively phosphorylated sites of endogenous δ-catenin in the rat brain. Interestingly, tryptic fragment of δ-catenin containing Thr337 was recovered by them, and they showed no phosphorylation on the Thr337 residue as we reported. We could not find any common constitutively phosphorylated residues on mouse δ-catenin, possibly because of differences either in the recovered tryptic fragments or in species (mouse versus rat). The sequence identity between human and mouse δ-catenin is 94%, whereas that between human and rat is 84%. The sequences of mouse and rat share 88% identity. Among 14 constitutively phosphorylated rat δ-catenin residues that Abu-Elneel et al. (29) reported, 12 residues were distributed between amino acids 200 and 450. The δ-catenin sequences in these areas were significantly varied between mouse and rat, showing only 46% identity, whereas those between human and mouse were 93% identical. In comparison, human and rat δ-catenin share only 49% identity between amino acids 200 and 450, suggesting that the mouse δ-catenin sequence is more closely related to human δ-catenin than is rat δ-catenin. Therefore, different kinases and phosphatases can be involved in the regulation of δ-catenin stability and/or function among different species.
The present study also demonstrates for the first time that δ-catenin undergoes ubiquitination and proteosome-dependent proteolysis. The question we set out to answer was whether the phosphorylation of δ-catenin by GSK-3 triggers its ubiquitination and subsequent proteosome-dependent proteolysis. Overexpressed GSK-3β WT significantly increased the levels of ubiquitinated and phosphorylated δ-catenin, whereas treatments with LiCl reduced levels of ubiquitinated δ-catenin. Thus, our results suggest that the phosphorylation of δ-catenin by GSK-3 triggers its ubiquitination and subsequent proteosome-dependent proteolysis. Recently published articles showed that phosphorylation promotes the association of SCF(β-TrCP) with its substrate (30–32). For example, phosphorylation of β-catenin at sites Ser22 and Ser37 on the DSGXXS motif is required for the interaction of β-catenin with the ubiquitin ligase SCF(β-TrCP) (30). There is a report that cAMP-dependent protein kinase triggers a cascade of Gli3 phosphorylation by GSK-3β and CK1 that leads to direct β-TrCP binding and ubiquitination by SCF(β-TrCP) (31). SCF(β-TrCP) is also known to bind wild type STAT1 but not nonphosphorylatable mutant STAT1 (S727A) (33). Even though our analyses of δ-catenin sequences showed that mouse δ-catenin lacks the DSG destruction motif, δ-catenin can be targeted by F-box proteins, including β-TrCP, through an unconventional recognition site as demonstrated in the case of cyclin D1 and β-TrCP association (34). Future experiments are required to identify the component(s) of δ-catenin ubiquitination, including the F-box protein.
In this study, we used two GSK-3 isoforms in parallel in most experiments. Both GSK-3α and GSK-3β interact with and phosphorylate δ-catenin. We observed slight differences between these GSK-3 isoforms in terms of their binding affinities to δ-catenin and their phosphorylating abilities. Furthermore, even though these GSK-3 isoforms have many substrates in common, several authors have suggested that they have unique functions in cells. For example, Phiel et al. (35) reported that GSK-3α siRNA specifically inhibits γ-secretase activity for APP but that GSK-3β siRNA does not. Our siRNA experiments showed that siRNA GSK-3α significantly increased the level of δ-catenin. Even though GSK-3β is relatively more abundant in human brain than GSK-3α (36), future experiments are required to clarifying their different effects on the functional aspects of δ-catenin in normal and aberrant expression states.
GSK-3 is abundantly expressed in the human brain, and its substrates have been implicated in the progression of Alzheimer disease. For example, GSK-3 induces the hyperphosphorylation of Tau and affects the formation of neurofibrillary tangles (37). GSK-3 also affects γ-secretase activity for APP and the subsequent production of Aβ (35). Furthermore, GSK-3β is known to form a tetrameric complex with PS-1 and β-catenin (38). Interestingly, familial Alzheimer disease PS-1 has been associated with β-catenin destabilization, possibly via GSK-3 (39). Moreover, because δ-catenin was first cloned as a binding partner of PS-1, our identification of δ-catenin as a new target of GSK-3 provides insight as to how GSK-3 affects brain function and the progression of Alzheimer disease. Many δ-catenin-binding proteins, including PS-1, cadherins, GRIP, and PSD-95, are known to play pivotal roles in synaptic plasticity and cognitive function. The expression of δ-catenin in adequate amounts and in the correct locations seems to be critical, because the natural or artificial down-regulation of δ-catenin results in an impaired brain function and severe form of mental retardation, e.g. Cri-du-Chat syndrome (5). Interestingly, δ-catenin overexpression has been reported in various cancers, including breast, esophageal, and prostate carcinoma (22). Therefore, it is important that we identify the molecules and/or signaling pathways that regulate δ-catenin expression. In this report, we provide the first biochemical evidence that GSK-3 phosphorylates δ- catenin and negatively regulates its stability via ubiquitination/proteosome-mediated proteolysis. Future experiments are needed to define its degradation pathways, i.e. to identify the ubiquitin ligases responsible, and to understand how aberrant expression of δ-catenin affects cellular functions.
Supplementary Material
Acknowledgments
We thank J. Woodgett for providing GSK-3β+/+ and GSK-3β−/− fibroblast cells.
This work was supported, in whole or in part, by National Institutes of Health Grants AG026630 (to Q. L.) and CA111891 (to Q. L.). This work was also supported by Korea Healthcare Technology R & D Project Grant A084462, funds from the Korean Ministry for Health, Welfare & Family Affairs (to K. K.), and a grant from the New Technology R & D Project (Gwang-Ju Technopark, Korea).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PS
- presenilin
- GSK
- glycogen-synthase kinase
- KD
- kinase dead
- MEF
- mouse embryonic fibroblasts
- WT
- wild type
- siRNA
- small interfering RNA
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- HA
- hemagglutinin
- DMEM
- Dulbecco's modified Eagle's medium
- CHX
- cycloheximide
- GFP
- green fluorescent protein
- STAT
- signal transducers and activators of transcription
- Ub
- ubiquitin
- ALLN
- N-acetyl-Leu-Lue-Nle-CHO.
REFERENCES
- 1.Zhou J., Liyanage U., Medina M., Ho C., Simmons A. D., Lovett M., Kosik K. S. (1997) Neuroreport 8, 2085–2090 [DOI] [PubMed] [Google Scholar]
- 2.Ide N., Hata Y., Deguchi M., Hirao K., Yao I., Takai Y. (1999) Biochem. Biophys. Res. Commun. 256, 456–461 [DOI] [PubMed] [Google Scholar]
- 3.Paffenholz R., Franke W. W. (1997) Differentiation 61, 293–304 [DOI] [PubMed] [Google Scholar]
- 4.Ho C., Zhou J., Medina M., Goto T., Jacobson M., Bhide P. G., Kosik K. S. (2000) J. Comp. Neurol. 420, 261–276 [PubMed] [Google Scholar]
- 5.Medina M., Marinescu R. C., Overhauser J., Kosik K. S. (2000) Genomics 63, 157–164 [DOI] [PubMed] [Google Scholar]
- 6.Israely I., Costa R. M., Xie C. W., Silva A. J., Kosik K. S., Liu X. (2004) Curr. Biol. 14, 1657–1663 [DOI] [PubMed] [Google Scholar]
- 7.Jones S. B., Lanford G. W., Chen Y. H., Morabito M., Kim K., Lu Q. (2002) Neuroscience 115, 1009–1021 [DOI] [PubMed] [Google Scholar]
- 8.Kosik K. S., Donahue C. P., Israely I., Liu X., Ochiishi T. (2005) Trends Cell Biol. 15, 172–178 [DOI] [PubMed] [Google Scholar]
- 9.Tanahashi H., Tabira T. (1999) Neuroreport 10, 563–568 [DOI] [PubMed] [Google Scholar]
- 10.Lu Q., Paredes M., Medina M., Zhou J., Cavallo R., Peifer M., Orecchio L., Kosik K. S. (1999) J. Cell Biol. 144, 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Q., Mukhopadhyay N. K., Griffin J. D., Paredes M., Medina M., Kosik K. S. (2002) J. Neurosci. Res. 67, 618–624 [DOI] [PubMed] [Google Scholar]
- 12.Kim J. S., Bareiss S., Kim K. K., Tatum R., Han J. R., Jin Y. H., Kim H., Lu Q., Kim K. (2006) Biochem. Biophys. Res. Commun. 351, 903–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodgett J. R. (1990) EMBO J. 9, 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeflich K. P., Luo J., Rubie E. A., Tsao M. S., Jin O., Woodgett J. R. (2000) Nature 406, 86–90 [DOI] [PubMed] [Google Scholar]
- 15.Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. (1997) EMBO J. 16, 3797–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaytor M. D., Orr H. T. (2002) Curr. Opin. Neurobiol. 12, 275–278 [DOI] [PubMed] [Google Scholar]
- 17.Fiol C. J., Mahrenholz A. M., Wang Y., Roeske R. W., Roach P. J. (1987) J. Biol. Chem. 262, 14042–14048 [PubMed] [Google Scholar]
- 18.Kim K., Sirota A., Chen Yh Y. H., Jones S. B., Dudek R., Lanford G. W., Thakore C., Lu Q. (2002) Exp. Cell Res. 275, 171–184 [DOI] [PubMed] [Google Scholar]
- 19.Kim H., Ki H., Park H. S., Kim K. (2005) J. Biol. Chem. 280, 22462–22472 [DOI] [PubMed] [Google Scholar]
- 20.Russell W. K., Park Z. Y., Russell D. H. (2001) Anal. Chem. 73, 2682–2685 [DOI] [PubMed] [Google Scholar]
- 21.Xia X., Mariner D. J., Reynolds A. B. (2003) Biochemistry 42, 9195–9204 [DOI] [PubMed] [Google Scholar]
- 22.Lu Q., Dobbs L. J., Gregory C. W., Lanford G. W., Revelo M. P., Shappell S., Chen Y. H. (2005) Hum. Pathol. 36, 1037–1048 [DOI] [PubMed] [Google Scholar]
- 23.Mackie S., Aitken A. (2005) FEBS J. 272, 4202–4210 [DOI] [PubMed] [Google Scholar]
- 24.Silverman J. B., Restituito S., Lu W., Lee-Edwards L., Khatri L., Ziff E. B. (2007) J. Neurosci. 27, 8505–8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodova M., Kelly K. F., VanSaun M., Daniel J. M., Werle M. J. (2004) Mol. Cell. Biol. 24, 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marambaud P., Shioi J., Serban G., Georgakopoulos A., Sarner S., Nagy V., Baki L., Wen P., Efthimiopoulos S., Shao Z., Wisniewski T., Robakis N. K. (2002) EMBO J. 21, 1948–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 28.Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., Burlingame A. L. (2008) Mol. Cell Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 29.Abu-Elneel K., Ochiishi T., Medina M., Remedi M., Gastaldi L., Caceres A., Kosik K. S. (2008) J. Biol. Chem. 283, 32781–32791 [DOI] [PubMed] [Google Scholar]
- 30.Megy S., Bertho G., Gharbi-Benarous J., Baleux F., Benarous R., Girault J. P. (2006) FEBS Lett. 580, 5411–5422 [DOI] [PubMed] [Google Scholar]
- 31.Tempé D., Casas M., Karaz S., Blanchet-Tournier M. F., Concordet J. P. (2006) Mol. Cell. Biol. 26, 4316–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setoyama D., Yamashita M., Sagata N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18001–18006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soond S. M., Townsend P. A., Barry S. P., Knight R. A., Latchman D. S., Stephanou A. (2008) J. Biol. Chem. 283, 16077–16083 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Wei S., Yang H. C., Chuang H. C., Yang J., Kulp S. K., Lu P. J., Lai M. D., Chen C. S. (2008) J. Biol. Chem. 283, 26759–26770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phiel C. J., Wilson C. A., Lee V. M., Klein P. S. (2003) Nature 423, 435–439 [DOI] [PubMed] [Google Scholar]
- 36.Lau K. F., Miller C. C., Anderton B. H., Shaw P. C. (1999) J. Pept. Res. 54, 85–91 [DOI] [PubMed] [Google Scholar]
- 37.Maccioni R. B., Muñoz J. P., Barbeito L. (2001) Arch. Med. Res. 32, 367–381 [DOI] [PubMed] [Google Scholar]
- 38.Tesco G., Tanzi R. E. (2000) Ann. N. Y. Acad. Sci. 920, 227–232 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Hartmann H., Do V. M., Abramowski D., Sturchler-Pierrat C., Staufenbiel M., Sommer B., van de Wetering M., Clevers H., Saftig P., De Strooper B., He X., Yankner B. A. (1998) Nature 395, 698–702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.