Abstract
The trophoblast-specific gene PLAC1 (placenta-specific 1) is ectopically expressed in a wide range of human malignancies, most frequently in breast cancer, and is essentially involved in cancer cell proliferation, migration, and invasion. Here we show that basal activity of the PLAC1 promoter is selectively controlled by ubiquitous transcription factor SP1 and isoform 2 of CCAAT/enhancer-binding protein β that we found to be selectively expressed in placental tissue and cancer cells. Binding of both factors to their respective elements within the PLAC1 promoter was essential to attain full promoter activity. Estrogen receptor α (ERα) signaling further augmented transcription and translation of PLAC1 and most likely accounts for the positive correlation between PLAC1 expression levels and the ERα status we observed in primary breast cancer specimens. DNA affinity precipitation and chromatin immunoprecipitation assays revealed that transactivation of the PLAC1 promoter by ligand-activated ERα is based on a nonclassical pathway independent of estrogen-response elements, by tethering of ERα to DNA-bound CCAAT/enhancer-binding protein β-2, and SP1. Our findings provide first insight into a novel and hitherto unknown regulatory mechanism governing selective activation of trophoblast-specific gene expression in breast cancer.
More than 100 years ago Scottish embryologist John Beard proposed a “trophoblastic theory of cancer” based on phenotypical similarities between the pregnancy trophoblast and cancer cells (1). In particular, these shared features are invasion in surrounding tissues (2), neovascularization (3), immunological escape (4), telomerase activity (5), aneuploidy (6), and epigenetic changes such as selective DNA hypermethylation (7). Based on these observations, it has long been speculated that cancer cells acquire characteristic traits by reactivation of embryonic or fetal gene programs, most prominently depicted by the ectopic production of oncofetal antigens, such as α-fetoprotein and carcinoembryonic antigen, and the aberrant production of chorionic gonadotropin and other trophoblastic hormones (8–10). The knowledge of the molecular mechanisms underlying ectopic activation of placenta-specific genes in cancer, however, is sparse.
In a recent study, we introduced PLAC1 as novel member of cancer-associated placental genes (11). The PLAC1 gene encodes a membrane-associated protein that is speculated to serve a receptor-like function modulating specific cell-cell or ligand-receptor interactions unique to the maternal-placental interface (12). PLAC1 is strictly confined to differentiated cells of the syncytiotrophoblast, in which it is expressed throughout human gestation, but underlies tight transcriptional repression in all other normal tissues (11–13). In the course of malignant transformation, however, PLAC1 is frequently activated and highly expressed in a variety of tumor types, in particular breast cancer. Applying RNA interference technology, we found that PLAC1 has a tumor-promoting role and is a critical factor for proliferation, migration, and chemotactic invasion of cancer cells (11). Most interestingly, treatment of human breast cancer cells with a PLAC1-specific antibody was sufficient to suppress cell proliferation and phosphorylation of AKT kinase, proposing PLAC1 as a target candidate for therapeutic antibody development against cancer.
PLAC1 in trophoblast cells seems to be modulated by growth factors important for normal trophoblast differentiation (13), but the mechanisms mediating ectopic activation of PLAC1 in neoplastic cells have not been specified so far. Identification of these mechanisms is likely to further shed light on the biological functions of PLAC1, its role in breast cancer carcinogenesis, and its suitability as therapeutic target. Moreover, such knowledge might help to gain a general understanding of the factors involved in activation of trophoblast-specific transcriptional programs in cancer. Having already shown that ectopic activation of PLAC1 is not linked to DNA hypomethylation of the promoter region (11), we now aimed to identify the cis-elements and trans-factors that regulate PLAC1 promoter activity in breast cancer cells.
EXPERIMENTAL PROCEDURES
Tissues and Cell Lines
Recombinant DNA work was done with the official permission and according to the rules of the State Government of Rhineland-Palatinate. Tissues were stored at −80 °C until use. Breast cancer cell lines SK-BR-3, MCF-7, and MDA-MB-231 were cultured in Dulbecco's modified Eagle's medium, 10% fetal calf serum. Prior to E23 treatment studies, MCF-7 cells were cultured in phenol red-free medium supplemented with charcoal-filtered fetal calf serum for 72 h. Cells were treated with 100 nm E2 (Sigma) and/or 5 μm ICI 182,780.
Transcription Factor Binding Assays
Binding of C/EBPβ and SP1 to the predicted binding sites was assessed using NoShift transcription factor assay kit (Novagen). Briefly, biotinylated double-stranded oligonucleotides covering the wild type (C/EBPβ, 5′-ACC ACC ATG TTG TAA CCC TCC-3′; SP1, 5′-CCT CCT CCC CGC CCT TCT TCC-3′) or the mutated (C/EBPβ, 5′-ACC ACC ATA CCA CGG TCC TCC-3′; SP1, 5′-CCT CCT AAA AGA AAT TCT TCC-3′) PLAC1 promoter-binding sites were incubated with nuclear extracts of MCF-7 cells. The extract/probe mixtures were transferred to a Streptavidin-coated 96-well plate and incubated for 1 h at 37 °C. Antibodies specific for C/EBPβ and SP1 (Abcam) were added to the samples and incubated for 1 h at 37 °C. The secondary antibody conjugated with horseradish peroxidase was incubated for 30 min at 37 °C. After final washing, TMB substrate was added to the samples to develop colorimetric signal before quenching the reaction with 1 n HCl. Sample absorbance was read at 450 nm on a Wallac Victor2 multiple label counter (PerkinElmer Life Sciences).
Small Interfering RNA Duplexes
The C/EBPβ small interfering RNA (siRNA) duplexes (Qiagen) targeted nucleotides 983–1001 (siRNA1) and 962–980 (siRNA2) of the C/EBPβ mRNA sequence (NM_005194.2) and SP1 siRNA duplexes targeted nucleotides 1984–2002 (siRNA1) and 5449–5467 (siRNA2) of the SP1 mRNA sequence (NM_138473.2). As control a nonsilencing siRNA duplex (sense 5′-r(UAA CUG UAU AAU CGA CUA G)dTdT-5′, antisense 5′-r(CUA GUC GAU UAU ACA GUU A)dGdA-3′) was used. Cells were transfected with 10 nm siRNA duplex using HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions.
RNA Extraction and Real Time RT-PCR
RNA extraction, first-strand cDNA synthesis and real time reverse transcription (RT)-PCR) were performed as previously described (11, 14). Real time quantitative expression analysis was performed in triplicates in a 40 cycle RT-PCR. After normalization to HPRT (sense 5′-TGA CAC TGG CAA AAC AAT GCA-3′; antisense 5′-GGT CCT TTT CAC CAG CAA GCT-3′, 62 °C annealing), expression of PLAC1 (sense 5′-AAA TTT GGC AGC TGC CTT CAC-3′; antisense 5′-TGA TGC CAC ATT CAG TAA CAC-3′, 60 °C annealing) was quantified using ΔΔCT calculation.
Luciferase Assays
The PLAC1 promoter region (−1022 to +12) was cloned into the basic pGL4 luciferase reporter vector (Promega). Variants containing mutated C/EBPβ- and SP1-binding sites were generated using QuickChange site-directed mutagenesis kit (Stratagene). 24 h prior to transfection, 4 × 105 cells were seeded into 6-well plates. Promoter constructs were co-transfected with an eGFP reporter construct as transfection and normalization control using Lipofectamine (Invitrogen). 48 h after transfection cells were collected and lysed, and luciferase activity was measured on an Infinite M200 microplate reader (Tecan).
Western Blots
Whole cell and tissue lysates were prepared using RIPA lysis buffer. For preparation of nuclear extracts, NucBuster protein extraction kit (Novagen) was used. Extracts were diluted in reducing sample buffer (Roth), subjected to SDS-PAGE, and subsequently blotted onto polyvinylidene difluoride membrane (Pall). Immunostaining was performed with antibodies reactive to PLAC1 (11), C/EBPβ, SP1, ERα, ERβ, and β-actin (all from Abcam), followed by detection of primary antibodies with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Jackson ImmunoResearch).
Generation of in Vitro-transcribed RNA and Electroporation of Cells
The coding sequence of PLAC1 was cloned into the pST1-A120 plasmid described recently (15–17). Plasmids were linearized, purified by phenol/chloroform extraction and sodium acetate precipitation, and used as templates for in vitro transcription (ivt) with Message-Machine kit (Ambion). ivtRNA concentration and quality were assessed by spectrophotometry and agarose/formaldehyde gel electrophoresis. 5 × 106 cells were suspended in 250 μl of X-VIVO-15 medium (Cambrex), transferred into a 4-mm gap sterile electroporation cuvette (Bio-Rad), electroporated (225 V, 250 microfarads) with 5 μg of ivtRNA using a Gene-Pulser II apparatus (Bio-Rad), and cultured for 12 h prior to experiments.
DNA Affinity Protein Assays (DAPA)
Biotinylated double-stranded oligonucleotides are described under “Transcription Factor Binding Assays.” 50 μg of nuclear extracts were preincubated with 40 μl of a 50% streptavidin-agarose slurry (Invitrogen) for 1 h at 4 °C with rotation. The supernatant collected by centrifugation was incubated with 0.2 μg of wild type or mutant biotin-labeled probe in binding buffer (60 mm KCl, 12 mm HEPES, pH 7.9, 4 mm Tris-HCl, 5% glycerol, 0.5 mm EDTA, 1 mm dithiothreitol, and 1× protease inhibitor mixture) overnight at 4 °C with rotation. DNA-protein complexes were then incubated with 40 μl of a 50% slurry of streptavidin-agarose (pre-equilibrated in the binding buffer for 1 h) overnight at 4 °C with gentle rotation. DNA-protein complexes were washed five times with the binding buffer. The pellet was resuspended in 25 μl of protein sample buffer (Roth) and then boiled for 5 min to dissociate the complexes. The proteins were resolved by PAGE followed by Western blot detection with specific antibodies.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed using the ChIP assay kit (Millipore) according to the manufacturer's instructions. Briefly, 1 × 107 MCF-7 cells under different treatments indicated in the corresponding experiments were used for each ChIP reaction. Cells were cross-linked with 1% formaldehyde and lysed in SDS lysis buffer. Soluble chromatin was prepared by sonication followed by immunoclearing. Immunoprecipitation was carried out at 4 °C overnight utilizing specific antibodies to transcription factors SP1 and C/EBPβ and transactivator ERα. Following immunoprecipitation, protein A-Sepharose beads and salmon sperm DNA were added. After several rounds of washing the precipitates were extracted three times with 1% SDS, 0.1% NaHCO3. After reversal of cross-linking with 5 m NaCl at 65 °C for 6 h, precipitates were treated with proteinase K, followed by phenol extraction and ethanol precipitation. The purified DNA was analyzed by conventional PCR and real time PCR for the presence of the human PLAC1 promoter fragment −348/−198 bp (sense 5′-CAA CAG CAA GCA CTA CAA GTG-3′; antisense 5′-GAA GCT CAA CTC GGT GCA CTT GTT C-3′) and for the fragment −1219/−1064 bp upstream to the promoter (sense 5′-AAG CAC TTA GGA CAG CAT CTG-3′; antisense 5′-TGA ATG ATA CCT ACT GTC ATG). PCR amplification of 1% of the soluble chromatin prior to immunoprecipitation was used as input control. The ChIP-precipitated DNA and input DNA were quantified by real time PCR using SYBR Green (Qiagen) in an ABI 7300 sequence detection system.
ChIP Re-immunoprecipitation
Complexes were eluted from the primary immunoprecipitation by incubation with 5 mm DTT at 37 °C for 20 min and diluted with buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl, pH 8.1) followed by re-immunoprecipitation with a different relevant antibody. Subsequent steps of ChIP re-immunoprecipitations were as for the initial immunoprecipitations.
RESULTS
C/EBPβ and SP1 Are Required for Activation of the PLAC1 Promoter in Breast Cancer Cells
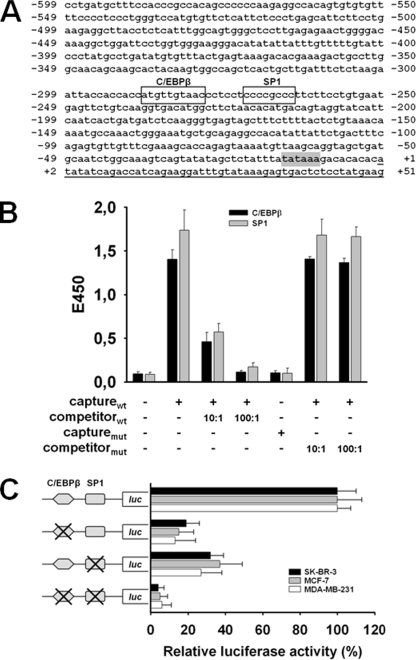
As a first step for exploration of PLAC1 gene regulation, we analyzed the sequence of the PLAC1 promoter region. This led to the identification of a typical TATA box element (TATAAA) at position −15 to −10 and the prediction of several other cis-acting elements (Fig. 1A). A sequence at position −286 to −278 closely resembling the consensus C/EBPβ-binding site attracted our attention, as C/EBPβ has been reported to transactivate several trophoblast-specific genes (18–22). The cytosine at position −278 did not match the consensus sequence T(T/G)NNGNAA(T/G); however, a considerable number of functional sites differ from the published consensus sequence at this position (23). It was particularly interesting to find a consensus sequence for specificity protein 1 (SP1) at position −271 to −264 in close vicinity to the C/EBPβ site, as this ubiquitous transcription factor is known to collaborate with cell-specific transcription factors serving as essential co-regulator of inducible or developmentally controlled genes (24–26).
FIGURE 1.
C/EBPβ and SP1 are required for activation of the PLAC1 promoter. A, proximal 5′-flanking region of the PLAC1 gene. C/EBPβ- (−286 to −278) and SP1 (−271 to −264)-binding elements are boxed; the TATA box element (−15 to −10) is shown in gray, and transcript start is underlined. B, enzyme-linked immunosorbent assay-based transcription factor binding assays were performed using nuclear lysates of MCF-7 cells. Transcription factors bound to the biotinylated capture probe harboring the respective binding element (capturewt) were detected by specific antibodies. competitorwt, nonbiotinylated capturewt probe; capturemut, biotinylated capture probe harboring the respective mutated binding element; competitormut, nonbiotinylated capturemut probe. C, activation of the PLAC1 promoter by C/EBPβ and SP1 was measured in luciferase (luc) activity assays 24 h after transfection of SK-BR-3, MCF-7, and MDA-MB-231 cells with luciferase reporter genes linked either to the wild type PLAC1 promoter domain (−1022 to +12) or domains with mutated C/EBPβ- and/or SP1-binding sites. Luciferase activity was normalized to fluorescence obtained by co-transfection with eGFP reporter plasmid. Activity of the wild type PLAC1 promoter domain was set to 100% for each individual cell line to allow direct comparison of mutated promoter domain activities between the cell lines analyzed. All assays were done in triplicate; mean values ± S.D. are shown.
Next, an enzyme-linked immunosorbent assay-based transcription factor binding assay was performed to verify that both transcription factors were able to bind to their respective elements. Incubation of nuclear cell extract from MCF-7 breast cancer cells as source of transcription factors with biotinylated oligonucleotide capture probes representing the putative C/EBPβ- and SP1-binding sequences within the PLAC1 promoter resulted in the formation of specific protein-DNA complexes (Fig. 1B). Mutation of the biotinylated oligonucleotide probes of either transcription factor binding site, however, suppressed binding of the respective transcription factors. Complex formation was inhibited by excess amounts of the nonbiotinylated capture probes as competitors, but not by competitor probes with mutated binding sites, confirming specificity of these interactions.
Having shown that C/EBPβ and SP1 were able to bind to their predicted nuclear binding sites, we wanted to assess whether binding of the two transcription factors was sufficient to activate the PLAC1 promoter. Luciferase reporter genes directed by the wild type PLAC1 promoter domain (−1022 to +12) or by domains with mutated C/EBPβ and/or SP1 protein binding sites were expressed in PLAC-positive SK-BR-3, MCF-7, and MDA-MB-231 breast cancer cells (11). In all three cell lines single mutation of the C/EBPβ-binding site decreased the promoter activity by 80–90%, and single mutation of the SP1 site caused marked reduction of the promoter activity by 60–70% (Fig. 1C). Double mutation of both elements nearly completely abrogated promoter activity in all cell lines, indicating that both factors are required to attain full PLAC1 promoter activity. These results showed that basal PLAC1 promoter activity in breast cancer cells is essentially regulated by C/EBPβ and SP1.
PLAC1 Is Selectively Activated by C/EBPβ Isoform 2
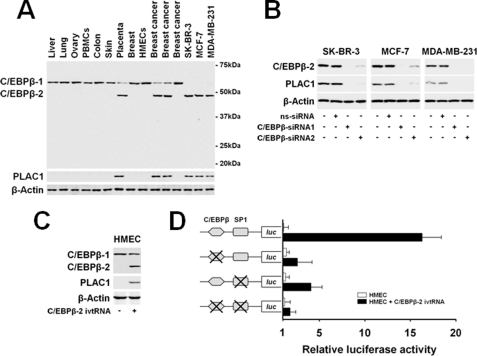
Constitutive expression of C/EBPβ has been reported in several human tissues, including liver, lung, ovary, placenta, and breast epithelial cells (27–30). These data required reanalysis as they did not comply with the highly specific expression of C/EBPβ-regulated PLAC1 in the trophoblastic lineage of placental tissue. Being aware of the existence of three C/EBPβ isoforms (C/EBPβ-1, 56 kDa; CEBPβ-2, 48 kDa; and C/EBPβ-3, 21 kDa) generated by differential initiation of translation, we resorted to an anti-C/EBPβ antibody directed against the C terminus of C/EBPβ capable of binding to all three protein isoforms. Analysis of an extended set of human tissues and cell lines disclosed C/EBPβ-2 as being selectively expressed in placenta and PLAC1-positive breast cancer samples and cell lines, whereas it was absent from all PLAC1-negative tissues (Fig. 2A). The broader expression pattern of C/EBPβ-1, in contrast, did not correlate at all with the tissue distribution of PLAC1. C/EBPβ-3 was not detected in any of the samples analyzed.
FIGURE 2.
PLAC1 is selectively activated by C/EBPβ isoform 2. A, protein expression of C/EBPβ isoforms and PLAC1 in tissues and cell lines analyzed by Western blot. β-Actin was used as loading control. B, protein expression in tumor cells 48 h after transfection with C/EBPβ-specific siRNA duplexes. ns-siRNA, nonsilencing siRNA. C, protein expression in HMECs 24 h after electroporation with C/EBPβ-2 ivtRNA (+) or eGFP ivtRNA as control (−). D, for measuring activation of the PLAC1 promoter by C/EBPβ-2 and SP1; HMEC cells were electroporated with C/EBPβ-2 or control ivtRNA, were transfected with luciferase (luc) reporter genes directed by the wild type PLAC1 promoter domain (−1022 to +12) or domains with mutated C/EBPβ- and/or SP1-binding sites, and subjected to luciferase activity assays after 24 h. Luciferase activity was normalized to fluorescence obtained by co-transfection with eGFP reporter plasmid. Assays were done in triplicate; mean values ± S.D. are shown.
To assess the impact of C/EBPβ-2 on PLAC1 expression more directly, we silenced C/EBPβ-2 expression in SK-BR-3, MCF-7, and MDA-MB-231 cells by transient transfection with two independent sets of siRNA duplexes, which resulted in loss of PLAC1 expression in all three cell lines (Fig. 2B). Analogously, introduction of C/EBPβ-2 into human mammary epithelial cells (HMECs) that only express C/EBPβ-1 by electroporation of in vitro transcribed RNA (ivtRNA) was sufficient to induce PLAC1 protein expression (Fig. 2C). As shown by luciferase reporter gene assays, the PLAC1 promoter was activated in HMECs in the presence of C/EBPβ-2 ivtRNA but was silent in the absence of this isoform or mutational disruption of at least one of the two binding sites for C/EBPβ-2 and SP1 within the promoter sequence (Fig. 2D). In summary, these investigations revealed C/EBPβ-2 to be essentially involved in selective activation of the PLAC1 gene.
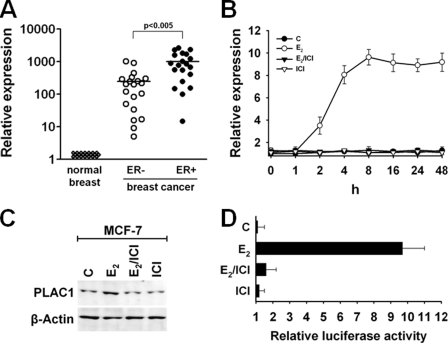
PLAC1 Is an Estrogen-responsive Gene
By correlation analyses, we sought to determine additional factors involved in transcriptional control of PLAC1 gene. Previously, we reported that PLAC1 expression in breast cancers was not associated with particular histological subtypes, tumor stage, or tumor grade (11). However, we found a striking correlation between PLAC1 expression and the ERα status of breast cancer samples (Fig. 3A). ERα-positive tumors display significantly higher PLAC1 expression levels compared with ERα-negative tumors. To analyze whether this reflects estrogen-induced transactivation, we quantified endogenous PLAC1 mRNA expression in ERα-positive MCF-7 breast cancer cells in response to treatment with E2. This was compared with cells, which were either not treated or treated with ER antagonist ICI 182,780 (ICI), which is known to efficiently inhibit the actions of endogenous and exogenous estradiol. Induction of PLAC1 expression was detectable as early as 2 h after addition of E2. Maximum induction of PLAC1 transcript levels to 9–10-fold higher as compared with control cells was reached after 8 h (Fig. 3B). This also resulted in increased PLAC1 protein levels in E2-treated cells but not in control cells (Fig. 3C). Importantly, PLAC1 protein was still detectable in the absence of E2, proving that basal expression of PLAC1 is not dependent on ERα signaling.
FIGURE 3.
PLAC1 is a 17β-estradiol-responsive gene. A, quantitative real time RT-PCR analysis of PLAC1 in breast cancers and matched normal breast tissues. Correlation of PLAC1 expression levels with ERα receptor status of breast cancer specimens (20 samples per group) was analyzed using unpaired t test. ER+, estrogen receptor positive; ER−, estrogen receptor negative. B, real time RT-PCR quantification of PLAC1 induction in MCF-7 cells treated with 100 nm E2, 100 nm E2, and 5 μm ER antagonist ICI 182,780 or 5 μm ICI 182,780 alone. Expression levels were normalized to untreated, estradiol-depleted control cells (c). Assay was done in triplicates and repeated twice; mean values ± STD are shown. C, Western blot analysis of PLAC1 expression in MCF-7 cells 12 h after treatment. D, luciferase activity was measured in MCF-7 cells transfected with the PLAC1 wild type promoter luciferase reporter gene 12 h after treatment. Luciferase activity was normalized to fluorescence obtained by co-transfection with eGFP reporter plasmid. Assay was done in triplicate; mean values ± S.D. are shown.
Transfection of MCF-7 cells with a luciferase reporter gene driven by the wild type PLAC1 promoter (−1022 to +12) resulted in 10-fold induction of luciferase activity after E2 treatment (Fig. 3D). This was not the case upon treatment with controls, indicating that the E2 regulatory elements are located within the cloned PLAC1 promoter sequence.
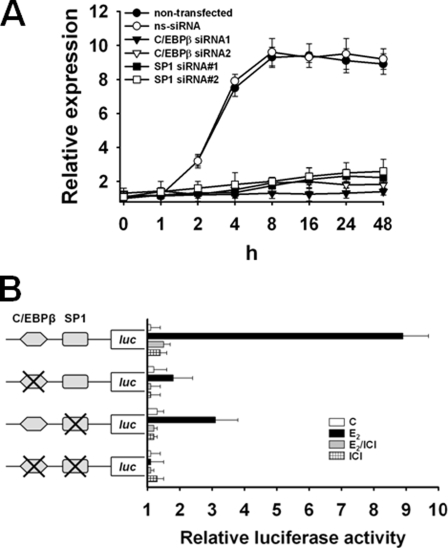
ERα-mediated Transactivation of PLAC1 Is Dependent on C/EBPβ-2 and SP1
Classically, activated ERα binds to estrogen-response elements (EREs) in target gene promoters and recruits cofactors to induce or repress transcription (31). We were not able to identify a classical ERE in the proximal 5′-flanking region of PLAC1, which implied that E2-mediated transactivation of PLAC1 may be directed by an ERE-independent ERα signaling pathway. ERα is capable of a nonclassical mode of action altering the transcription of genes in the absence of direct binding to DNA by engaging in protein-protein interaction with other transcription factors bound to their cognate DNA elements in the promoter of respective target genes (32, 33). We evaluated C/EBPβ-2 and SP1 as potential interaction partners of ERα for transactivation of PLAC1. siRNA-mediated silencing of C/EBPβ-2 and SP1 (supplemental Fig. 1) in MCF-7 cells abrogated inducibility of PLAC1 by treatment with E2, whereas E2 responsiveness of PLAC1 induction was not altered in untreated control cells or cells transfected with nonsilencing siRNA (Fig. 4A). Assays with luciferase reporter genes directed by the wild type PLAC1 promoter sequence, but not by variants with mutated C/EBPβ and/or SP1-binding sites, revealed the need for occupancy and functionality of both binding sites to allow for maximal E2-induced transactivation of PLAC1 (Fig. 4B).
FIGURE 4.
ERα-mediated transactivation of PLAC1 is dependent on C/EBPβ-2 and SP1. A, real time RT-PCR quantification of PLAC1 expression in MCF-7 cells in response to 100 nm E2 48 h after transfection of cells with C/EBPβ-specific siRNA duplexes. ns-siRNA, nonsilencing siRNA. Assay was done in triplicate and repeated twice; mean values ± S.D. are shown. B, luciferase reporter gene assay in MCF-7 cells measuring activity of the wild type PLAC1 promoter domain (−1022 to +12) or variants with mutated C/EBPβ- and/or SP1-binding sites 12 h after treatment with 100 nm E2, 100 nm E2 and 5 μm ER antagonist ICI 182,780, or 5 μm ICI alone. Luciferase activity was normalized to untreated, E2-depleted control cells (c), after normalization to fluorescence obtained by co-transfection with eGFP reporter plasmid. Assay was done in triplicate and repeated twice, shown are mean values ± S.D.
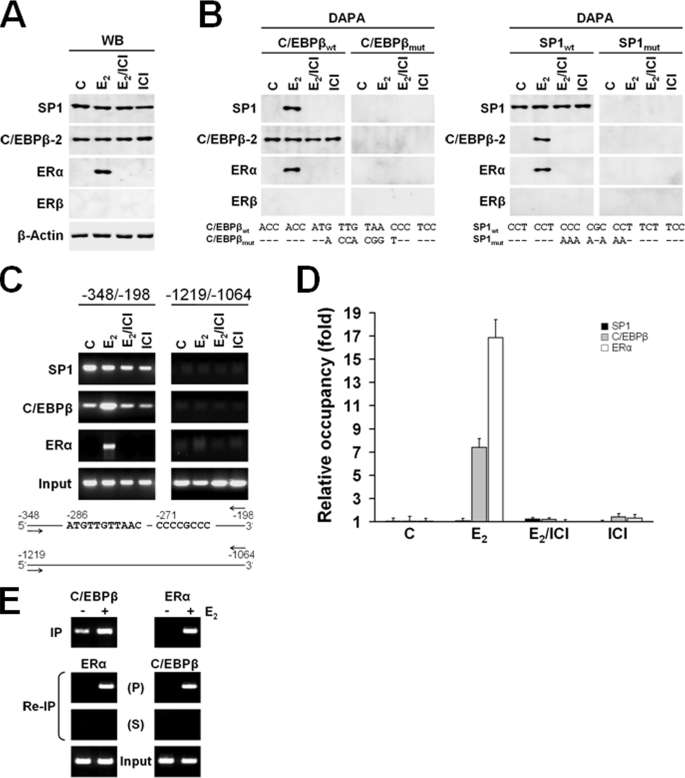
Activated ERα Directly Interacts with C/EBPβ-2 and SP1
To better understand the interaction of C/EBPβ-2 and SP1 with ERα, we conducted DAPA using biotin-labeled double-stranded oligonucleotides featuring wild type and mutant variants of C/EBPβ and SP1 sites derived from the PLAC1 promoter. These probes were incubated with nuclear extracts from untreated MCF-7 cells or MCF-7 cells treated with 100 nm E2, in the presence or absence of the estrogen receptor antagonist ICI, or with ICI alone. Immunoblots of these nuclear extracts showed that protein levels of SP1 and C/EBPβ-2 were not altered by these treatments (Fig. 5A). ERα levels increased significantly in response to E2 treatment, which was prevented by co-treatment with the E2 receptor antagonist ICI. Using an antibody specific to ERβ we confirmed previously published data that MCF-7 cells do not express ERβ protein (34, 35), proving that the observed E2-dependent effects were specifically mediated by activation of ERα. DNA affinity precipitation was conducted with these quality-controlled nuclear lysates to determine whether DNA-bound C/EBPβ-2 and SP1 target ERα to the complex (Fig. 5B). Most remarkably, probing with C/EBPβ-2 as well as SP1 wild type capture oligonucleotides allowed retrieval of a complex of ERα, C/EBPβ-2, and SP1 transcription factors from cells treated with E2 but not from cells that treated with E2/ICI or ICI. No binding or protein complex precipitation was shown with mutated probe variants of either transcription factor-binding site. These findings indicated that in addition to the mutual recruitment of ERα to SP1 and C/EBPβ, ERα could link both transfactors when only one of them is bound to DNA.
FIGURE 5.
Activated ERα directly interacts with C/EBPβ-2 and SP1. A, Western blot analysis of SP1, C/EBPβ-2, ERα, and ERβ expression in nuclear lysates of MCF-7 cells prepared 12 h after treatment with E2 and ICI 182,780. WB, Western blot. B, DAPA was performed using biotinylated wild type SP1 (SP1wt) and C/EBPβ (C/EBPβwt) probes or their mutated, inactive variants (SP1mut and C/EBPβmut), and nuclear lysates of MCF-7 cells were prepared 12 h after treatment with E2 and ICI 182,780. C, ChIP was performed with chromatin prepared from MCF-7 cells treated with E2 or without E2 (c) in the presence or absence of ICI or ICI alone. The promoter region containing the SP1 and C/EBPβ elements (−348/−198) and a region upstream to the promoter (−1219/−1064) as negative control were analyzed by PCR following immunoprecipitation with the antibodies indicated. Results of amplification of soluble chromatin prior to precipitation are shown as control (input). D, quantitative analysis of the recruitment/occupancy shown in C determined by real time PCR. The results corrected by input are shown as fold increase with unstimulated cells as reference. Samples from two individual ChIP assays were analyzed in triplicate. Results are shown as mean ± S.D. E, re-ChIP assays with sequential use of first C/EBPβ and than ERα antibodies (left) and ERα followed by C/EBPβ antibodies (right). P, precipitate; S, supernatant; IP, immunoprecipitate.
Next, we analyzed endogenous formation of the tertiary protein complex on the PLAC1 promoter in MCF-7 cells. ChIP assays showed no apparent differences in the recruitment of SP1 to the promoter in the presence or absence of E2 with or without ERα antagonist ICI (Fig. 5, C and D). In contrast, recruitment of C/EBPβ and ERα was strongly and specifically induced by E2. The association of these transfactors in the complex was further shown in re-ChIP assays by the sequential use of first C/EBPβ and thereafter ERα antibodies or vice versa (Fig. 5E). These observations not only demonstrate non-ERE-dependent tethering of E2-activated ERα to C/EBPβ-2 as well as SP1 bound to their cognate binding sites within the endogenous PLAC1 promoter. They further imply that transactivation of PLAC1 by E2-activated ERα is primarily mediated by enhanced recruitment of C/EBPβ-2 to its element after complex formation. Furthermore, as enhanced recruitment of C/EBPβ-2 to its element was not observed in DAPA despite complex formation with activated ERα and SP1, the presence of both C/EBPβ- and SP1-binding sites in the endogenous PLAC1 promoter appears to be a prerequisite for the formation of a functional transactivator complex.
DISCUSSION
In this study we provide a detailed molecular analysis of the regulatory mechanisms mediating ectopic activation of the trophoblast-specific gene PLAC1 in breast cancer. Our findings reveal a tandem-induced model for basal activity of the PLAC1 promoter. Cooperation of the two transcription factors C/EBPβ-2 and SP1 is essentially required for full promoter activity, with the single factors contributing 80 and 60%, respectively, to the expression level. Cooperation of ubiquitously expressed SP1 and C/EBPβ has previously been shown for the transcriptional control of several other tissue-specific genes (24–26).
Disclosure of C/EBPβ isoform 2 as key regulator is particularly interesting as it provides a rationale for the peculiar tissue distribution of PLAC1. C/EBPβ is known to be critically involved in regulation of both epithelial cell differentiation of the mammary gland (36, 37) as well as development of placental tissue (28). Three different protein isoforms of C/EBPβ have been described, which are generated by alternative translation initiation from the C/EBPβ transcript at three in-frame ATG initiator codons (29, 38–40). The largest variant C/EBPβ-1 (LAP* in rodents) is transactivating, as is C/EBPβ-2 (LAP in rodents), which lacks the 23 N-terminal amino acids of C/EBPβ-1. The isoform C/EBPβ-3 (LIP in rodents), in contrast, is considered to act as a dominant negative repressor, as it completely lacks the N-terminal activation domain while retaining the DNA binding/dimerization domain. The majority of studies dissecting C/EBPβ tissue distribution and function do not differentiate between these variants. One of the rare reports comparing the individual isoforms has shown that C/EBPβ-1 is exclusively expressed in normal breast cells, whereas C/EBPβ-2 was detected in 70% of invasive primary breast tumor samples and human breast cancer cell lines (41). Our data confirmed this observation and further extended them by profiling a comprehensive number of normal tissues, showing for the first time strict restriction of C/EBPβ-2 to placenta, the only normal tissue expressing PLAC1. The fact that C/EBPβ-2 introduction into HMECs, which constitutively express only C/EBPβ-1, is sufficient to activate the PLAC1 promoter and initiate protein expression provides further evidence for the specific role of C/EBPβ-2 in PLAC1 regulation.
The mechanisms that drive selective expression of C/EBPβ-2 in human cancer cells remain unknown. Intriguingly, for another of its target genes, placental lactogen, aberrant expression in primary breast cancers with frequencies in the range of >80% has been reported (42). This is in analogy to our findings for PLAC1 (11) and suggests that C/EBPβ-2-transactivated transcriptional programs may provide a source for discovery of tumor-associated proteins with restricted distribution in normal tissues.
Given that C/EBPβ isoforms 1 and 2 only differ N-terminally by 23 amino acids, selective activation of PLAC1 by C/EBPβ-2 may appear surprising but is consistent with previous reports of divergent gene regulatory activities assigned to the individual isoforms. It has been shown that C/EBPβ-1, but not C/EBPβ-2, can cooperate with Myb to activate the differentiation-specific mim-1 gene in chromatin via interaction of the N-terminal amino acids of C/EBPβ-1 with the SWI/SNF chromatin remodeling complex (43). C/EBPβ-2 in turn, in contrast to C/EBPβ-1, has been found to selectively transactivate cyclin D1 (41), a major regulator of the G1/S progression of the cell cycle.
Based on such data, it has been proposed that C/EBPβ-1 primarily activates transcriptionally silenced differentiation-specific genes by overcoming the repressive effects of chromatin. C/EBPβ-2 on the other hand is suspected to target growth-promoting genes. In case of aberrant activity of PLAC1 in mammary epithelial cells such proliferogenic signals may eventually culminate in malignant conversion, as implicated by the observation that HMECs engineered to selectively overexpress C/EBPβ-2, become transformed, undergo an epithelial to mesenchymal transition, acquire an invasive phenotype, and grow independently of epidermal growth factor (44, 45). In concordance with these findings in experimental models, C/EBPβ overexpression is a frequent phenomenon in human breast cancer specimens, and cyclin D1 overexpression is a shared early event in ∼50% of sporadic breast cancers (46, 47). In light of such evidence supporting an essential role of C/EBPβ-2 in tumor biology of breast cancer, it is particularly interesting that PLAC1 is so far the only tightly controlled gene selectively activated by C/EBPβ-2. This finding is of high relevance, as inhibition of PLAC1 function by siRNA or neutralizing antibodies results in reduced proliferation and down-regulation of cyclin D1 and phospho-AKT kinase, two key molecules that are involved in malignant proliferation (11).
Most interestingly, we found estrogen receptor stimulation, a signaling pathway known to contribute to cancer cell growth, augments PLAC1 protein levels (48). Estrogens induce cell proliferation in target cells by stimulating progression through the G1 phase of the cell cycle. Based on our recent findings that PLAC1 is a critical factor for the G1/S cell cycle transition (11), it remains to be analyzed to what extent PLAC1 is a mediator of estrogen actions in cancer cells. PLAC1 does not feature an ERE in its promoter and is transactivated via indirect ERα-DNA association. Such nonclassical ERα signaling has been reported for roughly 35% of the categorized human primary E2-responsive genes. In the majority of these cases, SP1 is the predominant mediator of such ERα-DNA indirect binding. Increasing numbers of genes with GC-rich promoter sequences are found to be induced by E2 via this mechanism, including the low density lipoprotein receptor, endothelial nitric-oxide synthase, c-fos, and most interestingly also cyclin D1 (49). Expression of these genes is modulated in response to estrogenic stimulation as ERα enhances the binding of SP1 to its site and contributes to co-activator recruitment. The DNA-binding domain of ERα has been shown to be dispensable for such activation. Transactivation of the PLAC1 gene promoter however, appears to be mediated by an extended “transcriptional cross-talk” (50) with ligand-activated ERα tethering directly to both the C/EBPβ-2 as well as the SP1 transcription factor. ChIP analysis showed that binding of SP1 to the PLAC1 promoter was not influenced by E2-activated ERα, which is consistent with our findings derived from DAPA. This indicates that SP1 constitutively associates with the PLAC1 promoter thereby contributing to basal levels of PLAC1 transcription. In contrast, we found enhanced recruitment of C/EBPβ-2 to the PLAC1 promoter after E2 treatment. E2-activated ERα presumably associates with C/EBPβ-2, bridging this transfactor to SP1 and favoring interaction of C/EBPβ-2 with its cognate element. Noteworthy, in a recent study it was shown that E2-dependent transactivation of the prolactin receptor in MCF-7 cells was mediated likewise by direct interaction between activated ERα, C/EBPβ, and SP1 (24). Although the authors did not provide fine specification of the C/EBPβ isoforms involved in prolactin regulation, C/EBPβ-2 is the likely candidate as it was the only variant we were able to detect in MCF-7 cells.
Viewed collectively, our findings provide first insight into a novel and previously unknown regulatory mechanism governing selective activation of trophoblast-specific genes in breast cancer. The tight co-regulation of PLAC1 by C/EBPβ-2 and ERα might provide a missing link between these cancer-associated transcription factors and their impact on malignant cell processes, and further strengthens the role of C/EBPβ-2 and PLAC1 in the pathogenesis of breast cancer.
Supplementary Material
This work was supported by the Combined Project Grant SFB 432, by the Forschungsfond and the Center for Natural Sciences and Medicine of the Johannes Gutenberg University, and by the GO-Bio funding of the Federal Ministry of Education and Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- E2
- 17β-estradiol
- C/EBPβ
- CCAAT/enhancer-binding protein β
- ERα
- estrogen receptor α
- ERE
- estrogen-response element
- ChIP
- chromatin immunoprecipitation
- siRNA
- small interfering RNA
- RT
- reverse transcription
- DAPA
- DNA affinity precipitation assay
- ivt
- in vitro transcription
- HMEC
- human mammary epithelial cell
- ICI
- ICI 182,780.
REFERENCES
- 1.Beard J. (1902) Lancet 1, 1758–1761 [Google Scholar]
- 2.Strickland S., Richards W. G. (1992) Cell 71, 355–357 [DOI] [PubMed] [Google Scholar]
- 3.Cross J. C., Hemberger M., Lu Y., Nozaki T., Whiteley K., Masutani M., Adamson S. L. (2002) Mol. Cell. Endocrinol. 187, 207–212 [DOI] [PubMed] [Google Scholar]
- 4.Wilczyñski J. R. (2006) Chemotherapy 52, 107–110 [DOI] [PubMed] [Google Scholar]
- 5.Rama S., Suresh Y., Rao A. J. (2001) Mol. Cell. Endocrinol. 182, 233–248 [DOI] [PubMed] [Google Scholar]
- 6.Weier J. F., Weier H. U., Jung C. J., Gormley M., Zhou Y., Chu L. W., Genbacev O., Wright A. A., Fisher S. J. (2005) Dev. Biol. 279, 420–432 [DOI] [PubMed] [Google Scholar]
- 7.Chiu R. W., Chim S. S., Wong I. H., Wong C. S., Lee W. S., To K. F., Tong J. H., Yuen R. K., Shum A. S., Chan J. K., Chan L. Y., Yuen J. W., Tong Y. K., Weier J. F., Ferlatte C., Leung T. N., Lau T. K., Lo K. W., Lo Y. M. (2007) Am. J. Pathol. 170, 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dirnhofer S., Koessler P., Ensinger C., Feichtinger H., Madersbacher S., Berger P. (1998) Hum. Pathol. 29, 377–382 [DOI] [PubMed] [Google Scholar]
- 9.Iles R. K., Chard T. (1991) J. Urol. 145, 453–458 [DOI] [PubMed] [Google Scholar]
- 10.Laurence D. J., Neville A. M. (1972) Br. J. Cancer 26, 335–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koslowski M., Sahin U., Mitnacht-Kraus R., Seitz G., Huber C., Türeci O. (2007) Cancer Res. 67, 9528–9534 [DOI] [PubMed] [Google Scholar]
- 12.Fant M., Weisoly D. L., Cocchia M., Huber R., Khan S., Lunt T., Schlessinger D. (2002) Mol. Reprod. Dev. 63, 430–436 [DOI] [PubMed] [Google Scholar]
- 13.Massabbal E., Parveen S., Weisoly D. L., Nelson D. M., Smith S. D., Fant M. (2005) Mol. Reprod. Dev. 71, 299–304 [DOI] [PubMed] [Google Scholar]
- 14.Koslowski M., Sahin U., Huber C., Türeci O. (2006) Hum. Mol. Genet. 15, 2392–2399 [DOI] [PubMed] [Google Scholar]
- 15.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci O., Sahin U. (2006) Blood 108, 4009–4017 [DOI] [PubMed] [Google Scholar]
- 16.Kreiter S., Konrad T., Sester M., Huber C., Türeci O., Sahin U. (2007) Cancer Immunol. Immunother. 56, 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreiter S., Selmi A., Diken M., Sebastian M., Osterloh P., Schild H., Huber C., Türeci O., Sahin U. (2008) J. Immunol. 180, 309–318 [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarty A., Roberts M. R. (2007) BMC Mol. Biol. 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H., Lin B., Chen C. L., Johnson P. F., Chou J. Y. (1995) DNA Cell Biol. 14, 681–688 [DOI] [PubMed] [Google Scholar]
- 20.Holland M. P., Bliss S. P., Berghorn K. A., Roberson M. S. (2004) Endocrinology 145, 1096–1105 [DOI] [PubMed] [Google Scholar]
- 21.Stephanou A., Handwerger S. (1995) Biochem. Biophys. Res. Commun. 206, 215–222 [DOI] [PubMed] [Google Scholar]
- 22.Toda K., Yang L. X., Shizuta Y. (1995) J. Steroid Biochem. Mol. Biol. 53, 181–190 [DOI] [PubMed] [Google Scholar]
- 23.Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. (1990) EMBO J. 9, 1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong J., Tsai-Morris C. H., Dufau M. L. (2006) J. Biol. Chem. 281, 18825–18836 [DOI] [PubMed] [Google Scholar]
- 25.Hu Z. Z., Zhuang L., Meng J., Dufau M. L. (1998) J. Biol. Chem. 273, 26225–26235 [DOI] [PubMed] [Google Scholar]
- 26.Lee Y. H., Yano M., Liu S. Y., Matsunaga E., Johnson P. F., Gonzalez F. J. (1994) Mol. Cell. Biol. 14, 1383–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bamberger A. M., Makrigiannakis A., Schröder M., Bamberger C. M., Relakis C., Gellersen B., Milde-Langosch K., Löning T. (2004) Virchows Arch. 444, 149–152 [DOI] [PubMed] [Google Scholar]
- 28.Bégay V., Smink J., Leutz A. (2004) Mol. Cell. Biol. 24, 9744–9751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramji D. P., Foka P. (2002) Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahnow C. A. (2002) Breast Cancer Res. 4, 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai M. J., O'Malley B. W. (1994) Annu. Rev. Biochem. 63, 451–486 [DOI] [PubMed] [Google Scholar]
- 32.Björnström L., Sjöberg M. (2005) Mol. Endocrinol. 19, 833–842 [DOI] [PubMed] [Google Scholar]
- 33.Marino M., Galluzzo P., Ascenzi P. (2006) Curr. Genomics 7, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omoto Y., Eguchi H., Yamamoto-Yamaguchi Y., Hayashi S. (2003) Oncogene 22, 5011–5020 [DOI] [PubMed] [Google Scholar]
- 35.Paruthiyil S., Parmar H., Kerekatte V., Cunha G. R., Firestone G. L., Leitman D. C. (2004) Cancer Res. 64, 423–428 [DOI] [PubMed] [Google Scholar]
- 36.Robinson G. W., Johnson P. F., Hennighausen L., Sterneck E. (1998) Genes Dev. 12, 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seagroves T. N., Krnacik S., Raught B., Gay J., Burgess-Beusse B., Darlington G. J., Rosen J. M. (1998) Genes Dev. 12, 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calkhoven C. F., Müller C., Leutz A. (2000) Genes Dev. 14, 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 39.Descombes P., Schibler U. (1991) Cell 67, 569–579 [DOI] [PubMed] [Google Scholar]
- 40.Timchenko N. A., Welm A. L., Lu X., Timchenko L. T. (1999) Nucleic Acids Res. 27, 4517–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eaton E. M., Hanlon M., Bundy L., Sealy L. (2001) J. Cell. Physiol. 189, 91–105 [DOI] [PubMed] [Google Scholar]
- 42.Horne C. H., Reid I. N., Milne G. D. (1976) Lancet 2, 279–282 [DOI] [PubMed] [Google Scholar]
- 43.Kowenz-Leutz E., Leutz A. (1999) Mol. Cell 4, 735–743 [DOI] [PubMed] [Google Scholar]
- 44.Bundy L., Wells S., Sealy L. (2005) Mol. Cancer 4, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bundy L. M., Sealy L. (2003) Oncogene 22, 869–883 [DOI] [PubMed] [Google Scholar]
- 46.Caldon C. E., Daly R. J., Sutherland R. L., Musgrove E. A. (2006) J. Cell. Biochem. 97, 261–274 [DOI] [PubMed] [Google Scholar]
- 47.Sutherland R. L., Musgrove E. A. (2004) J. Mammary Gland Biol. Neoplasia 9, 95–104 [DOI] [PubMed] [Google Scholar]
- 48.Clarke R., Dickson R. B., Lippman M. E. (1992) Crit. Rev. Oncol. Hematol. 12, 1–23 [DOI] [PubMed] [Google Scholar]
- 49.Safe S., Kim K., Kim K. (2008) J. Mol. Endocrinol. 41, 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Göttlicher M., Heck S., Herrlich P. (1998) J. Mol. Med. 76, 480–489 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.