Abstract
Paraquat (PQ), a herbicide used worldwide, causes fatal injury to organs upon high dose ingestion. Treatments for PQ poisoning are unreliable, and numerous deaths have been attributed inappropriate usage of the agent. It is generally speculated that a microsomal drug-metabolizing enzyme system is responsible for PQ toxicity. However, recent studies have demonstrated cytotoxicity via mitochondria, and therefore, the cytotoxic mechanism remains controversial. Here, we demonstrated that mitochondrial NADH-dependent PQ reductase containing a voltage-dependent anion channel 1 (VDAC1) is responsible for PQ cytotoxicity. When mitochondria were incubated with NADH and PQ, superoxide anion (O2˙̄) was produced, and the mitochondria ruptured. Outer membrane extract oxidized NADH in a PQ dose-dependent manner, and oxidation was suppressed by VDAC inhibitors. Zymographic analysis revealed the presence of VDAC1 protein in the oxidoreductase, and the direct binding of PQ to VDAC1 was demonstrated using biotinylated PQ. VDAC1-overexpressing cells showed increased O2˙̄ production and cytotoxicity, both of which were suppressed in VDAC1 knockdown cells. These results indicated that a VDAC1-containing mitochondrial system is involved in PQ poisoning. These insights into the mechanism of PQ poisoning not only demonstrated novel physiological functions of VDAC protein, but they may facilitate the development of new therapeutic approaches.
Paraquat (PQ2; methyl viologen, 1,1′-dimethyl-4,4′-bipyridinium dichloride) is an effective herbicide used in more than 120 countries (1). Although it is classified as a low hazard compound, PQ is hazardous when used improperly and has been found responsible for thousands of deaths worldwide because of intentional overdose and high levels of occupational and accidental exposure especially in developing countries (1). Direct exposure to PQ causes severe irritation to the eyes and skin, and ingestion of concentrated products may result in fatal injury to lungs because of edema, hemorrhage, and subsequent fibrosis as well as damage to other organs (2). Additionally, PQ has emerged as a risk factor for Parkinson disease (3). The acute toxicity of PQ in mammals is mediated by reactive oxygen species (ROS) produced by a cyclic oxidation-reduction reaction (4). It is generally speculated that NADPH-cytochrome P450 reductase in microsomal drug-metabolizing enzyme systems is responsible for the production of ROS (5). However, we previously observed that the initial ultrastructural alterations associated with PQ exposure occurred only in mitochondria and not in the endoplasmic reticulum in pulmonary cells in vivo (6) and in vitro (7). In addition, several reports have suggested the cytotoxicity of PQ via mitochondrial dysfunction (8–10). Despite the development of a number of treatments for PQ poisoning, the efficacy and reliability of currently available treatments have remained limited because of an insufficient understanding of PQ cytotoxicity (2).
We recently discovered that active NADH-dependent oxidoreductase located on the mitochondrial outer membrane reduced PQ to a radical form that spontaneously formed superoxide anion (O2˙̄) and destroyed mitochondria (11–13). Furthermore, we demonstrated that 1) PQ was initially metabolized to monopyridone in the cytosol and subsequently hydroxylated by the microsomes and 2) the induction of drug-metabolizing enzymes and the administration of a ROS scavenger reduced PQ toxicity in mice (11, 14). These results indicate that the mitochondrial system, not the microsomal system, is responsible for PQ toxicity. We verified that enzymes in the electron transport chain and NADH-cytochrome b5 reductase, an NADH-dependent oxidoreductase in the outer membrane, were not involved in this reaction (11, 12). A voltage-dependent anion channel (VDAC), an abundant pore-forming protein in the outer membrane, exerts numerous physiological functions as a channel; it regulates both the metabolite flux of mitochondria and transmembrane potential, and plays a role in apoptosis. Recently, it was reported that NADH regulates VDAC function (15), and an isoform of VDAC localized in the plasma membrane possesses NADH-ferricyanide reductase activity (16). Therefore, we attempted to determine whether or not NADH-PQ oxidoreductase on mitochondria is responsible for PQ cytotoxicity and if VDAC participates in this activity.
EXPERIMENTAL PROCEDURES
Cell Line
HeLa cells were provided by RIKEN Cell Bank (Tsukuba, Japan). Cells were cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum at 37 °C in a humidified CO2 incubator.
Intracellular ROS Production
Mitochondrial superoxide production in HeLa cells was detected using MitoSOX® (Molecular Probes Inc., Eugene, OR), a red fluorescent mitochondrial superoxide indicator, according to the given protocol. Cells were pretreated with 1 mm PQ; Sigma-Aldrich) for 50 min at 37 °C and incubated with 5 μm MitoSOX for 10 min in the dark. The medium was exchanged for fresh medium, and the cells were observed by a fluorescence microscope (Olympus IX70, Olympus Corp., Tokyo, Japan). The effects of benzoquinone (BQ; 0.2 mm, Sigma-Aldrich) were evaluated after 10 min of incubation in BQ-added medium. Intracellular H2O2 production in HeLa cells by PQ was detected using 2′,7′-dichlorofluorescein-diacetate (DCFH; Molecular Probes) (17, 18). Briefly, cells were pretreated with 1 mm PQ for 1 h at 37 °C, and then the cells were incubated with 5 μm DCFH for 20 min in the dark. Afterward, the medium was exchanged for fresh medium; fluorescence images that appeared after the formation of 2′,7′-dichlorofluorescein (DCF) were observed by fluorescence microscopy.
Preparation of Mitochondria
Mitochondria were isolated from the livers of male Wistar rats or from HeLa cells by differential centrifugation (11, 12). Mitochondria were suspended in 0.25 m sucrose solution containing 0.05 m Tris-HCl, 20 mm KCl, 2.0 mm MgCl2, and 1.0 mm Na2HPO4 (pH 7.4). The mitochondria were starved for 20 min at 37 °C to consume endogenous substrates before use. The Kanazawa Medical University Animal Care and Use Committee approved all studies. All animals were cared for and treated in accordance with the Committee guidelines.
PQ-dependent Hydrogen Peroxide (H2O2) Production on Mitochondria
Mitochondria were attached onto a glass-based culture dishes coated with Cell-Tak® (BD Biosciences) (19). The dishes were incubated with 10 mm PQ and 2 mm NADH (Oriental Yeast Co., Ltd., Tokyo, Japan) in the sucrose solution containing 5 μm DCFH, 5 μm rotenone (Sigma-Aldrich), and 1 μm p-hydroxymercuribenzoate (Sigma-Aldrich) at 37 °C. Fluorescence images were captured by a digital CCD camera (Pixera Penguin 150 CL, Pixera Corp., Los Gatos, CA) attached to a microscope and were analyzed by Lumina Vision bio-imaging analysis system (Mitani Corp., Fukui, Japan). The fluorescence intensity per 1000 mitochondria was calculated, and the mean value of three areas from each sample was compared. BQ (0.3 mm), anti-VDAC1 monoclonal antibody (mAb; anti-porin 31 HL mAb, 9 μg/ml; Calbiochem), and 4,4′-diisothiocyanatostilbene-2,29-disulfic acid (DIDS; 100 μm, Sigma-Aldrich) were evaluated by addition to the reaction mixture.
Electron Microscopy
Mitochondria were transferred to a sucrose solution containing 3 mm PQ, 2 mm NADH, 5 μm rotenone, 1 μm p-hydroxymercuribenzoate, and the solution was reacted for 30 min at 37 °C (11, 12). Superoxide dismutase (SOD; 3000 units/ml, Sigma-Aldrich) effects were evaluated by the addition of SOD to the reaction mixture. Anti-VDAC1 mAb (3 μg/ml) effects were evaluated by preincubation with the mitochondria for 5 min at 37 °C. The reaction was stopped by the addition of cold buffer. Mitochondria were immediately centrifuged, and the packed sediments were covered with 2% glutaraldehyde in phosphate-buffered saline and fixed for 1 h. The fixed clots were prepared for electron microscopy (11) and then observed by a transmission electron microscope (JEM-1200EX, JEOL Co. Ltd, Tokyo, Japan). The percentage of intact mitochondria per area was counted, and the mean of three areas was calculated.
Growth Inhibition Assays
Growth inhibition assays were performed by the stepwise addition of PQ, according to the method described by Saotome (20). Subconfluent HeLa cells were harvested by trypsinization and were precultured on 96-well plates (3 × 103 cells per well) for 24 h. Cells were treated with 7–250 μm PQ and were then cultured for 72 h. The effects of Trolox® (a water-soluble analog of vitamin E; 1 mm, Sigma-Aldrich) were evaluated by its addition to the medium. The 50% growth inhibition toxicity (IC50) was estimated at 72 h.
Extraction of NADH-PQ Oxidoreductase from the Outer Membrane
To extract NADH-PQ oxidoreductase, two-step extraction with Triton X-100, deoxycholate followed by SDS/Igepal® CA-630 was performed (21). The outer membranes were isolated from the mitochondria by discontinuous sucrose gradient centrifugation (12). The isolated outer membranes were suspended in 20 mm Tris-HCl buffer (pH 7.6) containing 1% Triton X-100, 1% sodium deoxycholate, and 1 mm EDTA. The suspensions were left to stand on ice for 1 h. Suspensions were then centrifuged at 105,000 × g for 60 min. The precipitates were resuspended in a 20 mm Tris buffer with 0.06% SDS and 0.1% Igepal CA-630. The suspensions were left on ice for 1 h. The supernatants were collected by centrifugation at 105,000 × g.
Preparation of NADH-PQ Oxidoreductase Fraction
The supernatants were diluted with 20 mm Tris-HCl buffer (pH 8.0) containing 0.03% Triton X-100 and 10% glycerol, and the dilutions were loaded onto an anion exchange column (DEAE MemSep® 1000; Millipore Corp. Billerica, MA). The columns were washed with the Tris buffer, and proteins were eluted using a NaCl gradient. The fractions containing NADH-PQ oxidoreductase were collected from 0.25–0.3 m NaCl fractions and dialyzed against a 20 mm Tris-HCl buffer (pH 7.6) containing 0.03% Triton X-100 and 10% glycerol.
Assays of NADH-PQ Oxidoreductase Activity
NADH Oxidation
The extracts were incubated with 0.2 mm NADH in Tris-buffered saline (TBS) containing 5 μm rotenone and 1 μm p-hydroxymercuribenzoate at 37 °C followed by the addition of 10 mm PQ. Activities were calculated by the first-order velocity of NADH oxidation measured at λ340 nm (e = 6.3 × 103 m−1cm−1).
O2˙̄ Production
O2˙̄ production by NADH-PQ oxidoreductase activity was assayed using a Diogenes® luminescence system (National Diagnostics Inc., Atlanta, GA). The extracts were mixed with NADH (0.1 mm), PQ (0.0012–5 mm), and Diogenes (3-fold dilution) in TBS on a 384-well plate. The effects of DIDS (100 μm) and anti-VDAC1 mAb (30 μg/ml) were evaluated by the addition of these reagents to the mixture. The total volume of the reaction mixture was 15 μl. Chemiluminescence produced by superoxide was detected by an Envision® multilabel plate reader (PerkinElmer Life Sciences).
Immunoprecipitation
The extracts were incubated with anti-VDAC1 mAb or normal mouse IgG as a control. After incubation in TBS for 90 min at 4 °C, Protein A slurries (Amersham Biosciences) were added to the solution and gently stirred for 90 min at 4 °C. The suspensions were centrifuged, and the supernatants were obtained for the assay.
Zymography and Western Blot Analysis
The NADH-PQ oxidoreductase fraction was mixed with 0.125 m Tris-HCl buffer (pH 6.8) containing 20% glycerol and 0.02% bromphenol blue (1:1, v/v), and the mixture was loaded on native-polyacrylamide gel (5–10% gradient gel; Funakoshi, Tokyo, Japan). Electrophoresis was performed at 5 mA for 5 h on ice, and the gel was immersed in 20 mm Tris-HCl buffer (pH 7.4) containing 20% glycerol, 0.25 mm nitro blue tetrazolium, 5 μm rotenone, and 1 μm p-hydroxymercuribenzoate. The gel was incubated with 2 mm NADH and 10 mm PQ at room temperature for 30 min and washed with TBS, and the active bands were stained by diformazan. The electrophoresed gel was blotted, and detection was performed with anti-VDAC1 mAb. Additionally, the active band was excised and subjected to SDS-PAGE followed by Western blot analysis with anti-VDAC1 mAb.
Synthesis of Biotinylated PQ
The biotinylated paraquat was synthesized in moderate yield by condensation reaction of (+)-biotin and 3-(1′-methyl-4,4′-bipyridinium)propylammonium salt, which was prepared by successive N-alkylation of 4,4′-bipyridine with iodomethane and 3-bromopropylamine hydrobromide. See the supplemental methods for detailed procedures.
Plasmid Construction
Human vdac1 cDNA was isolated as an XhoI fragment by PCR and was subcloned into pUC-CAGGS expression vector (22).
Synthesis of VDAC1 Protein Using a Cell-free Protein Synthesis System
The cDNA of VDAC1 was used. For wheat cell-free protein production of VDAC proteins, the VDAC DNA templates were constructed by “split-primer” PCR (23). The first round of PCR was performed on the cDNA using 10 nm concentrations of each of the following primers: a specific primer (5′-CCACCCACCACCACCAATGGCTGTGCCACCCACGT and AODA2306 primer, 5′-AGCGTCAGACCCCGTAGAAA). Then a second round of PCR was carried out to construct the templates for protein synthesis using a portion (5 μl) of the first PCR mix: 100 nm SPu primer (5′-GCGTAGCATTTAGGTGACACT), 100 nm AODA2303 primer (5′-GTCAGACCCCGTAGAAAAGA), and 1 nm deSP6E02 (5′-GGTGACACTATAGAACTCACCTATCTCTCTACACAAAACATTTCCCTACATACAACTTTCAACTTCCTATTCCACCCACCACCACCAATG). Wheat cell-free protein synthesis of VDAC protein was carried out using a robotic synthesizer (24, 25), GenDecorder1000® (CellFree Sciences, Yokohama, Japan) as described below. First, the transcript was created from each of the DNA templates mentioned above using SP6 RNA polymerase. The synthetic mRNAs were then precipitated with ethanol and collected by centrifugation using a Hitachi R10H rotor. Each mRNA (usually 30–35 μg) was washed and transferred into a translation mixture. The translation reaction was performed in the bilayer mode (26) with slight modifications. The translation mixture that formed the bottom layer consisted of 60 A260 units of wheat germ extract (CellFree Sciences) and 2 μg of creatine kinase (Roche Diagnostics) in 25 μl of SUB-AMIX® (CellFree Sciences). The SUB-AMIX® contained (final concentrations) 30 mm Hepes/KOH at pH 8.0, 1.2 mm ATP, 0.25 mm GTP, 16 mm creatine phosphate, 4 mm dithiothreitol, 0.4 mm spermidine, 0.3 mm concentrations of each of the 20 amino acids, 2.7 mm magnesium acetate, and 100 mm potassium acetate. 125 μl of the SUB-AMIX was placed on the top of the translation mixture, forming the upper layer. After incubation at 26 °C for 17 h, the synthesized proteins were confirmed by SDS-PAGE.
Binding Assay
The synthesized VDAC1 protein was mixed with biotinylated PQ (0–1.0 μm) in TBS containing 10% EZ block (Atto Corp., Tokyo, Japan) for 1 h. The mixtures were added to Nunc Immobilizer® streptavidin plates (Nunc, Roskilde, Denmark), which were incubated for 2 h. The plates were then washed with TBS containing 0.1% Tween 20 (TTBS). VDAC1 protein bound to biotinylated PQ was detected by anti-VDAC1 mAb (Calbiochem) followed by the addition of horseradish peroxidase-conjugated second antibody. ECL plus® (GE Healthcare) was used as a substrate of horseradish peroxidase. For NADH binding assay, biotinylated NAD+ (R&D Systems, Inc., Minneapolis, MN) was mixed with outer membrane extract, and serial dilutions of non-labeled NADH were added for 1 h. The mixtures were incubated with anti-VDAC1 mAb, which was immobilized on Nunc Immobilizer 96-well plates in 10% EZ block (Atto Corp) for 2 h. The plates were washed with TTBS to which ExtrAvidin® peroxidase (Sigma) was added followed by a wash with TTBS. ECL plus was used for the detection of binding. Immobilized normal mouse IgG was used as a control.
DNA Transfection
HeLa cells were transfected with the VDAC1 plasmid using Effectene® (Qiagen GmbH, Hilden, Germany).
Small Interfering RNA (siRNA) Transfection
HeLa cells were transfected for 72 h with 5 nm control siRNA or Hs_VDAC1_1HP_siRNA (Qiagen) using HiPerFect® transfection reagent (Qiagen).
Statistics
Statistical analyses were conducted using analysis of variance for multiple comparisons and Student's t test for comparing two groups.
RESULTS
PQ Produces O2˙̄ in an NADH-dependent Manner
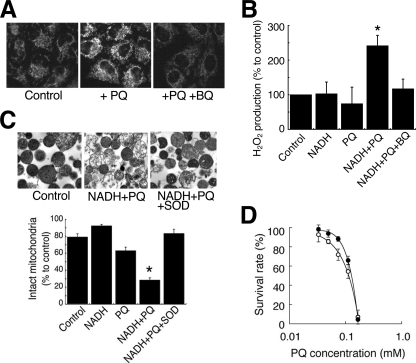
We first investigated whether PQ produced ROS on mitochondria in HeLa cells. We detected O2˙̄ on the mitochondria using MitoSOX fluorogenic dye (Fig. 1A). Whereas only slight fluorescence was detected on the mitochondria in cells exposed to normal conditions, highly intense levels of fluorescence were observed when the cells were exposed to PQ. Fluorescence was reduced to the control level with the addition of BQ, a scavenger of O2˙̄. In isolated rat liver mitochondria, we detected the NADH-dependent production of H2O2 by PQ using DCFH fluorescent dye (Fig. 1B). Although the fluorescence intensity did not change when PQ or NADH alone was added to the isolated mitochondria, the addition of PQ in combination with NADH raised the intensity of fluorescence in the mitochondria. BQ suppressed this augmentation. We also observed that the co-administration of PQ and NADH led to a loss of structural integrity of the isolated mitochondria, and SOD suppressed this damage (Fig. 1C). Furthermore, Torolox®, an O2˙̄ scavenger, significantly increased the survival rates of HeLa cells exposed to PQ (p < 0.05, Fig. 1D). These results indicated that PQ produced O2˙̄ in an NADH-dependent manner in mitochondria and damaged mitochondria followed by cell death.
FIGURE 1.
Damage to mitochondria caused by NADH-dependent O2˙̄ production induced by PQ. A, mitochondrial O2˙̄ production was visualized by MitoSOX in cells treated with PQ (1 mm) and BQ (0.2 mm). B, H2O2 production in isolated mitochondria was estimated by DCF florescence method. Mitochondria were incubated with 10 mm PQ, 2 mm NADH, and 0.3 mm BQ (*, p < 0.01, versus control). C, isolated mitochondria were incubated with 3 mm PQ, 2 mm NADH, and the 3000 IU/ml SOD. Upper panels, electron micrograph. Lower graph, percentages of intact mitochondria (*, p < 0.001, versus control). D, the survival rate of HeLa cells exposed to PQ (open circle) and PQ and 1 mm Trolox® (closed circle). Each point is the average of two to four experiments. Error bars represent S.E.
VDAC1 Is Responsible for NDAH-PQ Oxidoreductase Activity
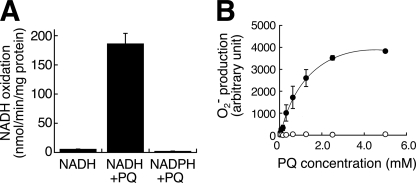
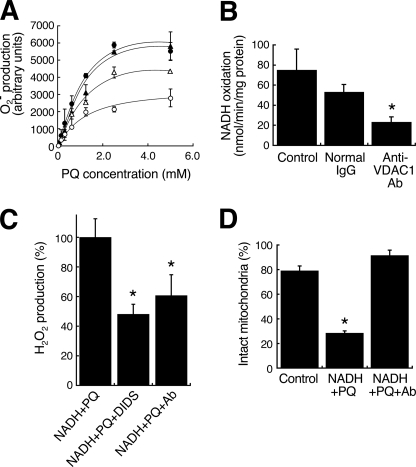
To reveal the components involved in this activity, we performed two-step extraction with Triton X-100, deoxycholate followed by SDS/Igepal CA-630 from the outer membrane and analyzed SDS/Igepal extract. The extract oxidized NADH, but not NADPH, by the addition of PQ (Fig. 2A), and O2˙̄ was produced in a PQ dose-dependent manner (Fig. 2B). The NADH oxidation activity was 4.4 times that of the Triton X-100/deoxycholate extract (data not shown). To ascertain whether or not VDAC protein is involved in NADH-PQ oxidoreductase activity, we examined the effects of VDAC inhibitors on this activity (Fig. 3A). O2˙̄ production by the extract from the outer membrane mixed with PQ and NADH was significantly inhibited by DIDS, an anion channel inhibitor, or anti-VDAC1 mAb, but such inhibition was not observed with exposure of the extract to normal mouse IgG. When the extract was immunoprecipitated with anti-VDAC1 mAb, the activity in the supernatant was lower than that observed with the administration of normal IgG (Fig. 3B). Furthermore, we confirmed that DIDS and anti-VDAC1 mAb inhibited the production of O2˙̄ and also inhibited the breakdown of isolated mitochondria exposed to PQ and NADH (Fig. 3, C and D). These results suggest that VDAC1 is responsible for NADH-PQ oxidoreductase activity.
FIGURE 2.
NADH-PQ oxidoreductase activity in the outer membrane extract. A, NADH (0.2 mm) was oxidized by the outer membrane extract in the presence of PQ (5 mm), but NADPH (0.2 mm) was not oxidized. B, PQ dose-dependent relationship to O2˙̄ production activity was observed by co-administration with NADH (0.1 mm) to the outer membrane extract (closed circle). In contrast, NADPH (0.1 mm) did not exert any such PQ effects (open circle). All error bars represent S.D. (n = 3).
FIGURE 3.
Participation of VDAC1 in the NADH-PQ oxidoreductase activity and mitochondrial damage. A, O2˙̄ production in the outer membrane extract (closed circle) was inhibited by DIDS (100 μm; open circle, p < 0.001, n = 3) and anti-VDAC1 mAb (30 μg/ml; open triangle, p < 0.05, n = 3). Closed triangle, treated with normal IgG (30 μg/ml). Error bars represent S.D. (n = 3). B, the extract was immunoprecipitated with anti-VDAC1 mAb or normal IgG, and the NADH-oxidation activity of the supernatants was measured. Control, no treatment. *, p < 0.01, versus control. Error bars represent S.D. (n = 3). C, H2O2 production in isolated mitochondria by PQ (10 mm) co-administered with NADH (2 mm) was estimated by DCF florescence method. DIDS (100 μm) and anti-VDAC1 mAb (9 μg/ml) were inhibited H2O2 production. *, p < 0.001 with respect to the control. Each point is the mean of triplicate experiments. Error bars represent S.E. D, effects of anti-VDAC1 antibody on the NADH-PQ-dependent breakage of mitochondria were estimated. Isolated mitochondria were ruptured by the co-administration of PQ (3 mm) and NADH (2 mm), whereas the addition of anti-VDAC1 mAb (9 μg/ml) protected the mitochondria from such breakage. *, p < 0.01 versus the control. Each point is the mean of triplicate experiments. Error bars represent S.E.
VDAC1 Is Component of NADH-PQ Oxidoreductase
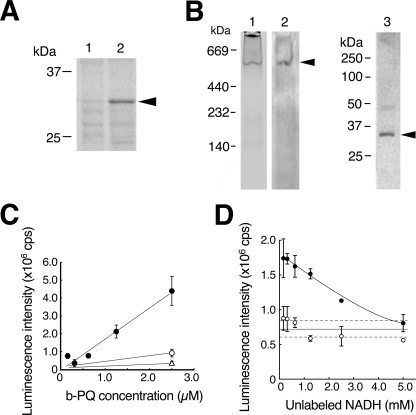
Because, VDAC1 protein was more highly concentrated in the SDS/Igepal extract than in the Triton X-100/deoxycholate extract (Fig. 4A), we investigated whether or not VDAC1 protein is contained in the oxidoreductase. We purified the active fraction from the SDS/Igepal extract using DEAE chromatography and carried out zymography on the fraction by native PAGE in blue tetrazolium solution with PQ and NADH. A major reactive band stained with dark blue diformazan, a form of blue tetrazolium reduced by O2˙̄, appeared at 500 kDa (Fig. 4B, lane 1); this band was recognized using anti-VDAC1 mAb (lane 2). Next, the excised band was examined by Western blot analysis with SDS-PAGE using anti-VDAC1 mAb. The antibody recognized a band at 31 kDa, the size of VDAC1 (lane 3). Because several proteins were detected in the reactive band by SDS-PAGE followed by silver staining (data not shown), the oxidoreductase may be a complex containing the VDAC1 protein. To confirm the direct interaction of VDAC1 with PQ, we performed a binding assay using recombinant VDAC1 protein and biotinylated PQ (Fig. 4C). We detected biotinylated PQ dose-dependently bound to the VDAC1 protein, and excess non-labeled PQ competed for the binding. Next, we examined the interaction of VDAC1 with NADH using biotinylated NAD+. Whereas the biotinylated NAD+ was not found to bind to the recombinant VDAC1 protein, which was immobilized by anti-VDAC1 mAb (data not shown), we did detect binding of the biotinylated NAD+ using the SDS/Igepal extract instead of the VDAC1 protein (Fig. 4D). These results were compatible with the absence of NADH-PQ oxidoreductase activity in the recombinant VDAC1 protein or purified VDAC from rat liver mitochondria (data not shown). The present results indicate that VDAC1 is involved in NADH-PQ oxidoreductase activity as a component of the PQ binding site.
FIGURE 4.
VDAC1 is a component of NADH-PQ oxidoreductase. A, extracts obtained from the mitochondrial outer membrane by treatment with Triton X-100/deoxycholate (lane 1) or SDS/ Igepal CA-630 (lane 2) were run on SDS-PAGE, and the results were analyzed by Western blotting with anti-VDAC1 mAb. VDAC1 protein was detected by the mAb (arrowhead). B, DEAE fractions from the extracts containing oxidoreductase activity were examined by zymography with NADH and PQ in blue tetrazolium solution (lane 1). The active band was consistent with the anti-VDAC1 mAb-detected band (lane 2, arrowhead), and this band was excised and subjected to Western blot analysis using anti-VDAC1 mAb (lane 3; the arrowhead indicates VDAC1 protein). C, direct binding to VDAC1 was assayed using biotinylated (b-) PQ (closed circle) in competition with non-labeled PQ (open circle, 25 μm, open triangle, 250 μm). Error bars represent S.D. (n = 3). D, assay of NADH binding to the outer membrane extracts was performed. The extracts were trapped by immobilized anti-VDAC1 antibody and incubated with biotinylated NAD+. Bound biotinylated NAD+ was reduced by exposure to non-labeled NADH (p < 0.01, closed circles). When normal IgG was used for trapping, no NADH competition was detected (open circles). Broken lines represent 95% confidence interval of the control value. Error bars represent S.D. (n = 3).
VDAC1 Is Responsible for the Cytotoxicity of PQ
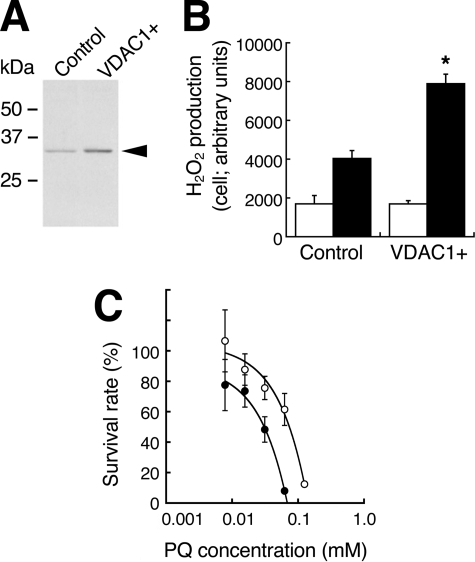
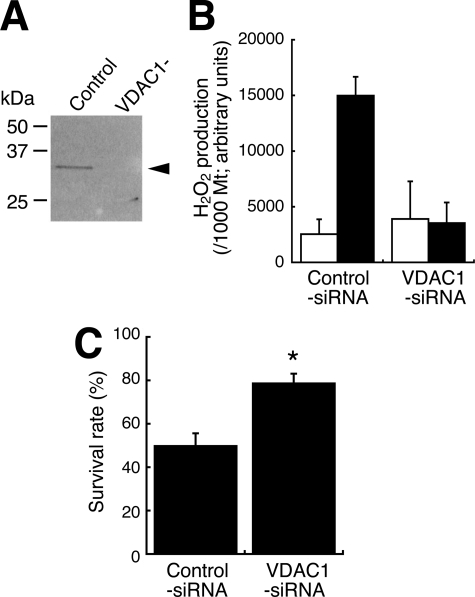
Finally, we determined whether the amount of VDAC1 protein in cells affects PQ sensitivity. We obtained stable transfectants of HeLa cells overexpressing VDAC1; these cells had 2.2 times the VDAC1 protein content of control cells (Fig. 5A). When treated with PQ, these VDAC1-overexpressing cells showed 2.0 times the intracellular production of H2O2 compared with that of control cells (Fig. 5B). The IC50 of control cells exposed to PQ was 72.3 μm, and this value fell to 30.7 μm in the VDAC1-overexpressing cells (Fig. 5C). When HeLa cells were transfected with VDAC1 siRNA, almost no VDAC1 protein was synthesized (Fig. 6A). Mitochondria were isolated from these cells, and the NADH-PQ dependent H2O2 production was estimated (Fig. 6B). The production of H2O2 on the mitochondria from knockdown cells was reduced to endogenous levels. The survival rate after exposure of the VDAC1 knockdown cells to 222 μm PQ for 24 h was 79% compared with 50% in controls (Fig. 6C). These results indicated that VDAC1 is responsible for the cytotoxicity of PQ as an NADH-dependent oxidoreductase.
FIGURE 5.
Effects of VDAC1 overexpression on the PQ-dependent H2O2 production and the cytotoxicity in HeLa cells. A, lysates from VDAC1-overexpressing cells were subjected to Western blot analysis with anti-VDAC1 mAb. Control, cells transfected with empty vector; VDAC+, cells transfected with the vector bearing vdac1 cDNA. B, H2O2 production by PQ in VDAC1-overexpressing cells (VDAC1+) was higher than that of control cells (*, p < 0.001). Light bars, no treatment; dark bars, exposure to 1 mm PQ. Error bars represent S.D. (n = 3). C, the survival rates of VDAC1-overexpressing HeLa cells (closed circle) were lower than those of controls (open circle; p < 0.001). Error bars represent S.E. of triplicate experiments.
FIGURE 6.
Effects of VDAC1 knockdown on the PQ-dependent H2O2 production and the cytotoxicity in HeLa cells. A, lysates from knockdown cells were subjected to Western blot analysis with anti-VDAC1 mAb. Control, cells transfected with the control siRNA; VDAC−, cells transfected with VDAC1 siRNA. The arrowheads indicate VDAC1 protein. B, NADH-PQ-dependent H2O2 production on mitochondria isolated from VDAC1-knockdown HeLa cells was estimated by DCF assay. Light bars, mitochondria incubated with 2 mm NADH only; dark bars, mitochondria were incubated with 10 mm PQ and 2 mm NADH. Error bars represent S.D. (n = 3). C, the survival rate of VDAC1-knockdown HeLa cells (VDA1-siRNA) after 24 h of exposure to 222 μm PQ was higher than that of control cells (p < 0.001). Error bars represent S.E. of triplicate experiments.
DISCUSSION
In this study we demonstrate that a mitochondrial system, not a microsomal system, is involved in PQ poisoning; PQ produces O2˙̄ by NADH-dependent oxidoreductase in the outer membrane of mitochondria and damages mitochondria, leading to cell death. Furthermore, we present that mitochondrial VDAC1 is responsible for this activity as a component of the PQ binding site.
We observed O2˙̄ production on the mitochondria after administering PQ to cells using MitoSOX, an O2˙̄-specific fluorescent dye. Additionally, we detected H2O2 production on isolated mitochondria in the presence of PQ and NADH using DCFH fluorescent dye, and BQ reduced the level of production. In a previous study we observed that cytochrome c, an O2˙̄ scavenger, diminished H2O2 production on mitochondria incubated with PQ and NADH (13). Because O2˙̄ is immediately (105 m−1s−1) converted into H2O2 in aqueous solution, DCF fluorescence demonstrating H2O2 is considered to be equivalent to a demonstration of O2˙̄ production (13). We also indicated that PQ destroyed isolated mitochondria in the presence of NADH and that SOD suppressed this damage. Furthermore, Trolox® suppressed the toxicity of PQ in cells. We formerly reported that PQ selectively destroyed the mitochondria of pulmonary type II cells and hepatocyte in vivo (6, 11) and also destroyed cultured type II cells (7). These results indicate that PQ attacks mitochondria by NADH-dependent O2˙̄ production in the course of its cytotoxicity.
In an ultrastructural study, we previously observed that NADH-dependent O2˙̄ production by PQ occurred in the outer membrane of mitochondria (13) and demonstrated that the NADH oxidation activity by PQ in the outer membrane fraction was five times that of the inner membrane fraction (12). O2 uptake on mitochondria took place with the addition of PQ and NADH (11, 12), and blue PQ radicals formed under anaerobic conditions (11). In the present study we again observed NADH oxidation and PQ radical formation in the outer membrane under anaerobic conditions (data not shown). These results indicated that an NADH-PQ oxidoreductase is localized in the outer membrane. We previously confirmed that NADH-cytochrome b5 reductase, an outer membrane-localized oxidoreductase, did not participate in the PQ reduction, based on its insensitivity to anti-NADH-cytochrome b5 reductase antibody and a different sensitivity to p-hydroxymercuribenzoate (12). We also reported that rotenone, an inhibitor of complex I in the electron transport chain, did not inhibit NADH-dependent PQ reduction (11, 12). Intriguingly, we find that VDAC1 is a constituent of the NADH-PQ oxidoreductase. VDAC1 is a small, abundant, pore-forming protein found in the outer membranes of all eukaryotic mitochondria and plays an important role in the passage of adenine nucleotides, Ca2+, and other metabolites through the outer membrane (27). In addition, VDAC1 located at contact sites between the outer and inner membranes forms permeability transition pores (PTPs) with the adenine nucleotide transporter, cyclophilin D, and other proteins (27). It is unknown whether PTP proteins besides VDAC1 participate in the NADH-PQ oxidoreductase activity; thus, it will still be necessary to investigate their involvement with PTP proteins.
Extramitochondrial oxidative stress induces PTP openings via VDAC protein without damage to the inner membrane (27). PQ does not penetrate mitochondrial membrane (28), and O2˙̄ production by PQ occurs on the outer surface of the outer membrane (13). Furthermore, we demonstrated the binding of PQ to VDAC1 protein by biotinylated PQ and the inhibition of PQ-dependent mitochondrial breakage by anti-VDAC1 mAb. These results indicated that the breakage of mitochondria by PQ occurred through VDAC1. The binding mechanism of PQ, a cation molecule, to VDAC1 remains unknown. It has been reported that NADH increased the voltage dependence of VDAC and reduced the conductance of the outer membrane (29, 30). The ion selectivity of VDAC changed from anions to cations when conductance decreased (31). NADH may, therefore, affect the binding of PQ to VDAC1. Baker et al. (16) reported that VDAC1 localized in the plasma membrane functions as NADH-ferricyanide reductase and that VDAC1 has a putative NAD+ binding motif. Yehezkel et al. (32) demonstrated that VDAC purified from rat liver mitochondria had nucleotide binding sites bound to ATP; however, NADH did not bind them. As in a previous study (Hirai et al. (11), the change in conformation from the orthodox to the condensed type occurred when NADH was added to starved intact mitochondria. In addition, NADH reduced the permeability of the outer membrane to ADP (15). These results indicated that NADH affects PTP even though NADH does not bind to VDAC1 directly. Although we did not observe the direct binding of NADH to VDAC1 alone, we did observe the binding of biotinylated NAD+ to the NADH-PQ oxidoreductase concentrated extract, which was trapped by anti-VDAC1 mAb. NADH-PQ oxidoreductase activity was inhibited by DIDS and anti-VDAC1 mAb, but recombinant VDAC1 protein or purified VDAC protein alone had no activity. Therefore, an NADH binding component is expected to be necessary to yield this activity.
Yagoda et al. (33) reported that VDAC2 or VDAC3 was implicated in the cytotoxicity of the anti-tumor agent erastin, which was shown to induce oxidative cell death; in particular, VDAC2 was found to bind directly to this agent. We have not yet identified the involvement of VDAC2 or VDAC3 in NADH-PQ oxidoreductase activity. It has been reported that VDAC1 is the most abundantly expressed of the three VDAC isoforms in mammalian mitochondria (34). In addition, we demonstrated a correlation between the production of O2˙̄ and VDAC1 expression, and we observed defective O2˙̄ production on the mitochondria isolated from VDAC1 knockdown cells. Therefore, it appears that VDAC1 participates primarily in NADH-PQ oxidoreductase activity. Recently, we found that several furanonaphthoquinones caused mitochondrial damage and the apoptosis of cancer cells by the production of ROS, and other studies revealed that VDAC1 induces ROS production by an NADH-dependent quinone reduction (17, 35). Additionally, we previously demonstrated that menadione, a naphthoquinone, was a substrate of NADH-PQ oxidoreductase (12). These previous and present results taken together suggest that the function of VDAC1 is not only to serve as a channel but also to function as part of an oxidoreductase enzyme.
Until now, management of PQ poisoning has been directed primarily at removing PQ from the gastrointestinal tract by the use of several absorbents (activated charcoal, Fuller's Earth, etc.) and increasing its excretion from the blood by hemoperfusion (2). However, the efficacy of these treatments is poor. Our results indicated that O2˙̄ production by a VDAC-containing mitochondrial system is responsible for PQ poisoning. DIDS and anti-VDAC1 antibody inhibited NADH-PQ oxidoreductase activity, mitochondrial O2˙̄ production, and the breakage of mitochondria by PQ. Furthermore, PQ cytotoxicity was suppressed in VDAC1-knockdown cells. These results suggest that specific VDAC inhibitors can be therapeutic agents of PQ poisoning.
Supplementary Material
Acknowledgment
We are grateful to Mayumi Mitani for secretarial assistance.
This work was supported by Grants-in-aid for Scientific Research 15591664, 17591899, 19390291, and 21791045 from the Japan Society for the Promotion of Science, Grants for Promoted Research S2003-12, S2004-12, C2007-4, S2007-9, C2008-1, S2008-10, C2009-3, and S2009-10 from Kanazawa Medical University, Grant for Project Research H2009-14 from High-Tech Research Center of Kanazawa Medical University, and in part by The Ministry of Education, Culture, Sports, Science, and Technology, Japan Grant S0801085.

The on-line version of this article (available at http://www.jbc.org) contains supplemental schemes.
- PQ
- paraquat
- BQ
- benzoquinone
- DCF
- 2′,7′-dichlorofluorescein
- DCFH
- DCF-diacetate
- DIDS
- 4,4′-diisothiocyanatostilbene-2,29-disulfic acid
- IC50
- 50% growth inhibition toxicity
- mAb
- monoclonal antibody
- PTP
- permeability transition pore
- TBS
- Tris-buffered saline
- VDAC
- voltage-dependent anion channel
- ROS
- reactive oxygen species
- SOD
- superoxide dismutase
- siRNA
- small Interfering RNA.
REFERENCES
- 1.Wesseling C., van Wendel de Joode B., Ruepert C., León C., Monge P., Hermosillo H., Partanen T. J. (2001) Int. J. Occup. Environ. Health 7, 275–286 [DOI] [PubMed] [Google Scholar]
- 2.Dinis-Oliveira R. J., Duarte J. A., Sánchez-Navarro A., Remião F., Bastos M. L., Carvalho F. (2008) Crit. Rev. Toxicol. 38, 13–71 [DOI] [PubMed] [Google Scholar]
- 3.McCormack A. L., Thiruchelvam M., Manning-Bog A. B., Thiffault C., Langston J. W., Cory-Slechta D. A., Di Monte D. A. (2002) Neurobiol. Dis. 10, 119–127 [DOI] [PubMed] [Google Scholar]
- 4.Baldwin R. C., Pasi A., MacGregor J. T., Hine C. H. (1975) Toxicol. Appl. Pharmacol. 32, 298–304 [DOI] [PubMed] [Google Scholar]
- 5.Bus J. S., Cagen S. Z., Olgaard M., Gibson J. E. (1976) Toxicol. Appl. Pharmacol. 35, 501–513 [DOI] [PubMed] [Google Scholar]
- 6.Hirai K., Witschi H., Côté M. G. (1985) Exp. Mol. Pathol. 43, 242–252 [DOI] [PubMed] [Google Scholar]
- 7.Wang G. Y., Hirai K., Shimada H. (1992) J. Electron Microsc. (Tokyo) 41, 181–184 [PubMed] [Google Scholar]
- 8.Yang W., Tiffany-Castiglioni E. (2005) J. Toxicol. Environ. Health A 68, 1939–1961 [DOI] [PubMed] [Google Scholar]
- 9.St. Clair D. K., Oberley T. D., Ho Y. S. (1991) FEBS Lett. 293, 199–203 [DOI] [PubMed] [Google Scholar]
- 10.Oliver P. D., Newsome D. A. (1992) Invest. Ophthalmol. Vis. Sci. 33, 1909–1918 [PubMed] [Google Scholar]
- 11.Hirai K., Ikeda K., Wang G. Y. (1992) Toxicology 72, 1–16 [DOI] [PubMed] [Google Scholar]
- 12.Shimada H., Hirai K., Simamura E., Pan J. (1998) Arch Biochem. Biophys 351, 75–81 [DOI] [PubMed] [Google Scholar]
- 13.Hirai K. I., Pan J., Shimada H., Izuhara T., Kurihara T., Moriguchi K. (1999) J. Electron Microsc. (Tokyo) 48, 289–296 [DOI] [PubMed] [Google Scholar]
- 14.Shimada H., Furuno H., Hirai K., Koyama J., Ariyama J., Simamura E. (2002) Arch. Biochem. Biophys. 402, 149–157 [DOI] [PubMed] [Google Scholar]
- 15.Lee A. C., Xu X., Colombini M. (1996) J. Biol. Chem. 271, 26724–26731 [DOI] [PubMed] [Google Scholar]
- 16.Baker M. A., Lane D. J., Ly J. D., De Pinto V., Lawen A. (2004) J. Biol. Chem. 279, 4811–4819 [DOI] [PubMed] [Google Scholar]
- 17.Simamura E., Hirai K., Shimada H., Koyama J., Niwa Y., Shimizu S. (2006) Cancer Biol. Ther. 5, 1523–1529 [DOI] [PubMed] [Google Scholar]
- 18.Ariyama J., Shimada H., Aono M., Tsuchida H., Hirai K. I. (2000) Intensive Care Med. 26, 981–987 [DOI] [PubMed] [Google Scholar]
- 19.Nakayama S., Sakuyama T., Mitaku S., Ohta Y. (2002) Biochem. Biophys. Res. Commun. 290, 23–28 [DOI] [PubMed] [Google Scholar]
- 20.Saotome K., Morita H., Umeda M. (1989) Toxicol. In Vitro 3, 317–321 [DOI] [PubMed] [Google Scholar]
- 21.Teraoka K., Matsui S. (1999) Nippon Rinsho. 57, (suppl.) 784–788 [PubMed] [Google Scholar]
- 22.Narita M., Shimizu S., Ito T., Chittenden T., Lutz R. J., Matsuda H., Tsujimoto Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14681–14686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawasaki T., Ogasawara T., Morishita R., Endo Y. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawasaki T., Kamura N., Matsunaga S., Saeki M., Tsuchimochi M., Morishita R., Endo Y. (2008) FEBS Lett. 582, 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawasaki T., Gouda M. D., Kawasaki T., Tsuboi T., Tozawa Y., Takai K., Endo Y. (2005) Methods Mol. Biol. 310, 131–144 [DOI] [PubMed] [Google Scholar]
- 26.Sawasaki T., Hasegawa Y., Tsuchimochi M., Kamura N., Ogasawara T., Kuroita T., Endo Y. (2002) FEBS Lett. 514, 102–105 [DOI] [PubMed] [Google Scholar]
- 27.Crompton M. (1999) Biochem. J. 341, 233–249 [PMC free article] [PubMed] [Google Scholar]
- 28.Gage J. C. (1968) Biochem. J. 109, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee A. C., Zizi M., Colombini M. (1994) J. Biol. Chem. 269, 30974–30980 [PubMed] [Google Scholar]
- 30.Zizi M., Byrd C., Boxus R., Colombini M. (1998) Biophys. J. 75, 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vyssokikh M., Brdiczka D. (2004) Mol. Cell. Biochem. 256–257, 117–126 [DOI] [PubMed] [Google Scholar]
- 32.Yehezkel G., Hadad N., Zaid H., Sivan S., Shoshan-Barmatz V. (2006) J. Biol. Chem. 281, 5938–5946 [DOI] [PubMed] [Google Scholar]
- 33.Yagoda N., von Rechenberg M., Zaganjor E., Bauer A. J., Yang W. S., Fridman D. J., Wolpaw A. J., Smukste I., Peltier J. M., Boniface J. J., Smith R., Lessnick S. L., Sahasrabudhe S., Stockwell B. R. (2007) Nature 447, 864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto T., Yamada A., Watanabe M., Yoshimura Y., Yamazaki N., Yoshimura Y., Yamauchi T., Kataoka M., Nagata T., Terada H., Shinohara Y. (2006) J. Proteome Res. 5, 3336–3344 [DOI] [PubMed] [Google Scholar]
- 35.Simamura E., Shimada H., Ishigaki Y., Hatta T., Higashi N., Hirai K. I. (2008) Anat. Sci. Int. 83, 261–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.