Abstract
Gram-negative binding protein 3 (GNBP3), a pattern recognition receptor that circulates in the hemolymph of Drosophila, is responsible for sensing fungal infection and triggering Toll pathway activation. Here, we report that GNBP3 N-terminal domain binds to fungi upon identifying long chains of β-1,3-glucans in the fungal cell wall as a major ligand. Interestingly, this domain fails to interact strongly with short oligosaccharides. The crystal structure of GNBP3-Nter reveals an immunoglobulin-like fold in which the glucan binding site is masked by a loop that is highly conserved among glucan-binding proteins identified in several insect orders. Structure-based mutagenesis experiments reveal an essential role for this occluding loop in discriminating between short and long polysaccharides. The displacement of the occluding loop is necessary for binding and could explain the specificity of the interaction with long chain structured polysaccharides. This represents a novel mechanism for β-glucan recognition.
The activation of the immune response is energetically costly and may be detrimental to the host, especially when inappropriately triggered. Therefore, the reliable detection of infections is a step of paramount importance in the immune response. To achieve the task of detecting potentially hazardous microorganisms, the innate immune system relies on several strategies. One of them is to sense both pathogenic and nonpathogenic microorganisms thanks to pattern recognition receptors (PRRs)4 that recognize intrinsic microbial molecular “signatures” (1). These immune receptors have been selected during evolution for their ability to bind to essential, conserved, structural components of the microorganisms such as flagellins, peptidoglycans of bacteria, lipopolysaccharides of Gram-negative bacteria, lipoteichoic acids of Gram-positive bacteria, and β-glucans of the fungal cell wall (2, 3). Examples of mammalian PRRs include Toll-like receptors (4), intracellular receptors of the NOD family (5), peptidoglycan recognition proteins (PGRPs) (6), and the membrane-bound Dectin-1 receptor, which detects fungal β-glucans (7).
One important arm of the innate immunity in Drosophila is a potent systemic response that relies on the synthesis in the fat body (a functional equivalent of the mammalian liver) of potent antimicrobial peptides (AMPs) that are secreted in the hemolymph where they attack invading microorganisms. Genetic analysis has delineated two major regulatory pathways of NF-κB type that control the expression of AMP genes (8). The immune deficiency (imd) pathway is mostly required in the host defense against Gram-negative bacteria (9) and is triggered by PRRs of the PGRP family, namely PGRP-LC (10) and PGRP-LE (11). The Toll pathway is essential for fighting fungal and some Gram-positive bacterial infections (12, 13). Toll, the funding member of the Toll-like receptor family, is not itself a PRR. Rather, it is activated by a ligand of the nerve growth factor family, the Spätzle cytokine. To bind to the Toll receptor, Pro-Spätzle needs to be proteolytically processed by a protease, the Spätzle-processing enzyme (SPE) (14), which is itself activated by upstream proteolytic cascades. One such cascade is activated in response to a Gram-positive bacterial challenge by a complex of PGRP-SA, PGRP-SD, and Gram-negative binding protein 1 (GNBP1) (13, 15, 16). Flies deficient for either PGRP-SA or GNBP1 are deficient in Toll pathway activation and are susceptible to infections by several Gram-positive bacterial species but not to fungal infections. In contrast, flies mutant for GNBP3, another gene encoding a GNBP family member, fail to activate the Toll pathway in response to killed fungi and succumb rapidly to fungal but not bacterial infections (17). GNBP3 is thought to activate a proteolytic cascade, which partially overlaps that triggered by the GNBP1·PGRP-SA complex (18). Even though they belong to the same family and activate the same pathway, GNBP1 and GNBP3 are required for sensing distinct classes of microorganisms.
The founding member of the GNBP family, a 50-kDa protein found in hemolymph of Bombyx mori and originally named p50, was characterized as a gram-negative (Escherichia coli) binding protein (19); hence, its name. However, it has become clear that GNBPs belong to the family of β-glucan recognition proteins (βGRP) that had first been purified on their ability to trigger the prophenol oxidase cascade (a wound response that leads to melanization at the injury site) in response to fungal infections (20). Members of the GNBP/βGRP family are extracellular proteins composed of a small N-terminal domain of about 100 residues and a longer C-terminal domain of about 350 residues (21, 22). In the insect Plodia interpunctella, both domains of βGRP bind to laminarin, a soluble β-1,3-glucan with a high affinity (KA in the 108 m−1 range) (23) which is in the same range as that of the Factor G of the Japanese horseshoe crab (24). The latter factor is used as a diagnostic reagent for the detection of glucans. The C-terminal domain displays sequence similarity to bacterial glucanases, yet the catalytic residues have not been conserved, suggesting that this domain has been selected during evolution for its ability to bind to glucans (21, 22). The N-terminal domain defines a novel β-1,3-glucan binding domain that binds to curdlan, an insoluble linear β-1,3-glucan polymer, a property that the C-terminal glucanase-like domain lacks (21). Full-length recombinant GNBP/βGRPs have been reported to bind to bacteria, lipopolysaccharides, or lipoteichoic acids (19, 22, 23, 25). Although the domain(s) that mediates these interactions has not been thoroughly mapped, it appears that the N-terminal P. interpunctella β-1,3-glucan domain is not required for binding to these bacterial compounds (23).
Numerous three-dimensional structures of PGRPs, in some cases complexed with their ligands, have been reported (26–29). In contrast, this knowledge is currently lacking as regarding GNBPs. As a first step toward elucidating the structure/function relationships of GNBPs, we report here that a recombinant polypeptide encoding the N-terminal domain of GNBP3 binds to fungi and to long β-1,3-glucan chains but not to short laminarioligosaccharides. The determination of the crystal structure of GNBP3 N-terminal domain reveals an immunoglobulin fold in which the β-glucan binding site is masked by a lid, which is likely to be displaced by long polysaccharide chains.
EXPERIMENTAL PROCEDURES
Strains
The following strains were used in this study: the Gram-positive bacteria Micrococcus luteus (CIP A270), the Gram-negative bacteria E. coli (1106), the Ascomycetes Candida albicans (Caf2.1), Candida glabrata (BG2), Saccharomyces cerevisiae (BY4741, EUROSCARF, kindly provided by Unité de Génétique Moléculaire des Levures, Institut Pasteur, Paris, France), Aspergillus fumigatus (CBS144-89, a clinical isolate), and the Basidiomycetes Cryptococcus neoformans (H99).
Materials
Curdlan (insoluble β-1,3-glucan, a kind gift from Dr. Hidemitsu Kobayashi), laminarin (rarely branched β-1,3-glucan; Sigma), Pustulan (linear β-1,6-glucan; Calbiochem), and Schizophyllan (a highly β-1,6-branched β-1,3-glucan from Kaken, Japan, kindly provided by Dr Kazutoshi Shibuya) were the polymers used, whereas colloidal chitin was prepared using chitin (Sigma) as described by Gómez Ramírez et al. (30). S. cerevisiae and A. fumigatus cell wall fractions were produced following the protocol of Fontaine et al. (31) using actively growing yeast cells and germinating mold conidia in a medium containing 2% glucose and 1% peptone at 37 °C for 15 h. Briefly, cell walls obtained after disruption of the fungal cells was boiled with Tris (50 mm), EDTA (50 mm), SDS (2%), β-mercaptoethanol (40 mm) reagent (pH 7.5) for 15 min, twice, to release cell-wall bound proteins and subsequently treated with 1 m NaOH, 0.5 m NaBH4 (70 °C, 1 h, twice). The supernatant (alkali-soluble (AS) fraction) obtained after centrifugation was neutralized and dialyzed against water, whereas the sediment (alkali-insoluble (AI) fraction) was washed till neutrality. Both the fractions were freeze-dried and stored at −20 °C. Laminarioligosaccharides with a degree of polymerization of 2–40 (mixture or in their pure form) were prepared according to the method described by Martín-Cuadrado et al. (32). Alternatively, laminaritetraose was obtained from Seikagaku. Preparation of curdlan beads was achieved following the protocol described by Ochiai and Ashida (33). All these materials were used for in vitro binding and pulldown assays or for direct and competition ELISA assays.
Expression, Purification, Crystallization, and Mutagenesis
Starting from the sequence alignment of full-length GNBP3 from the 12 known Drosophila genomes (supplemental Fig. 1), the N-terminal domain (that we called GNBP3-Nter) was defined from residues 1 to 128, including the signal peptide (residues 1–25). The protein was successfully expressed in Drosophila S2 cells. Details of expression, purification, and crystallization are described elsewhere (51). W77A and short-loop mutants of GNBP3-Nter were prepared using the QuikChange II site-directed mutagenesis kit (Stratagene). The mutation was confirmed by DNA sequencing (MWG).
Pulldown and Competition Assays
Overnight cultures of yeasts were collected by centrifugation, washed 3 times with PBS, and resuspended in PBS to an A600 = 1. Yeasts were either fixed with 4% paraformaldehyde overnight at 4 °C and then post-quenched with 0.2 m glycine or treated with 1.5 m NaOH solution twice for 30 min at 70 °C and washed with PBS until neutrality. p-Formaldehyde, sodium hydroxide-treated microorganisms, and curdlan beads were used for in vitro binding assays of GNBP3-Nter. 1 ml of killed microbes with an A600 of 1 or 50 μg of curdlan beads was added to 5 μg of purified GNBP3-Nter and incubated in 200 μl of binding buffer (10 mm Tris-HCl (pH 7.5), 500 mm NaCl) at room temperature with mild agitation for 1 h. The solution containing both recombinant protein and yeasts or curdlan particles was centrifuged (14,000 × g for 5 min), and the pellet was washed 3 times with 0.5 ml of washing buffer (10 mm Tris (pH 7.5), 500 mm NaCl, 0.02% Tween 20).
For competition assays (Western blotting coupled with immunodetection), S. cerevisiae AI fraction (100 μg) or curdlan was mixed with 0.5 μg of purified GNBP3-Nter-His tagged alone or pretreated with soluble laminaritetraose/laminarin (400 μg) in a total volume of 50 μl (in 10 mm Hepes (pH 7.5) containing 30 mm NaCl) at 37 °C for 1 h with mild intermittent agitation.
In both types of pulldown assays or competition assays, the unbound protein was recovered from the reaction mixture by centrifugation at 3000 × g for 5 min and analyzed by SDS-PAGE (15% gel) either directly or after acetone precipitation (90 μl of the sample). GNBP3-Nter bound to curdlan/Sc-AI-fraction was recovered after washing (6×) the centrifugation pellet with 100 μl of 10 mm Hepes containing 150 mm NaCl followed by boiling for 10 min in SDS sample buffer (15 μl). The protein thus released into the supernatant after subsequent centrifugation was analyzed by SDS-PAGE and Western blot using a mouse peroxidase-conjugated mAb-His (Sigma) following the manufacturer's instructions (Penta-His HRP Conjugate kit, Qiagen) or a rabbit polyclonal anti-GNBP3-Nter antibody as primary antibody.
Antibody Production
The purified recombinant GNBP3-Nter protein from S2 cells expression was used to produce polyclonal rabbit antisera. The anti-GNBP3-Nter antisera were screened for specific staining of GNBP3-Nter and Drosophila endogenous GNBP3 by Western blot analysis. The specificity of the antibody was assessed by comparing extracts of wild type flies to those of a null GNBP3 mutant strain (data not shown).
Immunolocalization
Recombinant His-V5-tagged GNBP3-Nter protein was incubated with paraformaldehyde-treated or NaOH-treated yeast for 1 h at room temperature in binding buffer. After coincubation, the mixture was centrifuged, and the supernatant was aspirated. The pellet was allowed to dry for 2 min. Cells were washed 3 times in washing buffer and blocked with 2% BSA in PBS for 1 h. GNBP3-Nter proteins were detected with a primary mouse anti-V5 antibody (Invitrogen). Primary antibodies were visualized with Cy3-conjugated goat anti-mouse (Zymed Laboratories Inc.). DNA was visualized with 4′,6-diamidino-2-phenylindole. Slides were mounted in Vectashield medium (Vector Laboratories) and were examined by confocal microscopy (Zeiss LSM510). Slides were kept at 4 °C, and the images were processed using Adobe PhotoShop CS (Adobe Systems) and analyzed using ImageJ plugin RVB profiler.
Direct and Competition ELISA Assays
Fungal cell wall fractions/commercial polymers (200 μg/ml) dispersed by ultrasonication in 50 mm Na2CO3 (pH 9.6) were added (100 μl) to microtiter wells on ELISA plates and incubated overnight at room temperature. Unbound material was removed, and the wells were blocked with 1% BSA and 2% Tween 20 (in PBS) for 1 h at room temperature. His-tagged GNBP3-Nter (0.5 μg/100 μl of binding buffer containing 1% BSA in PBS) was added to each well and incubated at 37 °C for 1 h followed by 3 washes with PBS containing 0.5% Tween 20. Peroxidase-conjugated mAb-His (Sigma) (1:10,000 dilution in PBS containing 1% BSA) was added, and the mixture was incubated for 1 h at 37 °C. Finally, the reaction was developed in the presence of 0.1 mg/ml O-phenylenediamine (Sigma) and 0.1% H2O2.
For the competition assays, microtiter wells on ELISA plates were coated with the AI fraction (100 μg/ml, 100 μl) A. fumigatus or curdlan as described above. At the same time 0.5 μg of GNBP3-Nter-His tagged was incubated with different concentrations of individual laminarioligosaccharides of DP 2–16 or a laminarioligo mixture of DP 12–20 and 20–40 or laminarin in 10 mm Hepes buffer (pH 7.0) in a total volume of 50 μl for 1 h at room temperature, after which 50 μl of PBS containing 2% BSA was added to all the tubes. Then these mixtures were added to each well, and ELISA readings were performed as described above. Statistical analyses were done on GRAPHPAD PRISM using the Student-Newman-Keuls test.
Isothermal Titration Microcalorimetry (ITC)
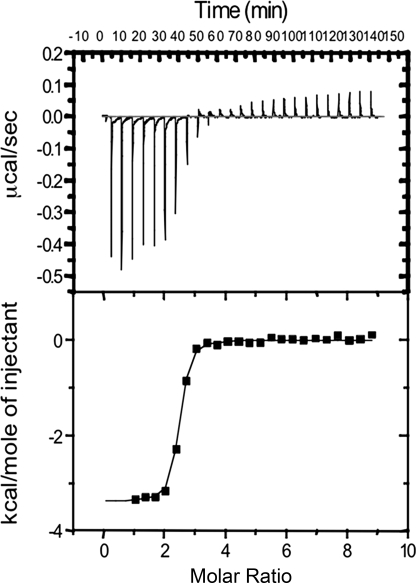
ITC experiments were performed using an iTC200 Isothermal Titration Calorimetry system (MicroCal; Northampton, MA) at a temperature of 30 °C. A typical titration profile is shown in Fig. 3. Protein and sugar samples were prepared in 20 mm Hepes and 150 mm NaCl (pH 7.5). Protein solution was taken in a syringe and loaded into the ITC sample cell (cell volume 200 μl). After the base line stabilized, 20 injections of 2 μl of the sugar ligand solution were added from the computer-controlled syringe into the protein solution, and exothermic heat changes accompanying the additions were recorded. The time period between the two consecutive injections was fixed at 340 s to allow the exothermic peak to return to the base line. The heat of mixing was measured by making identical injections into the cell containing buffer with no protein. The experimental data were fitted using software ORIGIN 7 supplied by Microcal, with ΔH (enthalpy change in kcal mol−1), KA (association constant in M−1), and n (number of binding sites/monomer) as adjustable parameters. Other thermodynamic parameters were calculated using the standard equation, ΔG = ΔH − TΔS = −RT log KA, where ΔG, ΔH, and ΔS are the changes in free energy, enthalpy, and entropy of binding, respectively. T is the absolute temperature in Kelvin, and R = 1.98 cal mol−1K−1.
FIGURE 3.
Interaction of GNBP3-Nter with laminarin in solution. The isothermal titration calorimetry of GNBP3-Nter was performed by injecting 2 μl of laminarin at 2 mm into 53 μm GNBP3-Nter at 340-s intervals. The upper panel shows a representative thermogram, and the lower part shows the integrated heats after fitting.
Structure Determination
The structure of GNBP3-Nter was determined by the single wavelength anomalous dispersion method using a samarium derivative. Diffraction data for the samarium derivative were collected at 100 K on a 300-mm Marresearch imaging plate mounted on a Rigaku RU200 rotating anode. They were indexed, integrated, and scaled using the XDS package (34). The space group was C2 with two molecules per asymmetric unit. Samarium sites were identified, refined, and used for phase calculation with the PHENIX suite (35). An initial model was then auto-built with PHENIX in which 69% of the total amount of residues was built. At this stage, the R value was 34%, and the Rfree value was 38%. This model was then refined against a high resolution diffraction data set (1.45 Å) collected on beamline ID23-1 at the European Synchrotron Radiation Facility (Grenoble, France). Refinement was performed using REFMAC5 (36), and manual rebuilding was carried out with the programs Coot (37) and Turbo-Frodo (38). The models of GNBP3-Nter lack interpretable electron density for the last residues 102–107. The final crystallographic model was refined to R and Rfree values of 16.5 and 19.8%, respectively. Statistics for all the data collections and refinement are summarized in Table 1. Figs. 4 and 5 were generated with PyMOL.
TABLE 1.
Data collection and refinement statistics
| GNBP3-Nter_Sm | GNBP3-Nter native | |
|---|---|---|
| Data collection statistics | ||
| Radiation source | In-house | ESRF ID23-1 |
| Wavelength (Å) | 1.5418 | 1.0332 |
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 135.32, 30.80, 51.72 | 135.53, 30.68, 51.65 |
| β (°) | 107.25 | 107.57 |
| Resolution range (Å) | 46.52-2.20 (2.32-2.20) | 46.52-1.45 (1.52-1.45) |
| Total observations | 132,581 | 129,875 |
| Unique reflections | 10,736 | 36,216 |
| Completeness (%) | 99.8 (99.0) | 98.8 (95.6) |
| Redundancy | 12.3 (11.9) | 3.6 (3.5) |
| Rmergea | 3.3 (8.6) | 6.7 (29.6) |
| Average I/σ(I) | 53.8 (30.1) | 12.2 (4.1) |
| Refinement and model statistics | ||
| Resolution range (Å) | 29.85-1.45 | |
| Proteins per asymmetric unit | 2 | |
| Number of reflections used | 34,308 | |
| Rwork (%)b/Rfree (%)c | 16.5/19.8 | |
| Average B values | ||
| All atoms (Å2) | 14.94 | |
| Protein atoms (Å2) | 13.28 | |
| Ethylene glycol atoms (Å2) | 35.87 | |
| Zinc atoms (Å2) | 4.29 | |
| Water atoms (Å2) | 28.40 | |
| Root mean square deviation from ideality | ||
| Bond lengths (Å) | 0.012 | |
| Bond angles (°) | 1.433 | |
| Torsion angles (°) | 6.756 | |
| Ramachandran analysis, favored regions/allowed regions/outliers (% of residues) | 99/1/0 | |
| No. of atoms | ||
| Protein | 1,612 | |
| Ethylene glycol | 8 | |
| Zinc | 6 | |
| Water | 192 | |
a Rmerge = ΣhΣi|Ih,i − 〈I〉h|/ΣhΣiIh,i, where 〈I〉h is the mean intensity of the symmetry-equivalent reflections.
b Rwork = Σh‖Fo| − |Fc‖/Σh|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively, for reflection h.
c Rfree is the R value for a subset of 5% of the reflection data, which were not included in the crystallographic refinement.
FIGURE 4.
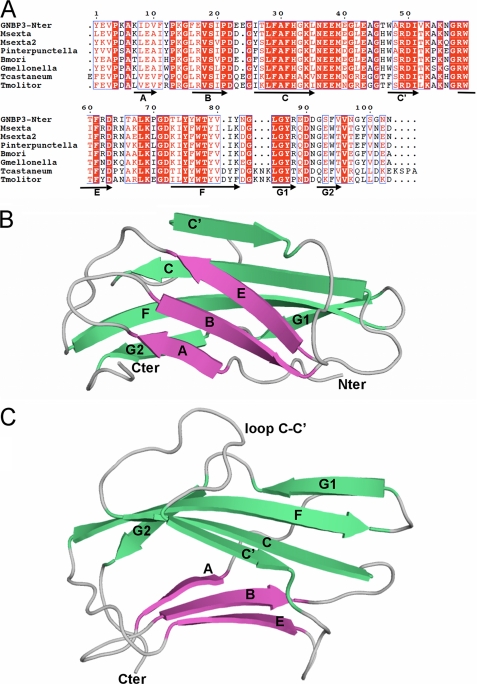
Sequence and overall structure of GNBP3-Nter. A, sequence alignment of N-terminal domains of βGRP (β-glucan recognition protein) from D. melanogaster (GNBP3-Nter), Manduca sexta (Msexta and Msexta2), P. interpunctella (Pinterpunctella), B. mori, Galleria mellonella (Gmellonella), Tribolium castaneum (Tcastaneum), and Tenebrio molitor (Tmolitor). The numbering is that of GNBP3-Nter of this study. Secondary structure elements (strands) are indicated below the sequences as arrows. Conserved residues are boxed, and strictly conserved residues are shown in white with a red background. Interestingly, the level of sequence identity is three times higher in the Nter domain (44%, 45/102) than in the C-terminal (Cter) domain (13%, 48/360). B, overall structure of GNBP3-Nter in ribbon representation with the strands A-B-E (sheet 1), colored in fuchsia, and C-C′-F-G1-G2 (sheet 2), colored in green, forming an immunoglobulin fold of fibronectin type III type. C, the same view with a 90° rotation along horizontal axis showing the C-C′ loop that folds back onto the β-sheet C′-C-F-G1-G2.
FIGURE 5.
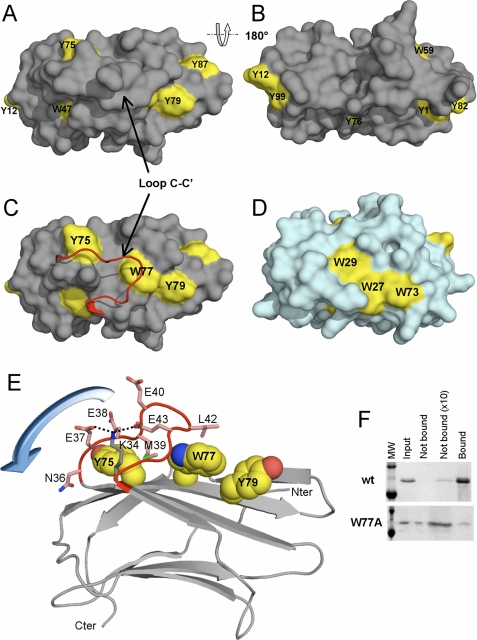
Binding site and the C-C′ loop of GNBP3-Nter. A, GNBP3-Nter is represented as a gray molecular surface, and tyrosine and tryptophan residues are colored in yellow. B, the back side of the molecule (after a 180° rotation), devoid of aromatic residues. C, the same view as in A with the C-C′ loop represented as a ribbon (in red). D, the molecular surface of the starch recognition domain of the pullulanase PulA from T. maritima in a similar orientation as in C and showing the aromatic residues (in yellow) involved in its binding site. E, the three aromatic residues (spheres colored according to the atom type) constituting the putative glucan binding site, covered by the C-C′ loop (in red) with its hypothetical movement (represented by a blue arrow). The side chains of the residues of the C-C′ loop are shown as sticks with carbon colored in pink, nitrogen in blue, oxygen in red, and sulfur in green. Discontinuous black lines represent the hydrogen bonds between Glu-37, Glu-43, and Lys-34. The remainder of GNBP3-Nter is represented as ribbon. The molecule is rotated of about 90° along horizontal axis as compared with C. F, SDS/PAGE analysis of the pulldown of GNBP3-Nter wild type (wt) and W77A mutant with curdlan. The first lane represents the molecular weight ladder (upper band = 20 KDa, lower band = 14 KDa). The next four lanes show one-tenth of the input, the unbound protein (10 μl of the supernatant), the unbound protein (90 μl of the supernatant after acetone precipitation), and the protein bound to curdlan (sediment).
RESULTS
The boundaries of GNBP3-Nter were delineated using an alignment of GNBP3 sequences from the genomes of 12 Drosophila species, as depicted in supplemental Fig. 1. The recombinant protein was overexpressed at a high level (>15 mg/liter of culture) in Drosophila S2 cells with the C-terminal extension V5-His6, which was used for detection and purification. The tag was proteolytically removed to allow crystallization of the recombinant protein (51).
GNBP3-Nter Binds to the Fungal Cell Wall
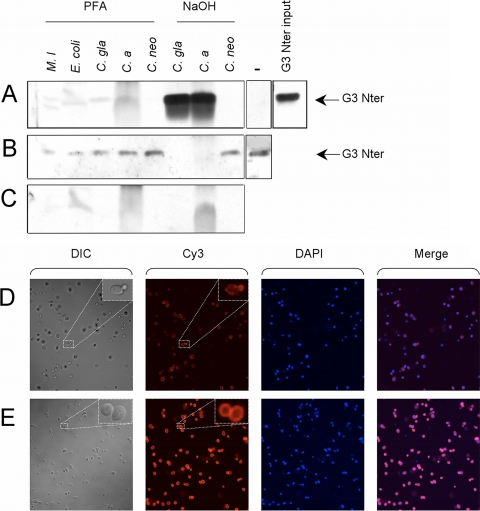
To determine whether the recombinant N-terminal domain of GNBP3 is able to bind to fungi, we first analyzed by pulldown experiments its binding to C. albicans, C. glabrata, and C. neoformans yeasts. We detected a mild binding to p-formaldehyde-fixed Candida yeasts and a strong binding to NaOH-treated Candida yeasts using either a tagged or a cleaved tag form of the recombinant protein (Figs. 1, A–C). The NaOH treatment strips the cell wall of its proteins and alkali-soluble polysaccharides, thus making the β-1,3-glucans more accessible. We did not, however, detect any binding to C. neoformans or bacteria, which have no β-glucan on their surface. We confirmed by immunohistochemistry the binding of the recombinant protein to C. albicans (Fig. 1D) and C. glabrata (data not shown). We found that the recombinant protein binds to discrete patches of the yeasts. Staining appeared strong in newly formed buds and bud scars. In contrast, GNBP3-Nter bound to the entire surface of NaOH-treated Candida yeasts (Fig. 1E).
FIGURE 1.
Binding of GNBP3-Nter to Candida. A–C, the recombinant protein was incubated with microorganisms, spun down by centrifugation, and washed, and the pellet (A) or one-tenth of the supernatant (B) was analyzed by Western blot using a specific antibody. A control of the pellets obtained after a similar treatment with no added recombinant protein is shown in C. Importantly, the recombinant protein in the absence of microorganisms did not precipitate during the procedure (−, ninth lane of A). One-tenth of the input protein is shown in the boxes on the right (C. gla, C. glabrata; C. a., C. albicans; C. neo, Cryptococcus neoformans; M. l, M. luteus). D–E, the V5-tagged recombinant protein was incubated with either paraformaldehyde (PFA)-fixed (D) and alkali-treated C. albicans (E) and detected by immunofluorescence (Cy3) using a V5 antibody. Similar results were obtained with C. glabrata-treated yeast. DIC, differential interference contrast; DAPI, 4′,6-diamidino-2-phenylindole nuclear staining.
GNBP3-Nter Binds Specifically to β-1,3-Glucans
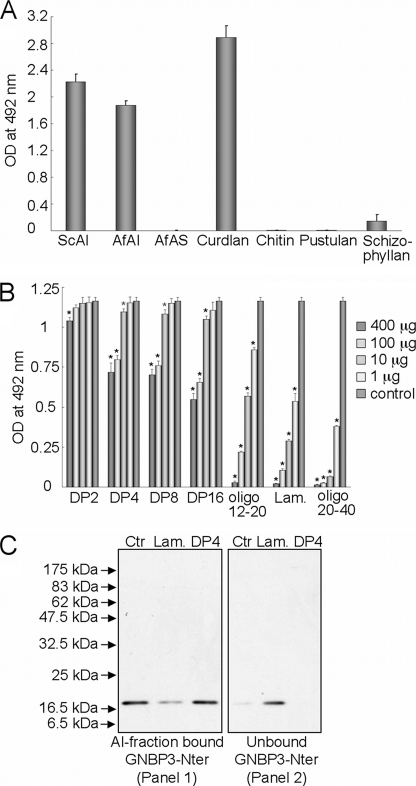
From the preceding experiments, we deduced that GNBP3-Nter binds to the fungal cell wall. However, as the latter is mainly a complex network of different polysaccharides, we performed binding assays on ELISA plates coated with different cell wall fractions or with commercially available polysaccharides. As depicted in Fig. 2A, GNBP3-Nter efficiently bound to the cell wall alkali-insoluble (AI) fraction from S. cerevisiae and A. fumigatus but not to the alkali-soluble (AS) fractions of A. fumigatus, which lacks β-1,3-glucans and contains mainly α-1,3-glucan and galactomannan. The structure common to the AI fraction of S. cerevisiae and A. fumigatus is a β-1,6-branched β-1,3-glucan covalently bound to chitin, suggesting that the polysaccharide recognized by GNBP3-Nter was either a β-1,3-glucan or chitin. However, we did not observe any binding with chitin, a linear polymer of N-acetylglucosamine. The binding efficacy to schizophyllan, a β-1,3-glucan with single β-(1,6)-bonded glucose at every third glucose molecule on the main chain, was less than 10% compared with the S. cerevisiae AI fraction. Also, there was no binding to pustulan, a linear β-1,6-glucan polymer. The highest ELISA values were obtained for curdlan, an insoluble linear β-1,3-glucan. Taken together, the binding assays indicate that GNBP3-Nter shows specific affinity toward β-1,3-glucan.
FIGURE 2.
Binding of GNBP3-Nter to polysaccharides versus oligosaccharides. A, direct ELISA assays showing the binding of GNPB3-Nter to the cell wall AI fractions of S. cerevisiae (ScAI) and A. fumigatus (AfAI), AS fraction of A. fumigatus (AfAS), and other commercially available fungal cell wall polysaccharides. Note that GNBP3-Nter does not bind to chitin, pustulan, or the AS fraction of A. fumigatus cell wall. Binding with schizophyllan (a highly branched β-glucan) is ∼1/10 of curdlan, confirming the affinity of GNBP3-Nter for linear β-(1,3)-glucan. Each bar represents mean ± S.D. of six repetitions. B, ELISA inhibition assays show that linear oligosaccharides of DP > 20 are the best inhibitor for binding of GNBP3-Nter to AI fractions of A. fumigatus at 1–400 μg/0.5 μg of GNBP3-Nter. Each bar represents the mean ± S.D. of four repetitions, black *, p < 0.001; gray *, p < 0.05. C, blot analysis of the competition between S. cerevisiae AI fraction and laminarin or laminaritetraose (DP4) for binding to GNBP3-Nter. GNBP3-Nter binds efficiently to the AI fraction (Panel 1, Ctr) even in the presence of high concentration of laminaritetraose (Panel 1, DP4). However, laminarin (soluble oligomeric mixture of DP > 20) competes with the AI fraction for GNBP3-Nter binding (Panel 2, Lam).
GNBP3-Nter Binding to β-Glucans Increases with Polysaccharide Chain Length
Competition assays for binding to GNBP3-Nter were performed between the cell wall AI fraction from A. fumigatus and soluble β-1,3-glucan oligosaccharides of different sizes (individually or in a mixture). After preincubation of GNBP3-Nter with laminarioligosaccharides of varying length (degree of polymerization (DP) of 2–16), there was weak or no reduction in the binding of GNBP3-Nter to the wells on the ELISA plates coated with the AI-fraction even when GNBP3-Nter was preincubated with laminarioligosaccharides at a 1/800 mass ratio (Fig. 2B). In contrast, water-soluble laminarioligosaccharide mixtures of higher DP (12–20, with maximum concentration of DP 14–18) and laminarin (a mixture of oligosaccharides of DP between 20 and 28 with 25-DP oligomer in the maximum concentration and having one branching point per oligosaccharide chain) competed efficiently with the AI fraction for the binding site(s) on GNBP3-Nter (Fig. 2B). However, the best inhibition was observed with a mixture of oligosaccharides of DP between 20 and 40 that was water-insoluble (Fig. 2B). These ELISA inhibition data were further confirmed by competition between laminaritetraose or laminarin and the AI fraction from S. cerevisiae for GNBP3-Nter binding using pulldown assays coupled with blotting immunodetection analyses (Fig. 2C). β-1,3-Linked tetraoses did not compete with the Sc-AI fraction for GNBP3-Nter binding, even at a protein/sugar molar ratio of 1/500, whereas laminarin reduced significantly the binding of GNBP3-Nter to Sc-AI fraction when used at a protein/sugar ratio of 1/10 (Fig. 2C). Thus, an increase in the binding affinity correlated with increasing oligosaccharide chain length and with concomitant decreasing aqueous solubility, whereas short linear or branched β-1,3-linked oligosaccharides were not efficient ligands for GNBP3-Nter. ITC was performed to quantify the interaction of GNBP3-Nter with laminarin in solution. ITC data for the binding fit a single-site binding model (Fig. 3). The stoichiometry for the interaction between GNBP3-Nter and laminarin was close to a ratio of 1–3 (n = 2.51), consistent with a triple helix organization of the laminarin in solution (39). The binding affinity is KA = 2.12 × 106 ± 0.4 × 106 m−1. The fitted data also yielded the interaction with negative enthalpy (ΔH = −3.34 kcal/mol) and entropy (ΔS = 17.9cal/mole/degree). In contrast and consistent with the competition assays, no interaction was detected between GNBP3-Nter and shorter sugars such as heptaose (DP7) or hexaose (DP6) even with repeated injections (2 μl) of highly concentrated sugar (10 mm) in the cell (data not shown). Taken together, these data indicate that GNBP3-Nter binds specifically to linear β-1,3-glucans with high DP.
Overall Structure of GNBP3-Nter
The structure of GNBP3-Nter was solved at 1.45 Å of resolution by single anomalous dispersion using a samarium derivative. The crystals contain two copies of the protein in the asymmetric unit. The two molecules were refined independently, and there are no significant differences (root mean square deviation = 0.53 Å for all Cαs). The protein is monomeric in solution up to a concentration of 0.5 mm as analyzed by gel filtration and dynamic light scattering, suggesting little functional significance for the crystallographic dimer.
The final refined model consists of residues 26–128 that were renumbered 1–102 (Fig. 4A). GNBP3-Nter is a globular domain of approximate 40 × 25 × 20 Å3 dimension. The overall structure consists of two antiparallel β sheets and belongs to the immunoglobulin fold family. The first sheet is made of strands A (residues 7–10), B (residues 17–21), and E (residues 58–63), whereas the strands C′ (residues 47–51), C (residues 26–35), F (residues 73–82), G1 (residues 85–88), and G2 (residues 92–95) constitute the second sheet (Fig. 4B). The two sheets are packed in a β-sandwich conformation enclosing a highly hydrophobic core organized around a cluster of three phenylalanines (Phe-16, Phe-31, and Phe-61). The closest structural homologue found using the DALI server (40) is a fibronectin type III domain of integrin α6β4 (PDB code 1QG3) (41) with a Z-score of 7.9. This molecule displays the same β-sheet organization, i.e. A-B-E and C′-C-F-G1-G2. Despite a very low level of sequence identity (9%), superimposition of the fibronectin III domain with GNBP3-Nter shows that 66 residues of 102 are structurally conserved, giving a root mean square deviation value of 1.54 Å. The main difference is the presence in GNBP3-Nter of a large negatively charged loop between strands C and C′ that folds back onto the β-sheet C′-C-F-G1-G2 (Fig. 4C). Interestingly, GNBP3-Nter also displays the same β-sheet organization as starch binding domains (42).
Carbohydrate Binding Site
Based on their binding characteristics, carbohydrate binding modules have been classified into three types named A (“surface binding,” for insoluble polysaccharides), B (“glycan chain binding,” which involves a groove), and C (“small sugar binding”) (43). The functional studies in this report show the preferential binding of GNBP3-Nter to long chain soluble or insoluble β-glucans, thus classifying GNBP3-Nter either as a type A or as a type B carbohydrate binding module. Higher affinity toward curdlan/cell wall AI fractions compared with soluble short chain sugars and the absence of any groove containing aromatic residues on its molecular surface led us to consider that GNBP3-Nter may belong to type A carbohydrate binding modules.
Type A carbohydrate binding modules display flat or platform-like binding sites made of three aromatic residues in most cases (43). The outer molecular surface of GNBP3-Nter does not display any obvious aromatic patch. The three Trp and the eight Tyr residues of GNBP3-Nter were, therefore, carefully examined (Fig. 5, A and B). Trp-47 participates in the previously described hydrophobic core located between the two β sheets. It makes van der Waals contacts with Phe-61, Phe-16, and Leu-35 and is partially buried by Thr-46. Trp-59 stands in a hydrophobic pocket and contacts Leu-26 (CG2), Ile-51 (CG2), Ala-54 (CB), Phe-29, and Phe-31. Thus, it is even more buried than Trp-47. The indole ring of Trp-77 is in stacking interaction with the His-32 imidazole group that lies beneath it. Trp-77 is only poorly accessible, as it is masked from the surface by Leu-42, which stands at the tip of the C-C′ loop. Five of the eight Tyr residues are distributed into two groups located at the two ends of the molecule. On one side Tyr-12 and Tyr-99 stand close to each other but are not stacked. They are accessible, with their OH groups pointing toward the surface of the molecule. On the opposite side, a stacking interaction occurs between Tyr-1 and Tyr-82. A third tyrosine, Tyr-87, positions its ring ∼90° from those of Tyr-1 and Tyr-82, leading to the formation of an imperfect aromatic cage. Tyr-76 is completely buried inside the molecule, whereas the Tyr-75 residue is masked from the surface by Glu-40 on the C-C′ loop. Finally, only Tyr-79 is fully solvent-exposed. This latter residue stands on the strand F, which is central to the β-sheet C′-C-F-G1-G2. Interestingly, strand F possesses two other aromatic residues, Tyr-75 and Trp-77, which are strictly conserved among β-glucan recognition domains. The spatial arrangement of the three aromatic side chains of Tyr-75, Trp-77, and Tyr-79 (Fig. 5C) is similar to the aromatic patch constituted by three neighboring residues described for starch binding domains, for example those of the starch recognition domain of the pullulanase PulA from Thermotoga maritima (44)(Fig. 5D). Nevertheless, the exposure of Tyr-75 and Trp-77 side chains to the surface is masked by the C-C′ loop. When this C-C′ loop is removed from GNBP3-Nter using a graphics display system, Tyr-75 and Trp-77 become accessible to solvent (Fig. 5C). Thus, we hypothesized that this loop acts as a mobile lid domain that covers the putative binding site (Fig. 5E). To verify our hypothesis, we mutated Trp-77 into Ala. Structural integrity of W77A mutant was assessed by circular dichroism spectroscopy. No structural difference between wild type and mutant proteins was detected (supplemental Fig. 3B). W77A mutant was expected to display a decreased affinity for β-1,3-glucan. Indeed, the binding to curdlan in a pulldown assay was severely decreased with the mutant protein (Fig. 5F). Moreover, in ITC experiments we failed to detect any binding of either long or short laminarioligosaccharides to the W77A mutant (data not shown), thus delineating a key role for Trp-77 in β-1,3-glucan recognition.
The C-C′ Loop of GNBP3-Nter
An unusual feature of GNBP3-Nter structure is the presence of a long loop between strands C and C′, which is composed of 10 residues and extends outwards from the compact body. Insertions between strands C and C′ have already been described for other members of the fibronectin type III-fold family and were assigned to be protein-protein interaction domains, as observed, for example, for fibronectin type III α6β4 integrin (41). The C-C′ loop of GNBP3-Nter is quite well conserved in terms of length and sequence among the N terminus domains of GNBP/βGRPs that have been shown to bind to β-glucans (Fig. 4A). The strict conservation of seven positions often gives the following consensus motif 36NEEMXGXEXG45. GNBP3-Nter carries four negatively charged amino acids (Glu-37, Glu-38, Glu-40, and Glu-43), which point outward from the surface (Fig. 5E). Two of them, Glu-37 and Glu-43, interact through side chain-side chain hydrogen bonds with the conserved Lys-34 residue. The NE2 nitrogen of Trp-77 interacts with the carbonyl oxygen of Glu-43 on the C-C′ loop. Finally, the side chains of the strictly conserved Met-39 and of Leu-42 contribute to the formation of a hydrophobic environment together with Tyr-75 and Trp-77 on the internal face of the C-C′ loop. As all these residues have been conserved throughout 350 million years of evolution (divergence of the Diptera and Lepidoptera lineages occurred during the early Carboniferous) (45), it is likely that these interactions have been selected to maintain the lid in a closed position in the absence of glucan ligands. A mutant protein in which the occluding loop was shortened and the conserved residues were mutated was cloned and produced in Drosophila cells (supplemental Fig. 3A). The structural integrity of the mutated protein was assessed by circular dichroism (supplemental Fig. 3B). The binding of the short-loop mutant to killed Candida yeasts was not detectable by immunohistochemistry. ELISA assays showed that the binding to the AI fraction of S. cerevisiae and curdlan was substantially reduced and corresponded to 30 and 10% that of the wild type, respectively (data not shown).
DISCUSSION
The discrimination between host and microbe-associated molecules is crucial to the function of PRRs. Short oligosaccharide chains may not constitute an ideal target for PRRs as they might also be displayed by host cells and, thus, may not represent a bona fide microbial signature. Therefore, it is likely that the host selected PRRs able to sense long glucan chains idiosyncratic to most fungal cell walls. In this manuscript we report that the glucan binding domain of GNBP3 binds preferentially to long β-1,3-glucan chains and shall discuss how the distinction between short and long chains of glucans is made by various PRRs.
The N-terminal domain of GNBP3 binds to the cell wall of C. albicans and most likely to β-glucans as indicated by the preferential binding to growing cell buds and bud scars, a pattern evocative of that of Dectin-1 (46). Indeed, the recombinant protein binds to the cell wall alkali-insoluble polysaccharide fraction of S. cerevisiae and A. fumigatus. The latter induces a GNBP3-dependent activation of the Toll pathway when injected into Drosophila (17). Our data indicate that the relevant biochemical moiety of these fungal cell wall AI extracts are β-1,3-glucan chains. These findings are confirmed by direct binding of GNBP3-Nter to curdlan and laminarin as assayed by ELISA, ITC, and pulldown experiments coupled to competition assays. The longer the glucan chain, the more efficient is the competition. Efficient binding to GNBP3-Nter is observed with polymeric chains that incorporate more than 16 glucan units. In keeping with this result, it has previously been shown that injection in Drosophila of the alkali-insoluble fraction of the A. fumigatus cell wall, which consists of long polysaccharides including β-1,3-glucans, induces a strong activation of the Toll pathway (17). At the same time, Gottar and colleagues in Strasbourg found that short laminarioligosaccharides with a DP ranging from 2 to 7 failed to induce Toll pathway activation when injected into flies.5 β-1,6-Branching in the linear chain of β-1,3-glucans does not appear to be required for recognition by GNBP3-Nter as we failed to observe strong binding with schizophyllan, a highly β-1,6-branched β-1,3-glucan from Schizophyllum commune (Fig. 2A). Interestingly, the glucan binding properties of the mammalian fungal receptor Dectin-1 have been reported to be fairly similar, with a minimum degree of polymerization of 11 required for β-glucan binding (47).
We have solved the GNBP3-Nter crystal structure that provides structural insight into the β-1,3-glucan recognition protein (βGRP) family. The overall structure displays an immunoglobulin-like fold similar to that of the fibronectin III superfamily. Although no solvent-exposed aromatic patch is present on GNBP3-Nter (Fig. 5, A and B), Tyr-75, Trp-77, and Tyr-79 are good candidates to constitute such a binding platform (Fig. 5C) after a structural rearrangement of the loop located between strands C and C′. In keeping with this hypothesis, we found that the binding to curdlan was strongly impaired, and the binding to laminarin was completely abolished with the W77A mutant, thus underscoring the importance of this initially buried residue for glucan binding. The essential role of the C-C′ loop in terms of binding and discrimination between short and long chains of β-glucan was confirmed by mutagenesis. To free access to the binding site, the C-C′ loop should fold back toward the C-terminal domain of GNBP3 (Fig. 5, C and E). Tyr-79 may act as a primary determinant that anchors β-glucan polymers. Then the negatively charged patch formed by the four glutamic acids on the top of the loop may be expulsed by the vicinity of a large sugar surface, unmasking the rest of the binding site (Tyr-75 and Trp-77). After the lid opening, the side chain of Tyr-75 is free to re-orientate toward Trp-77. Like this, the relative spacing between the three residues would not stretch beyond a distance required to accommodate a disaccharide and, thus, would be very comparable with that of starch binding domains (Fig. 5D). The internal hydrophobic surface of the lid could hardly be fully-exposed to solvent in the open conformation and may probably interact with the ligand. This putative interaction between the lid and the ligand may explain the results obtained for the short-loop mutant. Namely, this mutant does not appear to bind efficiently to long-chain oligosaccharides, possibly because the two conserved hydrophobic residues in the lid, which are missing in the mutant, no longer stabilize the interaction.
Both the sequence of the C-C′ loop and that of the putative binding site are conserved in βGRP family members that have been reported to bind to β-glucans. Noticeably, these sequences are not conserved in Drosophila melanogaster GNBP1 (and GNBP1 of other Drosophila species), a member of the family required in the host defense against Gram-positive bacteria (supplemental Fig. 2). We infer that the GNBP1 N-terminal domain will not bind significantly to β-1,3-glucans, even though some studies have reported some binding of full-length GNBP1 to curdlan (25). Thus, the sequences of the C-C′ loop and of the glucan binding site may be useful predictors of the function of uncharacterized GNBP/βGRP family members. Using this criterion, we predict that the function of the funding member of the GNBP family, B. mori p50, is not involved in defense against fungi, at least by a GNBP3/βGRP-like mechanism. In any case, GNBP full-length proteins, with their glucanase-like domains, are likely to have emergent properties not displayed by the Nter domain alone. These may include the activation of downstream proteolytic cascades (for Toll pathway and prophenol oxidase activation) and, obviously, agglutination, which requires two sugar binding domains in the protein.
An intriguing feature of the GNBP3-Nter domain is its capacity to discriminate between short and long chains of β-glucans. Many PRRs activate an immune response only when bound to long chains of carbohydrates through the use of spatially arranged multiple subunits or multimers. Yet, in striking contrast to GNBP3, the individual domains involved in carbohydrate recognition can bind to monomeric or short carbohydrate polymers. For instance, PGRP-SA, which binds to PGN muropeptide monomers as single molecules, requires the formation of PGRP-SA clusters on longer chains to trigger downstream proteolytic cascades (48). A similar case is presented by Factor G of the Japanese horseshoe crab where a laminariheptaose is required to activate the coagulation cascade even though it binds well to laminaribiose with an affinity that is only three times lower (49). Interestingly, the recognition domain that binds to β-glucans is actually made up of two carbohydrate binding subunits arranged in a tandem repeat. Only the tandem repeat, and not each individual subunit, is able to bind to the disaccharide. Another example of the importance of the spatial arrangements of multiple carbohydrate recognition domain (CRD) is provided by the mannose-binding lectin whereby each CRD head binds to a single sugar residue (mannose or fucose). Activation only occurs when the multiple heads arranged in a bouquet-like structure of trimers bind to an array of sugar residues present on the microbial but not the host cell surface (50). Here, we propose that the lid of the GNBP3-Nter domain that masks the carbohydrate binding site is displaced only by long chains of β-glucans. In this respect, the structures of laminarins and curdlan as triple helices in an aqueous environment may be an essential feature that triggers the opening of the carbohydrate binding site and the recognition of the fibrillar fungal structure.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility at Grenoble and in particular beamline ID23-1 staff for assistance. We are grateful to the ITC platform of the Institut de Biologie Moléculaire et Cellulaire in Strasbourg and in particular to Eric Ennifar. We acknowledge the expert technical help of Marie-Céline Lafarge. The Ferrandon laboratory is “Equipe FRM,” awarded by the Fondation pour la Recherche Médicale.
This work was supported, in whole or in part, by National Institutes of Health Grant PO1 AI44220. This work was also supported by an Action Thématique et Incitative sur Programme of the CNRS (2005–2008, to A. R.), by a grant from the Agence Nationale de la Recherche (ANR-MIME, to A. R. and C. H.), by the CNRS, and by a grant from the Ministère de l'Enseignement et de la Recherche (Programme de Recherche en Microbiologie).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
The atomic coordinates and structure factors (code 3IE4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
M. Gottar, personal communication.
- PRR
- pattern recognition receptor
- GRP
- glucan recognition protein
- PGRP
- peptidoglycan recognition protein
- AMP
- antimicrobial peptide
- GNBP1
- Gram-negative binding protein 1
- AS
- alkali-soluble
- AI
- alkali-insoluble
- ITC
- Isothermal titration calorimetry
- DP
- degree of polymerization
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- Nter
- N-terminal.
REFERENCES
- 1.Janeway C. A. (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13 [DOI] [PubMed] [Google Scholar]
- 2.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R., Janeway C. A., Jr. (2002) Science 296, 298–300 [DOI] [PubMed] [Google Scholar]
- 4.Takeda K., Kaisho T., Akira S. (2003) Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 5.Inohara N., Nuñez G. (2003) Nat. Rev. Immunol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 6.Royet J., Dziarski R. (2007) Nat. Rev. Microbiol. 5, 264–277 [DOI] [PubMed] [Google Scholar]
- 7.Brown G. D., Taylor P. R., Reid D. M., Willment J. A., Williams D. L., Martinez-Pomares L., Wong S. Y., Gordon S. (2002) J. Exp. Med. 196, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A. (2007) Nat. Rev. Immunol. 7, 862–874 [DOI] [PubMed] [Google Scholar]
- 9.Lemaitre B., Kromer-Metzger E., Michaut L., Nicolas E., Meister M., Georgel P., Reichhart J. M., Hoffmann J. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9465–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottar M., Gobert V., Michel T., Belvin M., Duyk G., Hoffmann J. A., Ferrandon D., Royet J. (2002) Nature 416, 640–644 [DOI] [PubMed] [Google Scholar]
- 11.Takehana A., Katsuyama T., Yano T., Oshima Y., Takada H., Aigaki T., Kurata S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13705–13710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A. (1996) Cell 86, 973–983 [DOI] [PubMed] [Google Scholar]
- 13.Michel T., Reichhart J. M., Hoffmann J. A., Royet J. (2001) Nature 414, 756–759 [DOI] [PubMed] [Google Scholar]
- 14.Jang I. H., Chosa N., Kim S. H., Nam H. J., Lemaitre B., Ochiai M., Kambris Z., Brun S., Hashimoto C., Ashida M., Brey P. T., Lee W. J. (2006) Dev. Cell 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 15.Gobert V., Gottar M., Matskevich A. A., Rutschmann S., Royet J., Belvin M., Hoffmann J. A., Ferrandon D. (2003) Science 302, 2126–2130 [DOI] [PubMed] [Google Scholar]
- 16.Bischoff V., Vignal C., Boneca I. G., Michel T., Hoffmann J. A., Royet J. (2004) Nat. Immunol. 5, 1175–1180 [DOI] [PubMed] [Google Scholar]
- 17.Gottar M., Gobert V., Matskevich A. A., Reichhart J. M., Wang C., Butt T. M., Belvin M., Hoffmann J. A., Ferrandon D. (2006) Cell 127, 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Chamy L., Leclerc V., Caldelari I., Reichhart J. M. (2008) Nat. Immunol. 9, 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee W. J., Lee J. D., Kravchenko V. V., Ulevitch R. J., Brey P. T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7888–7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida H., Ochiai M., Ashida M. (1986) Biochem. Biophys. Res. Commun. 141, 1177–1184 [DOI] [PubMed] [Google Scholar]
- 21.Ochiai M., Ashida M. (2000) J. Biol. Chem. 275, 4995–5002 [DOI] [PubMed] [Google Scholar]
- 22.Ma C., Kanost M. R. (2000) J. Biol. Chem. 275, 7505–7514 [DOI] [PubMed] [Google Scholar]
- 23.Fabrick J. A., Baker J. E., Kanost M. R. (2004) J. Biol. Chem. 279, 26605–26611 [DOI] [PubMed] [Google Scholar]
- 24.Muta T., Seki N., Takaki Y., Hashimoto R., Oda T., Iwanaga A., Tokunaga F., Iwanaga S. (1995) J. Biol. Chem. 270, 892–897 [PubMed] [Google Scholar]
- 25.Lee S. H., Carpenter J. F., Chang B. S., Randolph T. W., Kim Y. S. (2006) Protein Sci. 15, 304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M. S., Byun M., Oh B. H. (2003) Nat. Immunol. 4, 787–793 [DOI] [PubMed] [Google Scholar]
- 27.Chang C. I., Pili-Floury S., Hervé M., Parquet C., Chelliah Y., Lemaitre B., Mengin-Lecreulx D., Deisenhofer J. (2004) PLoS Biol. 2, E277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang C. I., Chelliah Y., Borek D., Mengin-Lecreulx D., Deisenhofer J. (2006) Science 311, 1761–1764 [DOI] [PubMed] [Google Scholar]
- 29.Leone P., Bischoff V., Kellenberger C., Hetru C., Royet J., Roussel A. (2008) Mol. Immunol. 45, 2521–2530 [DOI] [PubMed] [Google Scholar]
- 30.Gómez Ramírez M., Rojas Avelizapa L. I., Rojas Avelizapa N. G., Cruz Camarillo R. (2004) J. Microbiol. Methods 56, 213–219 [DOI] [PubMed] [Google Scholar]
- 31.Fontaine T., Simenel C., Dubreucq G., Adam O., Delepierre M., Lemoine J., Vorgias C. E., Diaquin M., Latgé J. P. (2000) J. Biol. Chem. 275, 27594–27607 [DOI] [PubMed] [Google Scholar]
- 32.Martín-Cuadrado A. B., Encinar del Dedo J., de Medina-Redondo M., Fontaine T., del Rey F., Latgé J. P., Vázquez de Aldana C. R. (2008) Mol. Microbiol. 69, 188–200 [DOI] [PubMed] [Google Scholar]
- 33.Ochiai M., Ashida M. (1988) J. Biol. Chem. 263, 12056–12062 [PubMed] [Google Scholar]
- 34.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 35.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 36.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 38.Roussel A., Cambillau C. (1991) in Silicon Graphics Geometry Partners Directory, Silicon Graphics, Mountain View, CA [Google Scholar]
- 39.Okobira T., Miyoshi K., Uezu K., Sakurai K., Shinkai S. (2008) Biomacromolecules 9, 783–788 [DOI] [PubMed] [Google Scholar]
- 40.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Pereda J. M., Wiche G., Liddington R. C. (1999) EMBO J. 18, 4087–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machovic M., Janecek S. (2006) Cell Mol. Life Sci. 63, 2710–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Bueren A. L., Boraston A. B. (2007) J. Mol. Biol. 365, 555–560 [DOI] [PubMed] [Google Scholar]
- 45.Gaunt M. W., Miles M. A. (2002) Mol. Biol. Evol. 19, 748–761 [DOI] [PubMed] [Google Scholar]
- 46.Gantner B. N., Simmons R. M., Underhill D. M. (2005) EMBO J. 24, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palma A. S., Feizi T., Zhang Y., Stoll M. S., Lawson A. M., Díaz-Rodríguez E., Campanero-Rhodes M. A., Costa J., Gordon S., Brown G. D., Chai W. (2006) J. Biol. Chem. 281, 5771–5779 [DOI] [PubMed] [Google Scholar]
- 48.Park J. W., Kim C. H., Kim J. H., Je B. R., Roh K. B., Kim S. J., Lee H. H., Ryu J. H., Lim J. H., Oh B. H., Lee W. J., Ha N. C., Lee B. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6602–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takaki Y., Seki N., Kawabata Si S., Iwanaga S., Muta T. (2002) J. Biol. Chem. 277, 14281–14287 [DOI] [PubMed] [Google Scholar]
- 50.Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. (1999) Science 284, 1313–1318 [DOI] [PubMed] [Google Scholar]
- 51.Mishima Y., Coste F., Bobezeau V., Hervouet N., Kellenberger C., Roussel A. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 870–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.