Abstract
Heparan sulfate proteoglycans (HSPGs) are important modulators for optimizing signal transduction of many pathways, including the Wnt pathways. We demonstrate that HSPG glycosaminoglycan levels increased with increasing metastatic potential of melanoma cells. Previous studies have demonstrated that Wnt5A increases the invasiveness of melanoma cells. We further demonstrate that HSPGs potentiate Wnt5A signaling, since enzymatic removal of the HSPG backbone resulted in a decrease in cellular Wnt5A levels, an increase in secreted Wnt5A in cell media, a decrease in downstream signaling, and ultimately, a decrease in invasiveness. Specifically, syndecan 1 and syndecan 4 expression correlated to Wnt5A expression and melanoma malignancy. Knockdown of syndecan 1 or 4 caused decreases in cell invasion, which could be restored by treating the cells with recombinant Wnt5A. These data indicate that syndecan 1 and 4 correlate to increased metastatic potential in melanoma patients and are an important component of the Wnt5A autocrine signaling loop, the activation of which leads to increased metastasis of melanoma.
The American Cancer Society estimates that in 2009 there will be 68,720 new cases of melanoma in this country with ∼8,650 deaths. Recent studies have demonstrated that the non-canonical Wnt pathway, also known as the Wnt/Ca2+ pathway, plays an important role in increasing the metastatic potential of melanoma cells (1–5). Studies from our laboratory demonstrated that increasing Wnt5A, which mediates the non-canonical Wnt/Ca2+ signaling pathway, increased melanoma metastasis (1–3), and silencing Wnt5A levels via siRNA3 decreased invasion (2, 3). In addition, we have shown that Wnt5A acts via protein kinase C (PKC) to mediate the motility of melanoma cells via the inhibition of metastasis suppressors and an initiation of the epithelial to mesenchymal transition, characterized by the loss of E-cadherin and the up-regulation of Snail (2).
Wnt signaling can be mediated by heparan sulfate proteoglycans (HSPGs) which are important signal transduction modulators. They mediate fibroblast growth factor, Hedgehog, epidermal growth factor, transforming growth factor-β, and WNT signaling pathways (6–11). HSPGs consist of two types, cell surface and basement membrane-associated HSPGs (12). Cell surface HSPGs are glycoproteins with covalently attached unbranched and modified sugar chains known as glycosaminoglycans (GAGs). There are two types of cell surface HSPGs, known as glypicans and syndecans (11, 13). Glypicans are attached to the cell surface via a glycosylphosphatidylinositol anchor, whereas syndecans are type 1 transmembrane proteins. HSPG GAG side chains are unbranched chains of modified repeating disaccharide units of N-acetylglucosamine and glucuronic acid. They are joined to the core protein via a tetrasaccharide linker attached to a serine residue. Following synthesis, these chains undergo modification with the addition of sulfates by N- and O-sulfation (14). The sulfation status determines to which specific portion of the GAG chains ligands, such as Wnt, will attach. The heparan sulfate endosulfatases Sulf1 and Sulf2 are cell surface enzymes that control growth factor signaling. The regulation of the 6-O-sulfation states by these endosulfatases changes the affinity of the GAG chains for ligand binding (15–17). Following sulfation modification, HSPGs can regulate signaling by dimerization (with other HSPGs or canonical signaling receptors), stabilization, or transport of the ligand to or away from the high affinity receptors (18–20). In addition, studies have suggested that the core proteins themselves may also play an important role in cell phenotype and function (21).
HSPGs have been implicated in a number of pathological conditions, such as Simpson-Golabi-Behmel overgrowth syndrome (22), fibrodysplasia ossificans progressiva (23), and Alzheimer disease (24). In addition, HSPGs are overexpressed in many forms of cancer, including prostate cancer and melanoma (25, 26). Importantly, in cancer, proteoglycans can have both tumor-promoting and tumor-suppressing activities. This depends on the type of protein core, the GAGs attached, and the localization of the proteoglycan and the molecules they associate with. In addition, the tumor subtype, stages, and degree of tumor differentiation also affect the function of HSPGs (27). HSPGs are cleaved by heparanases or heparin lyases (heparinases), which have been shown to have differing effects on tumor cell activity. For example, treating cancer cells with heparanase-1, which cleaves heparin-like regions (specifically HLGAG sites with O-sulfated l-iduronic acid residues), results in an increase in both tumor growth and metastatic dissemination (28). However, treating tumor cells with heparinase III, which more specifically cleaves HSPGs (i.e. unsulfated d-glucorinic acid, heparan-sulfate-like regions) results in an inhibition of their metastatic capacity (29). Importantly, heparanase I cleaves only certain side chains, where heparinase III treatment cleaves the entire backbone of the HSPG. It is likely that cleavage of specific side chains facilitates cell motility by releasing cells from adhesion to neighboring cells, whereas cleavage of the entire molecule decreases the availability of secreted ligands to their receptors, especially those involved in autocrine signaling, such as Wnt5A.
Historically, Wnt5A has been quite difficult to purify from cell culture media, despite the fact that it is a secreted protein. Further, in melanoma cells, Wnt5A appears to be signaling in an autocrine fashion (1, 2). These two observations, together with the fact that Wnt5A undergoes glycosylation (30), led us to hypothesize that HSPGs might be involved in increasing the availability of Wnt5A to its receptor, resulting in an increase in autocrine signaling and ultimately an increase in cellular invasion. In this study, we explore this hypothesis and investigate the role of HSPGs in the Wnt5A signaling cascade in metastatic melanoma cells.
EXPERIMENTAL PROCEDURES
Cell Culture
G361 cells were maintained in McCoy's (Invitrogen), and UACC903, M93-047, and UACC647 were maintained in RPMI (Invitrogen). All medium was supplemented with 10% fetal bovine serum (Invitrogen), 100 units/ml penicillin and streptomycin, and 4 mm l-glutamine. Cell lines were cultured at 37 °C in 5% CO2, 95% air, and the medium was replaced every 2–3 days. Cell lines were chosen based on their metastatic potential and Wnt5A levels, as reported previously (1, 2).
35S Binding
Cells were seeded in 96-well plates and grown to 60% confluence. Cells were then washed with PBS, and medium containing 0.5% fetal bovine serum and 100 μCi/ml Na235SO4 (PerkinElmer Life Sciences) was added. Cells were then incubated for 24 h and transferred to a microfiber filter using a TomTec Harvester 96. The filter was read using a Wallace 1205 Betaplate liquid scintillation counter to obtain counts/million. An identical plate was set up and used to quantitate protein levels. The counts/million were normalized to protein levels, determined with a BCA protein assay (Pierce) using bovine serum albumin as a standard.
1,9-Dimethylmethylene Blue (DMB) Assay
DMB binding assays were performed to quantify the levels of total GAG chains on the cell surface. Approximately 1 × 106 cells were collected in PBS or radioimmune precipitation buffer. For the cells in PBS, DMB (125 μl/well) was added to 40 μl of sample and quantified at 520 nm using a Bio-Rad 680XR spectrophotometer. For the cells in radioimmune precipitation buffer, protein was extracted and quantified. DMB binding levels were then compared with total protein levels.
Immunofluorescent Confocal Microscopy
Cells were seeded at 5 × 105 (G361) and 3 × 105 (UACC903, UACC647, and M93-047) into 1-well chamber slides (Nunc, Rochester, NY) and incubated overnight. Cells were then fixed in ice-cold 95% methanol at room temperature for 20 min, washed in PBS, and blocked using sterile filtered blocking buffer (0.2% Triton X-100, 0.2% bovine serum albumin, 0.2% casein, 0.2% gelatin, and 0.02% sodium azide) for 1 h at room temperature. Primary antibodies used were as follows: syndecan 1, syndecan 2, syndecan 3, and syndecan 4 (R&D Systems, Minneapolis, MN). Primary antibodies were added in blocking buffer, and cells were incubated overnight at 4 °C. Cells were then washed in PBS and treated with the appropriate secondary antibody (1:2000; Alexa fluor-488 or Alexa fluor-555). Cells were incubated for 1 h at room temperature, washed in PBS, mounted using Prolong Gold anti-fade reagent containing 4′,6-diamidino-2-phenylindole (Invitrogen), and imaged using a Zeiss Meta 510 confocal microscope (Zeiss, Thornwood, NY).
Heparinase III Treatments
Cells were serum-starved for 1 h and treated with 2 milliunits/ml heparinase III (Sigma) in serum-free media for 24 h.
Heparin Treatments
Cells were seeded at 2 × 105 and allowed to reach ∼80% confluence. Cells were then serum-starved for 2 h, and the appropriate concentration of heparin was added. After 6 h, cells were treated with either PBS or 200 ng/ml rWnt5A (R&D Systems) for a further 16 h. Media and cell lysates were then collected, concentrated in Microcon YM10 columns, and subjected to Western analysis.
SDS-PAGE and Western Blotting
Cell lysates were prepared using radioimmune precipitation buffer as described previously (2). 50 μg of protein was loaded into a 4–20% Tris/glycine gel (Invitrogen) and run at 120 V. Transfer to polyvinylidene difluoride membrane occurred using the i-Blot (Invitrogen), following the manufacturer's protocol. Membranes were washed in Tris-buffered saline containing 0.1% Tween 20 (TBS-T). Membranes were blocked using 5% milk in TBS-T for 1 h at room temperature with agitation. Primary antibodies were added at appropriate dilutions in 5% milk/TBS-T and incubated overnight at 4 °C with rotation. Primary antibodies used were biotinylated Wnt5A (1:100; R&D Systems), PO4-PKC (1:1000; Cell Signaling, Danvers, MA), anti-Δ-heparan antibody (Seikagaku America, Ijamsville, MD), β-catenin (1:1000; BD Biosciences), and β-tubulin (1:1000; Cell Signaling). Membranes were washed in TBS-T (3 × 5 min), and the appropriate secondary antibody was added in 5% milk/TBS-T for 1 h at room temperature with agitation. The membrane was washed in TBS-T and developed using ECL Plus (Amersham Biosciences).
Wound-healing Assay
Cells were seeded at 1 × 105 onto fibronectin-coated 24-well plates (BD Biosciences). Once confluent, a 200-μl pipette tip was heat-sealed, and two scratches were made, one horizontal and one vertical. Images were taken of the same field at 0, 9, or 12 h on a light microscope (Zeiss) at a gain of 1 using phased light. For post-transfection wound-healing assays, the transfection reagents were left on the cells for 5 h, removed, and replaced with media containing 10% fetal bovine serum. Cells were then incubated for 48 h, and then a wound-healing assay was performed as described above.
Quantatative Transwell Invasion Assays
Cells were serum-starved for 2 h and treated with 2 milliunits/ml heparinase III for 24 h and/or rWnt5A or rWnt3A for 16 h. Cells were then seeded at 5 × 105 cells/ml in the heparinase III/Wnt5A serum-free media in the transwell insert of a QCM plate (Millipore, Billerica, MA). RPMI containing 10% fetal bovine serum was placed in the bottom of the plate. Plates were then incubated for 48 h at 37 °C. The cells/media were removed, and the invasion chamber insert was placed into a clean well containing cell stain and incubated for 20 min at room temperature. The insert was washed in water, and while the insert was still moist, the non-invading cells and extracellular matrix layer were removed from the interior of the insert. The inserts were allowed to air-dry and then placed in extraction buffer for 15 min at room temperature. 100 μl of the dye mixture was transferred to a 96-well microtiter plate suitable for colorimetric measurement, and the optical density was measured at 570 nm.
siRNA Transfection
Negative control, SDC1, and SDC4 siRNA (200 nm; Qiagen, Valencia, CA) were transfected into cells using Lipofectamine Plus (Invitrogen) for 48 h as described previously (2). The siRNAs used were Invitrogen's “HP-validated siRNAs,” which have been validated to perform efficient knockdown with minimal off-target effects, as well as negative control siRNAs from the same company. These siRNAs are designed with unequal stabilities of the base pairs at the 5′-ends. This enables the antisense strand, which is less tightly bound at its 5′-end, to enter RISC while the sense strand is degraded. Asymmetry produces highly functional siRNAs and reduces the risk of off-target effects caused by the incorrect strand entering RISC. siRNA sequences are as follows: SDC1 sense, r(CGACUGCUUUGGACCUAAA)dTdT; SDC1 antisense, r(UUUAGGUCCAAAGCAGUCG)dTdT; SDC4 sense, r(GGGUGAGGUCAACCUAAUA)dTdT; SDC4 antisense, r(UAUUAGGUUGACCUCACCC)dTdT.
Real Time PCR
RNA was extracted using TRIzol (Invitrogen) and an RNeasy Mini kit (Qiagen) as described previously (2). RNA was quantified using an ND1000 spectrophotometer (Nanodrop, Wilmington, DE), and using 1 μg of RNA, cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad), and gene expression was quantified using the SYBR Green method of real time PCR. PCRs were performed in triplicate with a 100 nm concentration of each primer. Primers were designed to cross exon-intron junctions, using Primer Bank (available on the World Wide Web; see Table 1). Real time PCR was performed on an ABI Prism 7300 sequence detection system using standard (default) conditions. Samples were normalized against the 18 S gene using Universal 18 S primers (Ambion, Austin, TX). Expression was calculated using the standard curve method.
TABLE 1.
Primer sequences
| Gene | Sequence |
|---|---|
| Syndecan 1 | 5′-TGAAACCTCGGGGGAGAATAC-3′ |
| Syndecan 1 | 3′-GGTACAGCATGAAACCCACC-5′ |
| Syndecan 3 | 5′-GACATCCCTGAGAGGAGCAC-3′ |
| Syndecan 3 | 3′-TCTGGCTCATCCCGGATTGT-5′ |
| Syndecan 4 | 5′-ACTCCATGATCGGCCCTGAA-3′ |
| Syndecan 4 | 3′-TGGGGATAACCTCATTCTCCTC-5′ |
Cell Surface Biotinylation
Surface biotinylation was performed using a cell surface protein isolation kit (Thermo Scientific, Rockford, IL), following the manufacturer's protocol. Briefly, cells were grown in four T75 flasks to 90–95% confluence and labeled using sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate. Cells were lysed and then sonicated on low power and incubated for 30 min on ice. The labeled proteins were then isolated using NeutrAvidin-agarose columns. A trace amount of bromphenol blue was added to eluate. Samples were analyzed by Western blot.
Cell Membrane Extraction
Cell membrane proteins were isolated using a MEM-PER kit (Pierce) following the manufacturer's protocol. Briefly, 5 × 106 cells/sample were incubated on ice with the different components of the kit (reagents A, B, and C) and then incubated for 20 min in a 37 °C water bath. Tubes were centrifuged at room temperature for 2 min at 10,000 × g to isolate the hydrophobic fraction from the hydrophilic fraction. All fractions were diluted 2-fold to prevent band and lane distortion with the membrane fraction. Protein levels were quantified using a BCA protein kit, and 50 μg of protein was run on a 4–20% Tris/glycine gel.
Densitometry
Western images were saved as Tiff files and inverted, and integrated density was analyzed. Densitometry was performed on at least three independent experiments using ImageJ software (National Institutes of Health, Bethesda, MD), and values were normalized to β-tubulin.
Statistical Analysis
Student's t test was performed when required in at least three independent experiments. Significance was designated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
HSPG Levels Are Associated with Increased Metastatic Capacity of Melanoma Cells
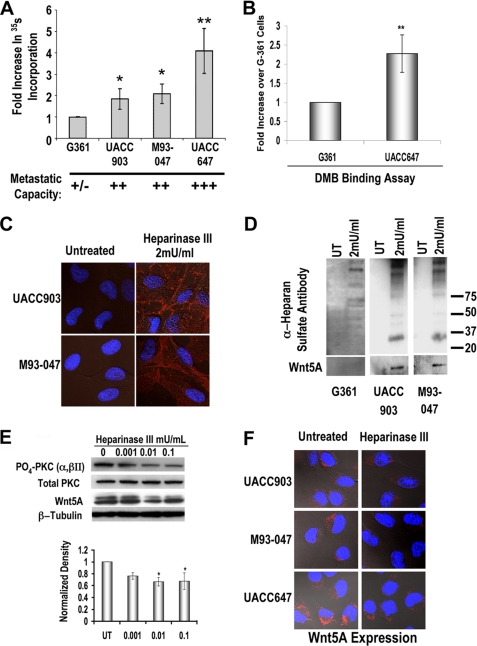
We have previously described the melanoma cells used in this study as having differing Wnt5A levels and differing metastatic ability (2). We used 35S labeling to examine the incorporation of sulfate into HSPG-specific GAG chains. More metastatic cells incorporated more sulfate (Fig. 1A), which may indicate several possible changes in HSPG core levels, side-chain synthesis, or elongation. To determine if total sulfated GAG levels increase in more metastatic cells, we also performed a DMB binding assay. DMB binding was increased in highly metastatic UACC647 cells as compared with less metastatic G361 cells (Fig. 1B).
FIGURE 1.
Total and HSPG-specific proteoglycans are elevated in highly metastatic melanoma cell lines, and GAG chain removal decreases Wnt5A signaling. A, increased 35S incorporation in metastatic melanoma cells (n = 5, error bars show S.D.; *, p < 0.05; **, p < 0.01). B, increased DMB binding in highly metastatic UACC647 melanoma cells as compared with less metastatic G361 melanoma cells (n = 4; **, p < 0.01). C, cells were treated with 2 milliunits/ml heparinase III for 24 h. Cells were then examined using immunofluorescence with a heparinase III Δ antibody that recognizes a neo epitope created by the treatment. D, Western analysis of the media of heparinase III-treated G361, UACC903, and M93-047 cells probed with both heparinase III δ and Wnt5A antibodies. E, Western analysis of the effects of heparinase III treatment show decreases in both cellular Wnt5A and Wnt5A downstream signaling. F, decreases in cellular Wnt5A are confirmed by immunofluorescent confocal microscopy. UT, untreated.
To investigate whether HSPGs could affect downstream Wnt5A signaling, heparinase III treatment was performed to cleave the HSPG backbone. Following treatment, cells were probed with an anti-Δ-heparan sulfate antibody that only recognizes a neo epitope created by heparinase III cleavage, and then analyzed by immunofluorescent confocal microscopy. As shown, only cells treated with heparinase III demonstrate antibody binding by immunofluorescence (Fig. 1C). This same antibody was also used to analyze media from heparinase III-treated cells in order to determine whether cleaved GAG chains could be seen in the media. As shown, increased levels of HSPG GAG chains can be seen in the treated samples as compared with the untreated control (Fig. 1D, upper panels). The lowly metastatic G361 cells have lower amounts of GAG chains. Upon heparinase III treatment, increased levels of Wnt5A can also be seen in the media (Fig. 1D, lower panels) in the Wnt5A high lines but not in the G361 cells, which have low Wnt5A expression. This increase of Wnt5A in the medium suggested one of two scenarios: 1) HSPGs may promote Wnt5A retention at the surface of the cell, in order to promote its internalization for autocrine signaling, and HSPG cleavage released Wnt5A into the medium, or 2) HSPGs may be affecting some other factor that, upon heparinase cleavage, increases Wnt5A production and secretion.
To determine which of these scenarios was the case, cells were treated with heparinase III, protein was collected, and Western analysis was performed. We have previously demonstrated that Wnt5A signaling increases PKC activity as well as cell motility (3). Thus, if HSPGs promote the retention and internalization of Wnt5A, increasing its signaling, then heparinase III treatment should release Wnt5A from the surface. This would result in a decreased ability of Wnt5A to signal and mediate motility. If, instead, HSPG cleavage results in increased production of Wnt5A and secretion into the medium, due to the release of a secondary factor, then we should see increased levels of cellular Wnt5A and increased PKC activity. As demonstrated, levels of PO4-PKC did not increase upon treatment with heparinase III (Fig. 1E) and in fact decreased by about 20 and 40%, respectively (based on densitometry). Cellular Wnt5A levels also decreased, confirming that the increased levels of Wnt5A in the media upon heparinase III treatment were not due to increased production of Wnt5A (Fig. 1E). This could be confirmed by immunofluorescent confocal microscopy, where cellular levels of Wnt5A were decreased after treatment with heparinase III (Fig. 1F), indicating a decrease in its internalization in the absence of HS. As we have previously demonstrated, Wnt signaling in melanoma is dependent on its internalization.4 Thus, these data indicate that in the absence of HS, Wnt5A signaling would be decreased due to the lack of internalization.
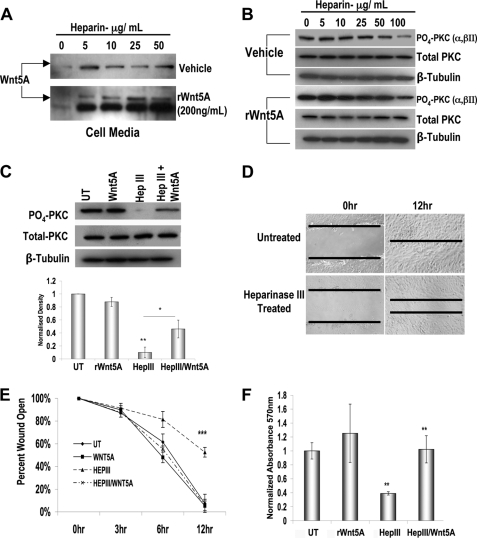
As a further confirmation that Wnt5A interacts with HS, cells were treated with heparin, which would be expected to compete for binding with Wnt5A, resulting in its accumulation in the medium. Indeed, in the presence of heparin, endogenously secreted Wnt5A accumulated in the medium of metastatic melanoma cells (Fig. 2A, top). The addition of recombinant Wnt5A (rWnt5A) to these highly metastatic cells resulted in its rapid uptake, leaving very little secreted into the medium (Fig. 2A, bottom, first lane). However, in the presence of heparin, rWnt5A accumulated in the medium, as evidenced by the presence of both the glycosylated and non-glycosylated Wnt5A bands (Fig. 2A, bottom, lanes 2–5). PKC signaling was also decreased upon heparin binding, and, at higher doses, this could not be overcome by the addition of rWnt5A (Fig. 2B). On the other hand, treatment of cells with heparinase, as shown above, decreased PKC signaling, but flooding the cell with rWnt5A could restore PKC signaling to some extent (Fig. 2C). This may indicate that although HS increases the efficacy of Wnt binding to its receptors and subsequent signaling, in the presence of an excess of ligand, there is still at least some receptor binding that occurs. It should be noted that the cells used in this figure have high levels of Wnt5A; thus, in control conditions, rWnt5A treatment has little or no effect on signaling or invasion.
FIGURE 2.
Heparin competes with Wnt5A binding of HS, and removal of HS decreases motility in melanoma cell lines. Cells were treated with increasing doses of heparin, and the medium of the cells was examined for Wnt5A release. A, in the presence of heparin, Wnt5A accumulates in the medium. The addition of rWnt5A to cells in the absence of heparin results in its rapid uptake and internalization, but in the presence of heparin, rWnt5A just accumulates in the medium. B, PKC signaling is affected by high doses of heparin, regardless of whether rWnt5A is added. C, cells were treated with 2 milliunits/ml heparinase III for 24 h and examined for PKC signaling. PKC signaling is decreased upon heparinase III treatment and partially reconstituted upon the addition of rWnt5A. D, representative images of the motility assays using M93-047 cells demonstrating that heparinase III treatment decreased the rate of wound closure. E, graphical representation of UACC903 scratch closure rates demonstrating that rWnt5A addition can overcome heparinase III treatment. F, quantitative extracellular matrix invasion assays also demonstrate that heparinase III treatment decreases the rate of melanoma cell invasion, and this can be restored upon the addition of exogenous Wnt5A to the media of the cells (n = 3; error bars show S.D.; **, p < 0.01). UT, untreated.
To further confirm that Wnt5A signaling was not increased upon heparinase III treatment, we performed migration assays, since we have previously demonstrated that Wnt5A signaling increases migration of melanoma cells (1–3). In a wound-healing assay, heparinase III treatment decreased the motility of melanoma cells (Fig. 2, D and E). M93-047 cells are shown in Fig. 2D, but data are similar across all Wnt5A high lines. In all cases, heparinase III treatment resulted in a reduction in the rate of closure compared with the untreated samples. To determine if increasing the amount of available Wnt5A by adding back rWnt5A to the medium could overcome the effects of the heparinase III treatment, cells were subjected to a quantitative extracellular matrix invasion assay. After heparinase III treatment, invasion was significantly inhibited, but the readdition of rWnt5A into the medium of heparinase III-treated UACC903 cells restored their invasive capacity (Fig. 2F).
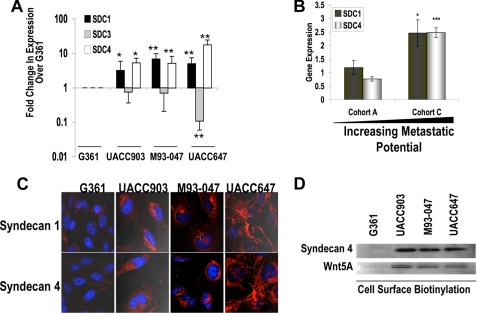
Syndecan 1 and Syndecan 4 Are the Most Abundantly Expressed HSPGs in Melanoma Cell Lines
Since total HSPG cleavage resulted in decreased motility and Wnt5A signaling in melanoma cells, we investigated which HSPG subtypes were expressed on Wnt5A high as compared with Wnt5A low melanoma cells. Gene and protein expression profiling was carried out for all glypicans and syndecans on melanoma cell lines. There was very little expression of all glypican subtypes by gene and protein expression (data not shown). Syndecan 1 and syndecan 4 gene expression demonstrated a correlation with metastatic potential, and syndecan 3 demonstrated an anti-correlation by real time PCR (Fig. 3A). Syndecan 2 expression was very low in all cell lines (data not shown). We then analyzed expression of syndecans in patient samples using the Mannheim data set (31). This is a microarray data base of melanoma patient samples that can be divided into low (Cohort A, n = 19) versus high (Cohort C, n = 16) metastatic potential (31). Analysis of this data set indicates that both syndecans 1 and 4 are elevated in patients with high metastatic ability (Fig. 3B). Immunofluorescent analysis was also carried out as an additional confirmation of these data (Fig. 3C). Although little change could be observed in syndecan 3 at the protein level (data not shown), syndecan 1 and 4 expression did increase as cells became more metastatic (Fig. 3C). Because the confocal experiments did not clearly indicate whether syndecans were bound at the surface of the cell, we performed cell surface biotinylation assays for syndecan 4 and Wnt5A. These assays indicate that syndecan 4 and Wnt5A are all present at the surface of the cell and at much higher levels in the metastatic cell lines (Fig. 3D).
FIGURE 3.
Syndecan 1 and 4 expression correlates with metastatic potential. A, syndecan mRNA expression by real time PCR (n = 3; error bars show S.D.; *, p < 0.05; **, p < 0.01) across melanoma cell lines. B, analysis of gene expression in a microarray data base of melanoma patients also confirms that cells from melanoma patients that have a lower metastatic capacity (Cohort A; n = 19) have lower levels of syndecan 1 and syndecan 4 than those with a high metastatic capacity (Cohort C; n = 16; error bars show S.E.; *, p < 0.05; ***, p < 0.001). C, immunofluorescent confocal microscopy also confirms that syndecan 1 and 4 protein levels are elevated in more metastatic melanoma cell lines. D, cell surface biotinylation studies indicate that syndecan 4 and Wnt5A are present on the surface of the cell.
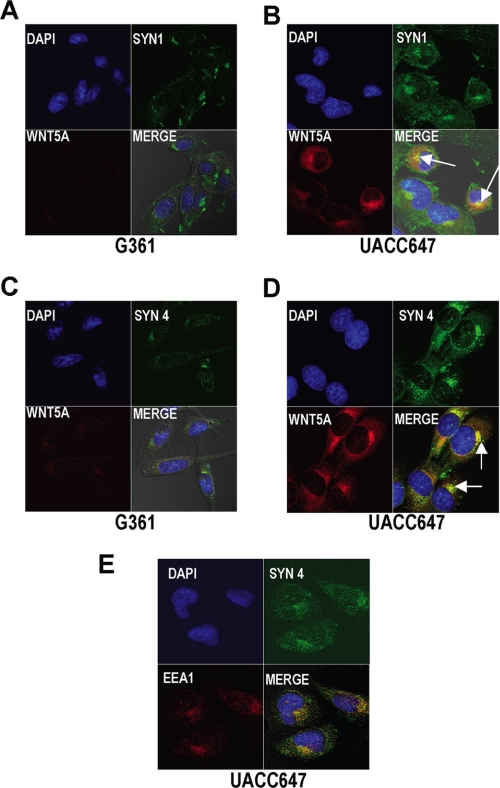
We then sought to determine whether syndecans bind Wnt5A in melanoma cells. Since glycosylated Wnt5A is probably bound to sugar chains, rather than the core protein, we hypothesized that co-immunoprecipitation experiments might be unsuccessful, and indeed our attempts at these assays were. Thus, we performed immunofluorescent confocal microscopy. In G361 cells, there are low levels of Wnt5A, syndecan 1, and syndecan 4 (Fig. 4, A and C). In highly metastatic cells (UACC647 cells are shown as an example, but this is true also for M93047 and UACC903 cells), syndecan 4 and, to a much lesser extent, syndecan 1 could be found all over the cell, including in foci near the nucleus. Wnt5A staining strongly co-localized with syndecan staining in these foci (Fig. 4, B and D, arrows). Both Wnts and syndecans have been shown to undergo internalization into endosomes, and we show here that this is also the case in melanoma cells, since syndecan 4 co-localized with the early endosome marker EEA1 (Fig. 4E). Immunofluorescent co-localization profiles and z-stack analysis confirmed that these proteins are co-localized (data not shown). These data indicate that Wnt5A and syndecans may be internalized into endosomes together, perhaps as part of a complex of Wnt, Wnt receptor, and syndecan.
FIGURE 4.
Syndecans and Wnt5A appear to be internalized together in Wnt5A high cells. A, G361 cells were stained for Wnt5A (red) and syndecan 1 (green) and show very little expression of either protein. B, in Wnt5A high UACC647 cells, Wnt5A co-localizes with syndecan 1, especially in perinuclear foci (yellow, arrows). C and D, the same is true for Wnt5A and syndecan 4 staining. Cells were stained for syndecan and Wnt5A co-localization with the early endosome, using an early endosome marker, EEA1. E, in the syndecan/EEA1 staining, syndecan 4 is detected using a green fluorophore, and EEA1 is stained red. DAPI, 4′,6-diamidino-2-phenylindole.
Syndecan 1 and 4 Knockdown Result in Decreases in the Motility and Invasiveness of Metastatic Melanoma Cells
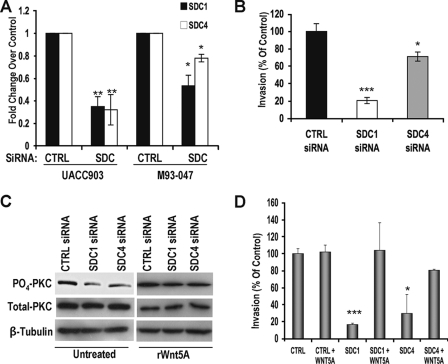
Since syndecan 1 and 4 correlated with increased melanoma cell metastasis, and generalized HSPG-GAG chain removal reduced motility in melanoma cells, syndecan 1 and syndecan 4 were knocked down using siRNA, to determine if reduction of the most abundantly expressed core proteins would also decrease the motility of the cells. Following 48 h of siRNA treatment, syndecan 1 (SDC1) and 4 (SDC4) mRNA expression was examined. As shown, SDC1 and SDC4 mRNA levels were reduced significantly as compared with the control (CTRL) siRNA-treated samples (Fig. 5A). UACC903 cells that were transfected with negative control, SDC1, or SDC4 siRNA were then placed in a Matrigel transwell invasion chamber to measure invasion. SDC1 and SDC4 transfection decreased the invasion of UACC903 melanoma cells as compared with control siRNA-transfected cells (Fig. 5B). Simultaneous transfection of both SDC1 and SDC4 siRNA did not further reduce invasion (data not shown). Additionally, PKC signaling was diminished in the presence of SDC1 or SDC4 siRNA but could be restored upon the addition of rWnt5A (Fig. 5C). To determine if the addition of excess exogenous Wnt5A could also restore the migratory capacity of melanoma cells, as in the case of the heparinase III treatment, syndecan expression was first reduced using siRNA, and cells were then treated with rWnt5A. UACC903 cells are already quite invasive and express high levels of Wnt5A, and treating the control siRNA-transfected cells with exogenous Wnt5A did not significantly affect invasion (Fig. 5D). As shown in Fig. 5, B and D, SDC1 and SDC4 knockdown inhibited invasion. The addition of excess rWnt5A to SDC1 and SDC4 siRNA-treated cells restored invasion. This may be due to the fact that the siRNA knockdown of the syndecans does not completely ablate them, and increasing the amount of available Wnt5A allows for signaling to be restored, or it may simply be an effect of excess ligand that can access receptors even in the absence of syndecans. These results suggest that SDC1, SDC4, and Wnt5A may work together to facilitate melanoma cell invasion.
FIGURE 5.
Syndecan 1 and 4 siRNA decrease motility and invasiveness of melanoma cell lines. A, real time PCR of SDC1 and SDC4 gene expression following siRNA transfection in UACC903 and M93-047 cells (n = 3; error bars show S.D.; **, p < 0.01; *, p < 0.05). B, Matrigel invasion assay following SDC1 and SDC4 siRNA in UACC903 cells (n = 3; error bars show S.D.; *, p < 0.05; ***, p < 0.001). Results were similar for M93-047 cells. C, Western analysis of PO4-PKC showing that PO4-PKC signaling is decreased upon syndecan knockdown but can be restored upon rWnt5A addition. D, Matrigel invasion assays demonstrating that exogenous Wnt5A addition can restore motility to UACC903 melanoma cells even after SDC1 and SDC4 knockdown. CTRL, control, *, p < 0.05; ***, p < 0.001.
DISCUSSION
The role of HSPGs in melanoma is not well documented. Previous studies have shown that sulfated GAGs are differently expressed in tumor cells characterized by different metastatic potential, and more metastatic cells contain a higher heparan sulfate/chondroitin sulfate ratio (32). Further, that study suggested that absolute or relative dominance of HSPGs over chondroitin sulfate proteoglycans at the cell surface of metastatic tumor cells can be considered a marker of a more metastatic phenotype. In other cell systems, HSPGs have been demonstrated to be important for Wnt signaling (33–35). It has been previously suggested that HSPGs can act as co-receptors that directly facilitate the formation of Wnt-Wg-Fz signaling complexes (36).
Our study demonstrated an increase in HSPGs, specifically syndecan 1 and 4 during melanoma progression, and shows that these proteins are important for Wnt5A-induced invasion of melanoma cells. We have previously shown that Wnt5A is critical for the induction of melanoma metastasis, via the induction of PKC and Ca2+, in cells that are not highly metastatic (1, 2, 5). It should be noted that in highly metastatic Wnt5A high cell lines, however, the addition of Wnt5A (above the already high levels of Wnt5A that exist) has no additional effect. Treating these cells with heparinase mimics the situation of low Wnt5A, as we show in Fig. 1, where Wnt5A is released away from the cell surface and into the media. Flooding the cell with excess recombinant Wnt5A negates the requirement for HS to bind it to the surface, since there is probably enough there to bind just by sheer availability of ligand, allowing for the restoration of both PKC activity and migration, although not as effectively as when there is HS present.
PKC up-regulation has been shown to stabilize Wnt5A mRNA, thereby increasing Wnt5A levels (37). Changes induced by Wnt5A activation of Ca2+/PKC include down-regulation of the metastasis suppressor Kiss-1 and up-regulation of CD44, Snail, and vimentin, leading to an epithelial to mesenchymal transition (2). Changes in the cytoskeleton accompany this transition and include filamin cleavage (5) and actin reorganization (1). Recently, we have shown that this occurs via Wnt5A activation of ROR2, an orphan tyrosine receptor.4 These previous observations indicate that Wnt5A exists in a positive feedback loop, where it binds to ROR2, activates PKC, which stabilizes Wnt5A, and increases its secretion. With the current data, we add one more piece of the puzzle, and hypothesize that, upon its secretion, HSPGs capture Wnt5A and present it to the receptor complex, increasing the efficacy of its signaling.
In vivo studies have shown that syndecan 4 can modulate the directionality of the migration of neural crest cells, via binding of Fzd7, resulting in PCP signaling and effects on Rac and the cytoskeleton. Loss of syndecan 4 decreases the migratory ability of these cells (38). In support of these in vivo data in Xenopus, our study demonstrates a correlation between levels of syndecan 1 and 4 and the metastatic potential of melanoma cells. Based on this observation and the fact that SDC1 or SDC4 siRNA results in a decrease in motility and invasiveness of melanoma cells, we propose that syndecan 1 and 4 also modulate Wnt5A signaling and cellular invasion in human melanoma. These data imply that HSPGs are required for maintaining the availability of endogenous Wnt5A for signaling, but the lack of HSPGs can be overcome by increasing the amount of available Wnt5A. This is probably true for other growth factors as well. An alternative interpretation of these results might suggest that the presence of HS is inhibitory, since it is clear from these data that the presence of HS promotes Wnt5A internalization. However, that would only hold true if internalization led to degradation, and data from our laboratory indicate that internalization of Wnt5A in our Wnt5A high cells does not correspond to either its lysosomal or proteasomal degradation. Further, based on other results that indicate that the internalization of Wnt5A and its receptor ROR2, via clathrin, is required for increased signaling and motility in melanoma cells,4 we believe that HS is required for Wnt5A internalization and subsequent signaling.
The role of syndecans specifically in melanoma progression is not well understood. In an early microarray study, syndecan 4 emerged, along with Wnt5A, as a gene that was up-regulated in highly metastatic melanoma (39), and more recent microarray studies have also identified both genes as being up-regulated in melanoma (40). In the current study, we confirm this observation at the protein level and demonstrate that syndecans, particularly syndecan 1 and 4, play an important role in melanoma invasion, via their role in the modulation of Wnt5A signaling. It is known that syndecans can bind to Fzd receptors to modulate Wnt signaling (38), and it has also been shown that clathrin-mediated endocytosis of frizzled receptors is a key component of Wnt signaling and that these clathrin-coated endosomes serve as a “signaling nexus” (41). Our previous data support this, demonstrating a clathrin-mediated internalization of ROR2, and Wnt5A, into the early endosome.4 In vivo studies have shown that syndecan 4 can modulate the directionality of the migration of neural crest cells, via binding of Fzd7, resulting in PCP signaling and effects on Rac and the cytoskeleton. Loss of syndecan 4 decreases the migratory ability of these cells (38). In support of these in vivo data in Xenopus, our study demonstrates a correlation between levels of syndecan 1 and 4 and the metastatic potential of melanoma cells. Based on this observation and the fact that syndecan 1 or 4 siRNA results in a decrease in motility and invasiveness of melanoma cells, we propose that syndecan 1 and 4 also modulate Wnt5A signaling and cellular invasion in human melanoma.
Data from other human cell systems also show a requirement for syndecans during cellular invasion. For example, syndecan 4 is essential for the invasion of fibroblasts into fibrin clots (42). Further, platelet-derived growth factor, the stimulus for migration, induces syndecan 4 core protein expression (42). Syndecan 1 is required for keratinocyte invasion across a laminin matrix and for the Rac-mediated formation of invadopodia (43). In melanoma specifically, the data are a little more confusing. Heparanase I has been shown to exist on the surface of metastatic melanoma cells, and its cleavage of syndecan 1 has been shown to promote metastasis, suggesting that syndecan 1 degradation may increase melanoma cell motility (44). However, as previously mentioned, heparanase I only cleaves certain side chains from the syndecans, and this may be a way for the cell to attempt to regulate the level of activity of syndecan 1. Our current study indicates that melanoma cells simply increase their rate of synthesis or core protein expression of these chains and, specifically, increase their expression of syndecan 1 and 4, perhaps as a way to try to overcome this heparanase I regulation. The role of heparanase I is complex, since a more recent study suggests that heparanase I may be playing a far more widespread role than simply the cleavage of syndecan 1 side chains. It appears that heparanase I promotes metastasis by releasing bioactive heparan sulfate fragments from the surface of melanoma cells as well as degrading components of the extracellular matrix, leading to increased invasion of the tumor cells lying adjacent to it (28). Syndecan 4 has been shown to mediate fibroblast growth factor 2 signaling, increasing adhesion to fibronectin but limiting invasion (45). However, because invasion is a delicate balance of three important steps that include adhesion, proteolytic dissolution of the basement membrane, and motility, it may be that the increased adhesion observed in that study may actually promote invasion on matrices that are not composed of fibronectin alone, as would be found in vivo.
In conclusion, this study demonstrates for the first time a correlation between syndecan 1 and 4 core protein levels, Wnt5A, and metastatic potential in human melanoma. It also shows a decrease in cellular invasion upon removal of HSPG core proteins, specifically syndecan 1 or 4. Finally, it demonstrates that HSPGs are essential for Wnt5A internalization and signal transduction in metastatic melanoma cells. Additional data suggest that this is specific to the non-canonical Wnt signaling pathway, since treatment of melanoma cells with high levels of heparinase III only serves to increase levels of β-catenin (supplemental Fig. S1A), whereas Wnt5A levels are decreased, consistent with previous reports that Wnt5A suppresses Wnt3A and β-catenin signaling (46, 47). In the presence of heparinase III, excess rWnt3A cannot restore invasion, unlike rWnt5A (supplemental Fig. S1C). Taken together, these results suggest that levels of HSPG subtypes may be indicative of metastatic potential, via their ability to promote Wnt5A-mediated invasion and that, specifically, syndecan 1 and 4 may be useful as markers of the metastatic ability of melanoma cells.
Supplementary Material
Acknowledgment
We thank Dr. Ian Morgan for providing the tri-acetyl-β-GlcNAc-serine.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the NIA (Baltimore, MD).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
O'Connell, M. P., Fiori, J. L., Xu, M., Carter, A. D., Frank, B. P., Camilli, T. C., French, A. D., Dissanayake, S. K., Indig, F. E., Bernier, M., Taub, D. D., Hewitt, S. M., and Weeraratna, A. T. (2009) Oncogene 28, in press
- siRNA
- small interfering RNA
- PKC
- protein kinase C
- HSPG
- heparan sulfate proteoglycan
- GAG
- glycosaminoglycan
- PBS
- phosphate-buffered saline
- DMB
- 1,9-dimethylmethylene blue.
REFERENCES
- 1.Weeraratna A. T., Jiang Y., Hostetter G., Rosenblatt K., Duray P., Bittner M., Trent J. M. (2002) Cancer Cell 1, 279–288 [DOI] [PubMed] [Google Scholar]
- 2.Dissanayake S. K., Wade M., Johnson C. E., O'Connell M. P., Leotlela P. D., French A. D., Shah K. V., Hewitt K. J., Rosenthal D. T., Indig F. E., Jiang Y., Nickoloff B. J., Taub D. D., Trent J. M., Moon R. T., Bittner M., Weeraratna A. T. (2007) J. Biol. Chem. 282, 17259–17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dissanayake S. K., Olkhanud P. B., O'Connell M. P., Carter A., French A. D., Camilli T. C., Emeche C. D., Hewitt K. J., Rosenthal D. T., Leotlela P. D., Wade M. S., Yang S. W., Brant L., Nickoloff B. J., Messina J. L., Biragyn A., Hoek K. S., Taub D. D., Longo D. L., Sondak V. K., Hewitt S. M., Weeraratna A. T. (2008) Cancer Res. 68, 10205–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Da Forno P. D., Pringle J. H., Hutchinson P., Osborn J., Huang Q., Potter L., Hancox R. A., Fletcher A., Saldanha G. S. (2008) Clin. Cancer Res. 14, 5825–5832 [DOI] [PubMed] [Google Scholar]
- 5.O'Connell M. P., Fiori J. L., Baugher K. M., Indig F. E., French A. D., Camilli T. C., Frank B. P., Earley R., Hoek K. S., Hasskamp J. H., Elias E. G., Taub D. D., Bernier M., Weeraratna A. T. (2009) J. Invest. Dermatol. 129, 1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamanna W. C., Frese M. A., Balleininger M., Dierks T. (2008) J. Biol. Chem. 283, 27724–27735 [DOI] [PubMed] [Google Scholar]
- 7.Yan D., Lin X. (2008) Nat. Cell Biol. 10, 761–763 [DOI] [PubMed] [Google Scholar]
- 8.Mahtouk K., Cremer F. W., Rème T., Jourdan M., Baudard M., Moreaux J., Requirand G., Fiol G., De Vos J., Moos M., Quittet P., Goldschmidt H., Rossi J. F., Hose D., Klein B. (2006) Oncogene 25, 7180–7191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñoz R., Moreno M., Oliva C., Orbenes C., Larraín J. (2006) Nat. Cell Biol. 8, 492–500 [DOI] [PubMed] [Google Scholar]
- 10.Hiraga T., Nakajima T., Ozawa H. (1995) Bone 16, 349–356 [DOI] [PubMed] [Google Scholar]
- 11.Filmus J., Capurro M., Rast J. (2008) Genome Biol. 9, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melrose J., Hayes A. J., Whitelock J. M., Little C. B. (2008) BioEssays 30, 457–469 [DOI] [PubMed] [Google Scholar]
- 13.Lopes C. C., Dietrich C. P., Nader H. B. (2006) Braz. J. Med. Biol. Res. 39, 157–167 [DOI] [PubMed] [Google Scholar]
- 14.Habuchi H., Habuchi O., Kimata K. (2004) Glycoconj. J. 21, 47–52 [DOI] [PubMed] [Google Scholar]
- 15.Ai X., Do A. T., Kusche-Gullberg M., Lindahl U., Lu K., Emerson C. P., Jr. (2006) J. Biol. Chem. 281, 4969–4976 [DOI] [PubMed] [Google Scholar]
- 16.Wang S., Ai X., Freeman S. D., Pownall M. E., Lu Q., Kessler D. S., Emerson C. P., Jr. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4833–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viviano B. L., Paine-Saunders S., Gasiunas N., Gallagher J., Saunders S. (2004) J. Biol. Chem. 279, 5604–5611 [DOI] [PubMed] [Google Scholar]
- 18.Dews I. C., Mackenzie K. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20782–20787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi J. Y., Han I., Oh E. S. (2006) Scientific World J. 6, 457–459 [Google Scholar]
- 20.Choi S., Lee E., Kwon S., Park H., Yi J. Y., Kim S., Han I. O., Yun Y., Oh E. S. (2005) J. Biol. Chem. 280, 42573–42579 [DOI] [PubMed] [Google Scholar]
- 21.Kramer K. L., Yost H. J. (2003) Annu. Rev. Genet. 37, 461–484 [DOI] [PubMed] [Google Scholar]
- 22.Pilia G., Hughes-Benzie R. M., MacKenzie A., Baybayan P., Chen E. Y., Huber R., Neri G., Cao A., Forabosco A., Schlessinger D. (1996) Nat. Genet. 12, 241–247 [DOI] [PubMed] [Google Scholar]
- 23.O'Connell M. P., Billings P. C., Fiori J. L., Deirmengian G., Roach H. I., Shore E. M., Kaplan F. S. (2007) J. Cell. Biochem. 102, 1493–1503 [DOI] [PubMed] [Google Scholar]
- 24.O'Callaghan P., Sandwall E., Li J. P., Yu H., Ravid R., Guan Z. Z., van Kuppevelt T. H., Nilsson L. N., Ingelsson M., Hyman B. T., Kalimo H., Lindahl U., Lannfelt L., Zhang X. (2008) Brain Pathol. 18, 548–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta S., Pierce M., Datta M. W. (2006) Int. J. Biochem. Cell Biol. 38, 1855–1861 [DOI] [PubMed] [Google Scholar]
- 26.Ikuta Y., Nakatsura T., Kageshita T., Fukushima S., Ito S., Wakamatsu K., Baba H., Nishimura Y. (2005) Clin. Cancer Res. 11, 8079–8088 [DOI] [PubMed] [Google Scholar]
- 27.Fjeldstad K., Kolset S. O. (2005) Curr. Drug Targets 6, 665–682 [DOI] [PubMed] [Google Scholar]
- 28.Roy M., Marchetti D. (2009) J. Cell. Biochem. 106, 200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y., MacLeod V., Dai Y., Khotskaya-Sample Y., Shriver Z., Venkataraman G., Sasisekharan R., Naggi A., Torri G., Casu B., Vlodavsky I., Suva L. J., Epstein J., Yaccoby S., Shaughnessy J. D., Jr., Barlogie B., Sanderson R. D. (2007) Blood 110, 2041–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dissanayake S. K., Weeraratna A. T. (2008) Methods Mol. Biol. 468, 157–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoek K. S., Schlegel N. C., Brafford P., Sucker A., Ugurel S., Kumar R., Weber B. L., Nathanson K. L., Phillips D. J., Herlyn M., Schadendorf D., Dummer R. (2006) Pigment Cell Res. 19, 290–302 [DOI] [PubMed] [Google Scholar]
- 32.Timar J., Ladanyi A., Lapis K., Moczar M. (1992) Am. J. Pathol. 141, 467–474 [PMC free article] [PubMed] [Google Scholar]
- 33.Colombres M., Henríquez J. P., Reig G. F., Scheu J., Calderón R., Alvarez A., Brandan E., Inestrosa N. C. (2008) J. Cell. Physiol. 216, 805–815 [DOI] [PubMed] [Google Scholar]
- 34.Baeg G. H., Lin X., Khare N., Baumgartner S., Perrimon N. (2001) Development 128, 87–94 [DOI] [PubMed] [Google Scholar]
- 35.Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P., Jr. (2001) Science 293, 1663–1666 [DOI] [PubMed] [Google Scholar]
- 36.Lin X., Perrimon N. (2000) Matrix Biol. 19, 303–307 [DOI] [PubMed] [Google Scholar]
- 37.Jönsson M., Smith K., Harris A. L. (1998) Br. J. Cancer 78, 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews H. K., Marchant L., Carmona-Fontaine C., Kuriyama S., Larraín J., Holt M. R., Parsons M., Mayor R. (2008) Development 135, 1771–1780 [DOI] [PubMed] [Google Scholar]
- 39.Bittner M., Meltzer P., Chen Y., Jiang Y., Seftor E., Hendrix M., Radmacher M., Simon R., Yakhini Z., Ben-Dor A., Sampas N., Dougherty E., Wang E., Marincola F., Gooden C., Lueders J., Glatfelter A., Pollock P., Carpten J., Gillanders E., Leja D., Dietrich K., Beaudry C., Berens M., Alberts D., Sondak V. (2000) Nature 406, 536–540 [DOI] [PubMed] [Google Scholar]
- 40.Tímár J., Mészáros L., Ladányi A., Puskás L. G., Rásó E. (2006) Cell. Immunol. 244, 154–157 [DOI] [PubMed] [Google Scholar]
- 41.Blitzer J. T., Nusse R. (2006) BMC Cell Biol. 7, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin F., Ren X. D., Doris G., Clark R. A. (2005) J. Invest. Dermatol. 124, 906–913 [DOI] [PubMed] [Google Scholar]
- 43.Bachy S., Letourneur F., Rousselle P. (2008) J. Cell. Physiol. 214, 238–249 [DOI] [PubMed] [Google Scholar]
- 44.Reiland J., Sanderson R. D., Waguespack M., Barker S. A., Long R., Carson D. D., Marchetti D. (2004) J. Biol. Chem. 279, 8047–8055 [DOI] [PubMed] [Google Scholar]
- 45.Chalkiadaki G., Nikitovic D., Berdiaki A., Sifaki M., Krasagakis K., Katonis P., Karamanos N. K., Tzanakakis G. N. (2008) Int. J. Biochem. Cell Biol. 41, 1323–1331 [DOI] [PubMed] [Google Scholar]
- 46.Westfall T. A., Brimeyer R., Twedt J., Gladon J., Olberding A., Furutani-Seiki M., Slusarski D. C. (2003) J. Cell Biol. 162, 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Topol L., Jiang X., Choi H., Garrett-Beal L., Carolan P. J., Yang Y. (2003) J. Cell Biol. 162, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.