Abstract
During early mammalian development, genesis of the first two cell lineages, inner cell mass (ICM) and trophectoderm (TE), is dependent upon functions of key transcription factors that are expressed in a regulated and spatially restricted fashion. In this study, we demonstrate that during early mouse development, mRNA expression of transcription factor GATA3 is induced at the 4-cell stage and is consistently present during pre-implantation embryonic development. Interestingly, at the blastocyst stage, Gata3 mRNA is selectively up-regulated within the TE lineage, and GATA3 protein is abundantly present only in the TE but not in the ICM. Using mouse trophoblast stem cells (TS cells) as a model, we found that, knockdown of GATA3 by RNA interference (RNAi) down-regulates expression of caudal-type homeobox 2 (CDX2), a key regulator of the TE lineage. Chromatin immunoprecipitation (ChIP) analyses revealed that, in TS cells, GATA3 directly regulates Cdx2 transcription from a conserved GATA motif at the intron 1 region of the Cdx2 locus. ChIP analyses with mouse blastocysts also detected GATA3 occupancy at intron 1 of the Cdx2 locus. In addition, down-regulation of GATA3 in pre-implantation mouse embryos reduces Cdx2 expression and inhibits morula to blastocyst transformation. Our results indicate a novel function of GATA3, in which it is selectively expressed in TE, regulates expression of key genes in TE lineage, and is involved in morula to blastocyst transformation.
Genesis of the trophectoderm (TE)2 and inner cell mass (ICM) lineages during early mouse development appears to occur in two stages (1–3). First, cells are allocated to different inside and outside positions via asymmetric divisions. Then, the cells in these different positions become specified, and they become committed to restricted developmental fates. Outside cells become committed to the TE, and inside cells become ICM. Development of ICM and TE is regulated by key transcription factors that specify TE and ICM cell fate, and CDX2 has been implicated in this process (4–6). Multiple studies indicated the importance of CDX2 in TS cell proliferation, proper function of TE, and successful implantation of blastocyst (5–8). However, molecular mechanisms that regulate Cdx2 expression in trophoblast cell lineages are poorly understood. Two other transcription factors, eomesodermin (Eomes) and TEA domain family member 4 (TEAD4), are also implicated in TE development. Mutation studies showed that the lack of Eomes also arrests blastocyst development (9). However, Cdx2 is still expressed in Eomes mutants (5). Tead4 mutants show more severe phenotypes than Cdx2 mutants and are characterized by loss of Cdx2 expression (10, 11). However, unlike Cdx2, Tead4 expression is not restricted to the TE lineage during pre-implantation development indicating that additional regulatory mechanisms are involved for the restricted expression of Cdx2 in TE lineage.
Earlier, we found that, among the six members (GATA1–6) of GATA family of transcription factors, only GATA3 is abundantly expressed in TS cells (12). GATA3 was first cloned as a T cell-specific transcript (13, 14), and germ line deletion of Gata3 results in embryonic lethality due to a multitude of phenotypic abnormalities (15). Interestingly, expression analysis during early mouse development indicated that at embryonic day (E) 8.5, Gata3 expression is only detectable at the ectoplacental cone and trophoblast cells surrounding the embryonic cavity (16). Furthermore, it is also shown that in vitro differentiation of human embryonic stem (ES) cells toward trophoblast cell lineage is associated with GATA3 up-regulation (17). These results strongly indicate that GATA3 is involved in the transcription factor networks that drive trophoblast cell linage development.
Although expression of several trophoblast cell-specific genes is impaired in Gata3 mutant mice (18), they die at mid-gestation (E11.5) without any apparent defect in early peri- and post-implantation development. As GATA factors have both distinct and overlapping biological activities (19–24), it is predicted that redundant function of another GATA factor, GATA2 might be the cause for this lack of early developmental phenotype in Gata3 mutant mice (18). Our observation that GATA3 negatively regulates Gata2 expression in TS cell population, and loss of GATA3 induces GATA2 expression (12) in a rat trophoblast stem cell line, further supports this prediction. However, trophoblast cell-specific GATA3 expression during early mouse development as well as its abundant expression in mouse TS cells led us to hypothesize that GATA3 might be an important regulator in TE lineage-specific gene expression. To test this hypothesis, in this study, we tested the expression pattern of GATA3 during early mouse development, and using TS cells as a model system, we tested its function on the regulation of key genes that are important for TE lineage development. Herein, we demonstrate that, at the blastocyst stage, GATA3 expression is selectively restricted in the TE lineage. Furthermore, we also demonstrate that GATA3 directly regulates Cdx2 transcription by occupying the intron 1 region of the Cdx2 chromatin domain. Our results indicate that GATA3 is an important transcription factor for gene expression in TE lineage during early mammalian development.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse TS cells were initially cultured on a feeder layer of primary mouse embryonic fibroblasts (MEFs) in the presence of 25 ng/ml fibroblast growth factor 4 (FGF4, Sigma-Aldrich) and heparin (1 μg/ml) in TS cell medium as mentioned earlier (12). For experiments, mouse TS cells were expanded in a proliferative state without MEF feeders by culturing in the presence of MEF-conditioned medium. E14 mouse ES cells were cultured as mentioned earlier (25).
Collection and Culture of Mouse Embryos
3–4-week-old CD-1 females were superovulated by intraperitoneal injection of 5 international units P.G. 600 (Intervet, Millsboro, DE), followed by 5 international units hCG (Sigma) 48 h later. Females were mated with C57BL/6 males and euthanized the following morning (d 0.5). Oviducts were removed, 1-cell embryos were harvested in M2 medium (Millipore, Billerica, MA) after treatment with hyaluronidase (300 μg/ml M2, Sigma) and cultured in KSOM (Millipore) at 37 °C in a humidified chamber at 5% CO2. Embryos were visualized daily, photographed, and sampled at each cell stage for RNA extraction.
Quantitative RT-PCR
For mRNA expression analyses in MEF, TS cells, and ES cells, total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA). For expression analysis in different stages of pre-implantation embryos, total RNA was isolated from embryos at different developmental stages using PicoPure RNA isolation kit (MDS Analytical Technology, Sunnyvale, CA) following the manufacturer's protocol. In each individual experiment, RNA was extracted from seventy 1-cell, thirty-five 2-cell, eighteen 4-cell, and nine 8-cell embryos. For the morula (≥16 cells) and blastocyst stages, five embryos at each stage were used for RNA extraction. cDNA was prepared by annealing RNA with a 5:1 mixture of random and oligo(dT) primers. This was followed by incubation with moloney murine leukemia virus reverse transcriptase (50 units) (Invitrogen) combined with 10 mm dithiothreitol, RNasin (Promega, Madison, WI), and 0.5 mm dNTPs at 42 °C for 1 h. Reactions, lacking reverse transcriptase, were used as control. Using gene-specific primers, amplified cDNAs were analyzed by quantitative PCR as mentioned earlier (12). Primers are mentioned in the supplemental information.
Protein Analysis
Western blot analysis was performed as mentioned earlier (12). Anti-GATA3 (sc268) and anti-Eomes (sc98555) antibodies were obtained from Santa Cruz Biotechnology, anti-CDX2 (catalogue 2475-1) was obtained from Epitomics Inc. (Burlingame, CA), anti-TEAD4 (ab58310) was obtained from Abcam (Cambridge, MA), and anti-β-actin was obtained from Sigma. Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse antibodies from Santa Cruz Biotechnology were used as secondary antibodies.
Immunosurgery of Blastocysts
Immunosurgery of mouse blastocysts was performed according to the method developed by Solter and Knowles (26) with some modifications. Zonae pellucidae were removed from blastocysts (15 blastocysts for each experiment) using 0.5% Pronase (Sigma) for 5 min. The zona-free blastocysts were then treated with rabbit anti-mouse serum (Sigma) at 1:100 dilution for 30 min, washed with phosphate-buffered saline, and incubated with guinea pig complement (Innovative Research, Novi, MI) at 37 °C in 5% CO2 for 30 min. After washing, ICMs were collected for RNA preparation by the PicoPure RNA isolation kit, and subsequent RT-PCR analysis was performed as mentioned earlier.
Immunostaining of Blastocysts
For immunostaining, early blastocysts were fixed with 4% paraformaldehyde (30 min), permeabilized in 0.25% Triton X-100 (15 min), and blocked with 10% fetal bovine serum and 0.1% Triton in phosphate-buffered saline for 1 h at room temperature. Embryos were incubated with anti-GATA3 antibody (1:50 dilution) in blocking solution for overnight at 4 °C, and washed three times in 0.1% Triton X-100 in phosphate-buffered saline. Blastocysts were incubated with anti-mouse secondary antibody (Alexa Fluor 488-conjugated, (Invitrogen) at 1:400 dilution) at room temperature for 1 h, washed three times, and mounted using anti-fade mounting medium (Invitrogen) and viewed in LSM 510 Laser Scanning Microscope (Carl Zeiss, Maple Grove, MN).
RNA Interference
Lentiviral vectors containing short-hairpin RNAs (shRNAs) targeting mouse Gata3 mRNA were cloned in pLKO.1 (Addgene, Cambridge, MA). Lentiviral supernatants were produced in HEK-293T cells as described earlier (12, 25). Lentiviral supernatants were collected after 24 and 48 h of transfection. Undifferentiated mouse TS cells were grown at ∼70% confluence without MEF feeder layer and infected with lentiviral supernatants. After 48 h, infected TS cells were selected by addition of puromycin (Sigma). After 3 days, samples were prepared for mRNA and protein analysis. The Gata3 target sequence 5′-AAGAGTGCCTCAAGTATCAGG-3′ successfully knocked down expression of the target gene. For control experiments, cells were infected with viral vectors expressing shRNA against the Gata3 target sequence 5′-CCCGAAACCGGAAGATGTCTAGCAAATCGA-3′ and 5′-GCTGTACTACAAGCTTCATAA-3′, which did not knockdown GATA3 expression.
To knockdown GATA3 in pre-implantation embryos, GATA3 shRNA was cloned in pLKO.1 lentiviral vector (Addgene). The vector was linearized with BamHI so that the shRNA expression cassette remains intact. pLKO.3G (Addgene), which contains an enhanced green fluorescence protein (EGFP) gene under the control of the phosphoglycerate kinase (PGK) promoter, was also linearized with BamHI and used as control. The linearized vectors were suspended in EmbryoMax Injection Buffer (Millipore) at a concentration of 1 ng/μl. 2-cell embryos were subjected to pronuclear injection in both the nucleus with 1–2 pl of suspended vectors. The embryos were grown in KSOM (Millipore) at 37 °C in a humidified chamber at 5% CO2. The cells were visualized and photographed daily under a fluorescent and phase contrast microscope for 3 days. 25 embryos were used for each set of vector, while 25 embryos, not subjected to pronuclear injection, were also cultured as an unmanipulated control. After day 3, the embryos were used to prepare total RNA using the Picopure RNA isolation kit.
Quantitative ChIP Assay
Quantitative ChIP analysis with mouse TS cells was performed as described earlier (12). For micro-ChIP analysis with blastocysts, for each experiment, 200 blastocysts were cross-linked with 1% formaldehyde, and ChIP analysis was performed following a protocol described earlier (27) with some modifications. In short, sonicated, cross-linked chromatin fragments were immunoprecipitated with anti-GATA3 antibody. Mouse IgG1 was used as control antibody. Immunoprecipitated chromatins were reverse-cross-linked, digested with proteinase K, and purified. Immunoprecipitated DNA fragments were amplified using the whole genome amplification procedure (WGA2 kit from Sigma), and were analyzed by quantitative PCR for GATA3 occupancy. Primers are mentioned in the supplemental information.
Transient Transfection Assay
For transient transfection analysis, RP23–355P12 BAC DNA was used to amplify regulatory elements of mouse Cdx2 locus. Promoter region (−663 bp to −18 bp) of mouse Cdx2 gene was cloned into BglII/HindIII sites in pGL3 Basic vector (Promega). A region of Cdx2 intron 1 (2601–3469 bp) containing the conserved element was cloned into MluI/BglII sites upstream of the Cdx2 promoter in the pGL3 Basic-Cdx2 promoter (Cdx2(pro)Luc) construct. To delete the conserved WGATAR motif, upstream of the WGATAR motif was cloned into MluI/XhoI sites of Cdx2(pro)Luc construct and downstream of WGATAR motif was cloned into XhoI/BglII sites of the new construct. Thus, conserved GATA binding WGATAR motif was deleted and replaced by an XhoI restriction site. For transient transfection analysis, TS cells were transfected with an equal amount of each plasmid (3 μg). Plasmids were added to 150 μl of Opti-MEM (Invitrogen) reduced serum medium, incubated with Lipofectamine reagent (Invitrogen) for 20 min at room temperature, and then added to the cells. After 3 h of incubation, the transfection mixture was replaced with culture medium. Cell lysates were harvested 48 h post-transfection, and luciferase activity was measured in a Veritas Microplate Luminometer using the luciferase assay buffer (Promega). The luciferase activity for each sample was normalized to the protein concentration of the lysate. At least three independent preparations of each plasmid were analyzed.
RESULTS
GATA3 Expression Is Selectively Induced at the TE Lineage during Early Mouse Development
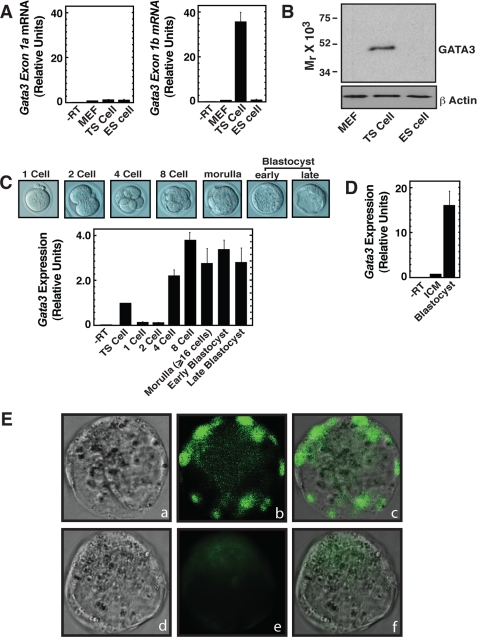
Earlier, we found that GATA3 is abundantly expressed in mouse TS cells (12). To determine whether GATA3 is selectively expressed in TS cells versus ES cells, we performed a quantitative RT-PCR analysis to measure Gata3 mRNA. It has been reported that Gata3 can be transcribed from two distinct promoters, distal and proximal (28, 29), thereby generating two transcripts with alternative untranslated first exons, exon-1a and exon-1b, respectively. The activity of these two promoters selectively drives Gata3 expression in a tissue-specific manner (29). So, to determine whether these two Gata3 transcripts are selectively expressed in TS versus ES cells, we performed an exon-specific quantitative PCR analysis. We found that in TS cells, only exon-1b-containing transcripts are expressed (Fig. 1A), indicating that, in TS cells, the proximal promoter drives Gata3 expression. In contrast to TS cells, and similar to mouse embryonic fibroblasts (MEF, negative control for GATA3 expression), very low levels (basal transcriptional level) of both exon-1a and exon-1b containing transcripts are present in ES cells. Quantitative RT-PCR using primers specific for Gata3 exon 3/4 also showed Gata3 mRNA expression in TS cells but not in the MEF and ES cells (data not shown). Similar to the RT-PCR analysis, Western blot analysis also detected GATA3 expression only in TS cells (Fig. 1B). These results indicate that Gata3 is expressed in the TE-derived stem cells but is absent in ICM-derived stem cells.
FIGURE 1.
GATA3 is selectively expressed in the TE of blastocyst. A, quantitative RT-PCR analysis of Gata3 transcripts with exon-specific primers (means ± S.E., three independent experiments). Graphs show Gata3 transcript levels with respect to that in MEF (used as negative control). B, Western blot analysis showing GATA3 protein in cells analyzed in A. C, different stages of early mouse embryo (top) that are analyzed by quantitative RT-PCR analysis (bottom) for Gata3 mRNA expression. The graph shows the relative Gata3 mRNA levels in pre-implantation embryos with respect to that in mouse TS cells (means ± S.E., three independent experiments). D, quantitative RT-PCR analysis showing the relative Gata3 mRNA levels in whole blastocyst with respect to that in ICM (means ± S.E., three independent experiments). E, localization of GATA3 protein at the early blastocyst stage as observed by immunofluorescence microscopy; a and d, phase contrast images of early blastocysts. b and e, fluorescence images after incubating blastocysts with anti-GATA3 antibody or only with the secondary antibody in the absence of anti-GATA3 antibody, respectively. c and f, merged fluorescence and phase-contrast images.
To obtain definitive information regarding the Gata3 mRNA expression at different developmental stages of the pre-implantation mouse embryo, we performed quantitative RT-PCR analyses (Fig. 1C). In our analyses, we detected very low levels of Gata3 transcripts in 1-cell and 2-cell embryos. However, Gata3 mRNA is highly induced beginning from 4-cell embryo and is consistently present up to the blastocyst stage. This result indicates that during pre-implantation development Gata3 is transcriptionally up-regulated at the 4-cell stage.
Although Gata3 transcription is induced as early as the 4-cell stage, as mentioned earlier, GATA3 is expressed selectively in TE-derived TS cells but not in the ICM-derived ES cells. So, we hypothesized that, at the blastocyst stage, GATA3 expression is restricted in the TE lineage cells. To test that, we performed two different experiments. In the first experiment, we selectively isolated mRNA from the ICM after separating the TE by immunosurgery and compared Gata3 mRNA expression with respect to the whole blastocyst. In the second experiment, we performed a whole-mount immunofluorescence microscopy to determine GATA3 protein expression in the blastocyst. For both of these experiments, we used embryos that are at the early blastocyst stage to eliminate the probability of signals coming from the primitive endoderm layer.
We found that, after immunosurgery, mRNA isolated only from the ICM contains significantly less Gata3 mRNA compared with that isolated from whole blastocysts (Fig. 1D). So, at the blastocyst stage, Gata3 mRNA expression is selectively induced in the TE lineage but not in the ICM lineage.
Finally, to determine the localization of GATA3 protein in the blastocyst, we performed immunofluorescence microscopy. We detected GATA3 protein expression specifically in the outer TE lineage cells but not in the ICM lineage cells (Fig. 1E). These results confirmed that at the blastocyst stage, GATA3 protein expression is restricted at the TE lineage.
GATA3 Directly Regulates Expression of the Cdx2 Gene in TS Cells and in Blastocysts
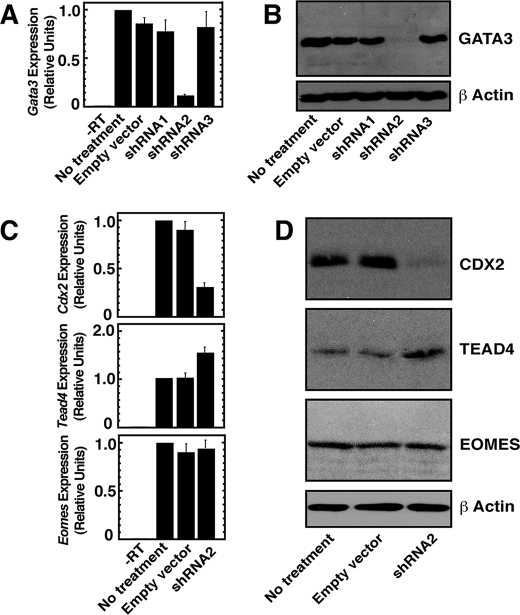
As GATA3 is selectively expressed in TE lineage cells and TE-derived TS cells, we next asked whether GATA3 regulates expression of key genes that are implicated in TE development. For this, we utilized an RNAi approach to knockdown GATA3 expression in TS cells and asked whether depletion of GATA3 affects expression of CDX2, Eomes, and TEAD4. We took an approach of expressing shRNA-utilizing lentiviral vectors to knockdown GATA3 and selected a shRNA (shRNA2, Fig. 2A) molecule that depleted Gata3 mRNA expression by ∼90% in TS cells. The efficient knockdown was also confirmed by Western blot analysis (Fig. 2B).
FIGURE 2.
GATA3 positively regulates Cdx2 expression in TS cells. A, quantitative RT-PCR analysis of Gata3 mRNA expression in TS cells, expressing shRNAs against Gata3 (means ± S.E., three independent experiments). Cells were infected with lentiviral vectors expressing shRNAs and analyzed as mentioned under “Experimental Procedures.” TS cells, without any treatment or infected only with empty vectors, were used as control. B, Western blot analysis of GATA3 protein expression in cells analyzed in A. C, quantitative RT-PCR analysis of Cdx2, Tead4, and Eomes in GATA3-knocked down TS cells (means ± S.E., three independent experiments). D, Western blot analysis of samples analyzed in C.
Under conditions when GATA3 was efficiently knocked down, we determined the mRNA expression of Cdx2, Eomes, and Tead4. We found that depletion of GATA3 selectively down-regulates Cdx2 mRNA expression by ∼68% (Fig. 2C). The Eomes mRNA expression did not change, and Tead4 mRNA expression was increased by ∼32%. Analyses of protein expression also validated the mRNA expression pattern. Western blot analysis (Fig. 2D) confirmed that CDX2 expression was significantly down-regulated, TEAD4 expression was slightly up-regulated, and Eomes expression did not change with GATA3 depletion. These results indicate that, in TS cells, GATA3 positively regulates expression of CDX2, a key regulator of the TE lineage.
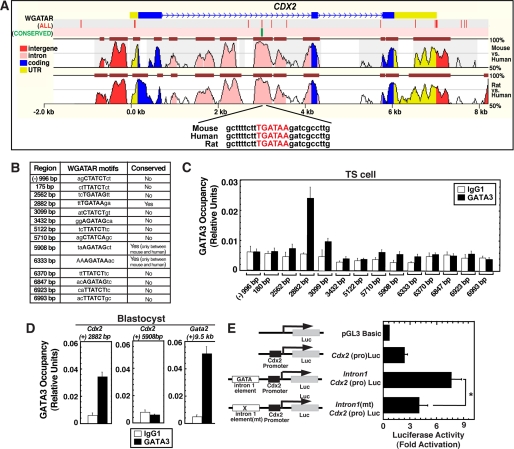
To test the hypothesis that GATA3 directly regulates Cdx2 transcription in TS cells, we tested GATA3 chromatin occupancy at the Cdx2 locus. Sequence analysis of ∼10 kb ((−) 2 kb to (+)8 kb region from the transcription start site) segment of mouse Cdx2 locus revealed the presence of 14 canonical GATA binding (W(A/T)GATAR(A/G)) (12) motifs (Fig. 3, A and B). Among those 14 motifs, only one GATA motif, which is located at (+)2882 bp within the intron 1 of the Cdx2 locus is conserved in multiple mammalian species (Fig. 3, A and B). Two other GATA motifs, located at (+)5908 and (+)6333 bp within the 3′-untranslated region (UTR) of mouse Cdx2 locus, are conserved between mouse and human only (Fig. 3B). The conserved GATA motif within intron 1 is located within a ∼500-bp evolutionary conserved region (Fig. 3A). Interestingly, earlier analysis in transgenic mice from two different laboratories revealed that the conserved region of intron 1 is an important regulatory element for Cdx2 expression in multiple tissues (30, 31). So, we tested whether GATA3 occupies the conserved GATA motif within the intron 1 of the Cdx2 locus. In addition, as non- or less-conserved GATA motifs might also serve as GATA binding sites, we analyzed GATA3 occupancy at all GATA motifs that are located within ∼10 kb of the mouse Cdx2 locus. However, quantitative ChIP analyses in TS cells revealed that, at the endogenous Cdx2 locus, GATA3 occupies only the conserved (+)2882 bp GATA motif of intron 1 (Fig. 3C) but not at other regions that lack conserved GATA motifs.
FIGURE 3.
GATA3 directly regulates Cdx2 expression in blastocysts by occupying a conserved GATA motif within intron 1 of the Cdx2 gene. A, alignment of ∼10 kb regions of mouse, human, and rat Cdx2 loci showing the presence of a conserved GATA motif at the Cdx2 intron 1 region. The red vertical bars on top indicate positions of all WGATAR motifs, and the green bar indicates the position of the conserved WGATAR motif in the ∼10-kb region of the mouse Cdx2 locus. B, table shows the coordinates of WGATAR motifs in the mouse Cdx2 locus. C, quantitative ChIP analysis showing GATA3 occupancy at the (+)2882-bp conserved WGATAR motif, located within intron 1 of the Cdx2 locus, in mouse TS cells (means ± S.E., four independent experiments). D, ChIP analysis showing that GATA3 occupies the conserved WGATAR motif of the Cdx2 intron 1 region in blastocysts but not at the GATA motif (5908 bp) of Cdx2 3′-UTR (means ± S.E., three independent experiments). Gata2 (+) 9.5-kb region was used as a positive GATA3 binding site. E, mouse TS cells were transiently transfected with plasmids in which the Cdx2 intron 1 element was fused to the Cdx2 promoter in front of a luciferase (Luc) reporter gene. In Intron1(mt)Cdx2(pro)Luc construct the conserved WGATAR motif was mutated. Plots depict luciferase activity of the cell lysates normalized by the protein concentration of the lysates (mean ± S.E., four independent experiments; *, p < 0.05). In each independent experiment, transfections were performed in triplicate.
As TS cells are derived from the TE, and GATA3 is selectively expressed in the TE of the peri-implantation mouse embryo, we wanted to determine GATA3 chromatin occupancy at the Cdx2 intron 1 within blastocysts. Our analysis showed that, similar to TS cells, GATA3 is occupied at the intron 1 of the Cdx2 locus in blastocysts (Fig. 3D, left panel). Gata2(+) 9.5-kb region, a GATA3 binding region in TS cells (12) is also occupied by GATA3 in blastocysts (Fig. 3D, right panel). However, similar to TS cells, the (+)5908 bp GATA motif within the 3′-UTR of Cdx2 locus did not show any GATA3 occupancy (Fig. 3D, middle panel). Collectively, these results along with the ChIP analysis and knockdown studies in TS cells indicate that GATA3 directly regulates Cdx2 expression in TE lineage.
To assess the functional importance of the intron 1 conserved GATA motif in trophoblast cells, we performed transient transfection assays. We found that when fused to the Cdx2 promoter in front of a luciferase reporter gene, the conserved intron 1 element of the mouse Cdx2 locus enhanced the promoter activity by ∼3-fold (Fig. 3E) in TS cells. However, mutation of the conserved WGATAR motif alone significantly reduced the enhancer activity of the intronic element. These data strongly implicate the WGATAR motif as being critical for Cdx2 intron 1 enhancer activity.
Down-regulation of GATA3 in Pre-implantation Embryos Inhibits Morula to Blastocyst Transformation
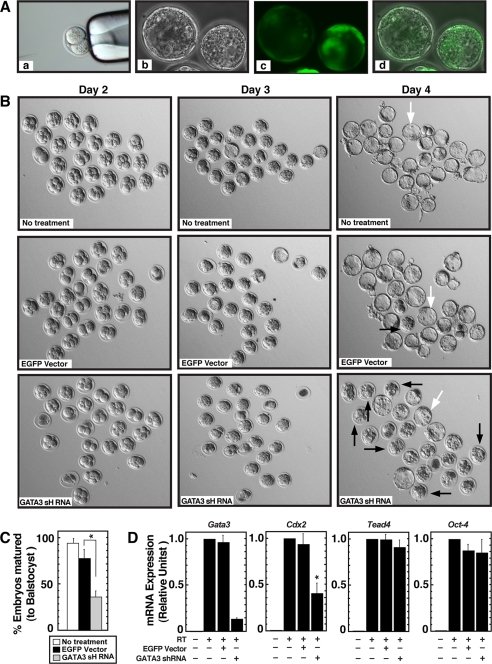
Our results show that GATA3 expression is induced as early as 4-cell stage in pre-implantation embryos and is restricted at the trophectoderm in peri-implantation embryos. In addition, ChIP analysis indicates that GATA3 directly regulates the Cdx2 gene in TS cells and blastocysts. However, GATA3 function and GATA3-mediated control of Cdx2 expression might be important at later rather than the early stage of trophectoderm development. The apparent lack of defect during pre-implantation development in GATA3 knockout mice further supports this possibility. However, to our knowledge, the role of GATA3 during the early stage of trophectoderm development has not been studied in a systematic way with GATA3 knockout mice. Thus, in an additional set of experiments, we attempted to determine the effects of GATA3 down-regulation in pre-implantation embryos on Cdx2 expression and on the pre-implantation development.
For this experiment, we injected the GATA3 shRNA-expressing lentiviral DNA in two cell mouse embryos and monitored pre-implantation development by culturing them in vitro. To confirm that the lentiviral vector can induce shRNA expression during the course of pre-implantation development, we injected an EGFP-expressing lentiviral vector in two cell embryos and tested expression of EGFP at the blastocyst stage (Fig. 4A). This control experiment also revealed that the injection of EGFP-expressing lentiviral vectors does not significantly inhibit pre-implantation development of mouse embryos when they are cultured in vitro for 3 more days to form matured blastocysts (Fig. 4B, top and middle rows). When injected with GATA3 shRNA-expressing vectors, two cell embryos develop normally to form morulas within 2 days, and like the untreated and EGFP-expressing morulas, the GATA3 shRNA-injected morulas showed no sign of noticeable degeneration. However, intriguingly, on day 3, ∼65% of the GATA3 shRNA-expressing morulas did not mature to form blastocysts (Fig. 4, B, bottom rows and C). A few additional morulas (∼15%) matured to the blastocyst stage when cultured for one additional day (data not shown). Quantitative RT-PCR analysis on day 3 with shRNA-injected embryos confirmed that Gata3 mRNA expression was inhibited by ∼85% (Fig. 4D, left panel). In addition, quantitative RT-PCR analyses also showed that in GATA3-knocked down morulas/blastocysts, Cdx2 mRNA expression was inhibited by ∼55%. However, no significant loss in Tead4 mRNA levels was observed in GATA3-knocked down morulas/blastocysts. In addition, mRNA expression of Oct-4, another important transcription factor involved in lineage specification and development of ICM lineage in early embryos (6), was not altered in GATA3-knocked down embryos (Fig. 4D, right panel). These results strongly implicate that GATA3 regulates Cdx2 expression during early trophectoderm development, and GATA3 function is required for efficient maturation of blastocysts from the morula stage.
FIGURE 4.
Inhibition of morula to blastocyst transformation in GATA3-depleted pre-implantation embryos. A, 2-cell mouse embryos were subjected to pronuclear injection (panel a) with lentiviral constructs expressing EGFP under the control of the phosphoglycerate kinase promoter. Embryos were cultured for 3 days, and images were captured under phase (panel b) or fluorescence (panel c) microscopy. Panel d shows merged phase and fluorescence images. B, 2-cell mouse embryos (25 embryos for each experimental set) were injected with EGFP-expressing (panels in middle row) or GATA3 shRNA-expressing (panels in bottom row) lentiviral constructs, pre-implantation embryonic development was monitored after 24 h (day 2 of development), 48 h (day 3 of development), and 72 h (day 4 of development), and compared with untreated embryos (panels of top row). Four independent experiments were performed, and representative pictures for each condition are shown. No significant defect is apparent in the development of GATA3 shRNA-expressing embryos up to day 3 of development. Most of the embryos developed to the morula stage. However, at day 4, development of the majority of GATA3 shRNA-expressing embryos was arrested at the morula stage (black arrows), whereas most of the untreated and EGFP-expressing embryos formed matured blastocysts (white arrows). C, graph shows the % total embryos (mean ± S.E., three independent experiments; *, p < 0.05) that formed matured blastocysts at day 4 of development under each experimental condition described in B. D, quantitative RT-PCR analysis of Gata3, Cdx2, Tead4, and Oct-4 mRNA expression in embryos analyzed in C (means ± S.E., *, p < 0.05).
DISCUSSION
The intricate transcriptional networks that dictate cellular and molecular events leading to the establishment and proper function of the TE and ICM lineages are still poorly understood. One of the strategies to understand these molecular mechanisms is to find out potential transcriptional regulators that are selectively expressed in TE versus ICM lineages followed by testing their functions.
Although multiple studies indicated that GATA3 is highly expressed in trophoblast cell lineages (12, 16, 18, 32) and regulates expression of multiple genes in trophoblast cells in vivo (18), major efforts have not been taken to determine its expression during early embryonic development. The lack of early developmental phenotype in Gata3 mutant mice might be one of the causes behind this lack of study. In this study (i) we have delineated the expression pattern of transcription factor GATA3 in pre-implantation embryos showing that it is selectively induced at the TE lineage at the blastocyst stage, and (ii) we have shown that down-regulation of GATA3 inhibits maturation of blastocyst from the morula stage (Fig. 4B).
We have also demonstrated here that GATA3 directly regulates Cdx2 expression in TS cells and in blastocysts (Figs. 3D and 4D). As CDX2 function is important for TE development, we speculate that GATA3 is an important regulator of molecular events that contributes to the early lineage commitment and TE development and/or TS cell self-renewal. In this respect GATA3 might have a role resembling transcription factors Elf5 and ETS2. It has been shown that during early lineage commitment Elf5 functions downstream of initial lineage determination, to reinforce commitment to the trophoblast lineage or to abort this pathway in epiblast cells (33). Another study indicated that ETS2 is essential for TS cell self-renewal (34). Like GATA3, both Elf5 and ETS2 regulate Cdx2 expression but Elf5- and Ets2-null mutations do not mimic Cdx2-null mutation (35, 36).
As mentioned earlier, the functional redundancy of GATA2 and GATA3 might rescue the lack of an early developmental or overt placental phenotype in Gata3−/− mice (18). The fact that GATA2 and GATA3 might function redundantly in certain contexts is evident from a recent study (37), in which both Gata2 and Gata3 transgenes, when expressed in the appropriate spatiotemporal pattern under the control of Gata1 regulatory elements, rescued the erythroid defect of Gata1-null mice. However, experiments have not been done in a context where both GATA2 and GATA3 are limiting to test their functional redundancy during early embryonic development.
GATA transcription factors regulate gene expression in multiple modes; (i) by activation of a repressed gene; (ii) by modulating the level of transcription; (iii) by maintaining transcriptionally active state; and (iv) by repressing an active gene. Our finding that GATA3 directly regulates Cdx2 transcription in a positive fashion raises the question via which mode GATA3 regulates Cdx2 expression in TE lineage. It is shown that, transcription factor TEAD4 regulates initial Cdx2 expression during pre-implantation mouse development (10, 11). Expression analysis in pre-implantation embryos indicated that initial expression of Cdx2 could be detected as early as 8-cell stage. However, later Cdx2 levels are differentially regulated across the inside/outside axis of the embryo, with progressively higher levels in outside cells, and low levels in inside cells (8). As GATA3 expression is also induced at the outside cells at the blastocyst stage (Fig. 1, D and E), we predict that GATA3 positively induces and maintains higher levels of Cdx2 transcription in outer cells after initial induction of Cdx2 expression by a TEAD4-dependent mechanism or other pathways. However, other possibilities like GATA3 and TEAD4 might coordinately regulate Cdx2 and other TE-specific gene expression cannot be ruled out.
Our findings also raise two other questions; (i) what are the molecular mechanisms that induce GATA3 expression during peri-implantation development and restricts its expression in the TE lineage at blastocyst stage? and (ii) does GATA3 activate parallel downstream pathways, independent of CDX2, to specify TE fate and development of trophoblast cell lineages? Regarding GATA3 expression, it is possible that TEAD4 might directly or indirectly regulate GATA3 expression in pre-implantation embryos. Our curious analysis revealed the presence of multiple conserved canonical TEAD binding sites within the Gata3 regulatory regions. The fact that TEAD4 expression is induced in GATA3-knocked down TS cells (Fig. 2, C and D) further indicates a possibility in which GATA3 might negatively regulate TEAD4 to properly orchestrate TE-specific gene expression.
Regarding the GATA3 downstream pathways, it is important to take new experimental approaches like determining global gene expression patterns after knocking down GATA3 expression in pre-and peri-implantation embryos. Along with this approach, performing a global analysis of GATA3 target genes within blastocysts will provide important insights regarding GATA3 function during early embryonic development.
Supplementary Material
Acknowledgment
We thank Dr. Michael J. Soares for important suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants P20 RRO16475 and P20 RRO24214.

The on-line version of this article (available at http://www.jbc.org) contains supplemental information.
- TE
- trophectoderm
- ICM
- inner cell mass
- EGFP
- enhanced green fluorescent protein
- ChIP
- chromatin immunoprecipitation assay
- Eomes
- eomesodermin
- shRNA
- short-hairpin RNA
- MEF
- mouse embryonic fibroblasts
- RT
- reverse transcriptase
- TS
- mouse trophoblast stem cells
- ES
- human embryonic stem cells
- UTR
- untranslated region
- CDX2
- caudal-type homeobox 2.
REFERENCES
- 1.Rossant J. (2004) Semin. Cell Dev. Biol. 15, 573–581 [DOI] [PubMed] [Google Scholar]
- 2.Zernicka-Goetz M. (2005) Nat. Rev. Mol. Cell Biol. 6, 919–928 [DOI] [PubMed] [Google Scholar]
- 3.Kunath T., Strumpf D., Rossant J. (2004) Placenta 25, Suppl. A, S32–S38 [DOI] [PubMed] [Google Scholar]
- 4.Chawengsaksophak K., James R., Hammond V. E., Köntgen F., Beck F. (1997) Nature 386, 84–87 [DOI] [PubMed] [Google Scholar]
- 5.Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. (2005) Development 132, 2093–2102 [DOI] [PubMed] [Google Scholar]
- 6.Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005) Cell 123, 917–929 [DOI] [PubMed] [Google Scholar]
- 7.Jedrusik A., Parfitt D. E., Guo G., Skamagki M., Grabarek J. B., Johnson M. H., Robson P., Zernicka-Goetz M. (2008) Genes Dev. 22, 2692–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ralston A., Rossant J. (2008) Dev. Biol. 313, 614–629 [DOI] [PubMed] [Google Scholar]
- 9.Russ A. P., Wattler S., Colledge W. H., Aparicio S. A., Carlton M. B., Pearce J. J., Barton S. C., Surani M. A., Ryan K., Nehls M. C., Wilson V., Evans M. J. (2000) Nature 404, 95–99 [DOI] [PubMed] [Google Scholar]
- 10.Yagi R., Kohn M. J., Karavanova I., Kaneko K. J., Vullhorst D., DePamphilis M. L., Buonanno A. (2007) Development 134, 3827–3836 [DOI] [PubMed] [Google Scholar]
- 11.Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K., Sasaki H. (2008) Mech. Dev. 125, 270–283 [DOI] [PubMed] [Google Scholar]
- 12.Ray S., Dutta D., Rumi M. A., Kent L. N., Soares M. J., Paul S. (2009) J. Biol. Chem. 284, 4978–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng W., Flavell R. A. (1997) Cell 89, 587–596 [DOI] [PubMed] [Google Scholar]
- 14.Flavell R. A., Li B., Dong C., Lu H. T., Yang D. D., Enslen H., Tournier C., Whitmarsh A., Wysk M., Conze D., Rincon M., Davis R. J. (1999) Cold Spring Harb. Symp. Quant. Biol. 64, 563–571 [DOI] [PubMed] [Google Scholar]
- 15.Pandolfi P. P., Roth M. E., Karis A., Leonard M. W., Dzierzak E., Grosveld F. G., Engel J. D., Lindenbaum M. H. (1995) Nat. Genet. 11, 40–44 [DOI] [PubMed] [Google Scholar]
- 16.George K. M., Leonard M. W., Roth M. E., Lieuw K. H., Kioussis D., Grosveld F., Engel J. D. (1994) Development 120, 2673–2686 [DOI] [PubMed] [Google Scholar]
- 17.Xu R. H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., Thomson J. A. (2002) Nat. Biotechnol. 20, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 18.Ma G. T., Roth M. E., Groskopf J. C., Tsai F. Y., Orkin S. H., Grosveld F., Engel J. D., Linzer D. I. (1997) Development 124, 907–914 [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara Y., Chang A. N., Williams A. M., Orkin S. H. (2004) Blood 103, 583–585 [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi-Osaki M., Ohneda O., Suzuki N., Minegishi N., Yokomizo T., Takahashi S., Lim K. C., Engel J. D., Yamamoto M. (2005) Mol. Cell. Biol. 25, 7005–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pevny L., Simon M. C., Robertson E., Klein W. H., Tsai S. F., D'Agati V., Orkin S. H., Costantini F. (1991) Nature 349, 257–260 [DOI] [PubMed] [Google Scholar]
- 22.Simon M. C., Pevny L., Wiles M. V., Keller G., Costantini F., Orkin S. H. (1992) Nat. Genet. 1, 92–98 [DOI] [PubMed] [Google Scholar]
- 23.Tsai F. Y., Keller G., Kuo F. C., Weiss M., Chen J., Rosenblatt M., Alt F. W., Orkin S. H. (1994) Nature 371, 221–226 [DOI] [PubMed] [Google Scholar]
- 24.Tsai F. Y., Orkin S. H. (1997) Blood 89, 3636–3643 [PubMed] [Google Scholar]
- 25.Dutta D., Ray S., Vivian J. L., Paul S. (2008) J. Biol. Chem. 283, 25404–25413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solter D., Knowles B. B. (1975) Proc. Natl. Acad. Sci. U. S. A. 72, 5099–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acevedo L. G., Iniguez A. L., Holster H. L., Zhang X., Green R., Farnham P. J. (2007) BioTechniques 43, 791–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asnagli H., Afkarian M., Murphy K. M. (2002) J. Immunol. 168, 4268–4271 [DOI] [PubMed] [Google Scholar]
- 29.Scheinman E. J., Avni O. (2009) J. Biol. Chem. 284, 3037–3048 [DOI] [PubMed] [Google Scholar]
- 30.Wang W. C., Shashikant C. S. (2007) J. Exp. Zoolog B Mol. Dev. Evol. 308, 308–321 [DOI] [PubMed] [Google Scholar]
- 31.Benahmed F., Gross I., Gaunt S. J., Beck F., Jehan F., Domon-Dell C., Martin E., Kedinger M., Freund J. N., Duluc I. (2008) Gastroenterology 135, 1238–1247, 1247.e1–3 [DOI] [PubMed] [Google Scholar]
- 32.Ng Y. K., George K. M., Engel J. D., Linzer D. I. (1994) Development 120, 3257–3266 [DOI] [PubMed] [Google Scholar]
- 33.Ng R. K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. (2008) Nat. Cell Biol. 10, 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen F., Tynan J. A., Cecena G., Williams R., Múnera J., Mavrothalassitis G., Oshima R. G. (2007) Dev. Biol. 312, 284–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donnison M., Beaton A., Davey H. W., Broadhurst R., L'Huillier P., Pfeffer P. L. (2005) Development 132, 2299–2308 [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto H., Flannery M. L., Kupriyanov S., Pearce J., McKercher S. R., Henkel G. W., Maki R. A., Werb Z., Oshima R. G. (1998) Genes Dev. 12, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira R., Wai A., Shimizu R., Gillemans N., Rottier R., von Lindern M., Ohneda K., Grosveld F., Yamamoto M., Philipsen S. (2007) Blood 109, 5481–5490 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.