Abstract
The mqsR gene has been shown to be positively regulated by the quorum-sensing autoinducer AI-2, which in turn activates a two-component system, the qseB-qseC operon. This operon plays an important role in biofilm formation in Escherichia coli. However, its cellular function has remained unknown. Here, we found that 1 base downstream of mqsR there is a gene, ygiT, that is co-transcribed with mqsR. Induction of mqsR caused cell growth arrest, whereas ygiT co-induction recovered cell growth. We demonstrate that MqsR (98 amino acid residues), which has no homology to the well characterized mRNA interferase MazF, is a potent inhibitor of protein synthesis that functions by degrading cellular mRNAs. In vivo and in vitro primer extension experiments showed that MqsR is an mRNA interferase specifically cleaving mRNAs at GCU. The mRNA interferase activity of purified MqsR was inhibited by purified YgiT (131 residues). MqsR forms a stable 2:1 complex with YgiT, and the complex likely functions as a repressor for the mqsR-ygiT operon by specifically binding to two different palindromic sequences present in the 5′-untranslated region of this operon.
It has been reported that quorum sensing is involved in biofilm formation (1–4). mqsR expression was found to be induced by 8-fold in biofilms (5) and also by the quorum-sensing signal autoinducer AI-2, which is a species-nonspecific signaling molecule produced by both Gram-negative and Gram-positive bacteria, including Escherichia coli (6). It has been reported that induction of mqsR activates a two-component system, the qseB-qseC operon, which is known to play an important role in biofilm formation (6). Thus, it has been proposed that MqsR (98 amino acid residues) is a regulator of biofilm formation because it activates qseB, which controls the flhDC expression required for motility and biofilm formation in E. coli (6). However, the cellular function of MqsR has remained unknown.
Interestingly, all free-living bacteria examined to date contain a number of suicide or toxin genes in their genomes (7, 8). Many of these toxins are co-transcribed with their cognate antitoxins in an operon (termed toxin-antitoxin (TA)2 operon) and form a stable complex in the cell, so their toxicity is subdued under normal growth conditions (9–11). However, the stability of antitoxins is substantially lower than that of their cognate toxins, so any stress causing cellular damage or growth inhibition that induces proteases alters the balance between toxin and antitoxin, leading to toxin release in the cell.
To date, 16 (12) TA systems have been reported on the E. coli genome, including relB-relE (13, 14), chpBI-chpBK (15), mazEF (16–18), yefM-yoeB (19, 20), dinJ-yafQ (21, 22), hipBA and hicAB (23, 24), prlF-yhaV (25), and ybaJ-hha (26). Interestingly, all of these TA operons appear to use similar modes of regulation: the formation of complexes between antitoxins and their cognate toxins to neutralize toxin activity and the ability of TA complexes to autoregulate their expression. The cellular targets of some toxins have been identified. CcdB directly interacts with gyrase A and blocks DNA replication (27, 28). RelE, which by itself has no endoribonuclease activity, appears to act as a ribosome-associating factor that promotes mRNA cleavage at the ribosome A-site (13, 29, 30). PemK (31), ChpBK (15), and MazF (32) are unique among toxins because they target cellular mRNAs for degradation by functioning as sequence-specific endoribonucleases to effectively inhibit protein synthesis and thereby cell growth.
MazF, ChpBK, and PemK have been characterized as sequence-specific endoribonucleases that cleave mRNA at the ACA, ACY (Y is U, A, or G), and UAH (H is C, A, or U) sequences, respectively. They are completely different from other known endoribonucleases such as RNases E, A, and T1, as these toxins function as protein synthesis inhibitors by interfering with the function of cellular mRNAs. It is well known that small RNAs, such as mRNA-interfering cRNA (33), microRNA (34), and small interfering RNA (35), interfere with the function of specific RNAs. These small RNAs bind to specific mRNAs to inhibit their expression. Ribozymes also act on their target RNAs specifically and interfere with their function (36). Therefore, MazF, ChpBK, and PemK homologs form a novel endoribonuclease family that exhibits a new mRNA-interfering mechanism by cleaving mRNAs at specific sequences. Thus, they have been termed “mRNA interferases” (2).
During our search for TA systems on the E. coli genome, we found that the mqsR gene is co-transcribed with a downstream gene, ygiT. These two genes appear to function as a TA system, as their size is small (98 residues for MqsR and 131 residues for YgiT) and their respective open reading frames are separated by 1 bp. In this study, we demonstrate that MqsR-YgiT is a new E. coli TA system consisting of a toxin, MqsR, and an antitoxin, YgiT. Moreover, we identify MqsR as a novel mRNA interferase that does not exhibit homology to MazF. This toxin cleaves RNA at GCU sequences in vivo and in vitro. The implication of this finding as to how this mRNA interferase is involved in cell physiology and biofilm formation will be discussed.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
E. coli BL21(DE3) and C43 were used. Both mqsR and ygiT genes in the mqsR-ygiT operon were separately amplified by PCR using the E. coli genomic DNA as template and first cloned into pET28a (Novagen). The mqsR-ygiT operon was also amplified by PCR with primers MqsR-Fw and YgiT-Rv using the E. coli genomic DNA as template and cloned into pET28a to express the MqsR-YgiT complex. Subsequently, the mqsR and ygiT genes were separately cloned into pBAD24, creating pBAD-mqsR and pBAD-ygiT, respectively. The promoter region of mqsR-ygiT was amplified by PCR with primers RT-proF and RT-proR and cloned into the pCR®2.1-Topo® vector (Invitrogen).
Assay of in Vivo DNA and Protein Synthesis
E. coli BL21(DE3) cells harboring pBAD-mqsR were grown in M9 medium with 0.5% glycerol (no glucose) and 1 mm each amino acids except for methionine. When the A600 value of the culture reached 0.3, arabinose was added to a final concentration of 0.2% to induce MqsR. Aliquots of the cell cultures (0.4 ml) were taken at the time intervals indicated in Fig. 2 and mixed with 30 μCi of [35S]methionine or 10 μCi of [3H]thymidine plus 80 μg of nonradioactive methionine and 30 μg of nonradioactive thymidine, respectively. After incubation at 37 °C for 30 s, the rates of protein and DNA synthesis were determined as described previously (32). For SDS-PAGE analysis of the total cellular protein synthesis, 400-μl samples were removed from the reaction mixture containing [35S]methionine at the time intervals indicated in Fig. 2F and transferred to chilled test tubes containing 100 μl of 100 μg/ml nonradioactive methionine solution. Cell pellets were collected by centrifugation, resuspended in 40 μl of Laemmli buffer, and subjected to SDS-PAGE, followed by autoradiography.
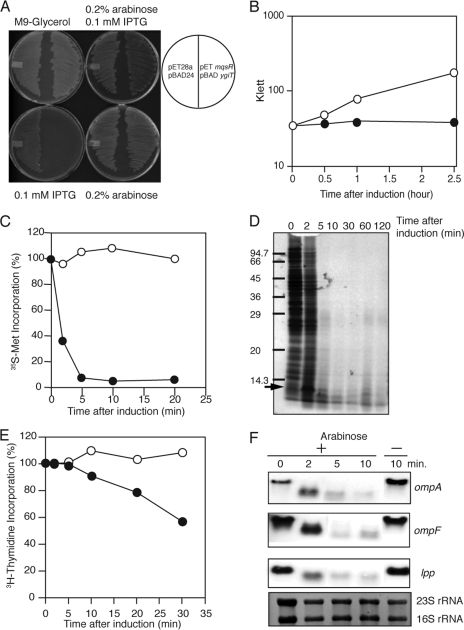
FIGURE 2.
Effect of MqsR induction on protein and DNA synthesis and mRNA stability. A, E. coli BL21 transformed with pET-mqsR and pBAD-ygiT and streaked on M9 (glycerol, CAA) plates with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG), with 0.2% arabinose, with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside plus 0.2% arabinose, or without either inducer. The plates were incubated at 37 °C for 18 h. B, growth curves of E. coli BL21 cells harboring pBAD-mqsR. The cells were cultured in M9-glycerol liquid medium at 37 °C in the presence (●) or absence (○) of 0.2% arabinose. C, effect of MqsR on [35S]methionine incorporation in vivo. At the different time intervals indicated, 0.4 ml of the culture was put into a test tube containing 30 μCi of [35S]methionine, and the mixture was incubated for 30 s at 37 °C. After the incubation, 50 μl of the reaction mixture was applied to a filter paper disk (Whatman No. 3MM, 2.3-cm diameter). The filter paper disks were treated with 10% trichloroacetic acid solution as described previously (32). The radioactivity on the filter was determined with a liquid scintillation counter. D, SDS-PAGE analysis of the products in C. The reaction mixture (400 μl) at the time points indicated was put into a chilled test tube containing 100 μg/ml nonradioactive methionine, and cells were collected by centrifugation. The pellets were dissolved in 40 μl of SDS-PAGE loading buffer. The samples were incubated in a boiling water bath for 10 min. After removal of insoluble materials by centrifugation, the supernatant fraction (12.5 μl) was applied to 15% SDS-polyacrylamide gel. E, effect of MqsR on [3H]thymidine incorporation in vivo. E. coli BL21 cells harboring pBAD-mqsR were grown at 37 °C. When the A600 value of the culture reached 0.3, MqsR was induced with arabinose (0.2%). At the different time intervals indicated, 0.4 ml of the culture was put into a test tube and incubated with 10 μCi of [3H]thymidine plus 30 μg of nonradioactive thymidine. The mixture was then incubated for 30 s at 37 °C. After the incubation, the incorporated radioactivity into the cells were determined as described previously (32). F, effect of MqsR on cellular mRNA stability. Total RNA was extracted from E. coli BL21 cells harboring pBAD-mqsR at various time points as indicated after the addition of arabinose (0.2%) and subjected to Northern blotting with labeled ompA, ompF, and lpp as probes. Before transferring RNA onto a membrane, the gel was stained with ethidium bromide to detect 23 S and 16 S rRNAs.
RNA Isolation and Northern Blot Analysis
E. coli BL21(DE3) cells containing pBAD-mqsR were grown at 37 °C in M9 medium with 0.2% glycerol (no glucose). When the A600 value reached 0.4, arabinose was added to a final concentration of 0.2%. The samples were taken at different intervals as indicated in Fig. 2. Total RNA was isolated using the hot phenol method as described previously (37). Northern blot analysis was carried out as described previously (38).
Primer Extension Analysis in Vivo
For primer extension analysis of mRNA cleavage sites in vivo, total RNAs were extracted from the E. coli BL21(DE3) cells containing pBAD-mqsR at different time points after MqsR induction as indicated in Fig. 3. Primer extension was carried out at 47 °C for 1 h with 10 units of avian myeloblastosis virus reverse transcriptase (Roche Applied Science) using 15 μg of total RNA and 1 pmol of primers (see Table 1) labeled with T4 polynucleotide kinase (Takara Bio Inc.) with [γ-32P]ATP. The reaction was stopped by the addition of 12 μl of sequencing loading buffer (95% formaldehyde, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol), heated at 95 °C for 2 min, and placed on ice. The products were analyzed on a 6% polyacrylamide-containing 8 m urea gel with a sequence ladder made with the same primer.
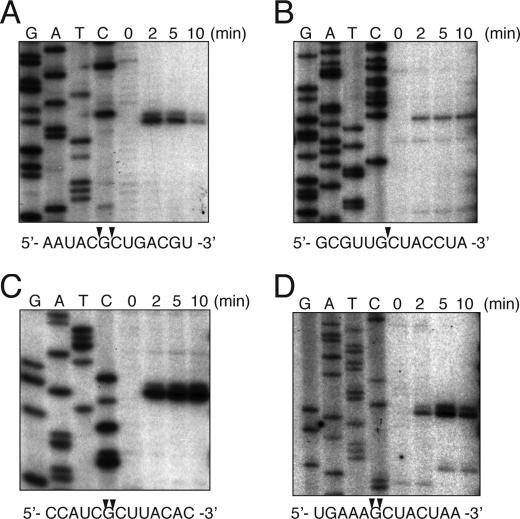
FIGURE 3.
Primer extension analysis of MqsR cleavage sites in ompF mRNA in vivo. Total RNA was prepared from E. coli BL21 cells harboring pBAD-mqsR at the indicated time points before and after the induction of MqsR. The sequence ladders were obtained with pCR®2.1-Topo®-ompF as template (32). The sequences around the cleavage sites are indicated below the panels, and the cleavage sites are indicated by arrowheads.
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| MqsR-Fw | 5′-TTTTTTTTTCATATGGAAAAACGCACACCACATACAC-3′ |
| mqsR-Rv | 5′-TTTGAATTCTTACTTCTCCTTAAACGAGACGATCAG-3′ |
| ygiT-Fw | 5′-TTTTTTTTTCATATGAAATGTCCGGTTTGC-3′ |
| ygiT-Rv | 5′-TTTGAATTCTTAACGGATTTCATTCAATAGTTCTGGATGC-3′ |
| RT-proF | 5′-TGCCTGACTCCAGCTTCCCTTA-3′ |
| RT-proR | 5′-TTAACGGATTTCATTCAATAGTTCTGGATGC-3′ |
| RT-Fw | 5′-ACGCACACCACATACACGTT-3′ |
| RT-Rv | 5′-GCGAAAACGCATTTACACCT-3′ |
| YT-Rv | 5′-TTAACGGATTTCATTCAATAGTTCTGGATGC-3′ |
| PX-RT | 5′-TGTATGTGGTGTGCGTTTTTCC-3′ |
| PX-F1 | 5′-TTGCCACCGTAACTGTTTTC-3′ |
| PX-F2 | 5′-TGTAACCCAGTGCATCATAAAC-3′ |
| PX-F3 | 5′-GCCAACACCGTCGCCGTTAGA-3′ |
| PX-F4 | 5′-TTTTTTACCGTTGCCAAGAGGT-3′ |
| PX-F5 | 5′-TGGCGAAGCCGCTGGTGTTTG-3′ |
| PX-F6 | 5′-GCCCACTTCAAAGTAGTTCA-3′ |
| PX-F7 | 5′-CATGTCGCCATTGCCACCGT-3′ |
| PX-F8 | 5′-CGAAAGAACCAACGTCAGCG-3′ |
| PX-F9 | 5′-GGTAGCAACGCCGCCAACAC-3′ |
| PX-F10 | 5′-GTTACGGGTTTCACCGTAG-3′ |
| PX-F11 | 5′-GCGCAACTAACAGAACGTCT-3′ |
| PX-F12 | 5′-TTCGGCATTTAACAAAG-3′ |
| PX-A | 5′-CAGTGTACCAGGTGTTATCTT-3′ |
| Px-Lp | 5′-AGCTGATCGATTTTAGCGTT-3′ |
| B3 | 5′-AGCACACCCACCCCGTTTAC-3′ |
| J | 5′-GGTTCAAGATACCTAGAGAC-3′ |
| D2 | 5′-TCTCTATTTATCTGACCGCG-3′ |
| E2 | 5′-TACAGGTTACTTTGTAAGCC-3′ |
| Palindrome 1F | 5′-CCCCTAACTAACCTTTTAGGTGCTTTTCCCC-3′ |
| Palindrome 1R | 5′-GGGGAAAAGCACCTAAAAGGTTAGTTAGGGG-3′ |
| Palindrome 2F | 5′-CCCAATTAACCTTTTAGGTTATAACCC-3′ |
| Palindrome 2R | 5′-GGGTTATAACCTAAAAGGTTAATTGGG-3′ |
Protein Purification
To purify N-terminally histidine-tagged MqsR (His-MqsR) and C-terminally histidine-tagged YgiT (YgiT-His), pET-mqsR-ygiT and pET-ygiT were introduced into E. coli BL21(DE3). Expression of the His-MqsR-YgiT complex and YgiT-His was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h. The His-MqsR-YgiT complex and YgiT-His were purified with Ni-NTA-agarose (Qiagen) following the manufacturer's protocol. Subsequently, the His-MqsR-YgiT complex was denatured with 6 m guanidine HCl. Denatured His-MqsR was then purified with Ni-NTA-agarose, and refolding of His-MqsR was carried out by stepwise dialysis as described previously for MazF (17).
Assay of Protein Synthesis in Vitro
Cell-free protein synthesis was performed with an E. coli T7 S30 extract system for circular DNA (Promega). The reaction mixture was prepared following the manufacturer's protocol. Different amounts of His-MqsR and YgiT-His were added in a final volume of 29 μl. The reaction was started by the addition of pET11a-mazG plasmid DNA (19, 39), and the mixture was incubated for 1 h at 37 °C. Proteins were precipitated with acetone and analyzed by 15% SDS-PAGE. The dried gel was analyzed by autoradiography.
mRNA Interferase Activity of MqsR
MS2 phage RNA (Roche Applied Science) was incubated with His-MqsR in 10 mm Tris-HCl (pH 8.0) containing 1 mm dithiothreitol at 37 °C for 10 min. To examine the antitoxin function of YgiT, His-MqsR was preincubated with YgiT-His for 10 min on ice and then further incubated with MS2 RNA for 10 min. After denaturation in urea, the products were separated on 1.2% agarose gel in 0.5× buffer containing 44.5 mm Tris borate and 1 mm EDTA (40).
Primer Extension Analysis in Vitro
MS2 RNA was incubated with or without purified His-MqsR in 10 mm Tris-HCl (pH 8.0) containing 1 mm dithiothreitol at 37 °C for 15 min, and the digested MS2 RNA (0.8 μg) was used for primer extension as described above.
Electrophoretic Mobility Shift Assays
Complementary strands (see Table 1) were annealed and purified to obtain palindrome 1 and 2 double-stranded DNAs, respectively. The double-stranded DNA fragments were end-labeled with [γ-32P]ATP by T4 kinase. The binding reactions were carried out at 4 °C for 30 min in 50 mm Tris-HCl (pH 7.2) containing 50 mm KCl, 5% glycerol, 100 ng of poly(dI-dC), labeled DNA fragment, and purified proteins. Electrophoresis was performed at 4 °C in 10 mm Tris-HCl and 1 mm EDTA (pH 7.2) at 110 V in 5% acrylamide/bisacrylamide (40:1.2) gel. After electrophoresis, the gel was dried and analyzed by autoradiography (41).
Reverse Transcription-PCR
Total RNA from E. coli was extracted at exponential phase (A600 = 0.8) as described above and treated with 100 units of RNase-free DNase I (Promega) in the presence of 0.5 μl (20 units) of RNase inhibitor (Roche Applied Science). The reverse transcription reaction was carried out at 47 °C for 1 h using total RNA (20 μg) and primer YT-Rv (20 pmol) with 10 units of avian myeloblastosis virus reverse transcriptase. PCR was carried out using the synthesized cDNA as template with primers RT-Fw and RT-Rv (see Table 1).
RESULTS
mqsR and ygiT Genes Are in an Operon
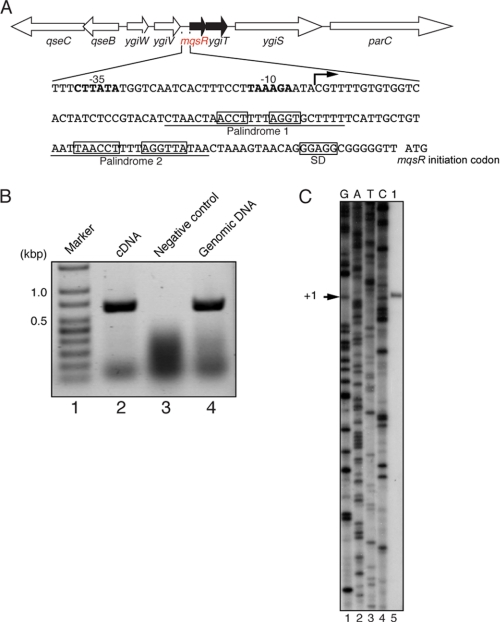
The location of the mqsR-ygiT operon at 68 min on the E. coli K12 chromosome is shown in Fig. 1A. MqsR is a 98-residue protein, and there is a predicted Shine-Dalgarno sequence (GGAGG) 8 bases upstream of the initiation codon for its open reading frame (boxed in Fig. 1A). The downstream YgiT is a 131-residue protein, and the initiation codon of ygiT is 1 base downstream of the translation stop codon of mqsR. To determine whether mqsR-ygiT is transcribed as an operon, reverse transcription-PCR was carried out using total RNA extracted from E. coli BL21(DE3). The cDNA was synthesized from total RNA using primer YT-Rv (Table 1), which is located 31 bp upstream of the ygiT stop codon, as described under “Experimental Procedures.” As shown in Fig. 1B (lane 2), the band was detected at the position of ∼600 bp by PCR using primers RT-Fw and RT-Rv (Table 1). When the E. coli genomic DNA was used as template for PCR with the same primers, the expected 576-bp band was detected (Fig. 1B, lane 3). This band was not detected in the reaction carried out without the addition of reverse transcriptase (Fig. 1B, lane 2). These results demonstrate that the mqsR gene is co-transcribed with the downstream gene, ygiT. To identify the transcription initiation site, we performed primer extension using the same RNA described above with primer PX-RT. This primer is located 2 bp downstream of the initiation codon of the mqsR gene. As shown in Fig. 1C, the transcription start site is located 109 bp upstream of the mqsR start codon, as indicated by the arrow. Thus, we identified the −10 region and the −35 region, a typical RNA polymerase promoter, in the upstream region of the transcription initiation site, as shown in Fig. 1A. It is important to note that no transcription start sites were detected in the region between mqsR and ygiT (data not shown), indicating that there is no independent transcriptional unit for the ygiT gene. It is interesting to note that there are two palindromic sequences in the 109-base 5′-untranslated region (5′-UTR) as indicated by boxes in Fig. 1A.
FIGURE 1.
Gene map of the mqsR-ygiT operon on the E. coli chromosome. A, arrows indicate the direction and size of the following genes: qseC, qseB, ygiW, ygiV, mqsR, ygiT, ygiS, and parC. The mqsR-ygiT promoter sequence is also shown, and the palindromic sequences (1 and 2) are boxed. The bent arrow represents the transcription initiation site of the mqsR-ygiT operon. The −10 and −35 regions of the mqsR-ygiT promoter are shown in boldface, and the Shine-Dalgarno (SD) sequence (GGAGG) is boxed. Underlined DNA sequences were used in electrophoretic mobility shift assay as shown in Fig. 5. B, shown are the results of reverse transcription-PCR analysis of the mqsR-ygiT operon. cDNA was synthesized with reverse transcriptase using total RNA from the E. coli BL21 strain grown at 37 °C to A600 = 0.8. Using the cDNA product as template, PCR was carried out with primers RT-Fw and RT-Rv. Lane 1, 100-bp DNA ladder (GenScript); lanes 2 and 4, cDNA and genomic DNA used as template for PCR, respectively; lane 3, PCR products without using reverse transcriptase. C, shown is the transcription start site of mqsR-ygiT. Primer extension analysis was carried out using the same RNA described for B and primer PX-RT. G, A, T, and C (lanes 1–4) comprise the sequence ladders using pCR®2.1-Topo®-mqsR-ygiT and the same primer. The transcription start site is indicated (+1).
Effect of MqsR on Cell Growth
The mqsR and ygiT genes were cloned into an isopropyl 1-thio-β-d-galactopyranoside-inducible pET28a plasmid (Novagen) and an arabinose-inducible pBAD24 plasmid (42), respectively. E. coli C43 cells harboring pET-mqsR and pBAD-ygiT could not form colonies on M9-glycerol-casamino acid-agar plates in the presence of arabinose (0.2%) (Fig. 2A). However, co-induction of YgiT in the presence of 0.2% arabinose neutralized the toxicity of MqsR, leading to the formation of colonies, indicating that MqsR is the toxin, whereas YgiT is the antitoxin for MqsR. We also examined the toxicity of MqsR in a liquid culture (Fig. 2B). When MqsR was induced by the addition of arabinose (0.2%), cell growth was completely inhibited after 30 min.
Next, we examined the effect of MqsR induction on protein synthesis as measured by [35S]methionine incorporation. Within 5 min of MqsR induction, protein synthesis was almost completely inhibited (Fig. 2C). These samples were analyzed by SDS-PAGE (Fig. 2D). Consistent with the results in Fig. 2C, MqsR completely blocked the incorporation of [35S]methionine into cellular proteins. The strong band present at the 2-min time point (indicated by the arrow) with an apparent molecular mass of 12 kDa is likely MqsR (11,232 Da). These results show that MqsR is a general inhibitor for the synthesis of all cellular proteins. Indeed, the incorporation of [3H]thymidine was not significantly affected upon MqsR induction (Fig. 2E), indicating that MqsR inhibits protein synthesis but not DNA synthesis. When the cellular mRNAs (ompA, ompF, and lpp) of E. coli BL21(DE3) cells carrying pBAD-mqsR were analyzed by Northern blotting at different time points after induction of MqsR by arabinose, the full-length mRNAs were observed only at the 0-time point in all cases (Fig. 2F). At 2 min, the full-size mRNAs were shortened by a certain length, indicating that all mRNAs tested have a preferential initial cleavage site located near the 5′- or 3′-end. The intensity of these bands was significantly reduced after 5 min. These data suggest that MqsR possesses endoribonuclease activity and inhibits protein synthesis through the cleavage of mRNA. It is important to note that 16 S and 23 S rRNAs were very stable in vivo even 10 min after mqsR induction, as no significant changes in their band intensities were observed (Fig. 2F). This was similar to the result seen with MazF mRNA interferase (32). The rRNAs appear to be protected from MqsR cleavage by the ribosomal proteins.
In Vivo Cleavage of the ompA, ompF, and lpp mRNAs by MqsR
Next, we examined the MqsR-mediated cleavage of the ompA, ompF, and lpp mRNAs by primer extension experiments. Primer extension analysis of ompA, ompF, and lpp using different primers identified distinct bands that appeared 2 min after induction of mqsR, corresponding to the specific cleavage sites in each mRNA (Fig. 3, A–D; and Table 2). Notably, these bands were not detected at 0 min. From the alignment of all cleavage sequences, the cleavage occurred before or after the G residue in the GCU sequences, indicating that MqsR cleaves mRNAs at the specific sequence GCU in vivo. It should be noted that all of the GCU sequences in the ompF mRNA were cleaved after mqsR induction without exception (Table 2).
TABLE 2.
Cleavage sites of MqsR in vivo and in vitro
|
In vivo |
In vitro |
|||
|---|---|---|---|---|
| ompA | ompF | lpp | MS2RNA | |
| ACA G↓CUAU | CCU G↓CU CU | AAA↓G↓CUAC | GAC↓G↓CUAG | |
| ATC G↓CGAU | CCU G↓CU CU | CUC↓G↓CUGC | ||
| CUG G↓CUGG | AAC↓G↓CU GC | AGC↓G↓CUAC | ||
| UUC G↓CUAC | GGC G↓CU GA | UUC↓G↓CUAA | ||
| UAC G↓CU GA | UUC↓G↓CUAC | |||
| GUU G↓CU AC | AGA↓G CUUC | |||
| UUC G↓CU UC | ||||
| UCA G↓CU AC | ||||
| AAG G↓CU UU | ||||
| GGU G↓CU UA | ||||
| GCA↓G↓CU GA | ||||
| GAA↓G CU CA | ||||
| AAA↓G CU GA | ||||
| UGG↓G CU AC | ||||
| AUC↓G↓CU UA | ||||
| AAC↓G CU AC | ||||
| GCG↓G CU UC | ||||
| Weak cleavage | AGC↓G CA AU | UGG G↓CGCG | ||
| CUG↓G↓CA GU | ||||
| GUA G↓CA GG | ||||
| AUG G↓CC UG | ||||
| CGA G↓CG AG | ||||
| UUG↓G CA AC | ||||
| AAA G↓CG AA | ||||
mRNA Interferase Activity of MqsR in Vitro
To obtain purified MqsR, His-MqsR was first expressed as the His-MqsR-YgiT complex from the E. coli BL21(DE3) cells harboring pET-mqsR-ygiT, and the complex was purified with Ni-NTA-agarose. The purified His-MqsR-YgiT complex was then denatured using 6 m guanidine HCl. Denatured His-MqsR was retrapped on Ni-NTA-agarose, eluted, and refolded by stepwise dialysis (17). YgiT-His was expressed in E. coli and purified as described under “Experimental Procedures.” The molecular masses of purified His-MqsR, YgiT-His, and His-MqsR-YgiT complex were determined to be 26, 32, and 90 kDa by gel filtration, respectively (data not shown). The results suggest that both MqsR and YgiT exist as dimers and that the MqsR-YgiT complex likely consists of two MqsR dimers and one YgiT dimer, which is also the case for the MazE-MazF complex (17).
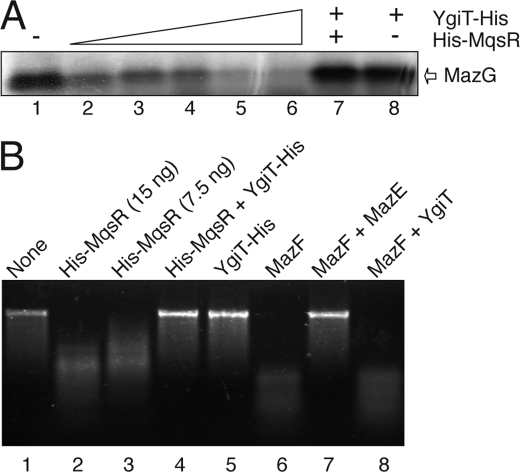
We next examined the effect of His-MqsR and His-MqsR-YgiT on cell-free protein synthesis using an E. coli T7 S30 extract system. The synthesis of MazG protein was almost completely inhibited by 40 nm or higher concentration of MqsR (Fig. 4A, lanes 5 and 6). Inhibition of protein synthesis in vitro was observed in the case of MazF (32) and YoeB (19). YgiT-His and the His-MqsR-YgiT complex did not inhibit protein synthesis (Fig. 4A, lanes 7 and 8).
FIGURE 4.
mRNA interferase activity of MqsR in vitro. A, effect of His-MqsR on protein synthesis in a cell-free system. MazG protein synthesis was carried out using an E. coli T7 S30 extract system for circular DNA with pET11a-mazG. Lane 1, without His-MqsR; lanes 2–6, 5, 10, 20, 40, and 80 nm His-MqsR, respectively; lane 7, 80 nm His-MqsR plus 40 nm YgiT-His; lane 8, 40 nm YgiT-His. B, mRNA interferase activity of purified His-MqsR in vitro. MS2 phage RNA (0.8 μg) was incubated with His-MqsR at 37 °C for 10 min in 10 mm Tris-HCl (pH 8.0) containing 1 mm dithiothreitol. The products were separated on a 1.2% agarose gel. The gel was stained with ethidium bromide.
To further prove that the in vivo cleavage of ompA, ompF, and lpp mRNAs observed above was due to the mRNA interferase activity of MqsR, MS2 phage RNA (3569 bases) was cleaved with purified His-MqsR in vitro. The purified MqsR preparation clearly showed endoribonuclease activity (Fig. 4B, lanes 2 and 3). The endoribonuclease activity was completely inhibited when purified YgiT-His was preincubated with His-MqsR (Fig. 4B, lane 4). It should be noted that purified YgiT-His by itself had no detectable effect on the mRNA (Fig. 4B, lane 5). The results confirm that YgiT functions as an antitoxin and blocks the MqsR mRNA interferase activity. To confirm that YgiT is the specific inhibitor for MqsR, we examined if YgiT inhibits MazF, which cleaves mRNA at ACA sequences. MazF cleaved MS2 RNA (Fig. 4B, lane 6), and its activity was completely inhibited when it was preincubated with purified MazE, the antidote for MazF (lane 7). However, when MazF was incubated with purified YgiT-His, its activity was not inhibited (Fig. 4B, lane 8). This result shows that YgiT specifically inhibits MqsR endoribonuclease activity.
It is important to note that the ability of MqsR to cleave RNA in the absence of ribosomes is distinctly different from that of RelE and YoeB, whose mRNA interferase activities are dependent on ribosomes (13, 19, 43). The activity of MqsR activity was inhibited by MgCl2 (data not shown), as described previously for MazF (32).
Site of In Vitro Cleavage of MS2 RNA by Purified MqsR
The in vitro MqsR activity on MS2 RNA was also analyzed by primer extension. The MS2 RNA was incubated at 37 °C for 10 min with MqsR. The product was used as template for primer extension. MqsR cleaved the MS2 RNA at five cleavage sites, and the sequences of all of the cleaved sites were determined to be GCU (Table 2). Taken together, the results of the in vivo and in vitro primer extension experiments (Fig. 3 and Table 2) indicate that MqsR is an mRNA interferase that specifically cleaves RNA at GCU sequences.
Binding of the MqsR-YgiT Complex to the mqsR-ygiT Promoter Region
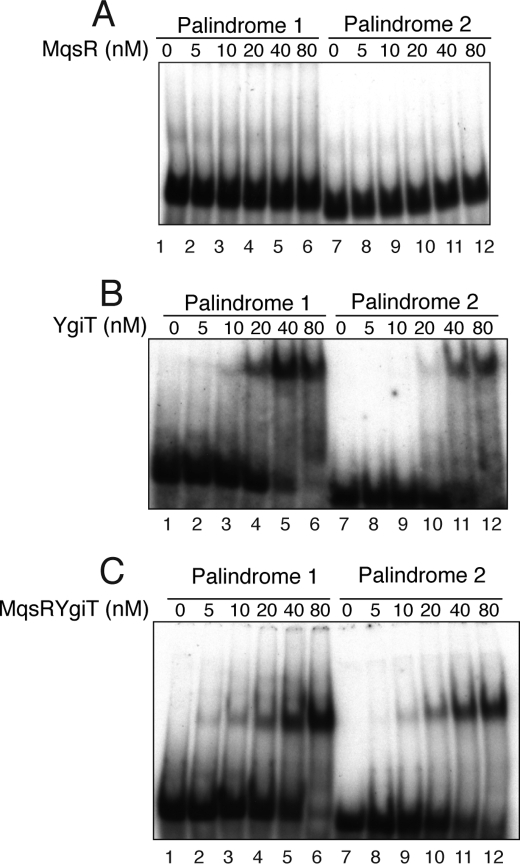
There are palindromic sequences in the promoter regions of many other TA systems, including ccdAB (44, 45), parDE (46), mazEF (47), and relBE (48). These antitoxins or TA complexes bind to their cognate palindromic sequence to negatively regulate their own operons. Because there are two palindromic sequences in the 5′-UTR of the mqsR-ygiT operon (Fig. 1A), we next examined if the MqsR-YgiT complex is able to bind them. Palindrome 1 and 2 DNA fragments were prepared as described under “Experimental Procedures” and labeled with [γ-32P]ATP by T4 kinase. YgiT and the MqsR-YgiT complex were mixed with labeled DNA to test their ability to bind the palindromic sequences. YgiT was able to shift the mobility of palindrome 1 and 2 fragments at 10 and 20 nm or higher concentrations (Fig. 5A, lanes 3–6 and 10–12, respectively). At 5 nm, no shifted bands were observed with either palindrome 1 or 2 fragments. Notably, the His-MqsR protein alone could not bind to either palindromic sequence, even at 80 nm (Fig. 5A). However, the addition of MqsR to YgiT enhanced YgiT binding to both palindromic sequences. MqsR was added to YgiT at a molar ratio of 2:1. The complex bound more strongly to both palindromic sequences compared with YgiT alone (Fig. 5C, lanes 2–6 and 9–12, respectively). Under these conditions, the positions of the bands representing the palindromic sequences were shifted at 5 and 10 nm MqsR-YgiT complex for the palindrome 1 and 2 fragments, respectively. The result suggests that both YgiT and the MqsR-YgiT complex bind to the palindromic sequences to negatively regulate the mqsR-ygiT operon like other TA systems.
FIGURE 5.
Binding of MqsR, MqsR-YgiT, and YgiT to the palindromic sequences in the mqsR-ygiT 5′-UTR. Electrophoretic mobility shift assay was carried out with 5′-end-labeled palindrome 1 (lanes 1–6) and 2 (lanes 7–12) DNA fragments (see Fig. 1A), which were incubated with different concentrations of proteins as described under “Experimental Procedures.” Lanes 1–6 and 7–12 represent 0, 5, 10, 20, 40, and 80 nm His-MqsR (A), YgiT-His (B), and His-MqsR-YgiT (C), respectively.
DISCUSSION
In this work, we have demonstrated that the mqsR and ygiT genes on the E. coli chromosome are co-transcribed and that MqsR-YgiT is a new TA system. In contrast to most other TA systems, the first gene in the operon encodes the toxin, MqsR, and the second gene encodes the antitoxin, YgiT. Although MqsR has no homology to the well characterized mRNA interferase MazF, which specifically cleaves at ACA sequences in mRNAs (29), MqsR was found to be an mRNA interferase that cleaves mRNAs at GCU sequences. Notably, MqsR is a ribosome-independent mRNA interferase like MazF, which is distinctly different from ribosome-dependent mRNA interferases such as RelE (13, 48), YoeB (19), and HigB (49).
It has been reported that MqsR is induced during biofilm formation (5) and by the addition of the quorum-sensing autoinducer AI-2 (6). In turn, the activation of MqsR activates a two-component system, qseBC, which is known to play an important role in biofilm formation (6). QseC is a sensor histidine kinase, and QseB is a transcription regulator that binds to the 5′-UTR of the qseBC operon and activates transcription of this operon (50, 51). The MqsR-YgiT complex is able to bind two palindromic sequences present in the 5′-UTR of the mqsR-ygiT operon and seems to repress transcription of mqsR-ygiT. We examined the possibility that the MqsR-YgiT complex may also regulate expression of the qseBC operon. However, the His-MqsR-YgiT complex was unable to bind the qseBC promoter region including the QseB-binding site (data not shown). Notably, both palindromic sequences (palindromes 1 and 2) (Fig. 1A) were found to be unique on the E. coli chromosome, as there are no other E. coli genes other than the mqsR-ygiT operon that have either of the two palindromic sequences. It should also be noted that purified QseB did not bind to the 5′-UTR of the mqsR-ygiT operon (data not shown). These results indicate that MqsR is not directly involved in the activation of the qseBC operon.
We analyzed all 4226 open reading frames on the E. coli genome (NCBI RefSeq accession number AC000091.1 for the existence of the GCU sequences and found that there are only 14 open reading frames that do not contain a single GCU sequence (Table 3). Of these 14 genes, six genes, pheL, tnaC, trpL, yciG, ygaQ, and ralR, have been shown to be induced during biofilm formation in E. coli (52). Of special interest is ygaQ (330 bp), which is induced by 32-fold in biofilms and has also been shown to be involved in the swarming mobility of E. coli (53). Because these genes are resistant to MqsR mRNA interferase activity, MqsR induction during biofilm formation may inactivate all E. coli mRNAs except for these 14 genes, which in turn may play an important role in biofilm formation. It is interesting to note that almost all cells die during biofilm formation in Pseudomonas aeruginosa (54). MqsR induction during biofilm formation may cause the cells to enter a quasi-dormant state similar to that caused by MazF (8, 55) and eventually lead to cell death.
TABLE 3.
MqsR resistance genes in the E. coli genome
| Location | Length | Gene | Product | Expected motif counts | Actual motif counts |
|---|---|---|---|---|---|
| bp | |||||
| 339017..339313 | 297 | yahH | Hypothetical protein | 4.64 | 0 |
| 736867..737121 | 255 | ybfQ | Predicted transposase | 3.98 | 0 |
| 795195..795344 | 150 | ybhT | Hypothetical protein | 2.34 | 0 |
| 1068503..1068676 | 174 | ymdF | Hypothetical protein | 2.72 | 0 |
| Complement (1317570..1317749) | 180 | yciG | Hypothetical protein | 2.81 | 0 |
| Complement (1324752..1324796) | 45 | trpL | trp operon leader peptide | 0.70 | 0 |
| Complement (1415447..1415641) | 195 | ralR | Restriction alleviation protein | 3.05 | 0 |
| Complement (1419722..1419943) | 222 | kilR | Inhibitor of FtsZ, killing protein | 3.47 | 0 |
| 2092133..2092183 | 51 | hisL | his operon leader peptide | 0.80 | 0 |
| 2736255..2736302 | 48 | pheL | pheA gene leader peptide | 0.75 | 0 |
| 2785053..2785385 | 333 | ygaQ | Hypothetical protein | 5.20 | 0 |
| Complement (3751906..3751980) | 75 | tnaC | Tryptophanase leader peptide | 1.17 | 0 |
| 4161624..4161824 | 201 | yheV | Hypothetical protein | 3.14 | 0 |
| 4357586..4357759 | 174 | yjdO | Hypothetical protein | 2.72 | 0 |
The identification of the MqsR-YgiT system as a new TA system in E. coli in this work increases the total number of the E. coli TA systems to as many as 16, including MazF-MazE (17, 32), RelE-RelB (13, 14), ChpBK-ChpBI (15), YafQ-DinJ (22), YoeB-YefM (19, 20), HipA-HipB (56, 57), HicA-HicB (23, 24), YhaV-PrlF (25), and YafO-YafN (12). Preliminary results suggest that there may be a few more TA systems in E. coli K12. It is highly intriguing to elucidate how each TA system plays a role in cellular physiology in E. coli under various stress conditions, including biofilm formation and quorum sensing. It remains to be determined if the functions of each of these TA systems are coordinated in E. coli by a network, raising the possibility of the existence of a TA network.
Acknowledgments
We thank for Dr. Sangita Phadtare and Jared Sharp for critical reading of the manuscript. We are also grateful to Drs. H. Nariya and Y. Zhang for critical discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant 1RO1 GM081567. This work was also supported by Takara Bio Inc.
- TA
- toxin-antitoxin
- Ni-NTA
- nickel-nitrilotriacetic acid
- 5′-UTR
- 5′-untranslated region.
REFERENCES
- 1.Balestrino D., Haagensen J. A., Rich C., Forestier C. (2005) J. Bacteriol. 187, 2870–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P. (1998) Science 280, 295–298 [DOI] [PubMed] [Google Scholar]
- 3.Hammer B. K., Bassler B. L. (2003) Mol. Microbiol. 50, 101–104 [DOI] [PubMed] [Google Scholar]
- 4.McNab R., Ford S. K., El-Sabaeny A., Barbieri B., Cook G. S., Lamont R. J. (2003) J. Bacteriol. 185, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren D., Bedzyk L. A., Thomas S. M., Ye R. W., Wood T. K. (2004) Appl. Microbiol. Biotechnol. 64, 515–524 [DOI] [PubMed] [Google Scholar]
- 6.González Barrios A. F., Zuo R., Hashimoto Y., Yang L., Bentley W. E., Wood T. K. (2006) J. Bacteriol. 188, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey D. P., Gerdes K. (2005) Nucleic Acids Res. 33, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi Y., Inouye M. (2009) Prog. Mol. Biol. Transl. Sci. 85, 467–500 [DOI] [PubMed] [Google Scholar]
- 9.Buts L., Lah J., Dao-Thi M. H., Wyns L., Loris R. (2005) Trends Biochem. Sci. 30, 672–679 [DOI] [PubMed] [Google Scholar]
- 10.Engelberg-Kulka H., Sat B., Reches M., Amitai S., Hazan R. (2004) Trends Microbiol. 12, 66–71 [DOI] [PubMed] [Google Scholar]
- 11.Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 12.Brown J. M., Shaw K. J. (2003) J. Bacteriol. 185, 6600–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., Ehrenberg M. (2003) Cell 112, 131–140 [DOI] [PubMed] [Google Scholar]
- 14.Takagi H., Kakuta Y., Okada T., Yao M., Tanaka I., Kimura M. (2005) Nat. Struct. Mol. Biol. 12, 327–331 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Zhu L., Zhang J., Inouye M. (2005) J. Biol. Chem. 280, 26080–26088 [DOI] [PubMed] [Google Scholar]
- 16.Kamada K., Hanaoka F., Burley S. K. (2003) Mol. Cell 11, 875–884 [DOI] [PubMed] [Google Scholar]
- 17.Zhang J., Zhang Y., Inouye M. (2003) J. Biol. Chem. 278, 32300–32306 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Zhang J., Hara H., Kato I., Inouye M. (2005) J. Biol. Chem. 280, 3143–3150 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Inouye M. (2009) J. Biol. Chem. 284, 6627–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamada K., Hanaoka F. (2005) Mol. Cell 19, 497–509 [DOI] [PubMed] [Google Scholar]
- 21.Motiejūnaite R., Armalyte J., Markuckas A., Suziedeliene E. (2007) FEMS Microbiol. Lett 268, 112–119 [DOI] [PubMed] [Google Scholar]
- 22.Prysak M. H., Mozdzierz C. J., Cook A. M., Zhu L., Zhang Y., Inouye M., Woychik N. A. (2009) Mol. Microbiol. 71, 1071–1087 [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen M. G., Pandey D. P., Jaskolska M., Gerdes K. (2009) J. Bacteriol. 191, 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova K. S., Grishin N. V., Koonin E. V. (2006) Bioinformatics 22, 2581–2584 [DOI] [PubMed] [Google Scholar]
- 25.Schmidt O., Schuenemann V. J., Hand N. J., Silhavy T. J., Martin J., Lupas A. N., Djuranovic S. (2007) J. Mol. Biol 372, 894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Kim Y., Wood T. K. (2009) ISME J., 10.1038/ismej.2009.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahassi E. M., O'Dea M. H., Allali N., Messens J., Gellert M., Couturier M. (1999) J. Biol. Chem. 274, 10936–10944 [DOI] [PubMed] [Google Scholar]
- 28.Kampranis S. C., Howells A. J., Maxwell A. (1999) J. Mol. Biol. 293, 733–744 [DOI] [PubMed] [Google Scholar]
- 29.Christensen S. K., Gerdes K. (2003) Mol. Microbiol. 48, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 30.Hayes C. S., Sauer R. T. (2003) Mol. Cell 12, 903–911 [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Zhang Y., Zhu L., Suzuki M., Inouye M. (2004) J. Biol. Chem. 279, 20678–20684 [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., Zhang J., Hoeflich K. P., Ikura M., Qing G., Inouye M. (2003) Mol. Cell 12, 913–923 [DOI] [PubMed] [Google Scholar]
- 33.Mizuno T., Chou M. Y., Inouye M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 1966–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambros V. (2001) Cell 107, 823–826 [DOI] [PubMed] [Google Scholar]
- 35.Billy E., Brondani V., Zhang H., Müller U., Filipowicz W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puerta-Fernández E., Romero-López C., Barroso-delJesus A., Berzal-Herranz A. (2003) FEMS Microbiol. Rev. 27, 75–97 [DOI] [PubMed] [Google Scholar]
- 37.Sarmientos P., Sylvester J. E., Contente S., Cashel M. (1983) Cell 32, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 38.Baker K. E., Mackie G. A. (2003) Mol. Microbiol. 47, 75–88 [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Inouye M. (2002) J. Bacteriol. 184, 5323–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y. C., Chou Y. C. (1990) BioTechniques 9, 558–560 [PubMed] [Google Scholar]
- 41.Yoshida T., Qin L., Egger L. A., Inouye M. (2006) J. Biol. Chem. 281, 17114–17123 [DOI] [PubMed] [Google Scholar]
- 42.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen S. K., Pedersen K., Hansen F. G., Gerdes K. (2003) J. Mol. Biol. 332, 809–819 [DOI] [PubMed] [Google Scholar]
- 44.Dao-Thi M. H., Charlier D., Loris R., Maes D., Messens J., Wyns L., Backmann J. (2002) J. Biol. Chem. 277, 3733–3742 [DOI] [PubMed] [Google Scholar]
- 45.Madl T., Van Melderen L., Mine N., Respondek M., Oberer M., Keller W., Khatai L., Zangger K. (2006) J. Mol. Biol. 364, 170–185 [DOI] [PubMed] [Google Scholar]
- 46.Oberer M., Zangger K., Gruber K., Keller W. (2007) Protein Sci. 16, 1676–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marianovsky I., Aizenman E., Engelberg-Kulka H., Glaser G. (2001) J. Biol. Chem. 276, 5975–5984 [DOI] [PubMed] [Google Scholar]
- 48.Li G. Y., Zhang Y., Inouye M., Ikura M. (2008) J. Mol. Biol. 380, 107–119 [DOI] [PubMed] [Google Scholar]
- 49.Hurley J. M., Woychik N. A. (2009) J. Biol. Chem. 284, 18605–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clarke M. B., Sperandio V. (2005) Mol. Microbiol. 58, 441–455 [DOI] [PubMed] [Google Scholar]
- 51.Sperandio V., Torres A. G., Kaper J. B. (2002) Mol. Microbiol. 43, 809–821 [DOI] [PubMed] [Google Scholar]
- 52.Domka J., Lee J., Bansal T., Wood T. K. (2007) Environ. Microbiol. 9, 332–346 [DOI] [PubMed] [Google Scholar]
- 53.Inoue T., Shingaki R., Hirose S., Waki K., Mori H., Fukui K. (2007) J. Bacteriol. 189, 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb J. S., Thompson L. S., James S., Charlton T., Tolker-Nielsen T., Koch B., Givskov M., Kjelleberg S. (2003) J. Bacteriol. 185, 4585–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inouye M. (2006) J. Cell. Physiol. 209, 670–676 [DOI] [PubMed] [Google Scholar]
- 56.Keren I., Shah D., Spoering A., Kaldalu N., Lewis K. (2004) J. Bacteriol. 186, 8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korch S. B., Henderson T. A., Hill T. M. (2003) Mol. Microbiol. 50, 1199–1213 [DOI] [PubMed] [Google Scholar]