Abstract
CCAAT/enhancer-binding Protein β (C/EBPβ) is a member of the bZIP transcription factor family that is expressed in various tissues, including cells of the hematopoietic system. C/EBPβ is involved in tissue-specific gene expression and thereby takes part in fundamental cellular processes such as proliferation and differentiation. Here, we show that the activity of C/EBPβ is negatively regulated by the transcriptional co-repressor Daxx. C/EBPβ was found to directly interact with Daxx after overexpression as well as on the endogenous level. Glutathione S-transferase pulldown assays showed that Daxx binds via amino acids 190–400 to the C-terminal part of C/EBPβ. Co-expression of C/EBPβ changed the sub-nuclear Daxx distribution pattern from predominantly POD-localized to nucleoplasmic. Daxx suppressed basal and p300-enhanced transcriptional activity of C/EBPβ. Furthermore, Daxx decreased the C/EBPβ-dependent phosphorylation of p300, which in turn was associated with a diminished level of p300-mediated C/EBPβ acetylation. Co-expression of promyelocytic leukemia protein abrogated the repressive effect of Daxx on C/EBPβ as well as the direct interaction of Daxx and C/EBPβ, presumably by re-recruiting Daxx to PML-oncogenic domains. In acute promyelocytic leukemia (APL) cells, C/EBPβ activity is known to be required for all-trans-retinoic acid-induced cell differentiation and disease remission. We show that all-trans-retinoic acid as well as arsenic trioxide treatment leads to a reduced C/EBPβ fraction associated with Daxx suggesting a relief of Daxx-dependent C/EBPβ repression as an important molecular event leading to APL cell differentiation. Overall, our data identify Daxx as a new negative regulator of C/EBPβ and provide first clues for a link between abrogation of Daxx-C/EBPβ complex formation and APL remission.
CCAAT/enhancer-binding protein β (C/EBPβ)2 is a member of the C/EBP family, which is composed of a least six different proteins (C/EBPα, -β, -δ, -γ, -ϵ, and -ζ). Common to all C/EBP proteins is the highly conserved C-terminal basic region leucine zipper DNA binding domain, which also mediates a large number of protein-protein interactions (1). In addition, C/EBPβ contains an N-terminal transactivation domain and a central regulatory motif, which has an autoinhibitory function (2). C/EBPβ is expressed in various different cell types, including hepatic cells, fat cells, and cells of the hematopoietic system such as granulocytes and macrophages (1). Due to alternative translation initiation C/EBPβ is present in three isoforms referred to as LAP1 (full-length), LAP2 (residues 22–296), and the inhibitory LIP form (151–296) (3). By regulating tissue-specific gene expression C/EBPβ is implicated in essential cellular processes like differentiation and proliferation and also exerts crucial functions during tumorigenesis (4, 5). In the latter context, Duprez et al. recently demonstrated that C/EBPβ activity is required during all-trans-retinoic acid- (ATRA)-induced differentiation of APL cells (6). Transcriptional activity of C/EBPβ was shown to involve the recruitment of co-regulatory components leading to post-translational modifications of C/EBPβ such as phosphorylation, sumoylation, and acetylation (7–11). For instance, p300, CBP and GCN5 directly bind to C/EBPβ and via their intrinsic acetyltransferase activity trigger C/EBPβ acetylation leading to an increase in transcriptional C/EBPβ activity (12–14). Interestingly, C/EBPβ reciprocally enhances the co-activator capability of p300 by supporting the phosphorylation of p300 thus indicating a complex interdependent regulation of both proteins (15). Deacetylation of C/EBPβ is controlled by the histone deacetylase HDAC1, which results in repression of transcriptional activity (9). Additional to the basic function of transcription factors to support RNA synthesis of a given gene, we previously showed that C/EBPβ is also involved in the very early steps of transcription initiation by triggering chromatin opening (16).
Daxx was initially identified as a pro-apoptotic protein that binds to the death domain of the CD95 death receptor (17). By activating the c-Jun NH2-terminal kinase-pathway Daxx was shown to enhance CD95-mediated as well as transforming growth factor-β-dependent apoptosis (17, 18). Interestingly, Daxx down-regulation by RNA interference is also associated with an increased level of apoptosis (19). Moreover, targeted disruption of the murine Daxx gene results in embryonic lethality due to extensive global apoptosis, suggesting Daxx acts in a rather anti-apoptotic manner (20). Besides its controversial role during apoptosis, Daxx is a well established regulator of transcription. Daxx binds to the transcriptional co-regulators CBP and HDAC as well as to numerous transcription factors, including members of the Pax and p53 families, ETS1, glucocorticoid, and androgen receptor (21–29). In most cases, Daxx serves as a transcriptional repressor presumably through recruitment of HDAC proteins as demonstrated for the repression of p53, the co-activator CBP, and the impact of Daxx on the expression of c-met (22, 23, 29). Nevertheless, Daxx was also reported to act in an HDAC-independent manner and even to trigger the activity of certain transcription factors such as HSF1 and Pax5, thus indicating Daxx does not act exclusively as a transcriptional repressor (30–32). Moreover, beyond affecting transcription by binding to transcription factors recent findings point toward a role of Daxx in modulation of chromatin remodeling and DNA methylation, which indicates that Daxx may control gene expression also via epigenetic mechanisms (33–35). Consistent with the involvement in transcriptional regulation Daxx is predominantly a nuclear protein. Here, it mainly localizes to sub-nuclear structures called PML-oncogenic domains (PODs) by binding to SUMO-modified PML (36). According to this, recently a SUMO-interacting motif (SIM) within the C-terminal tail of Daxx was identified that is responsible for the association with PML and several other transcription factors which also require sumoylation for Daxx binding (37). Several lines of evidence indicate that the repressor activity of Daxx is controlled by subnuclear compartmentalization. For instance, PML expression relieves the repressive effect of Daxx on Pax3 and glucocorticoid receptor-dependent transcription by sequestering Daxx to PODs (24, 27). In a similar way MSP58 and ASK1 were shown to inhibit the Daxx-mediated transcriptional repression by recruiting Daxx to the nucleolus and the cytoplasm, respectively (38, 39). Therefore it might be reasonable to assume that nucleoplasmic Daxx resembles the active fraction of the protein while localization to other compartments could be regarded as “out of action Daxx,” at least with respect to transcriptional regulation. Consistent with this idea, the constitutive repressor activity of Daxx that was proposed to be involved in the formation of acute promyelocytic leukemia (APL) was shown to be related to its aberrant localization pattern (25, 40). In APL cells POD formation is disrupted due to the expression of the oncogenic PML-RARα fusion protein leading to de-localization of POD-associated proteins such as Daxx (25, 41, 42). Interestingly, treatment with ATRA or As2O3 both of which induce differentiation and cause disease remission results in re-organization of POD structure and a PML-dependent recruitment of Daxx to PODs (25). This in turn is again associated with a relief of Daxx-dependent transcriptional repression suggesting that Daxx (like C/EBPβ, as mentioned above) is involved in the pathology of APL (25).
In the present study we report on the identification of Daxx as a new negative regulator of the transcription factor C/EBPβ. We show that Daxx binds to C/EBPβ and represses the p300 induced acetylation and transcriptional activity of C/EBPβ. The interaction of C/EBPβ and Daxx is accompanied by a localization change of Daxx leading to its release from PODs. Accordingly, the association of Daxx and C/EBPβ is inhibited by PML expression resulting in the de-repression of C/EBPβ-dependent transcription. Finally, we show that, upon differentiation of APL cells, the complex between C/EBPβ and Daxx is released. This in turn suggests that the relief of Daxx-dependent C/EBPβ repression is an important contribution to the C/EBPβ activation, which was recently shown to be required for terminal APL differentiation.
EXPERIMENTAL PROCEDURES
Expression Constructs
To generate the GFP-Daxx expression construct the coding sequence of human Daxx was amplified from cDNA derived from HeLa cells using specific forward (5′-act-tcc-tcc-gtc-gac-ggg-att-gga-tcc-c-3′) and reverse primers (5′-tcc-ggt-gga-tcg-atg-cag-cta-atc-ag-3′) that contain SalII and ClaI restriction sites, respectively. The PCR product was then cloned into pLEGFP-C1 (Clontech) to obtain pLEGFP-Daxx. To create a HA-Daxx expression vector, a double-stranded DNA-cassette coding for the HA tag was generated by annealing the complementary oligonucleotides HA-I (5′- gat-cta-ccg-gtc-gcc-acc-atg-gct-tac-cca-tac-gat-gtt-cca-gat- tac-gcg-g-3′) and HA-II (5′-tcg-acc-gcg-taa-tct-gga-aca-tcg- tat-ggg-taa-gcc-atg-gtg-gcg-acc-ggt-a-3′), which contains an internal AgeI site and is flanked by XhoI and SalI “sticky ends.” The cassette was cloned into the pLEGFP-Daxx vector via the XhoI and SalI restriction sites. The DNA sequence coding for GFP was then excised off the resulting construct by AgeI cleavage and subsequent ligation resulted in pLHA-Daxx, coding for HA-tagged Daxx. The expression constructs coding for full-length p300 (pCMV-p300CHA) was kindly provided by R. Eckner. Expression vectors for truncated p300 (p300/1751–2370), FLAG-tagged chicken C/EBPβ and the reporter construct p240-Luc coding for firefly luciferase driven by the C/EBPβ-responsive mim-1 promoter have been described (13). pEBFP-PML coding for human PML fused to blue fluorescent protein (BFP) was a generous gift from P. P. Pandolfi (43) and the GST-Daxx deletion constructs were kindly provided by T. G. Hofmann (44). Expression vectors coding for different GST-C/EBPβ fusion proteins were generated by cloning the full-length or different N- or C-terminally deleted parts of the C/EBPβ coding region between the BamHI and EcoRI sites of expression vector pGex-3X. The respective parts of the coding region were amplified by PCR using appropriately designed primers, and the final constructs were verified by sequencing. The β-galactosidase expression vector pCMVβ was obtained from Clontech.
Cell Culture and Transient Transfections
The quail fibroblast cell line QT6 and HeLa cells were cultured in Iscove's modified Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. HL-60 and NB4 cells were grown in RPMI medium supplemented with 10% fetal calf serum. For drug treatment of NB4 cells, the growth medium was supplemented with 1 μm of all-trans-retinoic acid (ATRA, 10 mm stock dissolved in ethanol) and 1 μm of As2O3 (10 mm stock dissolved in 1 m NaOH) for the indicated time periods. Transfection of QT6 cells plated in 10-cm dishes was carried out by calcium phosphate co-precipitation, as described previously (13). HeLa cells were transfected using Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. Total DNA amounts transfected were kept constant by adding required amounts of the respective empty vector.
RNA Extraction and Northern Blot Analysis
Preparation of poly(A) RNA and Northern blotting was performed as described before (13).
Immunoprecipitation and Western Blot Analysis
For immunoprecipitation cells were lysed in ELB buffer (50 mm Tris/HCl, pH 7.5; 120 mm NaCl; 20 mm NaF; 1 mm benzamidine; 1 mm EDTA; 6 mm EGTA; 15 mm sodium pyrophosphate; 1 mm phenylmethylsulfonyl fluoride; 0.5% Nonidet P-40). After incubation on ice for 30 min, lysates were centrifuged at 14,000 × g for 30 min, and the supernatant was used as total protein extract. Immunoprecipitations were carried out using aliquots of the total protein extract supplemented with the appropriate antibodies. After 1 h of incubation at 4 °C protein-A-Sepharose beads were added and incubated further for 12 h at 4 °C under constant agitation. Immune complexes were then collected by centrifugation, washed three times with lysis buffer, and finally subjected to SDS-PAGE. Immunostaining of proteins transferred to nitrocellulose membranes was performed with the following antibodies: anti-FLAG (M2, Sigma-Aldrich), anti-HA (HA.11, Hiss Diagnostics), anti-Daxx (Novocastra Laboratories, Newcastle, NCL-Daxx), anti-C/EBPβ (13) or H7 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-GFP (Roche Applied Science), anti-p300 (UBI), anti-acetylated lysine (Ack-103, Cell Signaling Technology), and anti-β-actin (Sigma-Aldrich).
Luciferase and β-Galactosidase Reporter Assays
QT6 cells were plated in 10-cm dishes and transfected with 3 μg of the p240-Luc reporter construct together with the indicated amounts of expression vectors. In all transfections 0.5 μg of pCMVβ was included to control the transfection efficiency using β-galactosidase assays. 24 h post transfection cells were harvested, lysed in ELB buffer, and crude protein extracts were prepared as mentioned above. Luciferase and β-galactosidase assays were performed as described (13).
GST Pulldown Assay
GST fusion protein expression was induced in logarithmically growing cultures of transformed Escherichia coli BL21-pLysS bacteria by adding isopropyl-d-thiogalactopyranoside to a final concentration of 0.5 mm. After additional 3 h of growth at 37 °C the bacteria expressing GST or GST-C/EBPβ fusion proteins were harvested by centrifugation for 10 min at 5,000 × g. Bacteria expressing GST-Daxx proteins were induced and cultured at 18 °C overnight in the presence of 2% ethanol before harvesting. Bacterial pellets were resuspended in GST lysis buffer (50 mm Tris-HCl, pH 8,0; 150 mm NaCl; 1% Triton X-100; 1 mm dithiothreitol; 0.1 mm phenylmethylsulfonyl fluoride) and lysed by three freeze-thaw cycles and sonification. An extract of soluble protein was prepared by ultracentrifugation for 1 h at 100,000 × g. Extracts containing 5–10 μg of GST fusion protein were then mixed with 30 μl of glutathione-Sepharose and incubated at 4 °C for 1 h. The Sepharose beads were then washed three times with ELB buffer and used for GST pull-down assays as follows: QT6 cells transfected with the appropriate expression vectors were lysed in ELB buffer and aliquots of the lysate were then incubated under constant agitation for 1 h at 4 °C with bacterially expressed GST fusion protein coupled to glutathione-Sepharose. Subsequently, beads were washed three times with ELB buffer. Then bound proteins were eluted from the beads by boiling in SDS sample buffer and analyzed by SDS-PAGE and Coomassie Blue staining or Western blotting using appropriate antibodies.
Fluorescence Microscopy and Immunostaining
HeLa cells were seeded on coverslips and transfected with the desired plasmids. 24 h later, cells were washed with PBS and fixed with 2% paraformaldehyde in PBS for 10 min at room temperature. The cells were then permeabilized with PBST (PBS containing 0.1% Triton X-100) and incubated with blocking buffer (PBST containing 5% bovine serum albumin). Primary antibodies were diluted in blocking buffer (anti-FLAG antibody, 1:3000 (Sigma); anti-PML antibody, 1:50 (Santa Cruz Biotechnology) and incubated for 1 h at room temperature. Subsequently, cells were washed five times with PBST followed by incubation with TRITC-coupled goat-anti mouse secondary antibody (Sigma-Aldrich) diluted 1:50 (detection of endogenous protein) or 1:3000 (detection of overexpressed protein) in blocking buffer for 1 h at room temperature in the dark. Finally, cells were washed five times with PBS, mounted in Aqua Poly/Mount (Polysciences), and analyzed by confocal laser scanning microscopy.
RESULTS
C/EBPβ Interacts with Daxx in Vivo
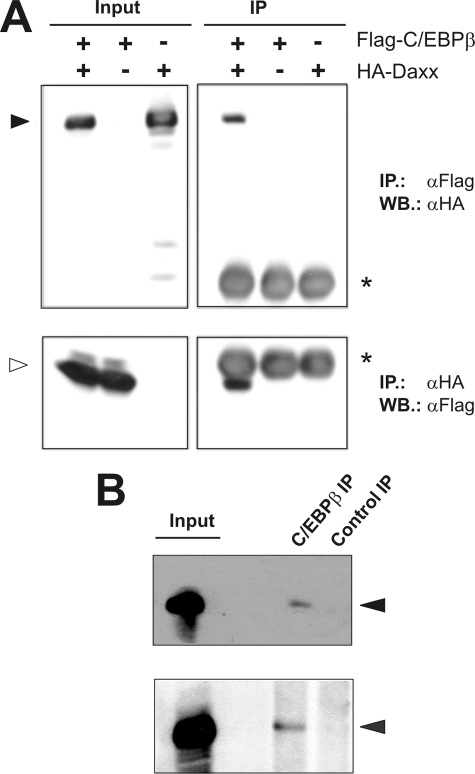
Regulation of transcription involves a complex interplay of transcription factors, transcriptional co-activators as well as co-repressors. Co-regulatory factors such as p300, CBP, or GCN5 have been extensively shown to enhance C/EBPβ-mediated transcription (12–14). On the other hand, factors that negatively influence C/EBPβ activity have been rarely detected. By performing co-immunoprecipitation experiments, we identified the transcriptional co-repressor Daxx as a C/EBPβ-interacting protein. As shown in Fig. 1A, after co-expression of HA-Daxx and FLAG-C/EBPβ, precipitation of Daxx via anti-HA antibody lead to the co-precipitation of FLAG-C/EBPβ. Similarly, Daxx could be co-precipitated together with C/EBPβ via anti-FLAG antibody. Single transfections of vectors encoding HA-Daxx or FLAG-C/EBPβ, respectively, served as controls and revealed no binding of the proteins to the reciprocal antibody. Next we checked whether the physical interaction of Daxx and C/EBPβ also occurs on the endogenous level. To this end, protein extracts derived from HL60 as well as NB4 cells were subjected to immunoprecipitation using an anti-C/EBPβ antibody. Indeed, in both cell extracts Daxx could be co-precipitated, whereas precipitation with an unrelated antibody failed to do so (Fig. 1B). These results demonstrated that C/EBPβ and Daxx interact as endogenous proteins. We also analyzed the subcellular localization of endogenous Daxx and C/EBPβ by immunofluorescence. Both proteins co-localized in the nucleoplasm. Daxx, additionally, showed a prominent association with in PODs (supplemental Fig. S1).
FIGURE 1.
In vivo interaction of C/EBPβ and Daxx. A, QT6 cells were transfected with the indicated combinations of plasmids encoding FLAG-C/EBPβ (5 μg) and HA-Daxx (5 μg). Cells were lysed after 24 h, and protein extracts were immunoprecipitated with anti-FLAG or anti-HA antibodies, followed by SDS-PAGE and Western blotting (right panels). Analyses of the crude protein extracts (input) demonstrate comparable expression levels of the proteins in the different samples. HA-Daxx and FLAG-C/EBPβ are marked by black and white arrowheads, respectively. The asterisks marks the immunoglobulin heavy chain of the FLAG antibody. B, protein extracts of HL-60 cells (upper panel) and NB4 cells (lower panel) were used for immunoprecipitation with an antibody against endogenous C/EBPβ (13) or with an unrelated antibody as control. Crude protein extracts (input) and precipitated proteins were analyzed by SDS-PAGE followed by Western blotting using a Daxx-specific antibody. Daxx is marked by a black arrowhead.
The C-terminal Domain of C/EBPβ Mediates the Interaction with Daxx
To identify the regions of C/EBPβ and Daxx responsible for their interaction GST-pulldown assays were performed using various GST-C/EBPβ and GST-Daxx deletion constructs. As shown in Fig. 2A, bacterially expressed GST-DaxxB (amino acids 190–400) specifically interacted with FLAG-C/EBPβ expressed in QT6 cells. C/EBPβ failed to bind to GST as well as to other domains of Daxx fused to GST. This indicated that, in contrast to most other Daxx-interacting proteins such as p53, Smad4, Pax3, and Pax5 or PML, which bind to the C-terminal S/P/T domain of Daxx (amino acids 635–740) (21, 22, 25, 26, 31, 43, 45, 46), C/EBPβ binding to Daxx involves amino acids 190–400 covering the two coil-coiled regions and the first NLS of Daxx (Fig. 2D). In addition, by using various recombinant GST-C/EBPβ proteins we mapped the Daxx interaction domain to the C-terminal basic-region-leucine-zipper (bZIP) domain of C/EBPβ. Fig. 2 (B and C) shows that overexpressed HA-Daxx was unable to bind to GST-C/EBPβ proteins comprising the first 200 amino acids of C/EBPβ. On the other hand GST-bZIP containing only the bZIP domain of C/EBPβ was able to bind Daxx. Together, this showed that Daxx does not bind to the N-terminal transactivation domain of C/EBPβ but rather requires the bZIP motif of C/EBPβ for binding (Fig. 2C). Binding was also demonstrated using GST-Daxx1–571 and in vitro translated C/EBPβ consistent with the idea that both proteins interact directly (supplemental Fig. S2).
FIGURE 2.
Mapping of the protein domains responsible for the C/EBPβ-Daxx interaction. A, GST-pull down experiments were performed with the indicated GST and GST-Daxx fusion proteins and lysates of QT6 cells transfected with 5 μg of a FLAG-C/EBPβ expression construct. Bound proteins were analyzed by SDS-PAGE followed by Western blotting. The interaction between GST-Daxx proteins and FLAG-C/EBPβ was detected using a FLAG-specific antibody. Crude protein extract of the transfected cells was used as control (upper panel). Coomassie Blue staining of GST proteins used in the pulldown experiments demonstrate comparable protein amounts (lower panel). The asterisk indicates a Daxx-unrelated co-purifying bacterial protein. B and C, GST pulldown experiments using different GST-C/EBPβ proteins and lysates of QT6 cells transfected with 5 μg of an HA-Daxx expression vector (+ lanes) or of untransfected cells (− lanes). Western blotting with an HA-specific antibody was performed to detect Daxx bound to the GST-C/EBPβ proteins. Crude protein extracts served as input control (upper panel). Coomassie Blue staining of GST proteins used in the pulldown experiments is shown in the lower panel of B and the right panel in C. D, schematic illustration of the C/EBPβ and Daxx constructs used and their respective binding capacity. The structural characteristics of the proteins are depicted by shaded boxes. Abbreviations: PAH, paired amphipathic helices; CC, coiled-coiled domain; D/E, acid-rich domain; NLS, nuclear localization signal; S/P/T, domain rich in serine, threonine, and proline; SIM, SUMO-interacting domain; TAD, transactivation domain; NRD, negative regulatory domain; BR, basic residue-rich domain; and LeuZ, leucine zipper. The numbers refer to human Daxx and chicken C/EBPβ.
Previous work has established a link between sumoylation and the interaction of Daxx with several transcription factors, including Smad4 (45) and glucocorticoid and androgen receptors (28, 37). Although the SIM, which resides at the C terminus of Daxx (37), is not involved in the interaction between Daxx and C/EBPβ, we wished to obtain experimental evidence that the interaction of C/EBPβ and Daxx is sumoylation-independent. We expressed Daxx and C/EBPβ together with the sumo-specific isopeptidase SENP1 followed by co-immunoprecipitation analysis. As shown in Fig. 3. C/EBPβ was co-precipitated via Daxx equally well in the absence or presence of increasing amounts of SENP1, indicating that the interaction of both proteins is SUMO-independent. In vitro GST pulldown experiments lead to a similar conclusion (supplemental Fig. S3). It therefore appears that the interaction of Daxx and C/EBPβ is sumoylation-independent.
FIGURE 3.
In vivo interaction of C/EBPβ and Daxx is not disrupted by SENP1. QT6 cells were transfected with the indicated combinations of plasmids encoding FLAG-C/EBPβ (5 μg), HA-Daxx (5 μg), and increasing amounts of FLAG-SENP1. Cells were lysed after 24 h, and protein extracts were immunoprecipitated with anti-HA antibodies, followed by SDS-PAGE and Western blotting with FLAG antibodies (top panel). The asterisk marks the immunoglobulin heavy chain of the anti-HA antibody. Co-precipitated C/EBPβ is marked by an arrowhead. Analyses of the crude protein extracts with HA- and FLAG-antibodies demonstrate comparable expression levels of the proteins in the different samples. HA-Daxx, FLAG-SENP1, and FLAG-C/EBPβ are marked by arrowheads.
Daxx Represses C/EBPβ-dependent Transcription
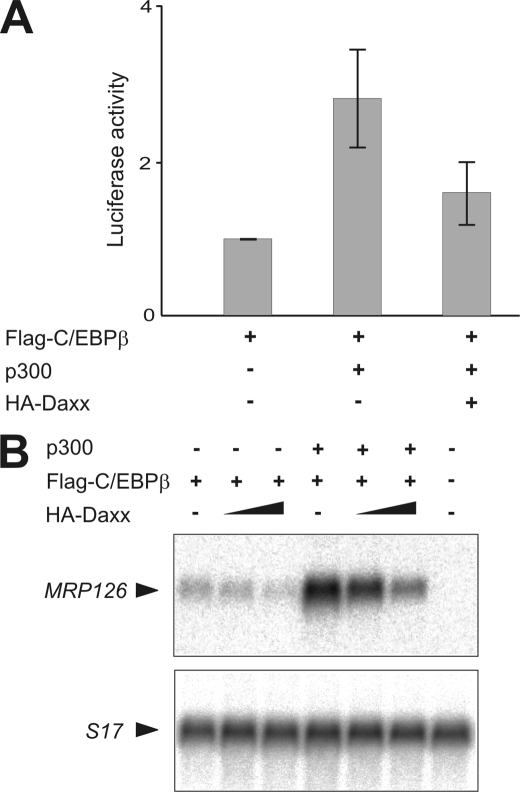
Daxx displays limited sequence similarity to the yeast transcriptional co-repressor Sin3 and its mammalian homologue Sin3a (21). Moreover, Daxx has been identified as a transcriptional co-repressor for a variety of transcription factors, including p53, Smad4, members of the Pax family, and androgen receptor (21, 22, 26, 28, 31, 45, 46). To investigate whether binding of Daxx to C/EBPβ is associated with repression of C/EBPβ-dependent transcription we performed reporter gene assays. To this end, combinations of FLAG-C/EBPβ, co-activator p300, and HA-Daxx expression constructs were co-transfected into QT6 cells together with a vector coding for firefly luciferase driven by the C/EBPβ-responsive mim-1 promoter (47). Consistent with our previous observations (13), co-expression of p300 with C/EBPβ resulted in a significantly higher transcriptional activity of C/EBPβ (Fig. 4A). However, after co-expression of HA-Daxx this effect was markedly decreased suggesting that Daxx was able to suppress C/EBPβ transcriptional activity. The repressive influence of Daxx was also evident without additional expression of p300 indicating that Daxx does not simply function by interfering with overexpressed p300 (data not shown). Next, we wanted to determine whether Daxx also inhibits C/EBPβ-dependent transcription of an endogenous, chromatin-embedded gene and analyzed the effect of Daxx on the expression of the C/EBPβ target gene MRP126. It had previously been shown that this gene is silent in fibroblasts but can be activated by ectopic expression of C/EBPβ (2). As expected, Northern blot analysis showed that expression of C/EBPβ was sufficient to induce endogenous MRP126 mRNA expression (Fig. 4B). p300 strongly increased the expression of this gene, confirming previous observations (13). Consistent with the results obtained by the reporter gene assay Daxx substantially suppressed basal as well as p300-enhanced transcription activity of C/EBPβ in a concentration-dependent manner (Fig. 4B). These experiments thus demonstrated that Daxx acts as a transcriptional inhibitor for p300-C/EBPβ-mediated transcription.
FIGURE 4.
Daxx inhibits the p300-enhanced C/EBPβ-dependent transcription. A, QT6 cells were transfected with the indicated combinations of expression vectors for FLAG-C/EBPβ (1 μg), HA-p300 (5 μg), and HA-Daxx (5 μg) together with the C/EBPβ-responsive p240-Luc reporter construct (3 μg) and pCMVβ (0.5 μg). Cells were harvested after 24 h, and luciferase and β-galactosidase activities were determined. The columns show the average luciferase activity normalized to the β-galactosidase activity. Thin lines show standard deviations. Data presented are derived from at least four independent experiments. The β-galactosidase-normalized luciferase activity mediated by exclusive expression of FLAG-C/EBPβ was arbitrarily set as one. B, Northern blot analysis of polyadenylated RNA from QT6 cells transfected with pCMVβ (0.5 μg) and the indicated combinations of FLAG-C/EBPβ (0.75 μg), HA-p300 (5 μg), and HA-Daxx (2 and 5 μg) expression vectors. β-Galactosidase assays of aliquots of the cells were used to confirm similar the transfection efficiencies in each case. The blot was hybridized sequentially with probes specific for the chicken MRP126 gene and the ribosomal protein S17 gene which was used as internal control.
p300-mediated Acetylation of C/EBPβ Is Suppressed by Daxx
Because our data presented in Fig. 2B showed that Daxx binds to the C-terminal part of C/EBPβ, which harbors its DNA-binding domain we initially considered the possibility that Daxx interferes with sequence-specific DNA binding of C/EBPβ. However, this was not the case, as demonstrated by electrophoretic mobility shift assays ( supplemental Fig. S4).
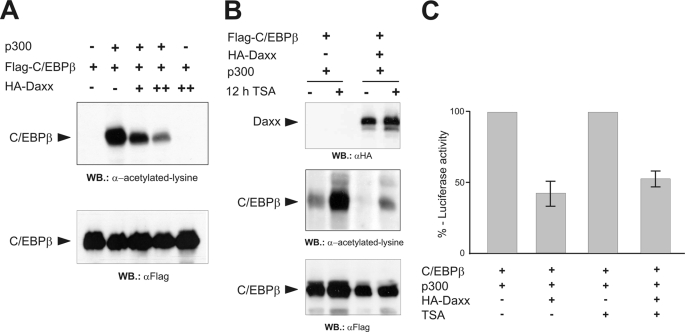
Previous studies have linked the control of C/EBPβ transcriptional activity to several different post-translational modifications, including acetylation (7–11). C/EBPβ acetylation was shown to be dynamically regulated by HDAC1 leading to deacetylation, and by acetyltransferases such as p300 or P/CAF, which support acetylation (8, 9), and to increase the transcriptional activity of C/EBPβ (8). Because Daxx is known to recruit HDAC proteins (8, 9) we suspected that the inhibitory activity of Daxx might be due to a suppression of the acetylation of C/EBPβ. To address this possibility, FLAG-C/EBPβ and full-length p300 were co-expressed without or with rising amounts of HA-Daxx. The extent of p300-mediated C/EBPβ acetylation was then monitored by Western blot analysis using an acetyl-lysine specific antibody. After co-expression of C/EBPβ with p300 a prominent band was detected by this antibody that co-migrated with C/EBPβ. This band was totally absent in samples expressing FLAG-C/EBPβ without p300 thus indicating this band as specific for acetylated forms of C/EBPβ (Fig. 5A, upper panel). More importantly, the p300-mediated C/EBPβ acetylation progressively decreased with rising levels of Daxx expression. Western blot analysis showed that this effect was not due to changes in the overall amount of C/EBPβ (Fig. 5A, lower panel) or p300 (data not shown) suggesting that Daxx does not influence protein stability but suppresses p300-mediated acetylation of C/EBPβ.
FIGURE 5.
Daxx inhibits the p300-mediated acetylation of C/EBPβ. A, QT6 cells were transfected with pCMVβ (0.5 μg) and expression constructs for FLAG-C/EBPβ (5 μg), HA-Daxx (5 and 10 μg), and HA-p300 (10 μg), as indicated. 24 h later protein extracts were prepared, and β-galactosidase-normalized protein amounts were analyzed by Western blotting using antibodies against acetyllysine or the FLAG tag. B, QT6 cells were transfected with pCMVβ and expression vectors for FLAG-C/EBPβ (1 μg), Ha-Daxx (5 μg), and full-length p300 (5 μg), as indicated at the top. Cells were treated with 400 nm TSA for 12 h before harvesting (+) or were left untreated (−). β-Galactosidase-normalized protein amounts were analyzed by Western blotting using antibodies against Daxx, acetyllysine, or the FLAG tag. Because smaller amounts of C/EBPβ expression vector were transfected in the experiment shown in B as compared with A, the signal intensities in the second lane of A and the first lane of B are different. C, QT6 cells were transfected with the indicated combinations of expression vectors for FLAG-C/EBPβ (1 μg), HA-p300 (5 μg), and HA-Daxx (5 μg) together with the C/EBPβ-responsive p240-Luc reporter construct (3 μg) and pCMVβ (0.5 μg). Cells were treated additionally with TSA as described in panel B or left untreated. Cells were harvested after 24 h, and luciferase and β-galactosidase activities were determined. The columns show the average luciferase activity normalized to the β-galactosidase activity. The activity of the reporter gene in the absence of Daxx was set to 100% in each case.
To investigate whether the suppression of the acetylation of C/EBPβ by Daxx was caused by Daxx-mediated recruitment of histone deacetylase to C/EBPβ we examined the inhibitory effect of Daxx in the presence of the HDAC inhibitor trichostatin (TSA). We reasoned that, if Daxx suppresses C/EBPβ acetylation by recruiting HDACs to C/EBPβ, TSA treatment should abrogate the inhibitory effect of Daxx. Fig. 5B shows that the acetylation of C/EBPβ was increased in the presence of TSA, consistent with previous work that showed that the level of acetylation of C/EBPβ is dependent on the balance between p300-mediated acetylation and HDAC1-mediated deacetylation (9). However, comparison of lanes 1 and 3 and lanes 2 and 4 of Fig. 5B showed that Daxx suppressed the acetylation of C/EBPβ to a similar extent in the absence or presence of TSA, suggesting that Daxx inhibits C/EBPβ acetylation in an HDAC-independent manner. To compare the inhibitory effect of Daxx in the absence or presence of TSA more quantitatively we used a reporter gene assay in which the inhibitory effect of Daxx on the C/EBPβ-p300-induced activity of the C/EBPβ-responsive mim-1 promoter was studied (Fig. 5C). These experiments confirmed that the inhibitory effect of Daxx was virtually identical in the absence or presence of TSA. This is consistent with the notion that Daxx does not exert its inhibitory effect to C/EBPβ by recruitment of histone deacetylase.
Daxx Reduces C/EBPβ-induced phosphorylation of p300
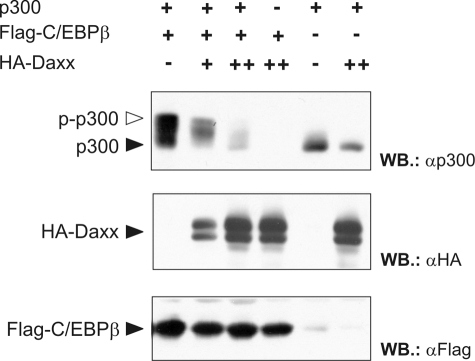
Recently, we have shown that C/EBPβ not only binds to p300 and recruits it to C/EBPβ target sequences but that C/EBPβ also triggers the phosphorylation at multiple sites in the C-terminal domain of p300. These phosphorylations, in turn, enhance the activity of p300 as a co-activator of C/EBPβ (13, 15). Because our data showed that Daxx inhibits p300-enhanced transcriptional activity of C/EBPβ, we wished to know whether the C/EBPβ-induced phosphorylation of p300 was affected by Daxx. To address this issue we used a truncated form of p300 (p300/1751–2370), which corresponds to the C-terminal part of p300 and displays a strong electrophoretic mobility shift in response to the C/EBPβ-induced phosphorylation (15). As illustrated in Fig. 6, co-expression of C/EBPβ resulted in the appearance of additional higher molecular weight forms of p300/1751–2370 which, as we have previously shown, correspond to phosphorylated forms of p300 (15). This mobility shift was markedly suppressed by co-expression of HA-Daxx in a dose-dependent manner, indicating that Daxx inhibits C/EBPβ-induced p300 phosphorylation. Previous studies have shown that the phosphorylation of p300 increases its HAT activity (48). Our results, therefore, suggest that Daxx affects the interdependent functional interaction of C/EBPβ and p300: by inhibiting the C/EBPβ-induced phosphorylation of p300 which, in turn, leads to a decreased p300-HAT activity and reduced levels of C/EBPβ acetylation. The overall result is that Daxx represses C/EBPβ-dependent transcription.
FIGURE 6.
Daxx suppresses the C/EBPβ-induced phosphorylation of p300. QT6 cells were transfected with expression vectors for FLAG-C/EBPβ (0.5 μg), HA-Daxx (1 and 3 μg), and a truncated version of p300 (encompassing amino acids 1751–2379; 3 μg) together with pCMVβ (0.5 μg). After 24 h the cells were lysed, the β-galactosidase was measured, and transfection efficiency-normalized protein amounts were analyzed by SDS-PAGE and Western blotting using antibodies directed against FLAG tag, HA tag, and p300. The black and white arrows mark the unphosphorylated and highly phosphorylated forms of p300, respectively.
C/EBPβ Affects the Subcellular Localization of Daxx
Previously, we have shown that the binding of C/EBPβ to p300 results in a change of the subnuclear localization of p300 from a discrete speckled distribution in absence of C/EBPβ to more evenly distributed localization throughout the nucleus when C/EBPβ is present (15). To determine whether C/EBPβ has any impact on Daxx localization or vice versa single as well as combined expression of GFP-fused Daxx and FLAG-tagged C/EBPβ in HeLa cells was examined by immunofluorescence analysis using confocal microscopy. As shown in Fig. 7, in the absence of Daxx C/EBPβ was spread throughout the nucleus with nucleoli being excluded. In the absence of C/EBPβ Daxx was predominantly present in the nuclear speckles, which were previously identified as PODs (36). Additionally, a minor fraction of Daxx was localized in the nucleoplasm. Co-expression of C/EBPβ and Daxx, however, led to a dramatic change of the nuclear distribution of both proteins. In the presence of C/EBPβ, the association of Daxx with PODs was diminished resulting in a diffusely dispersed distribution of Daxx throughout the nucleus. A quantitative examination revealed that co-expression of C/EBPβ shifted Daxx localization pattern from >80% POD-associated in absence of C/EBPβ to less that 35% in its presence (Fig. 7B). Counterstaining of endogenous PML of cells expressing only GFP-Daxx clearly demonstrated a perfect co-localization between Daxx and PML, thereby confirming Daxx to be a POD-associated factor (21, 25, 36, 43). Counterstaining of PML in cells transfected with GFP-Daxx and FLAG-C/EBPβ expression constructs indicated that the Daxx redistribution was not due to a general disturbance of POD formation (supplemental Fig. S5). Moreover, Western blot analyses demonstrated that expression levels of endogenous PML protein were not affected by overexpression of Daxx alone or Daxx together with C/EBPβ (data not shown). Taken together, our data show that the Daxx-dependent repression of C/EBPβ activity is accompanied by a marked change in sub-cellular localization of Daxx from PODs to a predominantly nucleoplasmic localization.
FIGURE 7.
C/EBPβ affects the sub-nuclear localization of Daxx. A, HeLa cells were transfected with 1.3 μg of expression vectors for FLAG-C/EBPβ (a–c), GFP-Daxx (d–f), or both (g–i). After 24 h, cells were fixed and FLAG-C/EBPβ was stained with anti-FLAG and TRITC-conjugated secondary antibody. Sub-cellular localization of the proteins was examined by confocal microscopy. White arrows indicate localization of GFP-Daxx to PODs. B, quantitative assessment of POD-associated Daxx localization in GFP-Daxx and GFP-Daxx/FLAG-C/EBPβ co-expressing cells. Data shown are the mean (± S.D.) of three independent experiments each with >100 cells analyzed.
PML Inhibits the Interaction of C/EBPβ and Daxx and Relieves the Repressive Effect of Daxx on C/EBPβ-dependent Transcription
In many cases the repressive effect of Daxx on the activity of other transcription factors like p53, Pax5, or glucocorticoid receptor is blocked by proteins such as PML, MSP58, or ASK1, which interrupt the Daxx association of the transcription factor and sequester Daxx to PODs, the nucleoli, or the cytoplasm (22, 24, 27, 38, 39). Having shown that Daxx is redistributed from PODs to the nucleoplasm in the presence of C/EBPβ, it appeared reasonable to assume that C/EBPβ is competing with PML for Daxx recruitment. Therefore, co-immunoprecipitation analyses were carried out to determine whether the C/EBPβ-Daxx interaction is, in turn, affected by expression of PML. As shown in Fig. 8A, increasing amounts of BFP-PML led to a drastic reduction of HA-Daxx co-precipitating with C/EBPβ. Western blot analysis of total cell extracts showed comparable Daxx and C/EBPβ protein levels irrespective of BFP-PML expression thus excluding varying protein amounts to be responsible for the reduced Daxx-C/EBPβ interaction. Taken together, these data suggest that C/EBPβ via direct interaction with Daxx induces the release of Daxx from PODs and that this interaction can be reverted by rising amounts of PML. This presumably results in re-recruitment of Daxx to PODs, as shown previously (24, 25).
FIGURE 8.
PML disrupts the interaction of C/EBPβ and Daxx and abrogates the repressive effect of Daxx on C/EBPβ-dependent transcription. A, QT6 cells were transfected with expression vectors coding for FLAG-C/EBPβ (5 μg), HA-Daxx (5 μg), and BFP-PML (5 and 10 μg), as indicated. Cells were lysed after 24 h, and protein extracts were immunoprecipitated with FLAG-specific antibody. Precipitated proteins were separated by SDS-PAGE and amount of co-precipitated HA-Daxx was detected by Western blotting using an anti-HA antibody. Analyses of the crude protein extracts (input) show increasing BFP-PML expression and comparable amounts of HA-Daxx and FLAG-C/EBPβ in the respective samples. B, QT6 cells were transfected with 3 μg of the p240-Luc reporter construct and 0.5 μg of pCMVβ together with plasmids coding for HA-Daxx (5 μg), FLAG-C/EBPβ (1 μg), HA-p300 (5 μg), and BFP-PML (1 and 3 μg) in the indicated combinations. Cells were harvested after 24 h, and luciferase and β-galactosidase activities were determined. The columns show the average β-galactosidase-normalized luciferase activity of at least three independent experiments. Thin lines indicate standard deviations. The luciferase activity in the presence of FLAG-C/EBPβ was arbitrarily designated as one.
To further investigate whether PML also antagonizes the Daxx-mediated repression of C/EBPβ activity, we performed luciferase reporter gene assays. QT6 cells were transiently transfected with a combination of expression vectors encoding FLAG-C/EBPβ, p300, HA-Daxx, and BFP-PML along with the C/EBPβ-responsive reporter construct. Consistent with the results shown before, co-expression of Daxx again significantly decreased the p300-dependent transcriptional activity of C/EBPβ. By contrast, additional expression of BFP-PML restored the C/EBPβ activity in a dose-dependent manner indicating that PML is able to relieve the inhibitory effect of Daxx on C/EBPβ-dependent transcription (Fig. 8B). Of note, co-expression of high BFP-PML amounts in absence of Daxx was also associated with a slightly stronger C/EBPβ-dependent transcription. This might be attributed to the ability of PML to relieve the repressive impact of endogenous Daxx. In summary, these data indicate that PML inhibits the association of C/EBPβ with the transcriptional repressor Daxx and, in turn promotes C/EBPβ activity.
Reduction of Daxx-C/EBPβ-interaction during APL Differentiation
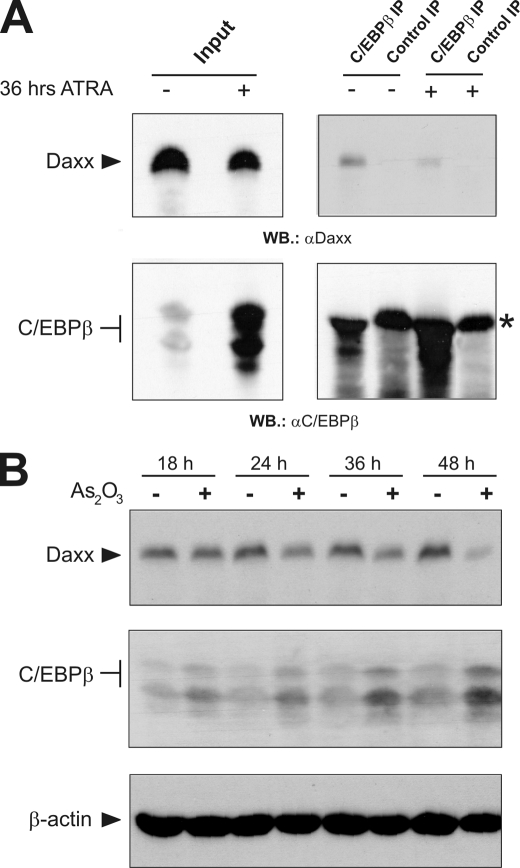
In APL the t(15;17) chromosomal translocation leads to the expression of the oncogenic PML-RARα fusion protein (41, 42). This is associated with the disruption of normal POD formation and results in de-localization of POD-linked proteins, including Daxx. Treatment with ATRA, which induces differentiation of leukemic cells and disease remission in patients, induces reorganization of intact POD structures and, consequently, relocation of POD-associated proteins such as Daxx (49–51). Several lines of evidence indicate that Daxx is crucially involved in APL biology by acting as a constitutive transcriptional co-repressor of PML-RARα (25, 40). Interestingly, in a recent report, Duprez et al. showed that C/EBPβ is up-regulated during ATRA treatment and, more importantly, that C/EBPβ activity is required for ATRA-induced APL differentiation (6). These observations together with our results that identified C/EBPβ as a Daxx-controlled transcription factor prompted us to analyze the association of the endogenous proteins during differentiation of APL cells. To this end, APL-derived NB4 cells were incubated with ATRA for 36 h or left untreated. The cells were then harvested and subjected to co-immunoprecipitation analysis. As shown in Fig. 9A, in untreated NB4 cells Daxx could be efficiently co-precipitated by an antibody directed against C/EBPβ. In contrast, after incubation with ATRA the interaction of C/EBPβ and Daxx was diminished, although the C/EBPβ protein level was markedly up-regulated, as reported previously (6). Consequently, Western blot analysis revealed a significantly higher amount of precipitated C/EBPβ protein after ATRA treatment thus indicating that antibody limitation was not responsible for the reduced Daxx co-precipitation. In conclusion this suggested that during ATRA-mediated APL differentiation the fraction of Daxx associated with C/EBPβ is specifically reduced.
FIGURE 9.
Induction of APL differentiation by ATRA and As2O3 treatment relieves the interaction of C/EBPβ and Daxx. A, NB4 cells were treated with 1 μm ATRA for 36 h or left untreated. Aliquots of crude protein extracts were immunoprecipitated with C/EBPβ-specific antiserum or an unrelated control antiserum. Precipitated proteins were separated by SDS-PAGE and the amount of precipitated C/EBPβ and co-precipitated Daxx was detected by Western blotting using C/EBPβ and Daxx-specific antibodies, respectively. Analyses of the crude protein extracts were used as input controls. The asterisk in the lower right panel marks the immunoglobulin heavy chain. B, NB4 cells were incubated with or without 1 mm of As2O3 for the indicated times. Subsequently, cells were analyzed by Western blotting with antibodies against Daxx, C/EBPβ, and β-actin.
Beyond ATRA treatment the application of As2O3 also produces clinical remission of APL. It was shown that As2O3 triggers the ubiquitin-mediated degradation of PML-RARα or PML via the ubiquitin ligase RNF4 in a SUMO-dependent manner and also induces POD re-organization (25, 52, 53). We therefore also investigated the interaction of Daxx and C/EBPβ after As2O3 treatment. We found that the amount of Daxx that was co-precipitated via C/EBPβ strongly decreased after 36 h of As2O3 treatment, however, we also noticed that the total amount of Daxx was diminished under these conditions (data not shown). To determine the change of the expression level of Daxx more systematically we analyzed NB4 cells after different times of As2O3 treatment Western blotting (Fig. 9B). It is apparent that the amount of Daxx progressively decreased to very low levels after 48 h of As2O3 treatment, whereas the amount of C/EBPβ increased. Taken together, the experiments illustrated in Fig. 9 indicate that treatment with ATRA as well as As2O3, both of which are commonly used for APL therapy, induce a significant release of the Daxx-C/EBPβ interaction. According to previous observations this is likely due to the PML-dependent re-location of Daxx to re-organized PODs (24, 25) and a decrease of the Daxx protein levels, particularly in the case of As2O3 treatment.
DISCUSSION
The transcription factor CCAAT/enhancer-binding protein β (C/EBPβ) is a member of the C/EBP family and is involved in the control of cell type-specific gene expression of various tissues, including hematopoietic cells (1). C/EBPβ activity is controlled by several co-stimulatory factors such as p300/CBP, G9a, or GCN5 leading to an increase in transcriptional activity (12–14). By contrast, factors that negatively affect C/EBPβ activity have been poorly detected. Here, we have demonstrated that Daxx is a novel negative regulator of C/EBPβ. Daxx was found to interact with C/EBPβ in transfected cells as well as on the level of the endogenous proteins. In vitro interaction experiments have mapped the regions of both proteins responsible for their interaction to the C-terminal bZIP domain of C/EBPβ and a central region (amino acids 190–400) of Daxx. Daxx inhibited C/EBPβ and p300-stimulated C/EBPβ transcriptional activity as assayed by the activation of a C/EBPβ-dependent reporter gene and of a chromatin-embedded endogenous C/EBPβ-inducible gene. Taken together, these data identify Daxx as an inhibitor of C/EBPβ transcriptional activity. Previous work has established strong links between sumoylation and Daxx-mediated repression of the activity of several transcription factors, such as Smad4 (45), PML (25, 36), the glucocorticoid and androgen receptors (28, 37), as well as the transcriptional co-activator CBP (23). The identification of an SIM at the C terminus of Daxx (37) and the finding that this motif was essential for the SUMO-dependent interaction of Daxx with several transcription factors have suggested that SUMO-dependent binding of Daxx and co-recruitment of HDACs comprise a common mechanism of Daxx-mediated transcriptional repression (37, 54). Although C/EBPβ is also negatively regulated by sumoylation (7, 55) Daxx recruitment by C/EBPβ does not appear to be SUMO-dependent. The binding of Daxx to C/EBPβ is not mediated by the C-terminal domain of Daxx, which harbors the SIM, but with the poorly characterized central region that encompasses a coiled-coil domain, as shown in Fig. 2C. Furthermore, the sumoylation site of C/EBPβ is located in the central negative regulatory domain of C/EBPβ around lysine 134 (of the rat homologue) and not within the C-terminal domain of the protein that binds to Daxx (7). Of note, the C-terminal domain of C/EBPβ is also required for the binding to Smad3, which is also associated with a transcriptional repression of C/EBPβ (56).
The interaction of Daxx and C/EBPβ resulted in obvious changes of the subnuclear localization of Daxx. In the absence of C/EBPβ, Daxx localized mainly to PODs, as also shown by others (25, 36). In the presence of C/EBPβ, Daxx was re-distributed and a significant fraction co-localized with C/EBPβ in the nucleoplasm. Our data indicate that the decrease in POD-associated Daxx induced by C/EBPβ was not due to a general disturbance of POD structures but rather might reflect a competition of POD-associated proteins (presumably PML) and C/EBPβ for recruitment of Daxx. This notion was also supported by the observation that increased expression of PML disrupts the C/EBPβ-Daxx interaction and additionally abrogates the repressive effect of Daxx on the C/EBPβ-dependent transcription.
How does Daxx inhibit the activity of C/EBPβ? Although Daxx binds to the part of C/EBPβ that harbors its DNA-binding domain, Daxx appears not to interfere with DNA binding of C/EBPβ but rather with the cooperation of C/EBPβ and p300. We and others have shown previously that the activity of C/EBPβ is enhanced by the recruitment of the coactivator p300/CBP (13, 57). Moreover, we showed that the interaction of p300 with C/EBPβ triggers the phosphorylation of p300 by an as yet unknown protein kinase, thereby stimulating the activity of p300 as co-activator of C/EBPβ (15). Our results clearly demonstrate that Daxx disrupts the cooperation of C/EBPβ and p300, as evidenced by inhibitory effects of Daxx on p300-stimulated transcriptional activity of C/EBPβ, on the C/EBPβ-induced phosphorylation of p300 and on the p300-dependent acetylation of C/EBPβ. How Daxx inhibits the C/EBPβ-induced phosphorylation of p300 and the acetylation of C/EBPβ and whether both inhibitory effects are linked is currently under investigation. Phosphorylation is known to increase the HAT activity of p300 (48); it is therefore possible that the inhibitory effect of Daxx on C/EBPβ acetylation is a consequence of the diminished phosphorylation of p300. Because Daxx is known to bind to different HDACs (23, 25, 33) the diminished acetylation of C/EBPβ might, in principle, also be due to the Daxx-mediated recruitment of HDACs to C/EBPβ. However, we found that inhibitory effect of Daxx on C/EBPβ acetylation was not abrogated by TSA, arguing for a HDAC-independent mechanism of Daxx-mediated repression. In any case, as acetylation is known to increase the transcriptional activity of C/EBPβ, the suppression of acetylation by Daxx probably accounts for the decreased activity of C/EBPβ as also demonstrated for Daxx-mediated inhibition of the NF-κB-dependent transcription (32). The mechanism by which Daxx inhibits the C/EBPβ-induced phosphorylation of p300 is subject of ongoing studies. One obvious possibility is that Daxx interferes with the C/EBPβ-p300 interaction and thereby displaces p300 from C/EBPβ. Alternatively, Daxx could interfere with the protein kinase that phosphorylates p300.
An important implication of our work concerns the role of the C/EBPβ-Daxx interaction in APL. In APL, the t(15;17) chromosomal translocation leads to the expression of the oncogenic PML-RARα fusion protein (41, 42). PML-RARα is a potent transcriptional repressor and initiates APL by repressing the myeloid differentiation program, which results in an accumulation of blasts blocked at the promyelocytic stage (58, 59). Interestingly, these cells are sensitive to treatment with ATRA and arsenic trioxide, both of which induce their differentiation and, in turn, lead to disease remission (60). The expression of PML-RARα also results in the disruption of normal POD formation and consequently the de-localization of POD-associated proteins, including Daxx, whereas treatment with ATRA or arsenic trioxide induces the re-organization of intact POD structures with subsequent re-localization of Daxx to re-formed PODs (25, 49–51). Recently, Duprez et al. have identified C/EBPβ as a major PML-RARα-responsive gene in APL cells that is drastically up-regulated upon ATRA treatment, thus demonstrating that PML-RARα is not exclusively a transcriptional repressor (6). Moreover, they have shown that the transcriptional activity of C/EBPβ is required for ATRA-induced APL differentiation. Our data strongly suggest that in addition to being transcriptionally up-regulated the function of C/EBPβ is also controlled on the protein level via interaction with Daxx during APL cell differentiation. We showed that Daxx binds to C/EBPβ in APL cells, presumably inhibiting its function, whereas treatment with ATRA or arsenic trioxide diminished the interaction, presumably as a result of re-localization of Daxx to the PODs, as shown previously (25, 27). In the case of As2O3 treatment the level of Daxx was strongly reduced upon differentiation. This suggests that Daxx is not only re-localized but also degraded under these conditions as already demonstrated for PML-RARα (52, 53). Lallemand-Breitenbach et al. showed that As2O3 triggers the degradation of PML-RARα and PML via the ubiquitin ligase RNF4 within the PODs in a SUMO/ubiquitin-dependent manner. Supported by their finding that As2O3 also promotes the accumulation of SUMO, ubiquitin, and the 20 S proteasome to the PODs they further envisioned that additional POD-associated proteins could be targeted to this degradation pathway (52) thus fitting with our observation that the Daxx amount is deceased after As2O3 treatment. Interestingly, the treatment of APL cells with HDAC inhibitors that induce differentiation and clinical remission as well were also reported to induce a significant reduction of Daxx expression levels (61). This suggests that an inactivation of Daxx correlates with the differentiation of APL cells and, in turn, that Daxx is critically involved in APL. Indeed, by acting as a co-repressor for PML-RARα-dependent transcriptional repression Daxx has been implicated in promoting the promyelocytic differentiation block (40). Our data indicate that the repressive effect of Daxx in the APL context is not restricted to PML-RARα but that Daxx also contributes to APL via the inhibition of C/EBPβ. Although future studies are needed to further clarify the functional role of the C/EBPβ-Daxx interaction in more detail, the data presented here identified Daxx as a novel physiologically relevant inhibitor of C/EBPβ.
Supplementary Material
Acknowledgments
We thank S. Steinmann and S. Chachra for experimental support and R. Eckner, P. P. Pandolfi, and T. G. Hofmann for providing expression vectors.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Deutsche Krebshilfe.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- C/EBPβ
- CCAAT/enhancer-binding protein β
- APL
- acute promyelocytic leukemia
- bZIP
- basic-region-leucine-zipper
- ATRA
- all-trans-retinoic acid
- SUMO
- small ubiquitin-like modifier
- CBP
- CREB (cAMP-response element-binding protein)-binding protein
- POD
- PML-oncogenic domain
- SIM
- SUMO-interacting motif
- RAR
- retinoic acid receptor
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- TSA
- trichostatin
- BFP
- blue fluorescent protein
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- TRITC
- tetramethylrhodamine isothiocyanate
- CMV
- cytomegalovirus
- HDAC1
- histone deacetyl transferase 1
- PML
- promyelocytic leukemia protein.
REFERENCES
- 1.Nerlov C. (2008) Curr. Opin. Cell Biol. 20, 180–185 [DOI] [PubMed] [Google Scholar]
- 2.Kowenz-Leutz E., Twamley G., Ansieau S., Leutz A. (1994) Genes Dev. 8, 2781–2791 [DOI] [PubMed] [Google Scholar]
- 3.Calkhoven C. F., Müller C., Leutz A. (2000) Genes Dev. 14, 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 4.Sundfeldt K., Ivarsson K., Carlsson M., Enerbäck S., Janson P. O., Brännström M., Hedin L. (1999) Br. J. Cancer 79, 1240–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu S., Yoon K., Sterneck E., Johnson P. F., Smart R. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duprez E., Wagner K., Koch H., Tenen D. G. (2003) EMBO J. 22, 5806–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berberich-Siebelt F., Berberich I., Andrulis M., Santner-Nanan B., Jha M. K., Klein-Hessling S., Schimpl A., Serfling E. (2006) J. Immunol. 176, 4843–4851 [DOI] [PubMed] [Google Scholar]
- 8.Ceseña T. I., Cardinaux J. R., Kwok R., Schwartz J. (2007) J. Biol. Chem. 282, 956–967 [DOI] [PubMed] [Google Scholar]
- 9.Ceseña T. I., Cui T. X., Subramanian L., Fulton C. T., Iñiguez-Lluhí J. A., Kwok R. P., Schwartz J. (2008) Mol. Cell Endocrinol. 289, 94–101 [DOI] [PubMed] [Google Scholar]
- 10.Piwien-Pilipuk G., MacDougald O., Schwartz J. (2002) J. Biol. Chem. 277, 44557–44565 [DOI] [PubMed] [Google Scholar]
- 11.Pless O., Kowenz-Leutz E., Knoblich M., Lausen J., Beyermann M., Walsh M. J., Leutz A. (2008) J. Biol. Chem. 283, 26357–26363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovács K. A., Steinmann M., Magistretti P. J., Halfon O., Cardinaux J. R. (2003) J. Biol. Chem. 278, 36959–36965 [DOI] [PubMed] [Google Scholar]
- 13.Mink S., Haenig B., Klempnauer K. H. (1997) Mol. Cell Biol. 17, 6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiper-Bergeron N., Salem H. A., Tomlinson J. J., Wu D., Haché R. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz C., Beck K., Mink S., Schmolke M., Budde B., Wenning D., Klempnauer K. H. (2003) EMBO J. 22, 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plachetka A., Chayka O., Wilczek C., Melnik S., Bonifer C., Klempnauer K. H. (2008) Mol. Cell Biol. 28, 2102–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Khosravi-Far R., Chang H. Y., Baltimore D. (1997) Cell 89, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlman R., Schiemann W. P., Brooks M. W., Lodish H. F., Weinberg R. A. (2001) Nat. Cell Biol. 3, 708–714 [DOI] [PubMed] [Google Scholar]
- 19.Chen L. Y., Chen J. D. (2003) Mol. Cell Biol. 23, 7108–7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michaelson J. S., Bader D., Kuo F., Kozak C., Leder P. (1999) Genes Dev. 13, 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollenbach A. D., Sublett J. E., McPherson C. J., Grosveld G. (1999) EMBO J. 18, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim E. J., Park J. S., Um S. J. (2003) Nucleic Acids Res. 31, 5356–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo H. Y., Chang C. C., Jeng J. C., Hu H. M., Lin D. Y., Maul G. G., Kwok R. P., Shih H. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16973–16978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehembre F., Müller S., Pandolfi P. P., Dejean A. (2001) Oncogene 20, 1–9 [DOI] [PubMed] [Google Scholar]
- 25.Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E. J., Chen J. D. (2000) Mol. Cell Biol. 20, 1784–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R., Pei H., Watson D. K., Papas T. S. (2000) Oncogene 19, 745–753 [DOI] [PubMed] [Google Scholar]
- 27.Lin D. Y., Lai M. Z., Ann D. K., Shih H. M. (2003) J. Biol. Chem. 278, 15958–15965 [DOI] [PubMed] [Google Scholar]
- 28.Lin D. Y., Fang H. I., Ma A. H., Huang Y. S., Pu Y. S., Jenster G., Kung H. J., Shih H. M. (2004) Mol. Cell Biol. 24, 10529–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morozov V. M., Massoll N. A., Vladimirova O. V., Maul G. G., Ishov A. M. (2008) Oncogene 27, 2177–2186 [DOI] [PubMed] [Google Scholar]
- 30.Boellmann F., Guettouche T., Guo Y., Fenna M., Mnayer L., Voellmy R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4100–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emelyanov A. V., Kovac C. R., Sepulveda M. A., Birshtein B. K. (2002) J. Biol. Chem. 277, 11156–11164 [DOI] [PubMed] [Google Scholar]
- 32.Park J., Lee J. H., La M., Jang M. J., Chae G. W., Kim S. B., Tak H., Jung Y., Byun B., Ahn J. K., Joe C. O. (2007) J. Mol. Biol. 368, 388–397 [DOI] [PubMed] [Google Scholar]
- 33.Hollenbach A. D., McPherson C. J., Mientjes E. J., Iyengar R., Grosveld G. (2002) J. Cell Sci. 115, 3319–3330 [DOI] [PubMed] [Google Scholar]
- 34.Puto L. A., Reed J. C. (2008) Genes Dev. 22, 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T. L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10635–10640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. (1999) J. Cell Biol. 147, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 38.Ko Y. G., Kang Y. S., Park H., Seol W., Kim J., Kim T., Park H. S., Choi E. J., Kim S. (2001) J. Biol. Chem. 276, 39103–39106 [DOI] [PubMed] [Google Scholar]
- 39.Lin D. Y., Shih H. M. (2002) J. Biol. Chem. 277, 25446–25456 [DOI] [PubMed] [Google Scholar]
- 40.Zhu J., Zhou J., Peres L., Riaucoux F., Honoré N., Kogan S., de Thé H. (2005) Cancer Cell 7, 143–153 [DOI] [PubMed] [Google Scholar]
- 41.de Thé H., Chomienne C., Lanotte M., Degos L., Dejean A. (1990) Nature 347, 558–561 [DOI] [PubMed] [Google Scholar]
- 42.Kakizuka A., Miller W. H., Jr., Umesono K., Warrell R. P., Jr., Frankel S. R., Murty V. V., Dmitrovsky E., Evans R. M. (1991) Cell 66, 663–674 [DOI] [PubMed] [Google Scholar]
- 43.Zhong S., Salomoni P., Ronchetti S., Guo A., Ruggero D., Pandolfi P. P. (2000) J. Exp. Med. 191, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofmann T. G., Stollberg N., Schmitz M. L., Will H. (2003) Cancer Res. 63, 8271–8277 [PubMed] [Google Scholar]
- 45.Chang C. C., Lin D. Y., Fang H. I., Chen R. H., Shih H. M. (2005) J. Biol. Chem. 280, 10164–10173 [DOI] [PubMed] [Google Scholar]
- 46.Gostissa M., Morelli M., Mantovani F., Guida E., Piazza S., Collavin L., Brancolini C., Schneider C., Del Sal G. (2004) J. Biol. Chem. 279, 48013–48023 [DOI] [PubMed] [Google Scholar]
- 47.Chayka O., Kintscher J., Braas D., Klempnauer K. H. (2005) Mol. Cell Biol. 25, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aikawa Y., Nguyen L. A., Isono K., Takakura N., Tagata Y., Schmitz M. L., Koseki H., Kitabayashi I. (2006) EMBO J. 25, 3955–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyck J. A., Maul G. G., Miller W. H., Jr., Chen J. D., Kakizuka A., Evans R. M. (1994) Cell 76, 333–343 [DOI] [PubMed] [Google Scholar]
- 50.Koken M. H., Puvion-Dutilleul F., Guillemin M. C., Viron A., Linares-Cruz G., Stuurman N., de Jong L., Szostecki C., Calvo F., Chomienne C. (1994) EMBO J. 13, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weis K., Rambaud S., Lavau C., Jansen J., Carvalho T., Carmo-Fonseca M., Lamond A., Dejean A. (1994) Cell 76, 345–356 [DOI] [PubMed] [Google Scholar]
- 52.Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 53.Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 54.Shih H. M., Chang C. C., Kuo H. Y., Lin D. Y. (2007) Biochem. Soc. Trans. 35, 1397–1400 [DOI] [PubMed] [Google Scholar]
- 55.Eaton E. M., Sealy L. (2003) J. Biol. Chem. 278, 33416–33421 [DOI] [PubMed] [Google Scholar]
- 56.Choy L., Derynck R. (2003) J. Biol. Chem. 278, 9609–9619 [DOI] [PubMed] [Google Scholar]
- 57.Oelgeschläger M., Janknecht R., Krieg J., Schreek S., Lüscher B. (1996) EMBO J. 15, 2771–2780 [PMC free article] [PubMed] [Google Scholar]
- 58.Grignani F., Ferrucci P. F., Testa U., Talamo G., Fagioli M., Alcalay M., Mencarelli A., Grignani F., Peschle C., Nicoletti I. (1993) Cell 74, 423–431 [DOI] [PubMed] [Google Scholar]
- 59.Lin R. J., Nagy L., Inoue S., Shao W., Miller W. H., Jr., Evans R. M. (1998) Nature 391, 811–814 [DOI] [PubMed] [Google Scholar]
- 60.Mistry A. R., Pedersen E. W., Solomon E., Grimwade D. (2003) Blood Rev. 17, 71–97 [DOI] [PubMed] [Google Scholar]
- 61.Amin H. M., Saeed S., Alkan S. (2001) Br. J. Haematol. 115, 287–297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.