Abstract
The Dbf4-Cdc7 kinase (DDK) is required for the activation of the origins of replication, and DDK phosphorylates Mcm2 in vitro. We find that budding yeast Cdc7 alone exists in solution as a weakly active multimer. Dbf4 forms a likely heterodimer with Cdc7, and this species phosphorylates Mcm2 with substantially higher specific activity. Dbf4 alone binds tightly to Mcm2, whereas Cdc7 alone binds weakly to Mcm2, suggesting that Dbf4 recruits Cdc7 to phosphorylate Mcm2. DDK phosphorylates two serine residues of Mcm2 near the N terminus of the protein, Ser-164 and Ser-170. Expression of mcm2-S170A is lethal to yeast cells that lack endogenous MCM2 (mcm2Δ); however, this lethality is rescued in cells harboring the DDK bypass mutant mcm5-bob1. We conclude that DDK phosphorylation of Mcm2 is required for cell growth.
The Cdc7 protein kinase is required throughout the yeast S phase to activate origins (1, 2). The S phase cyclin-dependent kinase also activates yeast origins of replication (3–5). It has been proposed that Dbf4 activates Cdc7 kinase in S phase, and that Dbf4 interaction with Cdc7 is essential for Cdc7 kinase activity (6). However, it is not known how Dbf4-Cdc7 (DDK)2 acts during S phase to trigger the initiation of DNA replication. DDK has homologs in other eukaryotic species, and the role of Cdc7 in activation of replication origins during S phase may be conserved (7–10).
The Mcm2-7 complex functions with Cdc45 and GINS to unwind DNA at a replication fork (11–15). A mutation of MCM5 (mcm5-bob1) bypasses the cellular requirements for DBF4 and CDC7 (16), suggesting a critical physiologic interaction between Dbf4-Cdc7 and Mcm proteins. DDK phosphorylates Mcm2 in vitro with proteins purified from budding yeast (17, 18) or human cells (19). Furthermore, there are mutants of MCM2 that show synthetic lethality with DBF4 mutants (6, 17), suggesting a biologically relevant interaction between DBF4 and MCM2. Nevertheless, the physiologic role of DDK phosphorylation of Mcm2 is a matter of dispute. In human cells, replacement of MCM2 DDK-phosphoacceptor residues with alanines inhibits DNA replication, suggesting that Dbf4-Cdc7 phosphorylation of Mcm2 in humans is important for DNA replication (20). In contrast, mutation of putative DDK phosphorylation sites at the N terminus of Schizosaccharomyces pombe Mcm2 results in viable cells, suggesting that phosphorylation of S. pombe Mcm2 by DDK is not critical for cell growth (10).
In budding yeast, Cdc7 is present at high levels in G1 and S phase, whereas Dbf4 levels peak in S phase (18, 21, 22). Furthermore, budding yeast DDK binds to chromatin during S phase (6), and it has been shown that Dbf4 is required for Cdc7 binding to chromatin in budding yeast (23, 24), fission yeast (25), and Xenopus (9). Human and fission yeast Cdc7 are inert on their own (7, 8), but Dbf4-Cdc7 is active in phosphorylating Mcm proteins in budding yeast (6, 26), fission yeast (7), and human (8, 10). Based on these data, it has been proposed that Dbf4 activates Cdc7 kinase in S phase and that Dbf4 interaction with Cdc7 is essential for Cdc7 kinase activity (6, 9, 18, 21–24). However, a mechanistic analysis of how Dbf4 activates Cdc7 has not yet been accomplished. For example, the multimeric state of the active Dbf4-Cdc7 complex is currently disputed. A heterodimer of fission yeast Cdc7 (Hsk1) in complex with fission yeast Dbf4 (Dfp1) can phosphorylate Mcm2 (7). However, in budding yeast, oligomers of Cdc7 exist in the cell (27), and Dbf4-Cdc7 exists as oligomers of 180 and 300 kDa (27).
DDK phosphorylates the N termini of human Mcm2 (19, 20, 28), human Mcm4 (10), budding yeast Mcm4 (26), and fission yeast Mcm6 (10). Although the sequences of the Mcm N termini are poorly conserved, the DDK sites identified in each study have neighboring acidic residues. The residues of budding yeast Mcm2 that are phosphorylated by DDK have not yet been identified.
In this study, we find that budding yeast Cdc7 is weakly active as a multimer in phosphorylating Mcm2. However, a low molecular weight form of Dbf4-Cdc7, likely a heterodimer, has a higher specific activity for phosphorylation of Mcm2. Dbf4 or DDK, but not Cdc7, binds tightly to Mcm2, suggesting that Dbf4 recruits Cdc7 to Mcm2. DDK phosphorylates two serine residues of Mcm2, Ser-164 and Ser-170, in an acidic region of the protein. Mutation of Ser-170 is lethal to yeast cells, but this phenotype is rescued by the DDK bypass mutant mcm5-bob1. We conclude that DDK phosphorylation of Ser-170 of Mcm2 is required for budding yeast growth.
EXPERIMENTAL PROCEDURES
Preparation of plasmids, proteins, and yeast strains are described in the supplemental material.
Size Exclusion Chromatography
The Superose 6 size exclusion column was pre-equilibrated in gel filtration solution containing 25 mm Tris-HCl, pH 7.5, 10% glycerol, 1 mm DTT, 0.1 mm EDTA, and 100 mm NaCl. 200 μg of Cdc7 alone or 200 μg of Cdc7 alone with 50 μg of Dbf4 were first incubated for 30 min at 15 °C. The samples were then subjected to Superose 6 size exclusion chromatography in gel filtration solution. Each fraction was then subjected to SDS-PAGE analysis followed by Coomassie staining. 7 μl of each column fraction was incubated with 500 ng of full-length Mcm2 and 10 μCi of [γ-32P]ATP in a final volume of 10 μl for 30 min at 30 °C, and the reactions were analyzed by SDS-PAGE followed by phosphorimaging.
DDK Analytical Kinase Assays
Kinase reactions were in a volume of 10 μl and contained 5 mm Tris-HCl, pH 8.5, 10 mm MgCl2, 1 mm DTT, 50 μm cold ATP, 10 μCi of [γ-32P]ATP, and 1.5 μg of DDK, as indicated for each reaction. The amount of Mcm2 in each reaction is described in the figure legends. Reactions were incubated at 30 °C for 1 h. Reactions were stopped by addition of 5 μl of 5× SDS sample buffer (25% glycerol, 200 mm Tris base, 125 mm DTT, 2.5% SDS, 0.1% bromphenol blue), and the products were resolved on either 10 or 15% SDS-PAGE.
GST Pulldown Assay
Mcm proteins were first radiolabeled in a reaction volume of 100 μl that contained 20 μm full-length or fragment Mcm2 protein in kinase reaction buffer (5 mm Tris-HCl, pH 8.5, 10 mm MgCl2, 1 mm DTT, 500 μm ATP, 100 μCi of [γ-32P]ATP) containing 5 μg of PKA. Reactions were incubated at 30 °C for 1 h.
The 100-μl GST-pulldown reaction contained 50 pmol of GST-tagged protein in GST-binding buffer (40 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.1 mm EDTA, 10% glycerol, 0.1% Triton X-100, 1 mm DTT, 0.7 μg/ml pepstatin, 0.1 mm phenylmethylsulfonyl fluoride, 0.1 mg/ml bovine serum albumin) and varying amounts of radiolabeled protein as described in each figure. Reactions were incubated at room temperature for 1 h. Following incubation, reactions were added to 40 μl of prepared glutathione-Sepharose and gently mixed. Binding of GST-tagged protein to the beads was performed for 20 min, with gentle mixing every few minutes. Once the binding was complete, the reaction mixture was aspirated, and the beads were washed two times with 0.5 ml of GST binding buffer. After the last wash, 30 μl of 5× SDS sample buffer were added to each reaction, and the samples were boiled for 10 min. Samples (20 μl) were then analyzed by SDS-PAGE.
Services
DNA sequencing and mass spectroscopy were performed by the Vanderbilt University Facilities.
RESULTS
Low Molecular Weight Form of Dbf4-Cdc7 Exhibits High Specific Activity for Mcm2
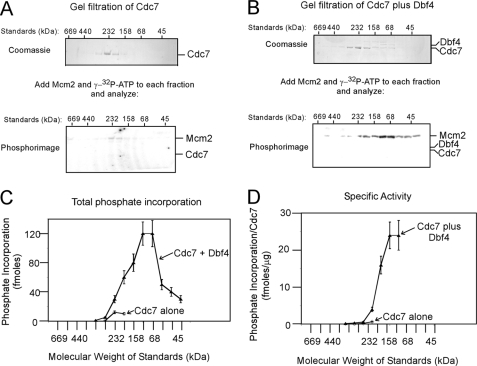
The molecular architecture of the active Dbf4-Cdc7 kinase is currently not known. To determine the functionally active state of Cdc7, the protein was analyzed by size exclusion chromatography (Fig. 1A). Cdc7 eluted in a peak at a Stokes radius that corresponded to that of a 232-kDa standard (Fig. 1A, top gel). The predicted molecular mass of Cdc7 is 58 kDa, suggesting that the majority of Cdc7 is in a tetrameric state. The elution of Cdc7 in neighboring fractions suggests that Cdc7 may be a mixture of tetramer and other forms. These results are consistent with in vivo data that Cdc7 alone exists as a multimer in the cell (27).
FIGURE 1.
Low molecular weight form of Dbf4-Cdc7 exhibits high specific activity for Mcm2. A, 200 μg of Cdc7 alone was subjected to Superose 6 size exclusion chromatography as described under “Experimental Procedures.” Each fraction was then subjected to SDS-PAGE analysis followed by Coomassie staining (top gel). The relative positions of molecular weight standards analyzed under identical conditions are shown above the gel. Each column fraction was incubated with 500 ng of full-length Mcm2 and [γ-32P]ATP for 30 min at 30 °C, and the reactions were analyzed by SDS-PAGE followed by phosphorimaging (bottom gel). The migration distances of Mcm2 and Cdc7, shown at the right of the gel, were determined by analyzing purified standards in the same gel. B, 200 μg of Cdc7 was incubated with 50 μg of Dbf4 for 30 min at 15 °C, and the mixture was then subjected to Superose 6 size exclusion chromatography as in A. The fractions were analyzed for protein abundance (top gel) and Mcm2 phosphorylation (bottom gel) as in A. C, quantitation of phosphate incorporation from the phosphorimages of A and B, and similar experiments are plotted as a function of size exclusion fraction (mean ± S.E., n = 3). The relative elution of molecular weight standards is shown at the bottom of the graph. D, amount of Cdc7 present in each reaction of C was quantified by densitometry of the Coomassie images. The phosphate incorporation into Mcm2 per μg of Cdc7 was calculated and plotted as function of gel filtration fraction.
To test if Cdc7 alone is active as a kinase at Mcm2, each fraction from the size exclusion column was incubated with Mcm2 and [γ-32P]ATP and then analyzed by SDS-PAGE and phosphorimaging (Fig. 1A, bottom gel). Cdc7 alone phosphorylates Mcm2 to low levels at a Stokes radius that corresponds to a 232-kDa protein (Fig. 1, C and D, open diamonds). These data suggest that a tetrameric from of Cdc7 alone is weakly active in phosphorylating Mcm2.
To determine the functionally active state of Dbf4-Cdc7, Dbf4 was incubated with excess Cdc7, and the protein mixture was then subjected to size exclusion chromatography. Once again, a peak of Cdc7 alone was present at a Stokes radius that corresponded to a 232-kDa protein. However, an additional low abundance peak of Cdc7 and Dbf4 is present in a fraction that peaks at a Stokes radius that is between 68 and 158 kDa (Fig. 1B, top gel). The molecular mass of one subunit of Dbf4 plus one subunit of Cdc7 is 139 kDa, suggesting that this complex is a heterodimer of Dbf4-Cdc7. Dbf4 was not observed in the higher molecular weight fractions, suggesting that Dbf4 does not bind to multimeric Cdc7. The elution fractions were then analyzed for phosphorylation of Mcm2, and peak phosphorylation levels were observed at a molecular mass between 68 and 158 kDa (Fig. 1, B, bottom gel, and C and D, filled triangles). Low levels of Cdc7 are present in the peak activity fractions, suggesting that a heterodimer of Dbf4-Cdc7 has a substantially higher specific activity for phosphorylating Mcm2 compared with the Cdc7 multimer (100-fold difference in specific activity, see Fig. 1D). Taken together, the data of Fig. 1 suggest that Cdc7 forms a multimer that is weakly active in phosphorylating Mcm2. Moreover, Dbf4 can bind to and activate a monomer of Cdc7, and the Dbf4-Cdc7 heterodimer has substantially higher specific activity than Cdc7 alone (100-fold higher specific activity).
Dbf4 or DDK, but Not Cdc7, Binds Tightly to Mcm2 and Mcm2-7
The data from Fig. 1 demonstrate that Dbf4 activates Cdc7 phosphorylation of Mcm2. We next investigated if Dbf4 alone, Cdc7 alone, or the complex of Dbf4-Cdc7 (DDK) binds to Mcm2. These data may help elucidate whether Dbf4 binds to Mcm2 to recruit Cdc7 or if Dbf4 activates Cdc7 that is pre-bound to Mcm2. To quantify Mcm2, an amino acid sequence bearing a kinase recognition site for protein kinase A was added to the N terminus of Mcm2. Protein kinase A (PKA) does not phosphorylate Mcm2 in vivo, and this sequence was added solely for the purpose of accurate quantitation. We mixed PKA-labeled full-length Mcm2 with GST-DDK, GST-Dbf4, or GST-Cdc7 and then isolated complexes bearing GST with glutathione beads (GST pulldown, Fig. 2A). When 6.6 pmol (66 nm), 20 pmol (200 nm), or 66 pmol (660 nm) of Mcm2 was added to the 100-μl binding reactions, more than half of the Mcm2 bound to GST-Dbf4-Cdc7 (GST-DDK) or GST-Dbf4 proteins (Fig. 2B). Thus, GST-DDK or GST-Dbf4 binds tightly to full-length Mcm2. In contrast, GST-Cdc7 binds weakly to Mcm2, because Mcm2 binding levels were only slightly greater than those achieved with GST alone (Fig. 2, A and B). The weak binding of Cdc7 to Mcm2 is consistent with the size exclusion data, which demonstrate that high concentrations of Cdc7 alone phosphorylate Mcm2 to very low levels (Fig. 1A).
FIGURE 2.
Dbf4 or DDK, but not Cdc7, binds tightly to Mcm2 and Mcm2-7. A, Mcm2 or Mcm2-7 bearing a site for protein kinase A phosphorylation (PKA site on Mcm3) was radiolabeled with protein kinase A as described under “Experimental Procedures.” 50 pmol of GST-DDK, GST-Dbf4, GST-Cdc7, or GST alone was incubated with varying concentrations of radiolabeled full-length Mcm2 (left) or Mcm2-7 complex (right) for 1 h at room temperature. For Mcm2, the total volume for each reaction was 100 μl, and the quantity of Mcm2 added was 66, 20, or 6.6 pmol. For Mcm2-7, the total volume for each reaction was 100 μl, and the quantity of Mcm2-7 complex added was 20, 6.6, or 2 pmol. After mixing, 40 μl of glutathione-Sepharose beads was added to each reaction, and the centrifuged pellet was washed and analyzed by SDS-PAGE followed by phosphorimaging. Input for the GST-pulldown was analyzed in the same gel to quantify the amount of protein bound. Results from experiments similar to those shown in A were quantified and plotted for Mcm2 in B and for Mcm2-7 in C. The mean ± S.E. is shown for each input amount.
We next determined whether the Mcm2-7 complex also binds to DDK, Dbf4, or Cdc7, because the Mcm2-7 complex provides the motor function for the replication fork helicase (29–31). We reconstituted Mcm2-7 complexes as described previously (31), except the Mcm3 subunit in this complex bears a PKA site. Using the same GST-pulldown approach, we found that the PKA-labeled Mcm2-7 complex binds tightly to Dbf4 or DDK but weakly to Cdc7 (Fig. 2, A and C). DDK phosphorylates multiple subunits within the Mcm2-7 complex; thus, the binding we observe for this complex likely reflects DDK or Dbf4 binding to different Mcm subunits within the Mcm2-7 complex.
The data in Fig. 2 suggest that Dbf4 does not activate Cdc7 that is pre-bound to Mcm2. It is more likely that Dbf4 binds to Mcm2 and then recruits Cdc7 to activate phosphorylation of Mcm2. It is also possible the Dbf4 first binds to Cdc7, and the DDK complex then binds to and phosphorylates Mcm2.
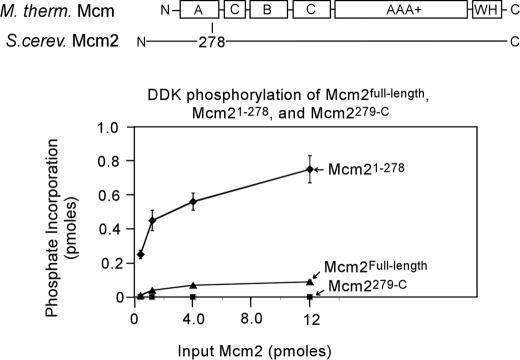
DDK Phosphorylates Mcm2-(1–278)
We next analyzed the region of Mcm2 that is phosphorylated by DDK. Based on structure-guided sequence alignment with Methanothermobacter thermoautrophicus Mcm (32), we constructed an Mcm2 region (1–278) that spans the poorly conserved N-terminal region and most of the A domain (Fig. 3A). The A domain is an α-helical bundle in the M. thermoautrophicus Mcm crystal structure (32). M. thermoautrophicus Mcm that lacks this region is active as a helicase, suggesting that the N-terminal region and A domain are not directly involved in Mcm helicase function (33). DDK phosphorylates Mcm2-(1–278) to high specific activity relative to full-length Mcm2 (Fig. 3A). In contrast, Mcm2-(279-C) is not phosphorylated by DDK. These data suggest that DDK phosphorylates only the N-terminal region and/or A domain of Mcm2 (Mcm2-(1–278)).
FIGURE 3.
DDK phosphorylates the N-terminal region of Mcm2. A structure-based sequence alignment with Sulfolobus solfataricus Mcm was used to divide Saccharomyces cerevisiae Mcm2 into two segments, an N-terminal fragment encompassing the nonconserved N-terminal region and most of domain A (Mcm2-(1–278)) and a C-terminal fragment encompassing the rest of the protein (Mcm2-(279-C)). S. cerevisiae Mcm2Full-length, Mcm2-(1–278), or Mcm2-(279-C) was incubated with 50 ng of DDK and [γ-32P]ATP in a volume of 10 μl for 30 min at 30 °C, and the reactions were then analyzed by SDS-PAGE followed by phosphorimaging. The results were quantified, and the fraction of phosphate incorporation is plotted as a function of input in picomoles.
Region 161–182 of Mcm2 Is Critical for DDK Phosphorylation
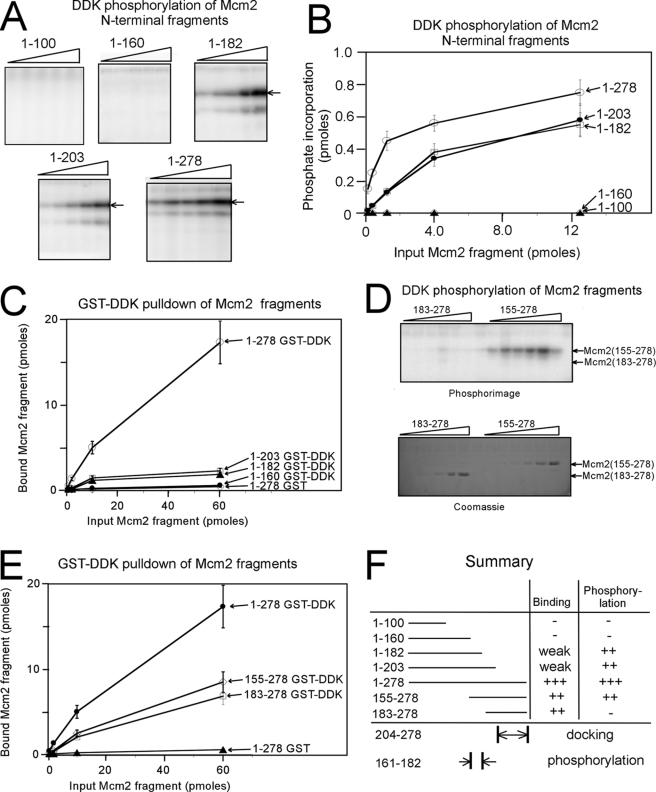
The DDK binding and kinase regions of budding yeast Mcm2 are unknown; thus, we investigated the action of DDK at Mcm2-(1–278) in further detail. We first constructed a series of N-terminal fragments of Mcm2 and tested the ability of DDK to phosphorylate these regions (Fig. 4, A and B). DDK was active in phosphorylating Mcm2-(1–203) and Mcm2-(1–182), but DDK did not phosphorylate Mcm2-(1–160) or Mcm2-(1–100) (Fig. 4A). These data suggest that amino acids 161–182 are critical for DDK phosphorylation of Mcm2. When DDK phosphorylation is plotted as a function of Mcm2 fragment concentration, the importance of amino acids 161–182 is evident (Fig. 4B). The plot also reveals that at low concentrations of Mcm2, region 204–278 is important for maximal DDK phosphorylation of Mcm2.
FIGURE 4.
Region 161–182 of Mcm2 is critical for DDK phosphorylation. A, N-terminal fragments of Mcm2 of varying amounts were incubated with 50 ng of DDK and [γ-32P]ATP in a volume of 10 μl for 30 min at 30 °C. The amount of Mcm2 fragment added was 13, 4, 1.3, 0.4, and 0.13 pmol. The reactions were then analyzed by SDS-PAGE followed by phosphorimaging. B, results from experiments similar to A were quantified, and the fraction of phosphate incorporation is plotted as a function of input in picomoles. The data are mean ± S.E. C, N-terminal fragments of Mcm2 with a site for protein kinase A phosphorylation was radiolabeled with [γ-32P]ATP and PKA as described under “Experimental Procedures.” 50 pmol of GST-DDK was incubated with varying amounts of radiolabeled Mcm2 fragments for 1 h at room temperature in a total volume of 100 μl. After mixing, the reactions were analyzed as described under “Experimental Procedures.” D, fragments 155–278 and 183–278 of Mcm2 of were tested for DDK phosphorylation as described in A. The amount of Mcm2 fragment added was 130, 40, 13, 4, 1.3, 0.4, and 0.13 pmol. The reactions were then analyzed by SDS-PAGE followed by phosphorimaging (top gel) or Coomassie staining (bottom gel). E, fragments of Mcm2 were analyzed by a GST-pulldown assay with GST-DDK as described in C. F, summary of data from A to E.
We next tested the ability of DDK to bind to Mcm2 fragments to identify the region for DDK docking on Mcm2. GST-DDK pulldown of PKA-radiolabeled Mcm2 fragments was accomplished, and the quantity of Mcm2 fragment bound as a function of input amount was plotted (Fig. 4C). A slight increase in binding is detected as Mcm2 fragment length is increased from 1–160 to 1–182, suggesting that residues 161–182 bind weakly to DDK. A substantial increase in binding is observed as the Mcm2 fragment length is increased from 1–203 to 1–278. These data suggest that region 204–278 binds tightly to DDK.
Regions 161–182 and 204–278 of Mcm2 are important for DDK activity. We next dissected the importance of these regions in further detail by investigating DDK phosphorylation and binding of two Mcm2 internal fragments Mcm2-(183–278) and Mcm2-(155–278). DDK phosphorylated Mcm2-(155–278) to high levels at low concentrations of Mcm2-(155–278) (Fig. 4D). In contrast, DDK was completely inactive in phosphorylating Mcm2-(183–278) (Fig. 4D). However, DDK was roughly equally efficient in binding to each of these Mcm2 fragments (Fig. 4E). Therefore, the substantial difference in phosphorylation between Mcm2-(155–278) and Mcm2-(183–278) may reflect the presence of a DDK phosphoacceptor sites between residues 155 and 182 of Mcm2. Hence, a model that is consistent with all of the data in Fig. 5 is that DDK binds tightly to region 204–278 of Mcm2 and DDK phosphorylates region 161–182 of Mcm2 (Fig. 4F).
FIGURE 5.
DDK phosphorylates Ser-164 and Ser-170 of Mcm2. A, residues 161–181 of S. cerevisiae Mcm2 are shown, and Ser-164 and Ser-170 were targeted for site-directed mutagenesis. B, mutants of Mcm2-(1–278) of varying amounts were incubated with 50 ng of DDK and [γ-32P]ATP in a volume of 10 μl for 30 min at 30 °C. The amount of Mcm2-(1–278) added was 12 or 4 pmol (left image) or 1.2 or 0.4 pmol (right image). The reactions were then analyzed by SDS-PAGE followed by phosphorimaging. C, results from experiments similar to B were quantified, and the fraction of phosphate incorporation is plotted as a function of input in picomoles. The data are mean ± S.E. D, mutants of Mcm2-(1–278) with a site for protein kinase A phosphorylation were radiolabeled with [γ-32P]ATP and protein kinase A (PKA) as described under “Experimental Procedures.” 50 pmol of GST-Dbf4 was incubated with varying amounts of radiolabeled Mcm2 fragments for 1 h at room temperature in a total volume of 100 μl. After mixing, the reactions were analyzed as described under “Experimental Procedures.” E, sequence alignment of budding yeast Mcm2 and budding yeast Mcm4. Serine and threonine residues are in boldface, and acidic residues are underlined. The aligned regions of these two proteins contain phosphoacceptor sites for DDK. F, alignment of budding yeast Mcm2 with Mcm2 from higher eukaryotes. DDK phosphoacceptor sites of human Mcm2 do not map to this region (10, 19, 20).
DDK Phosphorylates Ser-164 and Ser-170 of Mcm2
Fragment analysis suggests that DDK phosphorylates region 161–182 of Mcm2 (Fig. 4). There are two serine residues present in region 161–182 of Mcm2, Ser-164 and Ser-170 (Fig. 5A). To determine whether Ser-164 and Ser-170 are phosphorylated by DDK, these residues were mutated to alanine, and DDK phosphorylation and binding were examined. S614A exhibited a slight defect in DDK phosphorylation compared with wild-type Mcm2-(1–278), whereas S170A was modestly defective in DDK phosphorylation (Fig. 5, B and C). The double mutant S164A/S170A was not phosphorylated by DDK at all (Fig. 5, B and C).
The Dbf4 binding profiles of these mutants were also determined (Fig. 5D). Dbf4 bound to wild-type Mcm2-(1–278), S164A, S170A, and S164A/S170A with similar efficiency (Fig. 5D). Because S164A/S170A binds Dbf4 with nearly wild-type efficiency, but is not phosphorylated by DDK at all, the data suggest that DDK phosphorylates Ser-164 and Ser-170 of Mcm2.
Sequence alignment between budding yeast Mcm2 and budding yeast Mcm4 reveals that the DDK phosphorylation sites on Mcm2 are in the same region as the DDK phosphorylation sites on Mcm4 (Fig. 5E). In budding yeast Mcm4, seven serine and threonine residues between residues 161 and 178 are likely phosphoacceptor sites for DDK (26). In budding yeast Mcm2, the two serine residues between residues 161 and 178 are phosphorylated by DDK. Thus, although the number of residues phosphorylated by DDK is different, the region of Mcm that is phosphorylated by DDK is conserved between budding yeast Mcm2 and Mcm4. These regions are also acidic in both proteins. Sequence alignment between budding yeast Mcm2 and human Mcm2 reveals that the DDK phosphorylation sites are not conserved (Fig. 5F). Although there is some disagreement as to the particular residues of human Mcm2 that are phosphorylated by DDK (10, 19, 20), in every published study the human DDK phosphorylation sites do not align with those found here for budding yeast Mcm2.
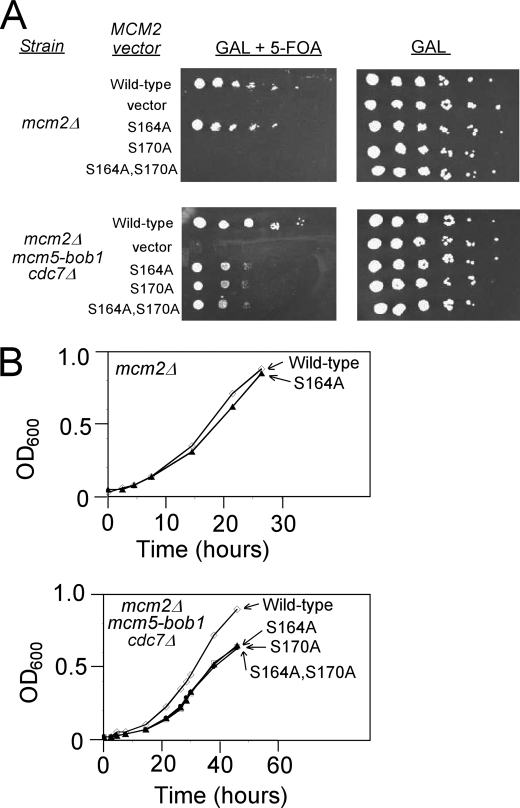
DDK Phosphorylation of Ser-164 of Mcm2 Is Not Required for Budding Yeast Cell Growth
To determine whether DDK phosphorylation of Ser-164 of Mcm2 is required for cell growth, we performed plasmid-shuffle assays in budding yeast cells lacking endogenous MCM2 (mcm2Δ) (Fig. 6). The deletion is complemented by wild-type MCM2 under expression control of its native promoter on a low copy centromere plasmid bearing a URA3 marker. The cells also contain a plasmid that expresses either wild-type of mutant MCM2 under control of the GAL1, galactose-inducible promoter. As expected, the cultures grow equivalently on media containing galactose (Fig. 6A, right). Growth on media containing 5-FOA and galactose selects for cells that have lost the URA3 complementing plasmid and are expressing only the MCM2 allele (or empty vector) indicated to the left of Fig. 6A. As seen in 10-fold serial dilutions, the mcm2S164A allele complements nearly as well as wild type in this background, a result confirmed by measuring the growth rates of cells taken from the 5-FOA plates and grown in media containing galactose (Fig. 6B, top graph). These results suggest that DDK phosphorylation of S164A is not required for yeast cell growth.
FIGURE 6.
DDK phosphorylation of Mcm2 Ser-170 is required for budding yeast growth. A, budding yeast cells deleted for mcm2 (mcm2Δ, top plates) or deleted for mcm2 and cdc7 and bearing the mcm5-bob1 mutation (mcm2Δ, mcm5-bob1, and cdc7Δ, bottom plates) were used. The cells harbor wild-type MCM2 on a plasmid with a URA3 selectable marker and mutant mcm2 on a LEU2 plasmid with expression controlled by a galactose-inducible promoter. In the presence of galactose and 5-FOA, only mcm2 under the control of the galactose-inducible promoter is expressed (left plates). In the presence of galactose (right plates), wild-type MCM2 is expressed as well. Cells were plated in serial 10-fold dilutions. B, colonies from A were grown in liquid media, and the rate of growth was measured as function of time. The top graph is for mcm2Δ cells, and the bottom graph is for mcm2Δ,mcm5-bob1,cdc7Δ cells.
DDK Phosphorylation of Ser-170 of Mcm2 Is Required for Budding Yeast Cell Growth
A similar experimental strategy was then used to determine whether DDK phosphorylation of Ser-170 of Mcm2 is required for yeast cell growth. When mcm2Δ cells were grown in the presence of galactose and 5-FOA, cells expressing mcm2S170A did not grow at all at any dilution, in marked contrast to the growth of cells expressing mcm2S164A or wild-type MCM2 (Fig. 6A, top left panel). These data suggest that Ser-170 of Mcm2 is required for yeast cell growth.
Serine 170 of Mcm2 is a target of DDK phosphorylation, but it may also be required for an additional unknown function. To determine whether the cell death caused by expression of mcm2S170A is related to DDK function, the plasmid-shuffle assay was performed in cells harboring the genetic bypass for DDK function, mcm5-bob1. These cells are also deleted for CDC7 (cdc7Δ). When mcm2Δ,mcm5-bob1,cdc7Δ cells were grown in the presence of galactose and 5-FOA, cells expressing mcm2S170A grew on agar plates in serial 10-fold dilutions, whereas cells harboring the vector alone were dead at every dilution (Fig. 6A, bottom left panel). Thus, the mcm5-bob1 DDK bypass rescues the cellular defect caused by the expression of mcm2S170A, suggesting that the lethality of mcm2S170A is primarily due to the absence of DDK phosphorylation of Ser-170 of Mcm2. It is not clear why mcm5-bob1 cells expressing mcm2S170A required higher concentrations to grow on agar plates compared with cells expressing wild-type MCM2, but it may be that in the mcm2Δ,mcm5-bob1,cdc7Δ strain, MCM2-Ser-170 is involved in a Cdc7-independent function. This is the first study of an mcm mutant with a lethal phenotype that is rescued by mcm5-bob1.
mcm2Δ,mcm5-bob1,cdc7Δ yeast colonies grown in galactose and 5-FOA were then grown in galactose-containing liquid media, and the rate of growth was determined (Fig. 6B, bottom graph). mcm5-bob1 cells expressing mcm2S164A grew, although the rate of growth was slightly decreased compared with cells expressing wild-type MCM2 for unknown reasons. Cells expressing mcm2S170A grew in liquid media, albeit slightly less fast than cells expressing wild-type MCM2, suggesting that the lethal phenotype of mcm2S170A in Δmcm2 cells is related to DDK phosphorylation of Ser-170 (Fig. 6B, bottom graph). Taken together, the data strongly support that DDK phosphorylation of Ser-170 of Mcm2 is required for budding yeast cell growth.
DISCUSSION
In this study we find that Cdc7 exists in solution primarily as a multimer, and this species at high concentrations is weakly active in phosphorylating Mcm2. Dbf4 binds to a low molecular weight form of Cdc7, forming what might be a heterodimer of Dbf4-Cdc7, and this species phosphorylates Mcm2 with substantially higher specific activity. Dbf4 alone binds tightly to Mcm2, whereas Cdc7 alone binds weakly to Mcm2, suggesting that Dbf4 recruits Cdc7 to phosphorylate Mcm2. DDK phosphorylates Mcm2-(1–278) but not Mcm2-(279-C). DDK phosphorylates Ser-164 and Ser-170 of budding yeast Mcm2, and these two serine residues are positioned in a highly acidic region. Although expression of mcm2S164A supports cell growth, expression of mcm2S170A does not. However, the lethality of mcm2S170A is rescued by the DDK-bypass mutation mcm5-bob1. We conclude that DDK phosphorylation of Ser-170 of Mcm2 is required for normal cell growth.
Dbf4 May Recruit a Monomer of Cdc7 to Target Mcm2 for Phosphorylation
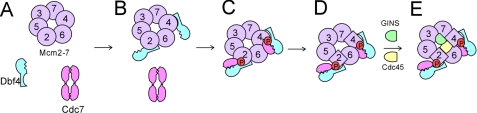
In this study we report that Cdc7 alone is a multimer that is weakly active in phosphorylating Mcm2. Cdc7 is present at high levels during G1 and S phase, whereas Dbf4 levels peak in S phase (18). It is unlikely that Cdc7 activity during G1 is physiologically relevant, given the low levels of Mcm2 phosphorylation observed in this study, and no activity was reported in other investigations (7, 8). In contrast, we report that Dbf4-Cdc7 is stable as a low molecular weight form, likely a heterodimer, and this species phosphorylates Mcm2 to substantially higher levels than Cdc7 alone. Thus, during S phase, Dbf4 may bind to a monomer of Cdc7, thereby forming a highly active heterodimer. The high molecular weight form of Cdc7 may be less active than the monomeric form because the high molecular weight form does not efficiently bind to Dbf4 or Mcm2. If this model is correct, multimerization of Cdc7 may be an auto-inhibitory mechanism to prevent high level Cdc7-phosphorylation of Mcm2 in the absence of Dbf4 (Fig. 7A).
FIGURE 7.
Model for DDK phosphorylation of Mcm2-7. A, Cdc7 is weakly active as a multimer on its own. B, Dbf4 binds tightly to residues 203–378 of Mcm2. Dbf4 likely binds to the homologous region of Mcm4 as well. C, Dbf4 recruits a monomer of Cdc7 to Mcm2 to activate the kinase, phosphorylating Ser-170 of Mcm2. The homologous region of Mcm4 is likely phosphorylated as well (26). D, phosphorylation of Ser-170 of Mcm2 induces a conformational change in the Mcm2-7 complex that is mimicked by the mcm5-bob1 mutation. E, conformational change in Mcm2-7 allows for the proper positioning of Cdc45 and GINS, and the Mcm2-7-Cdc45-GINS complex is activated to unwind DNA. DDK may also phosphorylate other Mcm proteins (data not shown).
We also find that Dbf4 and DDK bind tightly to Mcm2 and the Mcm2-7 complex but that Cdc7 alone does not bind tightly to these Mcm proteins. Thus, Dbf4 may recruit Cdc7 to Mcm2 and to the Mcm2-7 complex (Fig. 7B). Our in vitro data are consistent with a previously published in vivo report suggesting that Dbf4 recruits Cdc7 to origins of replication (23), and a recent study suggesting that Dbf4 recruits Cdc7 to Mcm2-7 complexes that are tightly bound to origins (24).
In Budding Yeast, DDK Phosphorylation of Mcm4 Is Important for Cell Growth, and DDK Phosphorylation of Mcm2 Is Required for Cell Growth
DDK phosphorylates the N termini of Mcm2 and Mcm4 in budding yeast cells (26). Furthermore, the DDK phosphorylation site region is conserved between these two proteins (Fig. 4) (26). The Dbf4 docking region for the kinase is positioned between residues 204 and 278 of Mcm2. This region corresponds to residues 201–275 of Mcm4. Region 175–333 of Mcm4 has been previously identified as the DDK-docking site of Mcm4 (26). Thus, it is likely that a conserved mode of binding occurs for Dbf4 docking to Mcm2 and Mcm4, with phosphorylation of serines and threonines in a region N-terminal to the conserved docking domain. Interestingly, the function of DDK phosphorylation of Mcm2 may be different from that of Mcm4. Whereas DDK phosphorylation of the Mcm4 residues is important for cell growth, it is not required (26). In contrast, DDK phosphorylation of Ser-170 of Mcm2 is required for yeast cell growth (Fig. 7C). Thus, although the mechanism of DDK binding and phosphorylation may be conserved between Mcm2 and Mcm4, the relationship to cell growth is somewhat different. Studies with S. pombe suggest some functional redundance between Mcm4 and Mcm6 (10). One possibility is that DDK phosphorylation of Mcm4 is redundant with DDK phosphorylation of another Mcm protein, such as Mcm6, whereas DDK phosphorylation of Mcm2 does not have a redundant counterpart.
Essential Requirement for DDK Phosphorylation of Mcm2 Is Likely to Be Conserved from Yeast to Humans
In this study, the lethal phenotype of Mcm2S170A is rescued by the mcm5-bob1 mutation, providing very strong evidence that DDK phosphorylation of Mcm2 is required for budding yeast cell growth. In a previous study of human cells, mutation of DDK phosphorylation sites to alanine resulted in inhibition of DNA replication, whereas mutation of these sites to glutamate did not (20). Thus, data from yeast and human support the notion that DDK phosphorylation of Mcm2 is essential for DNA replication and cell growth. These results suggest that the function of DDK at Mcm2 is conserved from yeast to human. Interestingly, the DDK phosphorylation sites of human Mcm2 do not align with the kinase sites of yeast Mcm2. We speculate that although the function of DDK phosphorylation of Mcm2 is conserved from yeast to human, the mechanistic details may be slightly different between these species.
Proposed Role for DDK Phosphorylation of Mcm Proteins
DDK phosphorylation of serine 170 of Mcm2 is required for cell growth, but what is the function of this phosphorylation? The N-terminal region and A domain of budding yeast Mcm2 encompasses the Dbf4 docking domain and Cdc7 kinase site. The homologous region of the archaeal Mcm is dispensable for helicase activity for archaeal Mcms (33, 34), suggesting that DDK phosphorylation of Mcm2 does not directly activate Mcm2-7 helicase function. Importantly, DDK phosphorylation of budding yeast Mcm4 or human Mcm4 is important for the loading of Cdc45 at origins of replication (10, 26). Thus, DDK phosphorylation of Mcm proteins likely influences the binding of key helicase accessory proteins such as Cdc45. However, the relationship between DDK phosphorylation of Mcm4 and Cdc45 binding is complex, because overexpression of Cdc45 is lethal to strains expressing mutations of mcm4 deficient in DDK phosphorylation (26). Thus, phosphorylation of Mcms by DDK does not appear to directly stimulate the interaction between Mcms and Cdc45. In this study, we report that DDK phosphorylation of Mcm2 can be suppressed by a mutation in mcm5 (mcm5-bob1). Changes in the conformation of Mcm5 as a result of the mcm5-bob1 mutation have recently been reported (35). This observation suggests that phosphorylation of Ser 170 of Mcm2 may be partially mimicked by a conformational change in a different Mcm subunit (Mcm5) (Fig. 7D). Interestingly, Mcm5 is positioned adjacent to Mcm2 in the Mcm2-7 complex (31, 36). We propose that DDK phosphorylation of Ser-170 of Mcm2 results in a conformational change in the Mcm2-7 complex that allows for the proper positioning of accessory proteins, such as Cdc45 and GINS, which ultimately leads to the activation of the replication fork helicase (Fig. 7E).
Supplementary Material
Acknowledgments
We thank Dr. Robert Sclafani for providing mcm5-bob1 yeast strains, Dr. Mike O'Donnell for providing expression constructs for Mcm proteins, and Dr. Susan Taylor for purified PKA protein. We also thank Katherine Friedman, Dave Cortez, and Ellen Fanning for helpful comments.
This work was supported by American Cancer Society Research Scholar Grant RSG-08-124-01-CCG (to D. K. and I. B.), a pilot grant from the Vanderbilt Ingram Cancer Center (to D. K.), and a Discovery Grant from Vanderbilt University (to D. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Fig. 1, and additional references.
- DDK
- Dbf4-Cdc7 kinase
- DTT
- dithiothreitol
- GST
- glutathione S-transferase
- 5-FOA
- 5-fluoroorotic acid
- PKA
- protein kinase A
- Mcm
- minichromosome maintenance.
REFERENCES
- 1.Bousset K., Diffley J. F. (1998) Genes Dev. 12, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donaldson A. D., Fangman W. L., Brewer B. J. (1998) Genes Dev. 12, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka S., Tak Y. S., Araki H. (2007) Cell Div. 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 5.Zegerman P., Diffley J. F. (2007) Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 6.Weinreich M., Stillman B. (1999) EMBO J. 18, 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown G. W., Kelly T. J. (1998) J. Biol. Chem. 273, 22083–22090 [DOI] [PubMed] [Google Scholar]
- 8.Masai H., Matsui E., You Z., Ishimi Y., Tamai K., Arai K. (2000) J. Biol. Chem. 275, 29042–29052 [DOI] [PubMed] [Google Scholar]
- 9.Jares P., Luciani M. G., Blow J. J. (2004) BMC Mol. Biol. 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J. M., Ishii A., Tanaka T., Kobayashi T., Tamai K., Ohtani K., Arai K. (2006) J. Biol. Chem. 281, 39249–39261 [DOI] [PubMed] [Google Scholar]
- 11.Moyer S. E., Lewis P. W., Botchan M. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labib K., Tercero J. A., Diffley J. F. (2000) Science 288, 1643–1647 [DOI] [PubMed] [Google Scholar]
- 13.Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C. (2006) Mol. Cell. 21, 581–587 [DOI] [PubMed] [Google Scholar]
- 14.Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 15.Pacek M., Walter J. C. (2004) EMBO J. 23, 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy C. F., Dryga O., Seematter S., Pahl P. M., Sclafani R. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3151–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei M., Kawasaki Y., Young M. R., Kihara M., Sugino A., Tye B. K. (1997) Genes Dev. 11, 3365–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshiro G., Owens J. C., Shellman Y., Sclafani R. A., Li J. J. (1999) Mol. Cell. Biol. 19, 4888–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. (2006) J. Biol. Chem. 281, 10281–10290 [DOI] [PubMed] [Google Scholar]
- 20.Tsuji T., Ficarro S. B., Jiang W. (2006) Mol. Biol. Cell 17, 4459–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nougarède R., Della, Seta F., Zarzov P., Schwob E. (2000) Mol. Cell. Biol. 20, 3795–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasero P., Duncker B. P., Schwob E., Gasser S. M. (1999) Genes Dev. 13, 2159–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowell S. J., Romanowski P., Diffley J. F. (1994) Science 265, 1243–1246 [DOI] [PubMed] [Google Scholar]
- 24.Francis L. I., Randell J. C., Takara T. J., Uchima L., Bell S. P. (2009) Genes Dev. 23, 643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino K., Takeda T., Matsui E., Iiyama H., Taniyama C., Arai K., Masai H. (2001) J. Biol. Chem. 276, 31376–31387 [DOI] [PubMed] [Google Scholar]
- 26.Sheu Y. J., Stillman B. (2006) Mol. Cell 24, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shellman Y. G., Schauer I. E., Oshiro G., Dohrmann P., Sclafani R. A. (1998) Mol. Gen. Genet. 259, 429–436 [DOI] [PubMed] [Google Scholar]
- 28.Cho W. H., Lee Y. J., Kong S. I., Hurwitz J., Lee J. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11521–11526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochman M. L., Schwacha A. (2008) Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]
- 30.Schwacha A., Bell S. P. (2001) Mol. Cell 8, 1093–1104 [DOI] [PubMed] [Google Scholar]
- 31.Davey M. J., Indiani C., O'Donnell M. (2003) J. Biol. Chem. 278, 4491–4499 [DOI] [PubMed] [Google Scholar]
- 32.Fletcher R. J., Bishop B. E., Leon R. P., Sclafani R. A., Ogata C. M., Chen X. S. (2003) Nat. Struct. Biol. 10, 160–167 [DOI] [PubMed] [Google Scholar]
- 33.Kasiviswanathan R., Shin J. H., Melamud E., Kelman Z. (2004) J. Biol. Chem. 279, 28358–28366 [DOI] [PubMed] [Google Scholar]
- 34.Barry E. R., McGeoch A. T., Kelman Z., Bell S. D. (2007) Nucleic Acids Res. 35, 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoang M. L., Leon R. P., Pessoa-Brandao L., Hunt S., Raghuraman M. K., Fangman W. L., Brewer B. J., Sclafani R. A. (2007) Mol. Cell. Biol. 27, 7594–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bochman M. L., Bell S. P., Schwacha A. (2008) Mol. Cell. Biol. 28, 5865–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.