Abstract
The cellular and molecular pathways that regulate platelet activation, blood coagulation, and inflammation are emerging as critical players in cancer progression and metastasis. Here, we demonstrate a novel signaling mechanism whereby protease-activated receptor 1 (PAR1) mediates expression of melanoma cell adhesion molecule MCAM/MUC18 (MUC18), a critical marker of melanoma metastasis, via activation of platelet-activating factor receptor (PAFR) and cAMP-responsive element-binding protein (CREB). We found that PAR1 silencing with small hairpin RNA inhibits MUC18 expression in metastatic melanoma cells by inhibiting CREB phosphorylation, activity, and binding to the MUC18 promoter. We further demonstrate that the PAF/PAFR pathway mediates MUC18 expression downstream of PAR1. Indeed, PAR1 silencing down-regulates PAFR expression and PAF production, PAFR silencing blocks MUC18 expression, and re-expression of PAFR in PAR1-silenced cells rescues MUC18 expression. We further demonstrate that the PAR1-PAFR-MUC18 pathway mediates melanoma cell adhesion to microvascular endothelial cells, transendothelial migration, and metastatic retention in the lungs. Rescuing PAFR expression in PAR1-silenced cells fully restores metastatic phenotype of melanoma, indicating that PAFR plays critical role in the molecular mechanism of PAR1 action. Our results link the two pro-inflammatory G-protein-coupled receptors, PAR1 and PAFR, with the metastatic dissemination of melanoma and suggest that PAR1, PAFR, and MUC18 are attractive therapeutic targets for preventing melanoma metastasis.
Recent studies have emphasized the importance of the inflammatory tumor microenvironment, blood coagulation, and platelet activation in cancer (1–6). Indeed, tumor cells can coagulate blood and activate platelets via mediators such as thrombin, factor X, tissue factor, fibrinogen, von Willebrand factor, and platelet-activating factor (PAF)2 (3–9). Mediators like thrombin are abundant in the tumor microenvironment; tumor cells, owing to their aberrant expression of tissue factor, catalyze thrombin production on their surface (4, 8). Growing evidence suggests that thrombin not only modifies tumor microenvironment, but also produces direct effect on tumor cells by stimulating G-protein-coupled receptors, also called protease-activated receptors (PARs). Thrombin receptor PAR1 plays a major role in orchestrating the interplay between coagulation and inflammation and stimulates cancer progression (6, 10–12). Recently, we have demonstrated that targeting PAR1 expression in melanoma using small hairpin (sh)RNA or the systemic delivery of small interfering (si)RNA can inhibit melanoma tumor growth and metastasis in nude mice (13).
Thrombin activates PAR1 by inducing proteolytic cleavage of the extracellular amino terminus of PAR1, which acts as a tethered ligand that interacts with the second extracellular loop of PAR1 (14). PAR1 activation stimulates various signaling pathways, including the phospholipase C/protein kinase C, mitogen-activated protein kinase, c-Jun N-terminal kinase, and NF-κB pathways (10, 12). In fibroblasts, epithelial cells, and inflammatory cells, PAR1 mediates the release of inflammatory mediators such as chemokine (C–C motif) ligand 2, IL-1β, IL-6, and IL-8 and the up-regulation of vascular endothelial growth factor, platelet-derived growth factor, and integrins (6, 10, 12).
PAR1 acts as an oncogene capable of transforming NIH3T3 cells in a thrombin-independent manner (15). PAR1 is overexpressed in various tumor types, including melanoma, breast, and prostate cancers (16–18). We have previously demonstrated that PAR1 expression is directly correlated with tumor progression in melanoma and that overexpression of PAR1 stimulates melanoma metastasis in mice (17, 19). Nierodzik et al. also demonstrated that PAR1 expression is a rate-limited factor in thrombin-mediated lung metastasis. In line with these observations, PAR1 overexpression was found to stimulate melanoma cell invasion through Matrigel (20). In breast cancer, PAR1 expression correlates with tumor progression (16) and targeted disruption of PAR1 expression reduces breast cancer cell invasion through the Matrigel (20). In prostate cancer, PAR1 was found to be overexpressed in cell lines derived from bone metastases (18). Additional mechanisms by which thrombin may facilitate tumor growth include its mitogenic effect on melanoma or colon carcinoma cells, as well as its ability to accelerate tumor angiogenesis (16, 20–25). However, the exact molecular mechanisms of how PAR1 contributes to the acquisition of the metastatic phenotype of melanoma are not yet defined.
Herein, we demonstrate that, in melanoma, PAR1 works in concert with another G-protein-coupled receptor, platelet-activating factor receptor (PAFR), to mediate the expression of melanoma cell adhesion molecule MCAM/MUC18 (MUC18) by activating CREB transcription factor. By using RNA interference approach to target PAR1, PAFR, or MUC18 expression, we further demonstrate that PAR1-PAFR-MUC18 pathway mediates melanoma cell adhesion to microvascular endothelial cells, transendothelial migration, and metastatic retention in the lungs. Identification of this new pathway that connects two pro-inflammatory G-protein-coupled receptors, PAR1 and PAFR, with the adhesion molecule MUC18 provides a molecular link between the inflammatory microenvironment and melanoma metastasis.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit polyclonal anti-phospho(Ser-133)-CREB antibody, rabbit polyclonal antibody against total CREB, and rabbit polyclonal antibody against Sp1 were from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal anti-PAFR antibody was from Cayman Biochemicals (Ann Arbor, MI). Fully human anti-MUC18 antibody ABX-MA1 was from Amgen (Thousand Oaks, CA). Mouse monoclonal anti-PAR1 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Lines
A375SM human melanoma cells were described elsewhere (26). C8161-c9 human melanoma cells were a gift from Dr. D. R. Welch (University of Alabama at Birmingham, Birmingham) (27). The human dermal microvascular endothelial cells (HDMECs) were from PromoCell (Germany).
Stable Silencing of PAR1, PAFR, MUC18, and CREB Using Lentiviral Delivery of shRNA
Stable silencing of PAR1 was performed using pLVTHM lentiviral vector coding for shRNA against the human PAR1 gene sequence AGATTAGTCTCCATCAATA as described previously (13). Briefly, sense and antisense shRNA oligonucleotides targeting PAR1 gene and the nontargeting (NT) sequence with no sequence homology to any known human mRNA (TTCTCCGAACGTGTCACGT (Qiagen, Valencia, CA)) were designed with a hairpin and sticky ends (Cla1 and Mlu1) for use with the lentiviral system developed and kindly provided by Didier Trono (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland (28)). The oligonucleotides were annealed into the lentiviral gene transfer vector, pLVTHM, using the Cla1 and Mlu1 restriction enzyme sites. Following sequence verification, lentivirus was produced by transfecting human embryonic kidney cells (293FT, Invitrogen) with the pLVTHM-shNT control or pLVTHM-shPAR1 vector, MD2G packaging plasmid (Invitrogen), and PAX2 envelope plasmid (Invitrogen). Seventy-two hours later, the viral supernatant was collected and filtered to remove cellular debris. The recombinant lentivirus was titrated according to the GFP positivity by flow cytometry in A375SM or C8161-c9 cells. The melanoma cell transduction was typically performed with the viruses at a multiplicity of infection of 5. The highly metastatic and PAR1-positive A375SM and C8161 cell lines were plated at 70% confluency in 6-well plates (5 × 105 cell/well) and, 16 h later, transduced with the virus. After 16 h, the virus-containing medium was removed and replaced with normal growth medium. Following transduction with pLVTHM-shNT control or pLVTHM-shPAR1 virus, PAR1/GFP double-positive (shNT) or PAR1-negative/GFP-positive (shPAR1) cells were selected by fluorescence-activated cell sorting after staining with phycoerythrin-conjugated anti-PAR1 antibody (13).
Stable silencing of PAFR, MUC18, and CREB was achieved by transducing cells with a pLVTHM lentiviral construct coding for shRNA against the human PAFR gene sequence GGGATATCTACTGTGGTCT, or against the human MUC18 gene sequence ACATCGATCTGAGGCATTA, or against the human CREB gene sequence GAGAGAGGTCCGTCTAATG (29), as described above. The transduced, GFP-positive cells were selected by fluorescence-activated cell sorting. PAFR, MUC18, or CREB expression was verified by Western blot.
Transient Silencing of PAFR with siRNA
PAFR siRNA purchased from Dharmacon (Lafayette, CO) was used to silence PAFR expression in melanoma cell lines (target sequence: CAACGUCACUCGCUGCUUU). NT siRNA (Qiagen), with no sequence homology to any known human mRNA, was used as control (target sequence: UUCUCCGAACGUGUCACGU). A375SM or C8161-c9 cells were grown to 60% confluency in 6-well plates and transiently transfected with PAFR siRNA or NT siRNA using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturers' instructions. PAFR protein expression was analyzed 72 h later.
Plasmids, Transduction, and Transfection
For stable PAR1 re-expression (rescue experiment) in C8161-c9 and A375SM PAR1-silenced cells, PAR1 expression construct with an N-terminal prolactin signal peptide and FLAG tag (kindly provided by Shaun R. Coughlin, University of California, San Francisco, CA) was combined with shRNA-non-targetable PAR1 coding region (7-bp silent mutations that will not be recognized by PAR1 shRNA). The resulting open reading frame insert was then ligated into the pLVX-DsRed-Monomer-C1 vector (Clontech, Mountain View, CA) replacing the red protein coding sequence of DsRed. The recombinant lentivirus was produced by transfecting 293FT cells with the sequenced-verified pLVX-PAR1 vector, MD2G packaging plasmid, and PAX2 envelope plasmid (Invitrogen) as described above. The recombinant lentivirus was titrated according to the GFP positivity by flow cytometry in A375SM or C8161-c9 cells. Melanoma cells were seeded in 6-well plates (5 × 105 cell/well) and, 16 h later, transduced with the virus at a multiplicity of infection of 5. Following cell transduction with the control pLVX-ev or pLVX-PAR1 virus, the population of puromycin-resistant cells was selected.
For transient PAFR re-expression in A375SM or C8161-c9 PAR1-silenced cells, the open reading frame of an intronless PAFR gene was amplified from C8161-c9 genomic DNA using forward primer GGGAATTCCATATGGAGCCACATGACTCCTCCCAC and reverse primer CCCAAGCTTCTAATTTTTGAGGGAATTGCCAGG. The PCR product was digested and cloned into a pcDNA3.1 vector. Melanoma cells were plated at a 70% confluency in 6-well plates (5 × 105 cell/well) and, 16 h later, transfected with 4 μg of control empty pcDNA3.1 or PAFR expression vectors. 48 h later, cells were either lysed for protein extraction or collected for the endothelial cell attachment or in vivo lung attachment assays. For the later assay, cells were labeled with [3H]thymidine (20 μCi) for the last 24 h of the post-transfection incubation period (as described below under “Animals and Tumor Cell Lung Seeding Assay”).
For stable MUC18 re-expression in C8161-c9 PAR1-silenced cells, the open reading frame of MUC18 gene was amplified from cDNA prepared by reverse transcription of C8161-c9-derived RNA. The PCR product was digested and cloned into lentiviral pLVTHM expression vector (28). The recombinant lentivirus was produced as described above and titrated according to the GFP positivity by flow cytometry in C8161-c9 cells. Melanoma cells were seeded in 6-well plates (5 × 105 cell/well) and, 16 h later, transduced with the virus at a multiplicity of infection of 5. Following transduction of C8161-c9 shPAR1 cells with control pLVTHM or pLVTHM-MUC18 virus, the population of puromycin-resistant cells was selected.
MUC18 promoter reporter was created by amplifying a 673-bp MUC18 promoter region (−644 to +29 relative to the transcription start site) from C8161-c9 genomic DNA using forward primer CTAGCTAGCACTTGCAGGAGCTTGAGTTTGAAGG and reverse primer GCCCAAGCTTGCTTCCCGGCCGGAGGAGAGCCAAG. The fragment was digested and ligated to a pGL4.12-Luc2-cp vector (Promega, Madison, WI). The cAMP response element (CRE)-driven luciferase reporter vector pGL3-CRE was described previously (26).
Dual Luciferase Reporter Assay
Transient transfections were performed using Lipofectamine 2000 reagent as previously described (26). For promoter reporter assays, 1 μg of the firefly luciferase reporter and 5 ng of CMV-driven Renilla luciferase reporter (pRL-CMV, Promega) plasmids were used for co-transfections of melanoma cells plated in 24-well plates. Luciferase activity was measured 48 h (MUC18 reporter) or 24 h (CRE reporter) post transfection using a dual-luciferase reporter assay system (Promega). The ratio of firefly luciferase activity to Renilla luciferase activity was used to normalize for differences in transfection efficiency. For transient PAFR overexpression, cells were analyzed after 48 h.
Western Blot Analysis
Western blot analysis was performed as previously described (26).
Immunoprecipitation
PAR1 immunoprecipitation was performed using monoclonal anti-PAR1 antibody and protein G-PLUS agarose beads (Santa Cruz Biotechnology) as previously described (26).
Scintillation Proximity Assay of Intracellular PAF
Lipid fractions were removed from cell pellets using a modified version of the Bligh and Dyer extraction method (30); formic acid was added to lower the aqueous phase pH to 3.0 (31). Lipids were purified using lipid purification columns (Amersham Biosciences). PAF levels were quantified using scintillation proximity assay for PAF (Amersham Biosciences) as instructed by the manufacturer. Scintillation proximity assay detects a decrease in binding of a [3H]PAF tracer (also used as a standard) to an anti-PAF antibody, immobilized on scintillant beads, in the presence of cell-derived PAF.
ChIP Assay
ChIP assay was performed using a ChIP-IT Express kit (Active Motif, Carlsbad, CA) as described previously (32). PCR amplification of a 258-bp fragment spanning the −204 to +54 region of the MUC18 promoter was performed using forward primer sequence 5′-AGTGGTAAGACATTTGCCCGAGGT-3′ and reverse primer sequence 5′-TTTCCCTCCCTGACAGCCTTCTTT-3′.
Attachment of Melanoma Cells to HDMECs
Attachment of melanoma cells to HDMECs was assayed in 24-well plates, after 16-h co-culture as described previously for HUVECs (33).
Transendothelial Migration of Melanoma Cells
Cell diapedesis across a monolayer of HDMECs was measured using the electric cell-substrate impedance sensing (ECIS) technique (ECIS Model 1600, Applied BioPhysics, Troy, NY), which detects a decrease in endothelial cell resistance due to retraction of endothelial cell monolayers after the addition of melanoma cells. Briefly, 8-well ECIS arrays were coated with rat-tail collagen type I (BD Biosciences), and HDMECs (5 × 105 cells per well) or media only were plated and allowed to form monolayers. A baseline impedance measurement for the endothelial monolayer was monitored for 3–4 h. Then, melanoma cells (5 × 105 cells/well) were added. During 24-h data acquisition, the impedance of HDMECs alone remained stable. Data plots are representative of triplicate experiments, with each graph showing impedance readings from a separate well. Each time point is the mean impedance, normalized to the initial value, at 10 distinct electrodes per well.
Animals and Tumor Cell Lung Seeding Assay
All experiments with female athymic BALB/c nude mice (NCI-Frederick Cancer Research Facility, Frederick, MD) were performed according to regulations approved by the Institutional Animal Care and Use Committee. For lung seeding assay, melanoma cells were labeled with [3H]thymidine (20 μCi) for 24 h. Aliquots were used to determine [3H]thymidine incorporation (cpm/1 × 106 cells). A total of 0.5 × 106 cells was injected into the lateral tail veins of nude mice. Twenty-four hours after injection, the mice were killed, and the lungs were resected, washed with water, and weighed. Tissues were digested in 10 n NaOH solution for 24 h. Radioactivity was counted, normalized to the injected cpm, and expressed as the amount of radioactivity retained per gram of tissue.
Statistical Analysis
Results were evaluated using a two-tailed Student's t test. p values < 0.05 were considered statistically significant.
RESULTS
PAR1 Silencing Down-regulates MUC18 Expression in Metastatic Melanoma Cells
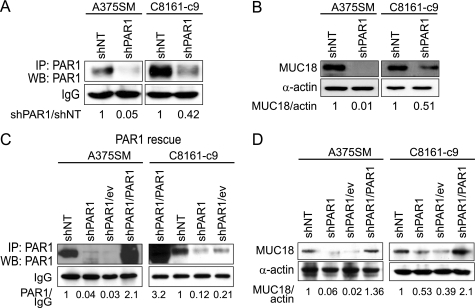
We have previously shown that PAR1 expression increases during melanoma progression (17). Furthermore, down-regulating PAR1 expression in metastatic melanoma cells significantly inhibits their metastatic potential in vivo (13). To investigate the molecular mechanisms of how PAR1 contributes to melanoma metastasis, we stably silenced PAR1 expression in two highly metastatic melanoma cell lines, A375SM and C8161-c9, using a small hairpin RNA (shRNA) and a lentiviral delivery system, described previously (13). Immunoprecipitation of cell lysates with anti-PAR1 antibody followed by Western blot analysis confirmed that PAR1 expression was down-regulated by 95% in A375SM cells and by 58% in C8161-c9 cells that had been transduced with shRNA targeting PAR1 (shPAR1), as compared with cells that had been transduced with nontargeting shRNA (shNT) (Fig. 1A).
FIGURE 1.
PAR1 regulates the expression of melanoma cell adhesion molecule MUC18 in metastatic melanoma. A, PAR1 silencing (shPAR1) using lentiviral shRNA results in down-regulation of PAR1 expression in A375SM and C8161-c9 cells as compared with control cells transduced with the nontargeting shRNA (shNT). Cell lysates were immunoprecipitated using anti-PAR1 antibody, and PAR1 expression was further detected by Western blot. The IgG band served as an indicator of equal loading. B, Western blot analysis demonstrates that PAR1 silencing results in down-regulation of MUC18 expression in A375SM and C8161-c9 cells as compared with control cells transduced with the nontargeting shRNA. C, stable re-expression of PAR1 (shPAR1/PAR1) in A375SM or C8161-c9 PAR1-silenced cells using lentiviral transduction as demonstrated by immunoprecipitation with anti-PAR1 antibody followed by Western blot. Note an increase in PAR1 levels in shPAR/PAR1 cells as compared with shPAR1 cells transduced with the empty expression vector (shPAR1/ev). The IgG band served as an indicator of equal loading. Please note that the loading order in C8161-c9 cells (right panel) is different from A375SM cells (left panel). D, re-expression of PAR1 (shPAR1/PAR1) in A375SM or C811-c9 PAR1-silenced cells rescues MUC18 expression as compared with PAR1-silenced cells transduced with the empty expression vector (shPAR1/ev).
The A375SM PAR1-silenced cells and cells transduced with non-targeting control shRNA were subjected to cDNA microarray to identify possible downstream target genes regulated by PAR1, as previously described (34). The melanoma cell adhesion molecule MCAM/MUC18 was found to be down-regulated by 9.6-fold following PAR1 silencing in A375SM cells. We and others have previously demonstrated that MUC18 plays a major role in melanoma metastasis by mediating heterotypic interactions between metastatic melanoma cells and endothelial cells (33, 35, 36). Therefore, we next tested the hypothesis that PAR1 regulates melanoma metastasis at list in part by stimulating MUC18 expression. Western blot analysis revealed that PAR1 silencing strongly diminished MUC18 expression in A375SM and C8161-c9 cell lines by 99 and 49%, respectively (Fig. 1B).
To confirm the specificity of shRNA against PAR1, we rescued PAR1 expression in PAR1-silenced cells using lentiviral shRNA-nontargetable PAR1 expression vector. Following cell transduction with the control pLVX-ev or pLVX-PAR1 virus and puromycin selection, the levels of PAR1 protein in PAR1 re-expressing cells (shPAR1/PAR1) increased dramatically, as compared with cells transduced with empty vector (shPAR1/ev) (Fig. 1C). (Please note that the loading order for C8161 cells (Fig. 1C, right panel) is different from the A375SM cells (Fig. 1C, left panel).) PAR1 re-expression rescued MUC18 expression both in A375SM and C8161-c9 cells (Fig. 1D). This experiment confirmed that MUC18 is indeed a specific downstream target gene of PAR1.
PAR1 Regulates MUC18 Expression via CREB Activation
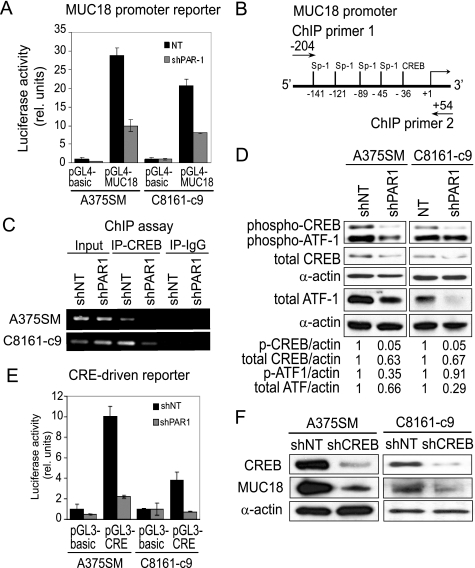
To understand the mechanism of PAR1-induced MUC18 expression, we cloned the MUC18 promoter (region from −644 to +29 bp relative to the transcription start site) and monitored the reporter-associated activity before and after PAR1 silencing using the dual luciferase assay. We found that PAR1 silencing down-regulated the activity of the MUC18 promoter-driven luciferase reporter gene in A375SM and C8161-c9 cells by 2.9- and 2.6-fold, respectively (Fig. 2A). This result suggested that PAR1 signaling mainly affected MUC18 expression at the transcriptional level. An analysis of the proximal MUC18 promoter region using MatInspector (Genomatix Software GmbH) revealed that this region contained a CREB binding site as well as four stimulating protein 1 (Sp1)-binding motifs (Fig. 2B). As demonstrated previously using promoter mutation analysis, both CREB and Sp1 can regulate MUC18 transcription (37, 38). In addition, we have previously found that dominant negative CREB inhibited the expression of MUC18 in metastatic melanoma cells (39). Therefore, we sought to determine whether PAR1 regulates MUC18 transcription via activation of CREB and Sp1. To monitor the in vivo binding of CREB to the promoter of MUC18 before and after PAR1 silencing, we performed a chromatin immunoprecipitation (ChIP) assay. ChIP analysis demonstrated that CREB was bound to the MUC18 promoter in control shNT cells, whereas minimal CREB binding was detected in C8161-c9shPAR1 cells, and no CREB binding was detected in A375SM cells after PAR1 silencing (Fig. 2C).
FIGURE 2.
PAR1 regulates MUC18 expression via CREB. A, dual luciferase assay demonstrates that PAR1 silencing inhibits activity of the MUC18 promoter reporter (673 bp, pGL4-MUC18) in A375SM and C8161-c9 cells as compared with control cells transduced with the nontargeting shRNA. B, a schematic of the 5′-flanking sequence of the MUC18 gene depicts possible binding sites for CREB and Sp1 transcription factors, as indicated by the position of the 3′-end of each site relative to the transcription start site. A schematic also demonstrates position of primers used in the ChIP assay. C, ChIP analysis demonstrates that PAR1 silencing (shPAR1) inhibits CREB binding to MUC18 promoter as compared with control cells transduced with the nontargeting shRNA (shNT). D, Western blot shows decrease in CREB and ATF-1 phosphorylation and expression following PAR1 silencing. E, as measured by dual luciferase assay, PAR1 silencing inhibits activity of the CRE-driven luciferase reporter. F, CREB silencing (shCREB) using lentiviral shRNA results in down-regulation of CREB expression in A375SM and C8161-c9 cells as compared with control cells transduced with the nontargeting shRNA (shNT). Western blot analysis further demonstrates that CREB silencing results in down-regulation of MUC18 expression.
The transcriptional activity of CREB is regulated by both its DNA binding and N-terminal phosphorylation, the latter of which facilitates CREB interaction with the p300/CREB-binding protein (40). We found that, after PAR1 silencing, CREB phosphorylation at serine 133 was strongly decreased (by 95%) in A375SM and C8161-c9 cells (Fig. 2D). Phosphorylation of activating transcription factor 1 (ATF-1), the CREB heteromerization partner, was also decreased (Fig. 2D). We also observed a slight decrease in total CREB levels in both shPAR1 cell lines (by 37% in A375SM cells and 33% in C8161-c9 cells), and a significant decrease in total ATF-1 levels (Fig. 2D). Consequently, PAR1 silencing decreased CRE-driven luciferase reporter activity in A375SM and C8161-c9 cells by 4.5- and 5-fold, respectively (Fig. 2E). Finally, we used lentiviral transduction to silence CREB expression using shRNA, as described elsewhere (29), and found that CREB silencing down-regulated MUC18 expression in A375SM and C8161-c9 cell (shCREB) as compared with cells transduced with shNT virus (Fig. 2F). Taken together, these data indicate that PAR1 silencing inhibits MUC18 expression at the transcriptional level by down-regulating the expression, phosphorylation, and DNA binding of CREB.
PAR1 Regulates MUC18 Expression via CREB-dependent Sp1 Modulation
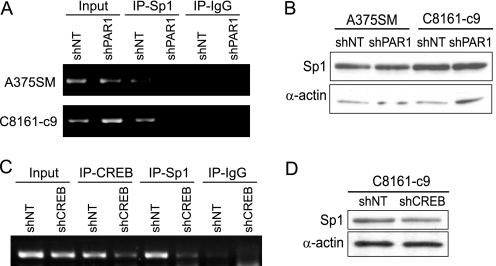
In addition to the CREB binding motif, the proximal MUC18 promoter sequence contains four Sp1 binding motifs (Fig. 2B). ChIP analysis revealed a decrease in the binding of Sp1 to the MUC18 promoter after PAR1 silencing (Fig. 3A). No changes in Sp1 protein levels were detected (Fig. 3B). To determine whether the loss of CREB binding affected Sp1 recruitment, we used ChIP to analyze Sp1 binding in CREB-silenced cells (C8161-c9 shCREB) and found that the loss of CREB inhibited the binding of Sp1 to the MUC18 promoter (Fig. 3C). Western blot analysis revealed that Sp1 protein levels remained unaltered after CREB silencing in C8161-c9 cells (Fig. 3D). Taken together, these results suggest that PAR1 regulates MUC18 transcription by regulating the binding of CREB and Sp1 to the MUC18 promoter and that the DNA binding of Sp1 is CREB-dependent.
FIGURE 3.
PAR1 and CREB regulate Sp1. A, PAR1 silencing down-regulates stimulatory protein-1 (Sp1) binding to the −204 to +54 promoter region of the MUC18 gene, as demonstrated by ChIP analysis. B, Western blot shows that PAR1 silencing does not affect the levels of Sp1 expression. C, CREB silencing (shCREB) down-regulates Sp1 binding to the −204 to +54 promoter region of the MUC18 gene as compared with cells transduced with the control non-targeting shRNA (shNT), as demonstrated by ChIP analysis. D, Western blot shows that CREB silencing does not affect the levels of Sp1 expression.
PAFR Mediates MUC18 Expression Downstream of PAR1
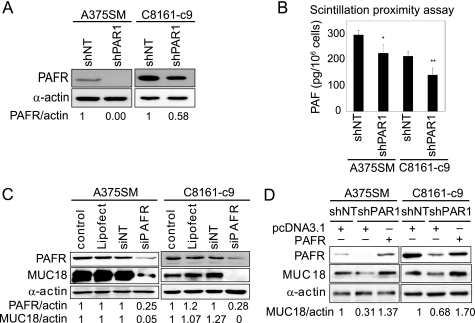
Recently, we demonstrated that biolipid PAF can stimulate the phosphorylation and activation of CREB and ATF-1 in melanoma cells (26). PAF is an essential mediator of platelet activation and inflammation that has been shown to promote tumor growth and angiogenesis of breast, prostate, non-melanoma, and melanoma skin cancers (41–43). Intriguingly, a number of biologically active compounds, including lipopolysaccharide, IL-1, or tumor necrosis factor-α rely on PAF-PAFR pathway to induce their downstream effects (44, 45). We therefore hypothesized that PAFR mediates MUC18 expression downstream of PAR1. To investigate this possible connection between PAR1 and PAFR in melanoma, we first monitored the levels of PAFR expression and PAF production after PAR1 silencing. Western blot analysis demonstrates that PAR1 silencing inhibited PAFR expression by >99% in A375SM cells and by 42% in C8161-c9 cells (Fig. 4A). The higher basal expression levels of PAR1 and PAFR may account for the weaker inhibition of their expression in the C8161-c9 cells. Notably, C8161-c9 cells also have higher metastatic potential in vivo than A375SM cells (13). Alternatively, the difference in degree of silencing could be explained by variations in transduction efficiency (shRNA incorporation and expression), or shRNA processing efficiency. Indeed, we have recently demonstrated variability in the expression of Dicer and Drosha, components of the RNA-interference machinery, in ovarian cancer (46).
FIGURE 4.
PAR1 regulates MUC18 expression in a PAFR-dependent manner. A, PAR1 silencing (shPAR1) down-regulates PAFR expression in A375SM and C8161-c9 metastatic melanoma cells as compared with cells transfected with the control non-targeting shRNA (shNT) (Western blot). B, PAR1 silencing reduces intracellular PAF concentrations in A375SM and C8161-c9 cells, as demonstrated by scintillation proximity assay. Data are mean ± S.D. from three experiments performed in triplicates. *, p < 0.1; **, p < 0.01. C, silencing PAFR expression with small interfering (si)RNA (siPAFR) significantly decreased MUC18 protein expression in A375SM and C8161-c9 melanoma cells as compared with parental cells, cells transfected with Lipofectamine 2000 (Lipofect), or cells transfected with the control non-targeting siRNA (siNT). D, MUC18 expression is rescued in shPAR1 cells transfected with PAFR as compared with shPAR1 cells transfected with control empty pcDNA3.1 vector, suggesting that PAFR mediates MUC18 expression downstream of PAR1.
To determine whether PAR1 silencing affected PAF production in melanoma cells, we used scintillation proximity assay for PAF. After PAR1 silencing, intracellular PAF levels in A375SM and C8161-c9 cells decreased by 24% (p < 0.1) and 35% (p < 0.01), respectively (Fig. 4B). Because PAF production was influenced only modestly, it suggested that PAR1 only partly contributed to generation of PAF by melanoma cells. Collectively, these findings suggest that PAR1 regulates basal PAF production and PAFR expression in metastatic melanoma cells.
To determine whether PAFR also regulates MUC18 expression, we used siRNA to transiently silence PAFR expression. Treating A375SM and C8161-c9 cells with siRNA down-regulated PAFR expression by 75% in both cell lines, which coincided with a strong down-regulation of MUC18 expression in A375SM cells (95%) and a complete loss of MUC18 expression in C8161-c9 cells (Fig. 4C).
Because silencing either PAR1 or PAFR was sufficient to down-regulate MUC18 expression, and because PAR1 silencing inhibited PAFR expression, we hypothesized that PAFR was required for the PAR1-dependent regulation of MUC18 expression. We therefore rescued PAFR expression in PAR1-silenced A375SM and C8161-c9 cells and found that MUC18 expression was restored following transient PAFR overexpression (Fig. 4D), indicating that PAR1 induces MUC18 expression via a PAFR-dependent mechanism. This is the first demonstration of a connection between the two G-protein coupled receptors, PAR1 and PAFR.
PAR1, PAFR, or MUC18 Silencing Inhibits the Attachment of Melanoma Cells to Human Dermal Microvascular Endothelial Cells and the Transendothelial Migration of Tumor Cells
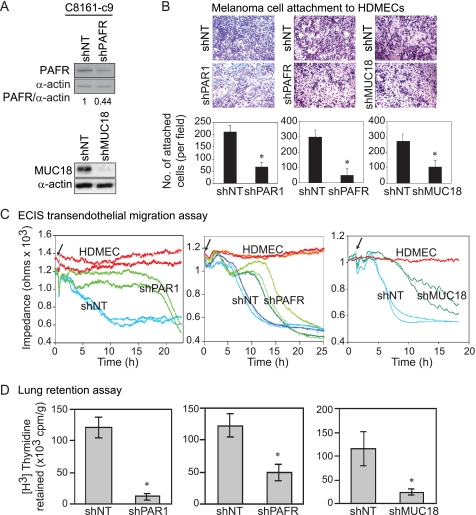
Because melanoma cells interact with HUVECs through MUC18-dependent adhesion (33, 35, 47), we next examined the role of the PAR1, PAFR, and MUC18 in the attachment of melanoma cells to HDMECs as a possible mechanism by which PAR1 contributes to melanoma metastasis. We used lentiviral delivery of shRNA to stably silence PAFR and MUC18 expression in C8161-c9 cells. Following lentiviral transduction and GFP-based selection, PAFR expression was down-regulated by 56%, and MUC18 expression was decreased by >95% (Fig. 5A). As in the case of transient PAFR silencing with siRNA, stable PAFR silencing in C8161-c9 cells resulted in down-regulation of MUC18 expression (data not shown). PAR1, PAFR, or MUC18 silencing significantly reduced the attachment of melanoma cells to HDMECs by 69% (p < 0.0001), 84% (p = 0.0002), and 62% (p = 0.005), respectively (Fig. 5B).
FIGURE 5.
Silencing PAR1, PAFR, or MUC18 in C8161-c9 cells decreased their adhesion to HDMECs, diapedesis, and retention in mouse lungs. A, Western blot analysis demonstrates down-regulation of PAFR or MUC18 proteins in C8161-c9 cells transduced with lentiviral PAFR-targeting shRNA (shPAFR) or MUC18-targeting shRNA (shMUC18). B, silencing PAR1, PAFR, or MUC18 in C8161-c9 cells decreased their adhesion to HDMECs. Representative images and quantitative results (bar graphs) demonstrate that PAR1, PAFR, or MUC18 silencing down-regulated endothelial cell attachment by 69%, 84%, and 62%, respectively. C, representative ECIS plots show a decrease in impedance of HDMEC monolayers on the addition of control C8161-c9 shNT cells (shNT), indicating transendothelial migration of melanoma cells (blue lines in each panel). Arrows indicate the addition of melanoma cells to HDMECs. PAR1 (shPAR1), PAFR (shPAFR), or MUC18 (shMUC18) silencing inhibited transendothelial cell migration (green lines). The impedance in samples with HDMECs supplemented with media alone remained stable during the entire experiment (red lines). D, PAR1, PAFR, or MUC18 silencing inhibited melanoma cell retention in the lung tissue of nude mice by 89%, 61%, and 79%, respectively. Tumor cells retention was measured 24 h after tail-vein injection and is expressed as the amount of radioactivity retained per gram of tissue. *, p < 0.01.
Tumor cell migration through the endothelial cells layer (diapedesis) is an integral step of the metastatic cascade. Therefore, we next used ECIS to assess the ability of melanoma cells to migrate across HDMEC monolayers (48). ECIS is a novel technique that allows real-time monitoring of tumor cell diapedesis. When HDMECs were grown in a monolayer on collagen-coated ECIS electrodes, the current flow was impeded with impedance being proportional to the adhesive properties of the cells (Fig. 5C). Layering C8161shNT cells (shNT) on top of the HDMECs decreased impedance (Fig. 5C), which reflected a loss of HDMEC monolayer integrity due to endothelial cell retraction (48). As evidenced by no decrease in impedance for up until 19 h after the addition of the C8161-c9 shPAR1 cells (shPAR1), diapedesis was strongly inhibited after PAR1 silencing (Fig. 5C, left panel). Silencing PAFR expression by 56% (Fig. 5A) resulted in a mean delay of 6 h in melanoma cell diapedesis (Fig. 5C, middle panel). Indeed, whereas 50% shNT cells migrated through the HDMEC monolayer ∼9.5 h after layering, it took 15.5 h for 50% of shPAFR cells to migrate (Fig. 5C, middle panel). MUC18 silencing produced a 7-h delay in melanoma cell diapedesis (Fig. 5C, right panel). Diapedesis inhibition after PAR1, PAFR, or MUC18 silencing could have been partly due to the decreased capacity of the melanoma cells to attach to HDMECs.
PAR1, PAFR, or MUC18 Silencing Inhibits Melanoma Cell Colonization in the Lungs
In a previous study, we found that PAR1 silencing inhibited melanoma metastasis after tail-vein injections of melanoma cells (13). The findings of the current study provide a mechanistic explanation for this effect and suggest that the PAR1, PAFR, and MUC18 are essential for melanoma cell adhesion to endothelial cells and transendothelial migration. To further investigate this mechanism, we examined the in vivo ability of melanoma cells to be retained in the lungs of nude mice following tail-vein injections of C8161-c9 melanoma cells before and after PAR1, PAFR, or MUC18 silencing. PAR1 silencing resulted in an 89% reduction in the ability of melanoma cells to seed in the lungs (Fig. 5D, left panel). Similarly, PAFR and MUC18 silencing resulted in decreases in the number of melanoma cells retained in the lungs of 61 and 79%, respectively (Fig. 5D, middle and right panels). These findings suggest that PAR1, PAFR, and MUC18 play a critical role in the metastatic arrest of melanoma cells in the lungs.
MUC18 Overexpression Partly Rescues PAR1 Functions, and PAFR Overexpression Fully Rescues PAR1 Functions in PAR1-silenced Cells
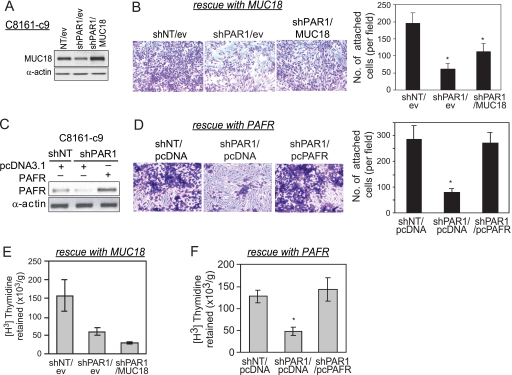
To determine the exact contribution of MUC18 or PAFR in PAR1-mediated melanoma metastasis, we rescued MUC18 or PAFR expression in C8161-c9 PAR1-silenced cells. Fig. 6A demonstrates that MUC18 protein levels increased by 2.3-fold after its stable re-expression in PAR1-silenced cells using lentiviral transduction (shPAR1/MUC18), as compared with shPAR1 cells transduced with empty vector virus (shPAR1/ev). To determine whether MUC18 mediates the attachment of melanoma cells to endothelial cells downstream of PAR1, we measured melanoma cell attachment to HDMECs after MUC18 rescue. Fig. 6B demonstrates that MUC18 re-expression partly restores the adhesion of PAR1-silenced cells to HDMECs (Fig. 6B). Indeed, PAR1 silencing down-regulated endothelial cell attachment by ∼70% (shPAR1/ev) as compared with control cell line (shNT/ev), whereas MUC18 re-expression (shPAR1/MUC18) restored melanoma cell adhesion to endothelial cells to 60% of the initial values seen in shNT/ev cells (Fig. 6B). Nevertheless, MUC18 re-expression alone was insufficient to repair the melanoma cells' ability to be retained in the lungs (Fig. 6E).
FIGURE 6.
MUC18 overexpression partly rescues PAR1 function, whereas PAFR overexpression completely rescues PAR1 function in C8161-c9 cells. A, Western blot analysis demonstrated stable overexpression of MUC18 in C8161-c9 shPAR1 cells (shPAR1/MUC18) as compared with shPAR1 cells transduced with control empty-vector lentiviral expression plasmid (shPAR1/ev). B, rescuing MUC18 expression in PAR1-silenced cells partly restored the ability of PAR1-silenced cells to attach to HDMECs. Representative images and quantitative results demonstrate that PAR1 silencing down-regulated endothelial cell attachment (shPAR1/ev) as compared with control cell line (shNT/ev). MUC18 overexpression (shPAR1/MUC18) partly restored melanoma cell adhesion to endothelial cells. Data are mean ± S.D. from three experiments performed in triplicates. C, Western blot analysis demonstrated transient overexpression of PAFR in C8161-c9 shPAR1 cells as compared with shPAR1 cells transfected with control empty-vector pcDNA3.1 plasmid. D, rescuing PAFR expression in PAR1-silenced cells restored the ability of PAR1-silenced cells to attach to HDMECs. Representative images and quantitative results demonstrate that PAR1 silencing down-regulated endothelial cell attachment by ∼70% (shPAR1/pcDNA) as compared with control cell line (shNT/pcDNA). PAFR overexpression (shPAR1/pcPAFR) restored melanoma cell adhesion to endothelial cells to 94% of the initial values seen in shNT/pcDNA cells. Data are mean ± S.D. from three experiments performed in triplicates. E, overexpression of MUC18 in PAR1-silenced cells (shPAR/MUC18) did not rescue the loss of melanoma cell retention in the lung after PAR1 silencing (shPAR1/ev). Tumor cell retention was measured 24 h after tail-vein injection and is expressed as the amount of radioactivity retained per gram of tissue. *, p < 0.01. F, overexpression of PAFR in PAR1-silenced cells (shPAR1/pcPAFR) rescued the loss of melanoma cell retention in the lung tissue after PAR1 silencing (shPAR1/pcDNA). Tumor cell retention was measured 24 h after tail-vein injection and is expressed as the amount of radioactivity retained per gram of tissue. *, p < 0.01.
We then rescued PAFR expression in PAR1-silenced cells to determine whether PAFR mediates the attachment of melanoma cells to endothelial cells and lung metastasis downstream of PAR1. Increased PAFR levels were detected after PAFR transient overexpression in shPAR1 cells as compared with shPAR1 cells transfected with empty vector (Fig. 6C). PAFR overexpression completely restored the adhesion of PAR1-silenced cells to HDMECs (Fig. 6D). Furthermore, PAFR re-expression in shPAR1 cells completely restored the melanoma cells' ability to be retained in the lungs (Fig. 6F).
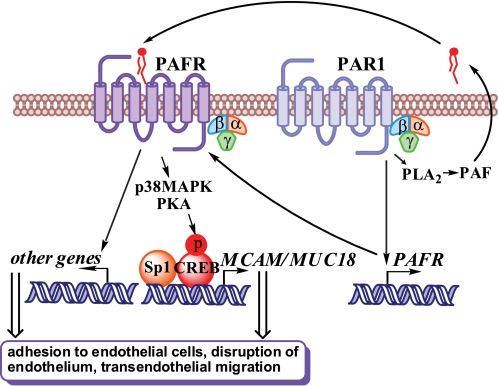
Taken together, our findings indicate that MUC18 is necessary but not sufficient in the absence of functional PAR1 to produce metastatic phenotype in melanoma. Our data further suggest that PAFR plays a central role in activating PAR1-induced metastatic phenotype and propose a novel mechanism linking the PAR1 and PAFR to melanoma metastasis. Fig. 7 depicts a putative model whereby constitutive activation of PAR1 in metastatic melanoma cells leads to the activation of PAFR signaling due to an increased expression of the PAF receptor and production of its ligand, PAF. Activation of PLA2 downstream of PAR1 may be responsible for the enzymatic generation of PAF: our preliminary analysis suggests that PAR1 silencing inhibited the phosphorylation of cPLA2 at serine 505, which is known to positively correlate with the enzyme's activity (data not shown). Our model further suggests that activation of PAFR stimulates phosphorylation of CREB (26) and the recruitment of CREB and Sp1 to the promoter of MUC18, thereby triggering MUC18 expression in metastatic melanoma. MUC18 acts as a rate-limiting factor in cancer cell adhesion to endothelium and cooperates with other downstream target genes of PAFR to enable the diapedesis and dissemination of melanoma cells into the distant organs.
FIGURE 7.
A putative model of cooperation between PAR1 and PAFR in melanoma metastasis. Constitutive PAR1 activation in melanoma cells causes PAFR expression, possibly through transcriptional activation, and PAF production, presumably through activation of PLA2. The PAF-PAFR signaling mediates PAR1-induced MUC18 expression via CREB phosphorylation. PAF can induce phosphorylation of CREB via activation of p38 MAPK and protein kinase A (PKA) (26). The binding of CREB to the promoter of MUC18 is accompanied by CREB-dependent Sp1 recruitment, resulting in the activation of MUC18 expression. MUC18 induces the adhesion of melanoma cells to endothelial cells and cooperates with other possible downstream targets of the PAR1-PAFR pathway to induce endothelial wall disruption, the transendothelial migration of melanoma cells, and the accumulation of melanoma cells in the lung.
DISCUSSION
We found that aberrantly activated PAR1 mediates PAF production, PAFR expression, CREB and Sp1 activation, and MUC18 expression in metastatic melanoma (Fig. 7). We also found that PAR1, PAFR, or MUC18 silencing strongly inhibited melanoma cell adhesion to endothelial cells, transendothelial migration, and retention in the lungs. Finally, we found that the re-expression of PAFR alone is sufficient to induce MUC18 expression and restore the effects of PAR1 on melanoma cell adhesion to HDMECs and accumulation in the lungs. Meanwhile, MUC18 is necessary but not sufficient in the absence of functional PAR1 to produce metastatic phenotype in melanoma. Collectively, these findings link PAR1 and PAFR with melanoma metastasis through identification of a novel common downstream target, an adhesion molecule MUC18 (Fig. 7).
MUC18 is a transmembrane glycoprotein that belongs to the immunoglobulin superfamily and functions as a Ca2+-independent adhesion molecule (47, 49, 50). MUC18 is thought to mediate the homotypic and heterotypic adhesion of melanoma cells to each other or to HUVECs by interacting with an unknown heterophilic ligand (35, 47). We previously found that MUC18 confers invasive and metastatic capability to melanoma cells by up-regulating MMP-2 expression (35). Blocking MUC18 by fully human antibodies inhibited tumor growth and metastasis and down-regulated MMP-2 expression and invasion of human melanoma (33). Using the genetic suppressor elements of MUC18 cDNA to down-regulate MUC18 expression inhibited melanoma cell aggregation, growth in soft agar, and tumorigenicity (51). The cytoplasmic domain of MUC18 can bind p59fyn, resulting in the phosphorylation of p125FAK and the binding of p125FAK to paxillin, implicating MUC18 in the focal adhesion assembly and cell migration (52). Together with the findings of the current study, these data indicate that MUC18 plays a rate-limiting role in melanoma metastasis. However, our data also suggest that MUC18 collaborates with other downstream targets of PAR1 to stimulate melanoma metastasis. Other surface adhesion proteins and vascular permeability factors that have been shown to influence tumor-to-endothelial cell adhesion and diapedesis include N-cadherin, Thomsen-Friedenreich glycoantigen and galectin-3, integrins α4β1, α5β1, and α3β2, P-selectin, vascular cell adhesion molecule-1, intercellular adhesion molecule-1, vascular endothelial growth factor, and transforming growth factor-β (22, 53–57). However, whether or not some of these mediators are downstream targets of PAR1 in melanoma has not been yet investigated.
Intriguingly, our results suggest that PAF plays a critical role in mediating functions of PAR1. PAF is an essential inflammatory biolipid. It is a mediator of platelet activation and inflammation that has been suggested to promote tumor growth and angiogenesis of breast, prostate, non-melanoma, and melanoma skin cancers (41–43). Although the molecular mechanisms of PAFR action in cancer remain to be investigated, in various normal cell types, PAF induces the expression of inflammatory and angiogenic molecules such as IL-6, IL-8, IL-10, cyclooxygenase-2, vascular endothelial growth factor, inducible nitric-oxide synthase, and tumor necrosis factor-α (58–61). Recently, we have demonstrated that PAFR antagonists inhibit human melanoma metastasis in nude mice, and that PAF contributes to melanoma metastasis by inducing phosphorylation of CREB and consequent expression of matrix metalloproteinase 2 (MMP-2) and membrane type 1 MMP (MT1-MMP) by melanoma cells (26).
Notably, we show that PAR1 mediates basal PAF production and PAFR expression in melanoma. A number of biologically active compounds including lipopolysaccharide, IL-1, or tumor necrosis factor-α rely on PAF-PAFR pathway to induce their downstream effects (44, 45). Our preliminary analysis suggested that decreased levels of PAF following PAR1 silencing were correlated with a strong inhibition of the phosphorylation of cPLA2 at serine 505, which is known to positively correlate with the activity of the enzyme (Fig. 7). Experiments are in progress to understand the exact mechanism of how PAR1 mediates PAFR expression on the transcriptional level. Our preliminary immunoprecipitation analysis revealed no evidence of a protein-protein interaction between PAR1-PAFR, therefore ruling out PAFR protein stabilization.
Analyzing the mechanism of MUC18 expression revealed a link between the PAR1-PAFR pathway and CREB and Sp1 activation. This is the first demonstration of PAR1-induced phosphorylation and activation of CREB. Intriguingly, we found that the binding of Sp1 to the MUC18 promoter depends on the level of CREB expression and CREB recruitment to the promoter. The mechanism of this effect is under investigation. Because we previously established that Sp1 mediates PAR1 expression (19), our new results suggest the existence of a positive feedback loop between PAR1 expression and Sp1 activation in melanoma.
Overall, our findings suggest that PAR1 plays a pivotal role in regulating melanoma cell adhesion to endothelial cells, transendothelial migration, and retention in secondary organs. The molecular mechanism of action of PAR1 involves MUC18 transactivation through the PAF-PAFR signaling and CREB and Sp1 activation. Therefore, the PAR1-PAFR-MUC18 axis is an attractive target for the prevention of melanoma metastasis.
Acknowledgment
We thank the University of Texas M. D. Anderson Cancer Center Dept. of Neuro-Oncology for the use of the ECIS apparatus.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1CA760980 (to M. B.-E.). This work was also supported by Career Development Award P50CA093459 from NIH SPORE in Skin Cancer and from Melanoma Research Foundation (to V. M.).
- PAF
- platelet-activating factor
- PAFR
- PAF receptor
- PAR
- protease-activated receptor
- shRNA
- small hairpin RNA
- siRNA
- small interference RNA
- IL-1
- interleukin-1
- NT
- nontargeting
- CRE
- cAMP response element
- CREB
- cAMP-responsive element-binding protein
- HDMEC
- human dermal microvascular endothelial cell
- GFP
- green fluorescent protein
- shNT
- nontargeting shRNA
- CMV
- cytomegalovirus
- ECIS
- electric cell-substrate impedance sensing
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type 1 MMP
- MAPK
- mitogen-activated protein kinase
- MUC18
- melanoma cell adhesion molecule MCAM/MUC18.
REFERENCES
- 1.DeNardo D. G., Johansson M., Coussens L. M. (2008) Cancer Metastasis Rev. 27, 11–18 [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 3.Bromberg M. E., Konigsberg W. H., Madison J. F., Pawashe A., Garen A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8205–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer E. G., Ruf W., Mueller B. M. (1995) Cancer Res. 55, 1629–1632 [PubMed] [Google Scholar]
- 5.Gasic G. J., Gasic T. B., Stewart C. C. (1968) Proc. Natl. Acad. Sci. U.S.A. 61, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nierodzik M. L., Karpatkin S. (2006) Cancer Cell 10, 355–362 [DOI] [PubMed] [Google Scholar]
- 7.Tímár J., Tóvári J., Rásó E., Mészáros L., Bereczky B., Lapis K. (2005) Oncology 69, 185–201 [DOI] [PubMed] [Google Scholar]
- 8.Ruf W., Mueller B. M. (2006) Semin. Thromb. Hemostasis 32, Suppl. 1, 61–68 [DOI] [PubMed] [Google Scholar]
- 9.Versteeg H. H., Schaffner F., Kerver M., Petersen H. H., Ahamed J., Felding-Habermann B., Takada Y., Mueller B. M., Ruf W. (2008) Blood 111, 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niessen F., Schaffner F., Furlan-Freguia C., Pawlinski R., Bhattacharjee G., Chun J., Derian C. K., Andrade-Gordon P., Rosen H., Ruf W. (2008) Nature 452, 654–658 [DOI] [PubMed] [Google Scholar]
- 11.Chen D., Carpenter A., Abrahams J., Chambers R. C., Lechler R. I., McVey J. H., Dorling A. (2008) J. Exp. Med. 205, 1739–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer P. F., Deng X., Chambers R. C. (2007) Ann. N.Y. Acad. Sci. 1096, 86–88 [DOI] [PubMed] [Google Scholar]
- 13.Villares G. J., Zigler M., Wang H., Melnikova V. O., Wu H., Friedman R., Leslie M. C., Vivas-Mejia P. E., Lopez-Berestein G., Sood A. K., Bar-Eli M. (2008) Cancer Res. 68, 9078–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coughlin S. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11023–11027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin C. B., Mahon G. M., Klinger M. B., Kay R. J., Symons M., Der C. J., Whitehead I. P. (2001) Oncogene 20, 1953–1963 [DOI] [PubMed] [Google Scholar]
- 16.Even-Ram S., Uziely B., Cohen P., Grisaru-Granovsky S., Maoz M., Ginzburg Y., Reich R., Vlodavsky I., Bar-Shavit R. (1998) Nat. Med. 4, 909–914 [DOI] [PubMed] [Google Scholar]
- 17.Tellez C. S., Davis D. W., Prieto V. G., Gershenwald J. E., Johnson M. M., McCarty M. F., Bar-Eli M. (2007) J. Invest. Dermatol. 127, 387–393 [DOI] [PubMed] [Google Scholar]
- 18.Chay C. H., Cooper C. R., Gendernalik J. D., Dhanasekaran S. M., Chinnaiyan A. M., Rubin M. A., Schmaier A. H., Pienta K. J. (2002) Urology 60, 760–765 [DOI] [PubMed] [Google Scholar]
- 19.Tellez C., McCarty M., Ruiz M., Bar-Eli M. (2003) J. Biol. Chem. 278, 46632–46642 [DOI] [PubMed] [Google Scholar]
- 20.Even-Ram S. C., Maoz M., Pokroy E., Reich R., Katz B. Z., Gutwein P., Altevogt P., Bar-Shavit R. (2001) J. Biol. Chem. 276, 10952–10962 [DOI] [PubMed] [Google Scholar]
- 21.Boire A., Covic L., Agarwal A., Jacques S., Sherifi S., Kuliopulos A. (2005) Cell 120, 303–313 [DOI] [PubMed] [Google Scholar]
- 22.Wojtukiewicz M. Z., Tang D. G., Nelson K. K., Walz D. A., Diglio C. A., Honn K. V. (1992) Thromb. Res. 68, 233–245 [DOI] [PubMed] [Google Scholar]
- 23.Nierodzik M. L., Chen K., Takeshita K., Li J. J., Huang Y. Q., Feng X. S., D'Andrea M. R., Andrade-Gordon P., Karpatkin S. (1998) Blood 92, 3694–3700 [PubMed] [Google Scholar]
- 24.Dardik R., Savion N., Kaufmann Y., Varon D. (1998) Br. J. Cancer 77, 2069–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klepfish A., Greco M. A., Karpatkin S. (1993) Int. J. Cancer 53, 978–982 [DOI] [PubMed] [Google Scholar]
- 26.Melnikova V. O., Mourad-Zeidan A. A., Lev D. C., Bar-Eli M. (2006) J. Biol. Chem. 281, 2911–2922 [DOI] [PubMed] [Google Scholar]
- 27.Welch D. R., Bisi J. E., Miller B. E., Conaway D., Seftor E. A., Yohem K. H., Gilmore L. B., Seftor R. E., Nakajima M., Hendrix M. J. (1991) Int. J. Cancer 47, 227–237 [DOI] [PubMed] [Google Scholar]
- 28.Wiznerowicz M., Trono D. (2003) J. Virol. 77, 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobroff A. S., Wang H., Melnikova V. O., Villares G. J., Zigler M., Huang L., Bar-Eli M. (July24, 2009) J. Biol. Chem. doi:10.1074/jbc.M109.019836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bligh E. G., Dyer W. J. (1959) Can J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 31.Bussolati B., Biancone L., Cassoni P., Russo S., Rola-Pleszczynski M., Montrucchio G., Camussi G. (2000) Am. J. Pathol. 157, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mourad-Zeidan A. A., Melnikova V. O., Wang H., Raz A., Bar-Eli M. (2008) Am. J. Pathol. 173, 1839–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills L., Tellez C., Huang S., Baker C., McCarty M., Green L., Gudas J. M., Feng X., Bar-Eli M. (2002) Cancer Res. 62, 5106–5114 [PubMed] [Google Scholar]
- 34.Villares G. J, Dobroff A. S., Wang H., Zigler M., Melnikova V. O., Huang L., Bar-Eli M. (2009) Cancer Res. 69, 6730–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie S., Luca M., Huang S., Gutman M., Reich R., Johnson J. P., Bar-Eli M. (1997) Cancer Res. 57, 2295–2303 [PubMed] [Google Scholar]
- 36.Shih I. M. (1999) J. Pathol. 189, 4–11 [DOI] [PubMed] [Google Scholar]
- 37.Mintz-Weber C. S., Johnson J. P. (2000) J. Biol. Chem. 275, 34672–34680 [DOI] [PubMed] [Google Scholar]
- 38.Rummel M. M., Sers C., Johnson J. P. (1996) Cancer Res. 56, 2218–2223 [PubMed] [Google Scholar]
- 39.Xie S., Price J. E., Luca M., Jean D., Ronai Z., Bar-Eli M. (1997) Oncogene 15, 2069–2075 [DOI] [PubMed] [Google Scholar]
- 40.Mayr B., Montminy M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 41.Robert E. G., Hunt J. D. (2001) Curr. Pharm. Des. 7, 1615–1626 [DOI] [PubMed] [Google Scholar]
- 42.Biancone L., Cantaluppi V., Del Sorbo L., Russo S., Tjoelker L. W., Camussi G. (2003) Clin. Cancer Res. 9, 4214–4220 [PubMed] [Google Scholar]
- 43.Sreevidya C. S., Khaskhely N. M., Fukunaga A., Khaskina P., Ullrich S. E. (2008) Cancer Res. 68, 3978–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo K. H., Lee H. S., Jung B., Ko H. M., Choi J. H., Park S. J., Choi I. H., Lee H. K., Im S. Y. (2004) Cancer Res. 64, 6482–6488 [DOI] [PubMed] [Google Scholar]
- 45.Kravchenko V. V., Pan Z., Han J., Herbert J. M., Ulevitch R. J., Ye R. D. (1995) J. Biol. Chem. 270, 14928–14934 [DOI] [PubMed] [Google Scholar]
- 46.Merritt W. M., Lin Y. G., Han L. Y., Kamat A. A., Spannuth W. A., Schmandt R., Urbauer D., Pennacchio L. A., Cheng J. F., Nick A. M., Deavers M. T., Mourad-Zeidan A., Wang H., Mueller P., Lenburg M. E., Gray J. W., Mok S., Birrer M. J., Lopez-Berestein G., Coleman R. L., Bar-Eli M., Sood A. K. (2008) N. Engl. J. Med. 359, 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih I. M., Speicher D., Hsu M. Y., Levine E., Herlyn M. (1997) Cancer Res. 57, 3835–3840 [PubMed] [Google Scholar]
- 48.Keese C. R., Bhawe K., Wegener J., Giaever I. (2002) BioTechniques 33, 842–844, 846,, 848–850 [DOI] [PubMed] [Google Scholar]
- 49.Johnson J. P. (1999) Cancer Metastasis Rev. 18, 345–357 [DOI] [PubMed] [Google Scholar]
- 50.Shih I. M., Elder D. E., Speicher D., Johnson J. P., Herlyn M. (1994) Cancer Res. 54, 2514–2520 [PubMed] [Google Scholar]
- 51.Satyamoorthy K., Muyrers J., Meier F., Patel D., Herlyn M. (2001) Oncogene 20, 4676–4684 [DOI] [PubMed] [Google Scholar]
- 52.Anfosso F., Bardin N., Francès V., Vivier E., Camoin-Jau L., Sampol J., Dignat-George F. (1998) J. Biol. Chem. 273, 26852–26856 [DOI] [PubMed] [Google Scholar]
- 53.Glinskii O. V., Huxley V. H., Glinsky G. V., Pienta K. J., Raz A., Glinsky V. V. (2005) Neoplasia 7, 522–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lauri D., Martin-Padura I., Biondelli T., Rossi G., Bernasconi S., Giavazzi R., Passerini F., Van Hinsbergh V., Dejana E. (1991) Lab. Invest. 65, 525–531 [PubMed] [Google Scholar]
- 55.Auguste P., Fallavollita L., Wang N., Burnier J., Bikfalvi A., Brodt P. (2007) Am. J. Pathol. 170, 1781–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Padua D., Zhang X. H., Wang Q., Nadal C., Gerald W. L., Gomis R. R., Massagué J. (2008) Cell 133, 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim M. P., Park S. I., Kopetz S., Gallick G. E. (2009) Cell Tissue Res. 335, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pei Y., Barber L. A., Murphy R. C., Johnson C. A., Kelley S. W., Dy L. C., Fertel R. H., Nguyen T. M., Williams D. A., Travers J. B. (1998) J. Immunol. 161, 1954–1961 [PubMed] [Google Scholar]
- 59.Deo D. D., Bazan N. G., Hunt J. D. (2004) J. Biol. Chem. 279, 3497–3508 [DOI] [PubMed] [Google Scholar]
- 60.Walterscheid J. P., Ullrich S. E., Nghiem D. X. (2002) J. Exp. Med. 195, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Countryman N. B., Pei Y., Yi Q., Spandau D. F., Travers J. B. (2000) J. Invest. Dermatol. 115, 267–272 [DOI] [PubMed] [Google Scholar]