Abstract
PCSK9 is a natural inhibitor of LDL receptor (LDLR) that binds the extracellular domain of LDLR and triggers its intracellular degradation. PCSK9 and LDLR are coordinately regulated at the transcriptional level by sterols through their promoter-imbedded sterol response elements (SRE) and co-induced by statins. Identification of regulatory networks modulating PCSK9 transcription is important for developing selective repressors of PCSK9 to improve statin efficacy by prolonging the up-regulation of LDLR. Interestingly, the plant-derived hypocholesterolemic compound berberine (BBR) up-regulates LDLR expression while down-regulating PCSK9. In our investigations to define mechanisms underlying the transcriptional suppression of PCSK9 by BBR in HepG2 cells, we have identified a highly conserved hepatocyte nuclear factor 1 (HNF1) binding site residing 28 bp upstream from SRE as a critical sequence motif for PCSK9 transcription and its regulation by BBR. Mutation of the HNF1 site reduced PCSK9 promoter activity >90%. A battery of functional assays identified HNF1α as the predominant trans-activator for PCSK9 gene working through this sequence motif. We further provide evidence suggesting that HNF1 site works cooperatively with SRE as HNF1 mutation significantly attenuated the activity of nuclear SREBP2 to transactivate PCSK9 promoter. Finally, we show that a coordinate modest reduction of HNF1α and nuclear SREBP2 by BBR led to a strong suppression of PCSK9 transcription through these two critical regulatory sequences. This is the first described example of SREBP pairing with HNF1 to control an important regulatory pathway in cholesterol homeostasis. This work also provides a mechanism for how BBR suppresses PCSK9 transcription.
Recent studies of human genetics and genome-wide screens have identified proprotein convertase subtilisin/kexin type 9 (PCSK9)2 as the third gene associated with autosomal dominant familial hypercholesterolemia, after LDL receptor (LDLR) and apoB100 (apoB) (1–3). In hypercholesterolemic humans, “gain of function” mutations were found (4, 5), whereas in hypocholesterolemic subjects “loss of function” mutations were detected (6). The function of PCSK9 as a secreted serine protease degrading hepatic LDLR (7–9) directly correlates with its tight association with plasma cholesterol levels and provides a new therapeutic target to combat hypercholesterolemia and coronary heart disease (10–12).
PCSK9 is predominantly expressed in adult liver hepatocytes and in small intestinal enterocytes (13). It is synthesized as a 72-kDa zymogen that undergoes autocatalytic cleavage in the endoplasmic reticulum into a heterodimer of a prosegment (122 amino acids) and a 60-kDa active form, which is secreted as a proteolytic inactive form (13, 14). Once secreted outside cells, PCSK9 can bind to the EGF-A extracellular domain of LDLR (15, 16) and trigger its intracellular degradation in lysosomes, thus increasing the level of circulating LDL-cholesterol (LDL-C).
At the transcriptional level, PCSK9 has been identified as a target gene of sterol regulatory element binding proteins (SREBPs) (17, 18). The proximal promoter of the PCSK9 gene contains a functional sterol regulatory element (SRE) that responds to changes in intracellular cholesterol levels (19). It has been shown that insulin induces PCSK9 transcription through the interaction of SREBP1c with the SRE motif in rodent primary hepatocytes (20). In HepG2 cells both SREBP1 and SREBP2 transcriptionally activate PCSK9 via this SRE site (21). In vivo however, it was suggested that the sterol-dependent regulation of PCSK9 is mediated predominantly by SREBP2 (17).
Statins reduce intracellular levels of sterols and activate the SREBP pathway by inhibiting hydroxymethylglutaryl-CoA, the rate-limiting enzyme in cholesterol biosynthesis. PCSK9 and LDLR both contain an SRE motif in their proximal promoters and thus are coordinately up-regulated by statins through activation of SREBP (20). Several studies have demonstrated the induction of PCSK9 by statins in cultured cells and in animal models (13, 22, 23). The abrogation of the effect of pravastatin on PCSK9 promoter harboring a mutated SRE further confirmed its regulation by the SREBP pathway (24). In human studies, it was reported that atorvastatin at a 40-mg dose significantly elevated circulating PCSK9 protein levels (25). This led to the speculation that the diminishing efficacy of statins to further reduce LDL-C levels might be caused by the induced degradation of LDLR protein by the concomitant up-regulation of PCSK9 at higher drug doses. These findings from in vitro and in vivo studies raised an important question as to whether PCSK9 gene transcription could be separately regulated from LDLR; such a mechanism, if it existed, might be applied to inhibit PCSK9 transcription to further enhance the actions of statins to lower plasma cholesterol through their effect on LDLR transcription. This could, for example lead to a lower and more effective dose of the statins and reduce the chance of unwanted side effects.
Recently, several studies have reported the inhibition of PCSK9 transcription by small molecules. In immortalized human hepatocytes, activation of PPARα by various fibrates decreased PCSK9 mRNA and protein levels (24). Co-incubation of fenofibrate acid with statins also significantly impaired the induction of PCSK9 protein expression by pravastatin. This effect was further confirmed at the transcriptional level by showing that fenofibrate repressed the wild-type PCSK9 promoter activity alone and with pravastatin. However, this study did not further address whether SRE is the sole cis-acting element responsible for this repression. In another report, it was shown that activation of farnesoid X receptor by chenodeoxycholic acid, a bile acid, or by a farnesoid X receptor synthetic agonist repressed PCSK9 expression (26). Similar to the effects of fibrates, coadministration of chenodeoxycholic acid counteracted the statin-induced PCSK9 expression, leading to a potentiation of LDLR ligand uptake activity. The observation that chenodeoxycholic acid treatment did not change PCSK9 mRNA half-life suggested a transcriptional regulation by an unknown mechanism.
Our laboratory has previously demonstrated that the natural cholesterol-lowering compound berberine (BBR) up-regulates LDLR expression through a post-transcriptional mechanism of mRNA stabilization (27–30). Interestingly, it was recently reported that BBR also exerts inhibitory effects on the expression of PCSK9 protein and mRNA in HepG2 cells (31). Thus, BBR could have dual actions on LDLR metabolism by prolonging its mRNA half-life as well as directly increasing the protein abundance through the blockage of PCSK9-mediated degradation. This would make BBR or BBR-like compounds attractive candidates for enhancing statin efficacy. Collectively, these studies provided evidence for divergent regulation of PCSK9 transcription from statins and warranted further investigations at the promoter level to fully characterize the regulatory mechanisms involved in PCSK9 transcription by BBR or other non-statin small molecules.
In the initial study of PCSK9 promoter, a computer-aided sequence analysis has identified putative binding sites for NF-Y (−613), Sp1 (−430), and SREBPs (−337) within a region ∼650 bp upstream from the translation start site (19). However, until this report, the SRE motif was the only proven functional cis-acting element that conferred cholesterol regulation through its interaction with SREBPs. The Sp1 binding site located 76 bp upstream of SRE was later shown to not be involved in cholesterol regulation and has a minor role in PCSK9 transcription as the PCSK9 promoter activity was moderately reduced when this Sp1 site was abolished (21).
In an effort to define the BBR-responsive element on PCSK9 promoter in the current study, we have identified a functional hepatocyte nuclear factor 1 (HNF1) binding site (−386 to −374) that is essential for PCSK9 transcription as a positive regulator through its interaction predominantly with HNF1α and to a lesser extent with HNF1β. The HNF1 site appears to work cooperatively with the SRE as mutation of HNF1 reduced the sensitivity of the promoter to sterols and attenuated the effect of SREBP2 to transactivate PCSK9 promoter. We further showed that the HNF1 motif along with the SRE is uniquely involved in BBR-mediated transrepression of PCSK9 as mutation of either site diminished the BBR effect.
Considering the fact that HNF1α is a liver-enriched transcription factor known to regulate many target genes in liver and intestine (32–34) and that the HNF1 site lies within the highly conserved section between Sp1 and SRE sites and its nucleotide sequence (GTTAATGTTTAAT) is 100% conserved among human, mouse, and rat PCSK9 promoters, our findings for the first time provide a mechanistic explanation for the abundant expression of PCSK9 in liver and intestine.
EXPERIMENTAL PROCEDURES
Cells and Reagents
The human hepatoma cell line HepG2 was obtained from American Type Culture Collection and cultured in minimum essential eagle medium supplemented with 10% fetal bovine serum (FBS). Antibodies specific to the following proteins were obtained from Santa Cruz Biotechnology: HNF1α (sc-6547x), HNF1β (Sc-22840x), HNF4α (sc-8987x), and HDAC-1 (sc-7872). Rabbit anti-SREBP2 and anti-PCSK9 antibodies were previously described (21). Berberine chloride, lovastatin, and simvastatin were obtained from Sigma. Fluvastatin was generously provided by Novartis (Ringaskiddy, Co., Cork, Ireland).
The constructs of PCSK9 promoter luciferase reporters (D1, D4, D5, D6, SRE-mu, and Sp1-mu) were generated as described before (21). Expression vectors of nuclear SREBP2 (pTK-nBP2), HNF1α (pTS-HNF1α), and HNF1β (pTS-HNF1β) have been previously described (21, 35, 36). In this study, the HNF1 mutant construct (HNF1-mu) was generated from D4 using the QuikChange site-directed mutagenesis kit (Stratagene) and the following sense oligonucleotide: 5′-AGTCCGGGGGTTCCtggAATGTTTAATCAGATAGGATC-3′. The small letters are mutated nucleotides. The HNF1/SRE dual mutated construct was generated using SRE-mu as the template and the above oligonucleotide. The correct sequence was verified by DNA sequencing.
Generation of Stable Clones Expressing a PCSK9 Promoter Luciferase Reporter
The plasmid pGL3-PCSK9-D1 (hereafter referred to as D1) contains 5′-flanking region of the PCSK9 gene from −1711 to −94, relative to the ATG start codon in front of the luciferase coding sequence (21). D1 and an empty vector pcDNA3.1 harboring a neomycin-resistant gene were cotransfected into HepG2 cells in a DNA ratio of 50:1 using a MicroPorator (Digital Bio, Seoul, Korea). Positive clones were selected by G418 (Invitrogen) at a concentration of 800 μg/ml. Three individual clones (CL26, CL28, and CL30) were tested for their responses to statin or BBR treatment, and clone 26 (CL26) was further used in this study.
Transient Transfections of Reporter Constructs
PCSK9 promoter reporters of wild-type, deletion, and mutation constructs were cotransfected with a Renilla luciferase vector constitutively expressed as a transfection control, and the dual luciferase activities were measured as described (21). The firefly luciferase activity was normalized to the Renilla luciferase activity (21). In experiments of sterol regulation, transfected cells were cultured in medium containing 10% lipoprotein-depleted serum (LPDS) or LPDS plus cholesterol (10 μg/ml cholesterol plus 1 μg/ml 25-hydroxycholesterol) for 24 h prior to cell lysis for the measurement of dual luciferase activities.
To measure the BBR effect, 1.5 × 104 HepG2 cells per well were seeded into 96-well plates. PCSK9 promoter constructs were cotransfected with pSV-β-galactosidase control vector. One day post-transfection the medium was changed to 0.5% FBS in minimum essential eagle medium overnight, and BBR was added for 24 h. Cells were lysed in 50 μl of reporter lysis buffer per well of which 20 μl of cell lysate were used to measure β-galactosidase activity according to the manufacturer's instruction by using β-Galactosidase Enzyme Assay System (cat. no. E2000, Promega, Madison, WI). The remaining 30 μl of lysate was used to measure the firefly luciferase activity by using Luciferase Assay System (cat. no. E1501, Promega). Absolute luciferase activity was normalized against β-galactosidase activity to correct for transfection efficiency. Triplicate wells were assays for each transfection condition, and at least four independent transfection assays were performed for each reporter construct.
Electrophoretic Mobility Shift Assays
HepG2 nuclear extracts were prepared as described (30). Oligonucleotide probes were annealed and end-labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. Each binding reaction comprised 10 mm Tris, pH 7.5, 5 mm MgCl2, 1 mm dithiothreitol, 50 mm KCl, 2.5% glycerol, 1 μg of poly(dI-dC), 0.05% Nonidet P-40, and 10 μg of HepG2 nuclear extract in a final volume of 20 μl. The nuclear extracts were incubated with 0.4–0.5 ng of 32P-labeled double-stranded synthetic oligonucleotide probe (1 × 105 cpm) for 10 min at room temperature. The reaction mixtures were loaded onto a 5% polyacrylamide gel and run in 0.5 × TBE buffer (89 mm Tris, 89 mm borate, and 2 mm EDTA, pH 8.4) at 30 mA for 2 h at 4 °C. Gels were dried and visualized on a PhosphorImager. For supershift assays, 2 μg of antibody was incubated with samples for 10 min at room temperature prior to the addition of the probe. The sense sequences of EMSA probes were as follows: PCSK9HNF1-UP, 5′-AGTCCGGGGGTTCCGTTAATGTTTAATCAGATAGGATC-3′ and PCSK9HNF1MU-UP, 5′-GTCCGGGGGTTCCGTTcgTGTTgccTCAGATAGGATC-3′. The HNF1 binding site is underlined.
Chromatin Immunoprecipitation
The ChIP assays were conducted according to the described protocol using aliquots of lysate obtained from 2 × 107 HepG2 cells (33). Briefly, cells were fixed in 1.42% formaldehyde for 15 min at room temperature. Cells were lysed and nuclear pellet was isolated. The chromatin was sheared to an average length of 0.5–1 kb by sonicating suspended nuclear pellets. Samples were immunoprecipitated at 4 °C overnight with 4 μg of goat anti-HNF1α (sc-6547x), rabbit anti-HNF1β (sc-22840x), normal goat or rabbit IgG as negative controls. Immunocomplexes were isolated by binding to protein A-agarose beads after extensive washing with the immunoprecipitation buffer. Chelex 100 slurry was added to the washed beads and boiled for 10 min. Precipitated DNA was isolated after ethanol precipitation. An aliquot of sheared chromatin was used in PCR reaction as a control for the amount of input DNA used in precipitations and was diluted 40× prior to PCR. The bound and the input DNA were analyzed by PCR with primers that amplify a 199-bp fragment of the human PCSK9 promoter region from −490 to −292, relative to the ATG start codon. The sequences of ChIP primers were as follows: PCSK9 ChIP primer 1-up, 5′-TCCAGCCCAGTTAGGATTTG-3′; PCSK9 ChIP primer 1-lo, 5′-CGGAAACCTTCTAGGGTGTG-3′.
The PCR conditions were 95 °C for 4 min, 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and 72 °C for 7 min. The 199-bp PCR product was visualized on a 2% agarose gel stained with ethidium bromide. The intensity of the PCR products was scanned and quantified with Kodak Image Station 4000R System.
siRNA Transfection
Three pre-designed siRNAs targeted to human HNF1α mRNA (cat. no. 16708A; ID 3544, 3450, and 3354), two siRNAs to HNF1β mRNA (cat. no. 16708; ID 3262 and 3170), and one silencer negative control siRNA (cat. no. 4618G) were obtained from Ambion. Pooled siRNAs for each gene were used in this study. For promoter analysis, HepG2 CL26 cells in suspension were mixed with siRNA in the SilencerTM siRNA transfection reagent for 30 min according to the vendor's instruction and were plated in 96-well plates at a density of 1 × 104 cells/well. Twenty-four hours later, cells were cultured in minimum essential eagle medium containing 0.5% FBS overnight and BBR at a concentration of 40 μm was added for 24 h before cell lysis for measuring luciferase activity.
For Western blotting or real-time PCR, suspended 1 × 106 cells were mixed with siRNA in the SilencerTM siRNA transfection reagent for 30 min and plated in 6-well plates. After 2 days, transfected cells were cultured in minimum essential eagle medium containing 0.5% FBS overnight and then treated with BBR for 24 h prior to cell lysis.
RNA Isolation and Real-time Reverse Transcription-PCR
Total RNA was extracted from HepG2 cells using UltraspecTM total RNA isolation reagent (Biotecx Laboratories). Two micrograms of total RNA was reverse transcribed with a high capacity cDNA reverse transcription kit (Applied Biosystems) using random primers according to the manufacturer's instructions. Real-time PCR was performed on the cDNA using an ABI Prism 7900-HT Sequence Detection System. Human LDLR, PCSK9, and GAPDH Pre-Developed TaqMan Assay Reagents (Applied Biosystems) were used to assess mRNA expression in HepG2 cells with or without BBR treatment. All values are reported as means ± S.D. of triple measurement of each cDNA sample.
Western Blot Analysis
Western blot analysis was performed to examine the protein expression in untransfected or siRNA-transfected cells as previously described (29).
Statistical Analysis
Each experiment is representative of at least three independent experiments with a minimum of triplicates per condition in promoter analysis. Significant differences between control and treatment groups or between wild-type and mutated vectors were assessed by one-way analysis of variance with post test of Bonferroni multiple comparison test or by two-tailed Student's t test. Values of p < 0.05 were considered statistically significant.
RESULTS
BBR Counteracts the Inducing Effects of Statins on PCSK9 Transcription
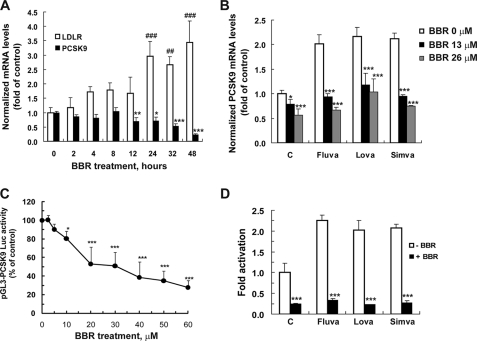
The time-dependent effects of BBR on the mRNA expression of LDLR and PCSK9 were examined in HepG2 cells that were treated with BBR at a dose of 20 μm for various times. BBR increased LDLR mRNA by ∼1.8-fold at 4 h and up to 3-fold at 24 h (p < 0.001), and this elevation was observed for at least 48 h. Conversely, the amount of PCSK9 mRNA was decreased by BBR treatment. A 30% reduction was detected at 12 h (p < 0.01), and the mRNA level of PCSK9 gradually declined in BBR-treated cells down to 23% of untreated control by 48 h (Fig. 1A). To further evaluate the effect of BBR on PCSK9 mRNA expression we treated cells with 1 μm of fluvastatin, lovastatin, or simvastatin in the absence or presence of two different concentrations of BBR. Although statins individually increased PCSK9 mRNA levels above 2-fold of control, their inducing effects were not observed in cells concomitantly treated with BBR at both doses (Fig. 1B). These results are in line with a previous observation of counteraction of BBR on mevastatin-induced PCSK9 mRNA expression (31).
FIGURE 1.
Berberine transcriptionally inhibits PCSK9 while upregulating LDLR mRNA quantity. A, HepG2 cells were exposed to BBR at a concentration of 20 μm for various times. PCSK9, LDLR, and glyceraldehyde-3-phosphate dehydrogenase mRNA levels were quantified by real-time PCR, and mRNA relative levels are presented. Triplicate RNA samples were measured per condition. *, p < 0.05 and ***, p < 0.001 as compared with time 0 h of PCSK9 mRNA level; ###, p < 0.001 as compared with time 0 h of LDLR mRNA level. B, HepG2 cells were treated with indicated doses of BBR in the absence or the presence of 1 μm of fluvastatin (Fluva), lovastatin (Lova), or simvastatin (Simva) for 24 h, and the PCSK9 mRNA abundance was assayed by real-time PCR. *, p < 0.05 and ***, p < 0.001 as compared with control. C, CL26 cells were incubated with BBR at various concentrations for 24 h. D, CL26 cells were incubated with BBR (40 μm) alone or with 1 μm of each statin for 24 h. Luciferase activities are expressed in relative to untreated control cells. Significant differences between control and treatment groups were assessed by one-way analysis of variance with post test of Bonferroni multiple comparison. *, p < 0.05 and ***, p < 0.001 as compared with untreated control. Data shown in A, B, and D are representative of two separate experiments with similar results. Data (mean ± S.D.) in C are derived from four separate assays.
To firmly demonstrate the transcriptional inhibition of PCSK9 mRNA expression by BBR, we established stable clones of HepG2 cells expressing a luciferase construct (Fig. 2A, D1) that contains the 5′-flanking region of the PCSK9 gene from −1711 to −94, relative to the ATG start codon (21). Preliminary screens of clones (CL26, CL28, and CL30) showed that luciferase activities of these clones were increased by simvastatin and were reduced by BBR, mimicking the response of endogenous PCSK9 transcriptional activity to the drug treatments. CL26 cells were studied to show a dose-dependent effect of BBR on PCSK9 promoter activity (Fig. 1C) and to demonstrate the antagonism of BBR on the statin-induced PCSK9 promoter construct (Fig. 1D). In parallel, we conducted a cell proliferation assay and demonstrated that BBR at a high dose (40 μm) did not affect cell growth under the assay conditions, indicating that the decreased luciferase activity in BBR-treated cells was not caused by reduction of viable cell number. Taken together, these results establish that BBR inhibits PCSK9 gene expression by suppressing its promoter transcriptional activity.
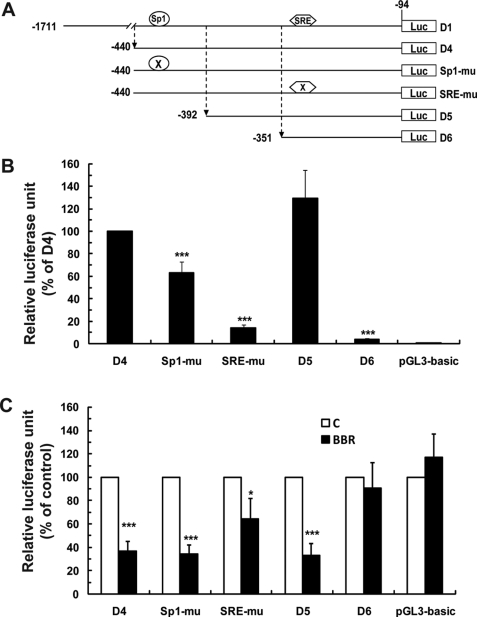
FIGURE 2.
Analysis of PCSK9 promoter constructs in transiently transfected HepG2 cells without and with BBR treatment. A, schematic presentation of the deletion and mutation constructs of human PCSK9 promoter-luciferase reporters. Position −1 was assigned to the nucleotide preceding the ATG start codon. B, the reporter constructs were transiently cotransfected with pSV-β-gal vector into HepG2 cells. One day post transfection, cells were incubated in 0.5% FBS medium overnight, followed by a 24-treatment of BBR. Cells were harvested, and the luciferase and β-galactosidase activities were measured as described under “Experimental Procedures.” After normalization, the relative luciferase unit of each vector is expressed as the percent of the luciferase activity of the D4 vector. C, relative luciferase activity of transfected cells that were untreated is expressed as 100%. Results shown are mean ± S.D. of four separate transfections. ***, p < 0.001 as compared with D4 in B or compared with untreated control in C.
Characterization of the PCSK9 Promoter to Define BBR-responsive Sequences
To define which region of PCSK9 promoter is responsive to BBR treatment we transfected a set of PCSK9 promoter constructs (Fig. 2A) into HepG2 cells together with plasmid pSV-β-gal for normalization. In these experiments, we included the promoter-less vector pGL3-basic as a negative control whose luciferase activity was not affected by BBR. The summarized results of four separate transfections are presented in Fig. 2 (B and C). Fig. 2B shows the basal luciferase activity of each construct, relative to the wild-type plasmid D4. Fig. 2C compares the inhibitory effect of BBR on different promoter constructs.
Plasmid D4 contains the functional proximal PCSK9 promoter (−440 to −94) wherein the Sp1 and SRE sites reside. BBR reduced D4 activity by 64%. Mutation of the Sp1 site modestly lowered the basal promoter activity to ∼63% of the wild type but had no effect on the BBR response. SRE mutation reduced the basal promoter activity by 86% and also partially prevented the BBR-mediated inhibition. Deletion of a small segment of 48 bp covering the Sp1 site from the 5′ proximal region of D4 slightly increased the basal activity of D5 (consistent with the previous report) (21), and the BBR response was fully retained in D5. Interestingly, PCSK9 promoter activity was drastically reduced by a further deletion of 41 bp from −392 to −352, and the resulting plasmid D6 became resistant to BBR treatment despite the presence of an intact SRE. These results suggest that, although the SRE motif is partially involved in the BBR-mediated suppression, the small segment of PCSK9 promoter upstream of SRE might contain a cis-regulatory element that is critically required for PCSK9 gene transcription and is likely involved in the action of BBR.
Identification of HNF1 Binding Site as an Essential Regulatory Element for PCSK9 Transcription
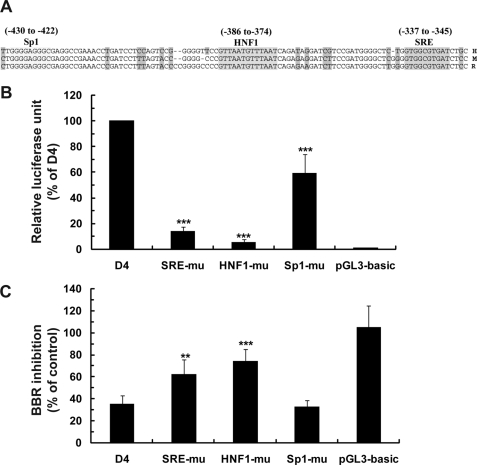
Analysis of nucleotide sequence within this 41-bp segment of PCSK9 promoter by the MatInspector software revealed the presence of a strong HNF1 binding site spanning the region −386 to −374. This HNF1 site resides 28 bp upstream from the SRE and 35 bp downstream of the Sp1 site. It is 100% conserved among human, mouse, and rat PCSK9 promoters (Fig. 3A). We mutated the core nucleotide sequence of the HNF1 site and transfected the mutated plasmid along with the wild type, SRE-mu, Sp1-mu, and the pGL3-basic vector individually into HepG2 cells. The data shown in Fig. 3B were derived from eight separate transfection experiments. Destroying the HNF1 binding site reduced PCSK9 promoter activity to ∼5% of the wild type (Fig. 3B), even more severe than SRE mutation (13.8% of the wild type), indicating that it is a vital cis-regulatory element for PCSK9 gene transcription. The combined mutation of HNF1 and SRE resulted in a complete loss of the promoter activity (data not shown). Although BBR-mediated suppression of luciferase activity was not affected by Sp1 mutation, mutation of SRE reduced BBR inhibitory activity from 35% to 62% of control (p < 0.01) and HNF1 mutation attenuated BBR activity to 73% of control (p < 0.001) (Fig. 3C). In contrast to the PCSK9 reporters, the negative control vector, pGL3-basic showed no response to BBR. Taken together, these data demonstrated the critical roles of HNF1 and SRE in PCSK9 transcription and their involvement in mediating the effect of BBR on transrepression of PCSK9.
FIGURE 3.
HNF1 binding site is critical for PCSK9 transcription and BBR-mediated suppression. A, sequence comparison of proximal regions of PCSK9 promoter of human, mouse, and rat. Gray-highlighted letters indicate divergent nucleotides among the three promoter sequences. B and C, summarized results (means ± S.D.) of eight independent transfections and treatments as described in Fig. 2. In B, the normalized basal luciferase activity of each vector is expressed as the percent of luciferase activity of the D4 vector. In C, the response of each mutant vector to BBR inhibition was compared with that of the wild-type D4 vector. **, p < 0.01; ***, p < 0.001.
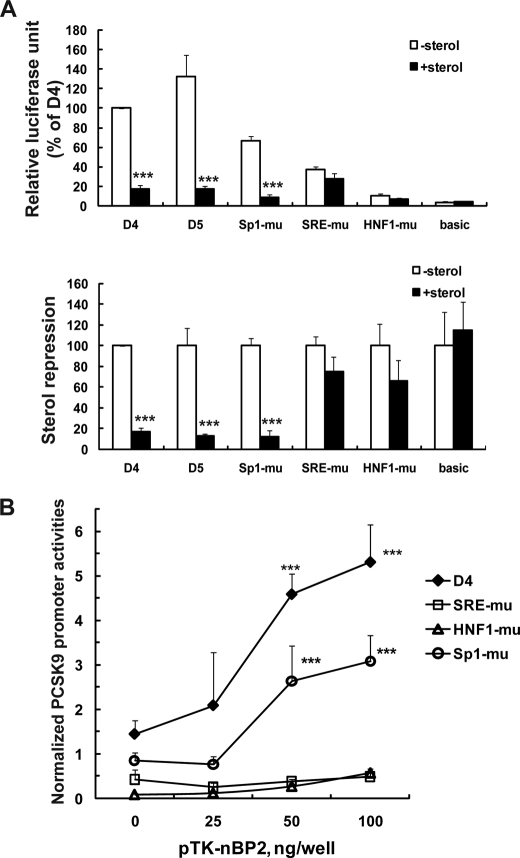
In typical promoters of cholesterol biosynthetic genes SRE motifs are adjacent to Sp1 or NF-Y binding sites (37–39). To our knowledge, PCSK9 is the first identified gene, involving in cellular cholesterol metabolism that utilizes an HNF1 binding site to cooperate with SRE. To determine whether the HNF1 motif is involved in cholesterol regulation, we tested promoter activities of various constructs in cells cultured in the absence or presence of sterols. Fig. 4A shows that the transcriptional activity of PCSK9 promoter was decreased by 83 and 88% in the presence of sterols in wild-type constructs D4 and D5. Mutation of Sp1 did not diminish the sterol effect at all (88% repression). In contrast, mutation of SRE as well as the HNF1 site in the D4 construct nearly abolished the sterol-dependent suppression of transcription. These results clearly demonstrate that Sp1 has no functional role in the sterol-mediated regulation of PCSK9 transcription but HNF1 site is critically involved.
FIGURE 4.
Examination of responses of the wild-type and mutated vectors to sterol suppression and SREBP2-mediated transactivation of PCSK9 promoter. A, reporter constructs were cotransfected with a Renilla expression vector (pRL-SV40) into HepG2 cells. One day post transfection, cells were cultured in medium supplemented with 10% LPDS or LPDS plus cholesterol (10 μg/ml cholesterol plus 1 μg/ml 25-hydroxycholesterol) for 24 h prior to cell lysis to measure dual luciferase activities. After normalization, the luciferase activity of each vector is expressed as the percent of the luciferase activity of the D4 vector without sterols (upper panel). In the lower panel of A, the luciferase activity of each vector in the absence of sterols is expressed as 100%. The relative luciferase of each vector in the presence of sterols was compared with that in the absence of sterols. Each value represents the mean ± S.D. of four independent transfection experiments in which triplicate wells were assayed. ***, p < 0.001. B, HepG2 cells were cotransfected with indicated PCSK9 luciferase reporters with different amounts of pTK-nBP2. Total amounts of transfected plasmids were adjusted to 0.2 μg/well with a mock vector. Dual Luciferase activities were measured as described in A. Each value represents the mean ± S.D. of six wells per condition. ***, p < 0.001 as compared with 0 ng/well pTK-nBP2. The data shown are representative of three separate transfections with similar results.
To further examine the relationship between SRE and HNF1 motifs, we examined the SREBP2-mediated trans-activation of PCSK9 promoters, including the wild-type, SRE-mutated, HNF1-mutated, and Sp1-mutated plasmids. Individual reporter constructs were cotransfected into HepG2 cells with a plasmid expressing the nuclear form of SREBP2 (pTK-nBP2) at different amounts. Fig. 4B shows that expressions of nSREBP2 dose-dependently increased the transcription of the wild-type and Sp1 mutated PCSK9 promoter activities. In contrast, the promoter activities of SRE-mu and HNF-mu were not significantly stimulated by nSREBP2 (Fig. 4B). These data further support the model that SREBP2 and HNF1 binding proteins may work cooperatively in control of PCSK9 gene transcription.
HNF1α Has a Predominant Role in PCSK9 Transcription
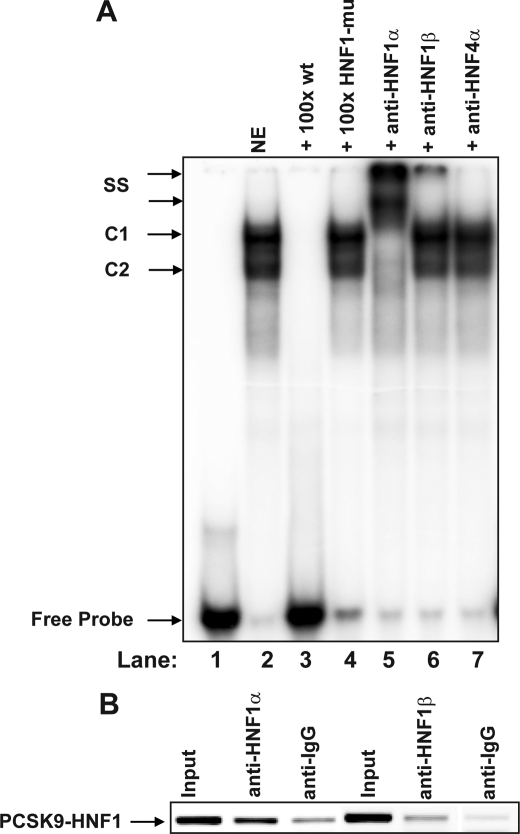
To detect transcription factors that bind to the HNF1 site of PCSK9 promoter, gel mobility shift assays using a 32P-labeled oligonucleotide probe (PCSK9-HNF1) containing PCSK9 promoter sequence −400 to −362 were performed with nuclear extracts prepared from HepG2 cells (Fig. 5A). Two complexes (C1 and C2) were detected with C1 being the abundant one. Formation of both complexes was inhibited by a 100-fold molar excess of the unlabeled wild-type oligonucleotides, but was not inhibited by a 100-fold molar excess of oligonucleotides containing the mutated HNF1 sequence, demonstrating the binding specificity. Moreover, the complexes were completely supershifted by anti-HNF1α antibody, whereas an irrelevant antibody anti- HNF4α had no effect on the mobility of the complexes. Anti-HNF1β antibody produced a supershifted band of a much lower intensity compared with anti-HNF1α antibody. These results suggest that PCSK9-HNF1 site predominantly interacts with HNF1α and to a lesser extend with HNF1β, likely through homodimerization of HNF1α and heterodimerization of 1α and 1β in the cellular context of HepG2.
FIGURE 5.
EMSA and ChIP analyses of HNF1α and HNF1β association with the PCSK9 promoter in HepG2 cells. A, a double-stranded oligonucleotide (designated as PCSK9-HNF1) corresponding to PCSK9 promoter region −400 to −362 was radiolabeled and incubated with 10 μg of HepG2 nuclear extract (lanes 2–7) for 10 min at 22 °C in the absence (lane 2) or presence of 100-fold molar amounts of unlabeled wild-type (lane 3) or mutated HNF1 (lane 4) probes. The binding reactions were also carried out in the presence of 2 μg of HNF1α antibody (lane 5) or HNF1β antibody (lane 6). Anti-HNF4α antibody was included as an irrelevant negative control (lane 7). B, antibodies to HNF1α and HNF1β were used in a ChIP analysis followed by PCR to amplify a 199-bp region surrounding the HNF1 binding site of PCSK9 promoter from genomic DNA isolated from HepG2 cells. Normal rabbit IgG and goat IgG were included in the assay as negative controls for nonspecific binding. The PCR product was separately on a 2% agarose gel, stained with ethidium bromide, and quantified by a Kodak Image Station 4000R. Input represents the starting material before immunoprecipitation. The data shown are representative of two separate EMSA and ChIP assays with similar results.
To examine the in vivo interactions of HNF1 factors with PCSK9 promoter, we performed ChIP assays to detect the direct binding of HNF1α or HNF1β to PCSK9 promoter HNF1 sequence in intact HepG2 cells. Fig. 5B shows that, although both isoforms of HNF1 were cross-linked to the HNF1 motif, the level of HNF1α bound to the HNF1 motif was ∼2.5-fold higher than HNF1β. Thus, the results of ChIP strongly support our in vitro finding that HNF1α is the major isoform of HNF1 factors that recognizes PCSK9-HNF1 sequence motif in HepG2 cells.
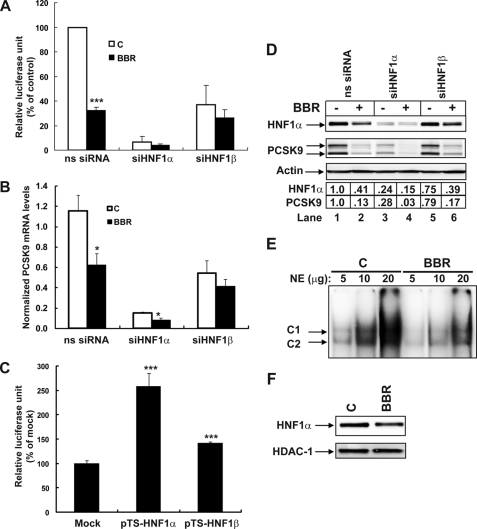
To determine the functional role of HNF1 isoforms in PCSK9 transcription, we first transfected siRNAs targeted to HNF1α or HNF1β into HepG2-CL26 cells that express the PCSK9 promoter construct D1. A nonspecific siRNA with scrambled nucleotide sequence was included in the experiments as a negative control. Compared with the nonspecific siRNA, transfections of siHNF1α and siHNF1β lowered luciferase activity by 94 and 63%, respectively. Depletion of HNF1 factors also compromised the BBR inhibitory effect on PCSK9 promoter.
Next, we examined the effects of siRNA transfection on the expression of endogenous PCSK9 mRNA in HepG2 cells with or without BBR treatment by real-time PCR. Consistent with the data of promoter analysis, the level of PCSK9 mRNA was drastically decreased to 15% of control in siHNF1α-transfected cells, and the effect of siHNF1β was again less severe (56% of control) (Fig. 6B).
FIGURE 6.
Effects of siHNF1α and siHNF1β knockdown on PCSK9 transcription and protein expression in HepG2 cells untreated and treated with BBR. A, analysis of PCSK9 promoter activity in CL26 cells transfected with targeted siRNA or control nonspecific siRNA. Cells were treated with BBR at a concentration of 40 μm or its vehicle (0.1% DMSO) as control for 24 h prior to cell lysis. Data represent the means ± S.D. derived from four separate transfection experiments. B, real-time reverse transcription-PCR analysis of PCSK9 mRNA levels in HepG2 cells after siRNA transfection of 2 days followed by BBR treatment of 24 h. Data represent the means ± S.E. derived from three separate transfection experiments. C, D4 was cotransfected with either pTS-HNF1a, pTS-HNF1b, or with a mock vector along with pRL-SV40 in a DNA ratio of 2:1:0.2 into HepG2 cells. Dual luciferase activities were measured. The normalized luciferase activity in D4-transfected cells with the mock vector is expressed as 100%, and luciferase activities in cells transfected with other expression vectors are plotted relative to that value. D, Western blot analysis of HNF1α, PCSK9, and β-actin in total cell lysates of siRNA-transfected HepG2 cells without and with BBR treatment. The HNF1α and PCSK9 bands were quantified using an imaging program of Kodak Image Station 4000R. Values were normalized to β-actin and were graphed relative to untreated cells without siRNA transfection. E, indicated amounts of nuclear extracts prepared from untreated or BBR-treated cells were incubated with the 32P-labeled PCSK9-HNF1 probe and analyzed by EMSA. F, HepG2 cells were treated with BBR (40 μm) or its vehicle 0.1% DMSO as control for 24 h. Nuclear extracts were isolated. 50 μg of nuclear proteins from each sample was analyzed by Western blotting with HNF1α antibody, followed by blotting the membrane with anti-HDAC1 as a loading control. The data shown in C–F are representative of 2–3 separate experiments with similar results.
In addition to knockdown, effects of exogenous overexpression of HNF1α or HNF1β on PCSK9 transcription were studied. Plasmid D4 was cotransfected with pTS-HNF1α, pTS-HNF1β, or the mock vector into HepG2 cells, and the luciferase activity was measured 48 h post transfection. PCSK9 promoter activity was increased 158 and 41% by pTS-HNF1α and pTS-HNF1β, respectively (Fig. 6C). All together, these data point to HNF1α as the critical factor in PCSK9 transcription, which is in line with the findings of EMSA and ChIP showing the predominant binding of HNF1α to the PCSK9-HNF1 site.
To further confirm the primary activator role of HNF1α in PCSK9 expression and its involvement in BBR action, we examined PCSK9 protein expression in control and BBR-treated cells without and with siRNA transfections (Fig. 6D). The abundance of HNF1α protein was greatly reduced in siHNF1α-transfected cells (compare lane 1 with lane 3), concomitant with an extensive reduction of PCSK9 protein expression. Depletion of HNF1β only slightly lowered PCSK9 signal.
Importantly, the results of Western blotting with anti-HNF1α further revealed that the amount of HNF1α protein in BBR-treated cells was decreased to ∼40% of control. This effect was also observed in siRNA-transfected cells. To validate this finding at the promoter level, we performed EMSA with nuclear extracts prepared from control and BBR-treated cells. As shown in Fig. 6E, the signal intensity of the complexes formed with 32P-labeled PCSK9-HNF1 probe was clearly reduced by BBR treatment at all three different doses. Further analysis of nuclear extracts by Western blotting (Fig. 6F) showed an equal abundance of the nuclear protein HDAC1 (histone deacetylase 1) in control and BBR-treated extracts, thereby validating the HNF1 EMSA result. These findings, combined with the promoter analysis, demonstrate that BBR suppresses PCSK9 transcription partially though its inhibitory effect on HNF1α expression.
Reduction of the Nuclear Form of SREBP2 by BBR Treatment
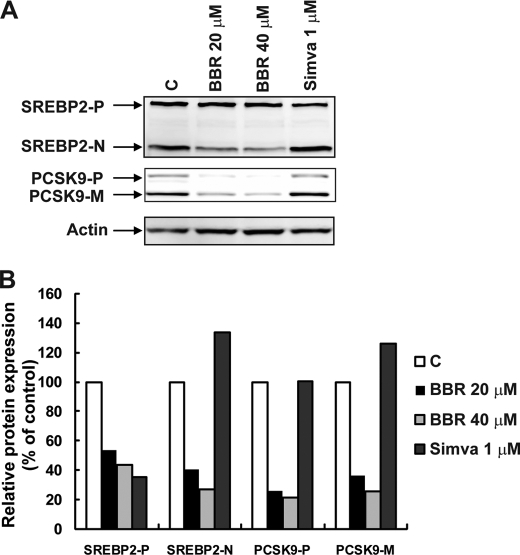
Finally, we sought to understand how SRE is involved in BBR-mediated suppression of PCSK9 transcription. We examined the expression of the nuclear and precursor forms of SREBP2 in HepG2 cells without and with treatments of BBR or simvastatin (Fig. 7). The results of Western blotting using anti-SREBP2 antibody showed that simvastatin increased the nuclear form of SREBP2 while reducing the membrane-bound precursor, a typical effect of statins on SREBP2 protein expression (21). BBR treatment resulted in a moderate reduction of nuclear SREBP2 and a lesser reduction of the precursor protein in a dose-dependent manner. The reduction of SREBP2 by BBR was accompanied by a more extensive inhibition of PCSK9 protein expression, which was in clear contrast to simvastatin that increased both nSREBP2 and PCSK9. These results, combined with the data in Fig. 6, showing a reduced expression of HNF1α in BBR-treated cells, suggest that BBR exerts an integrated action on these two transcription factors, which translates into a strong suppression PCSK9 transcription synergistically.
FIGURE 7.
Reduction of SREBP2 protein by BBR treatment is partially responsible for reduced PCSK9 expression. A, HepG2 cells were treated with BBR for 24 h at concentrations of 20 and 40 μm. Total cell lysate was isolated for Western blot analysis of SREBP2 and PCSK9. B, intensities of the bands of precursor SREBP2 (SREBP2-P), nuclear SREBP2 (SREBP2-N), precursor PCSK9 (PCSK9-P), and mature PCSK9 (PCSK9-M) were quantified using an imaging program of Kodak Imaging Station 4000R. Values were normalized to β-actin and were graphed relative to untreated cells. The figure shown is representative of three separate experiments with similar results.
DISCUSSION
We set out to understand the interesting conundrum of the differential action of the plant-derived hypocholesterolemic compound berberine on a gene shown to be important in cholesterol metabolism. Whereas cholesterol and statins regulated both LDLR and PCSK9 in a coordinated manner, BBR acted to down-regulate the transcription of PCSK9 while up-regulating LDLR mRNA level post-transcriptionally. This investigation led us to uncover a novel transcriptional regulatory network that modulated the expression of PCSK9, a promising new therapeutic target in LDL-C metabolism.
PCSK9, a recently discovered serine protease important in regulating the degradation of LDLR protein, is a new addition to the list of more than 30 genes whose transcription is controlled by SREBPs (40). When intracellular cholesterol levels are depleted, the mature SREBP translocates to the nucleus, where it activates PCSK9 and other genes' transcription by direct binding to nonpalindromic 9-bp sterol response elements in the promoter/enhancer regions (41, 42). Although in many cases, the key SRE motifs are adjacent to Sp1 or NF-Y binding sites and SREBPs work in concert with these coactivators to induce full transactivation (37–39), findings from our current study indicate that the PCSK9 promoter is rather unique in this aspect. Despite the initial characterization of PCSK9 promoter that identified putative NF-Y and Sp1 sites upstream of the SRE (19), both sequence motifs were later shown to not be critical for SRE function. The NF-Y site (−613) resides 5′ outside the functional proximal promoter region, whereas mutation of the Sp1 site (−430) modestly attenuated basal PCSK9 promoter activity in a manner that was not associated with sterol suppression (21) or berberine inhibition (present study). Additional putative Sp1 sites downstream of SRE also failed to exhibit a significant impact on PCSK9 transcription (21). These observations raised an important question as to what was the SREBP cofactor in controlling PCSK9 transcription. In this current study, by conducting several different lines of investigation, we provided solid evidence to demonstrate that the HNF1 binding site adjacent to SRE is the critical regulatory sequence motif and HNF1α is the predominant working partner for SREBP2 in the regulation of PCSK9 gene in the context of HepG2 cells.
It was previously reported that Sp1 and SRE sites, separated by ∼75 bp were perfectly conserved in the PCSK9 proximal promoter of human, mouse, and rat (19). In this study, we show that the HNF1 site residing between Sp1 and SRE is also 100% preserved in the PCSK9 promoter of these three species. This underscores the importance of this ∼100 bp proximal promoter region for PCSK9 gene transcription. Our promoter analysis revealed that deletion or mutation of this HNF1 site severely reduced PCSK9 promoter activity to levels even below those observed with the SRE mutation. Mutation of the HNF1 site also significantly mitigated the transactivation of PCSK9 promoter by nuclear SREBP2 through SRE, suggesting that these two sites work in concert. The liver-enriched trans-activator HNF1α shares dimerization and homeodomains with its related protein HNF1β and mediates sequence-specific DNA binding through the recognition of its cognate sequence 5′-GTTAATNATTAAC-3′ located in promoter regions of the target genes (32, 33). The PCSK9-HNF1 site (GTTAATGTTTAAT) shares 11 of 13 nucleotide identities with the consensus sequence (34). To identify the proteins interacting with the PCSK9-HNF1 promoter region, we combined approaches of EMSA, ChIP, siRNA knockdown, and exogenous overexpression. Nuclear extracts of HepG2 cells formed two discrete complexes with the labeled probe of PCSK9-HNF1. Both complexes were supershifted with anti-HNF1α antibody, indicating HNF1α was a major protein in these complexes. These complexes also modestly reacted with anti-HNF1β antibody as evidenced by a weak supershifted band. At present, we could not conclude whether the major complex represented the homodimer of HNF1α and the minor one was composed of HNF1α and HNF1β as a heterodimer. Further experiments are required to clearly define the protein components within these complexes. A stronger binding of HNF1α compared with HNF1β in intact HepG2 cells was also detected by ChIP assays. Taken together, these results firmly demonstrate the specific binding of HNF1α to this newly defined HNF1 site on PCSK9 promoter. The functional importance of this interaction is demonstrated by our siRNA transfection studies. Depletion of HNF1α by siRNA transfection reduced the promoter activity, endogenous mRNA, and protein expression of PCSK9 to near background levels. In comparison, siHNF1β transfection modestly reduced PCSK9 promoter activity and mRNA expression, and caused a small reduction in PCSK9 protein expression. The less significant role of HNF1β on PCSK9 transcription is further demonstrated by a small increase in PCSK9 promoter activity in HepG2 cells after transfection of pTS-HNF1β as compared with a substantial increase seen in pTS-HNF1α-transfected cells. Based upon these results we conclude that HNF1α is the major form of HNF1 factors that interacts with PCSK9-HNF1 site under our experimental conditions. It is possible that HNF1β may have a significant role in PCSK9 transcription in a different cellular context. Recently, in an integrated study of analysis of mRNA expression in Hnf1α−/− pancreatic islets and liver with a computational and experimental identification of HNF1α target genes, by conducting ChIP assays Servitja et al. showed that a promoter region containing a putative HNF1 binding motif of mouse PCSK9 gene was occupied by HNF1α in hepatocytes isolated from wild-type liver, whereas in hepatocytes isolated from Hnf1α−/− liver, HNF1β was found to bind to this promoter region (43). This observation is consistent with our speculation that HNF1β could have a compensational role in PCSK9 transcription under certain circumstances.
Since the discovery of PCSK9 and its implication in plasma LDL-C metabolism, different strategies have been explored to block the function of PCSK9 and its interaction with LDLR (44, 45). In this study, through elucidation of the mechanism of BBR-mediated suppression of PCSK9 we provide new evidence to support the notion that inhibition of PCSK9 expression at the transcriptional level by small molecules could be a viable means to increase LDLR expression and to minimally complement and potentially synergize with statin therapy. We showed that BBR inhibits PCSK9 mRNA and protein expression and counteracts the inducing effects of various statins. We have shown at the promoter level the inhibitory effect of BBR was partially abolished by each single mutation of SRE or HNF1 site; this led us to speculate that somehow BBR affected interactions of both SREBP and HNF1 factors to their recognition sequences. By Western blotting we showed that the amount of HNF1α was ∼60% less in BBR-treated cells compared with control. Similarly, the abundance of SREBP2 was modestly reduced by BBR treatment. In contrast to the moderate reductions of SREBP2 and HNF1α, the diminution of PCSK9 protein expression by BBR was pronounced. These data imply a synergistic effect occurred in BBR-treated cells by down-regulation of each transcription factor at modest levels. This observed synergy would be beneficial with regard to LDLR expression, because SREBP2 is absolutely required for LDLR transcription, and a strong inhibition of SREBP2 would eventually shut down LDLR expression. The facts that BBR treatment increases LDLR protein level in HepG2 cells (27, 46) and in livers of hyperlipidemic hamsters in vivo (27, 29) suggest that the balanced effects of BBR are in favor of LDLR expression and stability.
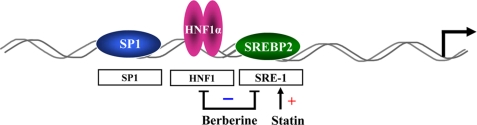
In summary (Fig. 8), we have identified a highly conserved HNF1 binding site located between SRE and Sp1 site as a critical cis-regulatory sequence of PCSK9 promoter. Our data suggest that the liver-enriched transcription factor HNF1α is the principal form of HNF1 factors that binds to the HNF1 sequence motif to stimulate PCSK9 transcription. Whereas statins induce PCSK9 transcription by enhancing the binding of SREBP2 to SRE-1, BBR lowers the cellular abundance of HNF1α and SREBP2, resulting in reduced interaction of these two critical trans-activators with their recognition sequences of PCSK9 promoter and leading to a transcriptional repression. These findings may provide novel insights into developing small molecule inhibitors of PCSK9 to improve the efficacy of statins in even further reducing LDL-C by overcoming one of the feedback loops that limits LDLR expression and longevity.
FIGURE 8.
Depiction of human PCSK9 promoter and its regulation by statin and BBR through the cis-regulatory elements and trans-acting factors. The plus sign indicates that statin induces SREBP2 to bind SRE-1 site of the PCSK9 promoter to stimulate transcription. The minus sign indicates the dual effects of BBR in reducing the binding of HNF1α to HNF1 site and the binding of SREBP2 to SRE-1 site.
This work was supported, in whole or in part, by National Institutes of Health Grants 1RO1 AT002543-01A1 and 1R21AT003195-01A2 from NCCAM. This work was also supported by the Department of Veterans Affairs (Office of Research and Development, Medical Research Service).
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- BBR
- berberine
- EMSA
- electrophoretic mobility shift assay
- HDAC1
- histone deacetylase 1
- HNF1
- hepatocyte nuclear factor 1
- LDL
- low density lipoprotein
- LDL-C
- LDL-cholesterol
- LDLR
- LDL receptor
- LPDS
- lipoprotein-depleted serum
- SRE
- sterol response element
- SREBP
- SRE-binding protein
- siRNA
- small interference RNA
- FBS
- fetal bovine serum
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Abifadel M., Varret M., Rabès J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derré A., Villegér L., Farnier M., Beucler I., Bruckert E., Chambaz J., Chanu B., Lecerf J. M., Luc G., Moulin P., Weissenbach J., Prat A., Krempf M., Junien C., Seidah N. G., Boileau C. (2003) Nat. Genet. 34, 154–156 [DOI] [PubMed] [Google Scholar]
- 2.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., Wahlstrand B., Hedner T., Corella D., Tai E. S., Ordovas J. M., Berglund G., Vartiainen E., Jousilahti P., Hedblad B., Taskinen M. R., Newton-Cheh C., Salomaa V., Peltonen L., Groop L., Altshuler D. M., Orho-Melander M. (2008) Nat. Genet. 40, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., Strait J., Duren W. L., Maschio A., Busonero F., Mulas A., Albai G., Swift A. J., Morken M. A., Narisu N., Bennett D., Parish S., Shen H., Galan P., Meneton P., Hercberg S., Zelenika D., Chen W. M., Li Y., Scott L. J., Scheet P. A., Sundvall J., Watanabe R. M., Nagaraja R., Ebrahim S., Lawlor D. A., Ben-Shlomo Y., Davey-Smith G., Shuldiner A. R., Collins R., Bergman R. N., Uda M., Tuomilehto J., Cao A., Collins F. S., Lakatta E., Lathrop G. M., Boehnke M., Schlessinger D., Mohlke K. L., Abecasis G. R. (2008) Nat. Genet. 40, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timms K. M., Wagner S., Samuels M. E., Forbey K., Goldfine H., Jammulapati S., Skolnick M. H., Hopkins P. N., Hunt S. C., Shattuck D. M. (2004) Hum. Genet. 114, 349–353 [DOI] [PubMed] [Google Scholar]
- 5.Leren T. P. (2004) Clin. Genet. 65, 419–422 [DOI] [PubMed] [Google Scholar]
- 6.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. (2005) Nat. Genet. 37, 161–165 [DOI] [PubMed] [Google Scholar]
- 7.Maxwell K. N., Breslow J. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S. W., Moon Y. A., Horton J. D. (2004) J. Biol. Chem. 279, 50630–50638 [DOI] [PubMed] [Google Scholar]
- 9.Qian Y. W., Schmidt R. J., Zhang Y., Chu S., Lin A., Wang H., Wang X., Beyer T. P., Bensch W. R., Li W., Ehsani M. E., Lu D., Konrad R. J., Eacho P. I., Moller D. E., Karathanasis S. K., Cao G. (2007) J. Lipid Res. 48, 1488–1498 [DOI] [PubMed] [Google Scholar]
- 10.Horton J. D., Cohen J. C., Hobbs H. H. (2007) Trends Biochem. Sci. 32, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seidah N. G. (2009) Expert Opin. Ther. Targets 13, 19–28 [DOI] [PubMed] [Google Scholar]
- 12.Cao G., Qian Y. W., Kowala M. C., Konrad R. J. (2008) Endocr. Metab. Immune Disord. Drug Targets 8, 238–243 [DOI] [PubMed] [Google Scholar]
- 13.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S. B., Stifani S., Basak A., Prat A., Chrétien M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., Trillard M., Abifadel M., Tebon A., Attie A. D., Rader D. J., Boileau C., Brissette L., Chrétien M., Prat A., Seidah N. G. (2004) J. Biol. Chem. 279, 48865–48875 [DOI] [PubMed] [Google Scholar]
- 15.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. (2007) J. Biol. Chem. 282, 18602–18612 [DOI] [PubMed] [Google Scholar]
- 16.McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., Lagace T. A. (2009) J. Biol. Chem. 284, 10561–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxwell K. N., Soccio R. E., Duncan E. M., Sehayek E., Breslow J. L. (2003) J. Lipid Res. 44, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 19.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N. G., Bernier L., Prat A. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1454–1459 [DOI] [PubMed] [Google Scholar]
- 20.Costet P., Cariou B., Lambert G., Lalanne F., Lardeux B., Jarnoux A. L., Grefhorst A., Staels B., Krempf M. (2006) J. Biol. Chem. 281, 6211–6218 [DOI] [PubMed] [Google Scholar]
- 21.Jeong H. J., Lee H. S., Kim K. S., Kim Y. K., Yoon D., Park S. W. (2008) J. Lipid Res. 49, 399–409 [DOI] [PubMed] [Google Scholar]
- 22.Attie A. D., Seidah N. G. (2005) Cell Metab. 1, 290–292 [DOI] [PubMed] [Google Scholar]
- 23.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y. A., Horton J. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5374–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kourimate S., Le May C., Langhi C., Jarnoux A. L., Ouguerram K., Zaïr Y., Nguyen P., Krempf M., Cariou B., Costet P. (2008) J. Biol. Chem. 283, 9666–9673 [DOI] [PubMed] [Google Scholar]
- 25.Careskey H. E., Davis R. A., Alborn W. E., Troutt J. S., Cao G., Konrad R. J. (2008) J. Lipid Res. 49, 394–398 [DOI] [PubMed] [Google Scholar]
- 26.Langhi C., Le May C., Kourimate S., Caron S., Staels B., Krempf M., Costet P., Cariou B. (2008) FEBS Lett. 582, 949–955 [DOI] [PubMed] [Google Scholar]
- 27.Kong W., Wei J., Abidi P., Lin M., Inaba S., Li C., Wang Y., Wang Z., Si S., Pan H., Wang Y., Wu J., Li Z., Liu J., Jiang J. D. (2004) Nat. Med. 10, 1344–1352 [DOI] [PubMed] [Google Scholar]
- 28.Abidi P., Zhou Y., Jiang J. D., Liu J. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 2170–2176 [DOI] [PubMed] [Google Scholar]
- 29.Abidi P., Chen W., Kraemer F. B., Li H., Liu J. (2006) J. Lipid Res. 47, 2134–2147 [DOI] [PubMed] [Google Scholar]
- 30.Li H., Chen W., Zhou Y., Abidi P., Sharpe O., Robinson W. H., Kraemer F. B., Liu J. (2009) J. Lipid Res. 50, 820–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cameron J., Ranheim T., Kulseth M. A., Leren T. P., Berge K. E. (2008) Atherosclerosis 201, 266–273 [DOI] [PubMed] [Google Scholar]
- 32.Mendel D. B., Hansen L. P., Graves M. K., Conley P. B., Crabtree G. R. (1991) Genes Dev. 5, 1042–1056 [DOI] [PubMed] [Google Scholar]
- 33.Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., Fraenkel E., Bell G. I., Young R. A. (2004) Science 303, 1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rey-Campos J., Chouard T., Yaniv M., Cereghini S. (1991) EMBO J. 10, 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowley C. W., Staloch L. J., Divine J. K., McCaul S. P., Simon T. C. (2006) Am. J. Physiol. Gastrointest. liver Physiol. 290, G466–G475 [DOI] [PubMed] [Google Scholar]
- 36.Qadri I., Hu L. J., Iwahashi M., Al-Zuabi S., Quattrochi L. C., Simon F. R. (2009) Toxicol. Appl. Pharmacol. 234, 281–292 [DOI] [PubMed] [Google Scholar]
- 37.Sanchez H. B., Yieh L., Osborne T. F. (1995) J. Biol. Chem. 270, 1161–1169 [DOI] [PubMed] [Google Scholar]
- 38.Magaña M. M., Koo S. H., Towle H. C., Osborne T. F. (2000) J. Biol. Chem. 275, 4726–4733 [DOI] [PubMed] [Google Scholar]
- 39.Egea M., Metón I., Córdoba M., Fernández F., Baanante I. V. (2008) Gen. Comp. Endocrinol. 155, 359–367 [DOI] [PubMed] [Google Scholar]
- 40.Horton J. D., Goldstein J. L., Brown M. S. (2002) J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown M. S., Goldstein J. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown M. S., Goldstein J. L. (1997) Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 43.Servitja J. M., Pignatelli M., Maestro M. A., Cardalda C., Boj S. F., Lozano J., Blanco E., Lafuente A., McCarthy M. I., Sumoy L., Guigo R., Ferrer J. (2009) Mol. Cell. Biol. 29, 2945–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham M. J., Lemonidis K. M., Whipple C. P., Subramaniam A., Monia B. P., Crooke S. T., Crooke R. M. (2007) J. Lipid Res. 48, 763–767 [DOI] [PubMed] [Google Scholar]
- 45.Frank-Kamenetsky M., Grefhorst A., Anderson N. N., Racie T. S., Bramlage B., Akinc A., Butler D., Charisse K., Dorkin R., Fan Y., Gamba-Vitalo C., Hadwiger P., Jayaraman M., John M., Jayaprakash K. N., Maier M., Nechev L., Rajeev K. G., Read T., Röhl I., Soutschek J., Tan P., Wong J., Wang G., Zimmermann T., De Fougerolles A., Vornlocher H. P., Langer R., Anderson D. G., Manoharan M., Koteliansky V., Horton J. D., Fitzgerald K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11915–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong W. J., Zhang H., Song D. Q., Xue R., Zhao W., Wei J., Wang Y. M., Shan N., Zhou Z. X., Yang P., You X. F., Li Z. R., Si S. Y., Zhao L. X., Pan H. N., Jiang J. D. (2009) Metabolism 58, 109–119 [DOI] [PubMed] [Google Scholar]